Abstract
The metallo-β-lactamase gene blaVIM-2 was identified in a strain of Pseudomonas aeruginosa isolated in India. The integron encoding blaVIM-2 was virtually identical to those recently found in the United States and Russia. These unusual structures are likely to have arisen from an ancestral integron predating the formation of the 3′ conserved sequence.
The increasing rates of antibiotic resistance among gram-negative bacteria, particularly Pseudomonas aeruginosa and Acinetobacter spp., is a serious cause for concern. The broad β-lactam resistance profile in these species may be mediated by metallo-β-lactamases (MBLs), which are capable of hydrolyzing most classes of β-lactams, and at present, there are currently no known effective clinical inhibitors. To date, there are five sub-major types of mobile MBL genes: blaIMP, blaVIM, blaSPM, blaGIM, and blaSIM (2, 12, 14). However, it would appear that blaVIM-2 has become the dominant genotype and has currently been reported from 23 counties, with the alleged “index” strain being a Portuguese P. aeruginosa isolate recovered in 1995 (1, 14). Most recently, blaVIM-2 has been reported from a few isolates in China and an outbreak in the United States (6, 15). Herein, we report on the first characterization of an MBL (blaVIM-2) from India that is carried on a unique integron but that shows genetic structures similar to those of integrons from the United States and Russia.
A clinical isolate (isolate 42) of Pseudomonas aeruginosa was collected from the Sri Ramachandra Medical College and Research Institute, Chennai, India, in 2003. The isolate displayed an MBL-like phenotype that was characterized by zone enhancement with EDTA-impregnated imipenem disks (750 μg) (16); and cell lysates also hydrolyzed imipenem and meropenem, as measured by spectrophotometry at 299 nm, as described previously (13). The resistance profile (MIC) of P. aeruginosa strain 42 from India was as follows: imipenem, >32 μg/ml; meropenem, >32 μg/ml; ceftazidime, 96 μg/ml; piperacillin, >256 μg/ml; piperacillin-tazobactam, >256 μg/ml; cefepime, >256 μg/ml; aztreonam, >256 μg/ml; colistin, 1 μg/ml; gentamicin, >1,024 μg/ml; amikacin, 8 μg/ml; and ciprofloxacin, >32 μg/ml. The isolate came from the bronchoalveolar lavage fluid of a 60-year-old man with ventilator-associated pneumonia. The patient was treated with meropenem in the intensive care unit of the hospital of the Sri Ramachandra Medical College and Research Institute but subsequently succumbed to the infection.
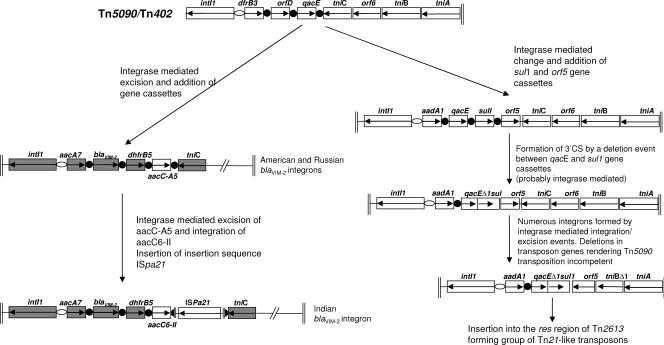
PCR with blaVIM-specific primers was positive by using the Expand high-fidelity master mix containing a mixture of Pfu and nonproofreading Taq polymerases and deoxynucleoside triphosphates (ABgene; Epsom United Kingdom) and primers, as reported previously (13). The amplicon was sequenced to confirm the presence of the blaVIM-2 gene cassette. Further PCR with class 1 integron conserved sequence (CS) primers 5′CS and 3′CS failed to amplify any class 1 integron genetic structures. However, subsequent PCRs with a combination of the 5′CS primer and a primer designed to detect the tniC gene of transposon Tn5090 (primer tniCF) (Table 1) was successful and amplified an integron that harbored the blaVIM-2 MBL gene but that lacked the normal 3′CS. This integron was sequenced in full by using a combination of primers 5′CS and tniCF and custom-made primers (Table 1). The integron had an unusual cassette structure consisting of a tandem array of aacC7, blaVIM-2, dhfrB5, and aacC6-II gene cassettes (Fig. 1). The cassette array and integron structure were strikingly similar to those of two other blaVIM-2-harboring integrons that have recently been sequenced from P. aeruginosa strains isolated in the United States and Russia (GenBank accession no. DQ522233) (6) (Fig. 1). In particular, all three integrons had the same three cassettes in positions 1 to 3 of their variable regions, i.e., aacA7, blaVIM-2, and dhfrB5 (previously called dhfrIIe), which confer resistance to aminoglycosides, β-lactams, and trimethoprim, respectively (4). Additionally, all three integrons lacked the 3′CS that is found in the vast majority of class 1 integrons in clinically relevant bacteria and that consists of fused qacE and sul1 gene cassettes, termed qacEΔ1/sul1. Instead, the tniC gene encoding the resolvase of transposon Tn5090 (also called tniR of Tn402) (7) was found 3′ adjacent to the variable region of each integron (Fig. 1). The Indian integron differs in only two respects from the integrons of the Russian and U.S. isolates. First, the fourth gene cassette is aacC6-II, an N-acetyltransferase gene that confers resistance to gentamicin, tobramycin, and netilmicin but not amikacin or isepamycin (9), rather than the aacCA5 gene found in the integrons of the Russian and U.S. isolates, which confers resistance only to gentamicin (Fig. 1) (3). Second, the integron of the Indian isolate contained an ISPa21-like insertion sequence that has inserted within the 59-base element of the aacC6-II gene, an event that would “fix” this gene in the integron, making it refractory to integrase-mediated excision events (Fig. 1).
TABLE 1.
Primer sequences used in this study
| Primer name | Primer sequence (5′-3′) |
|---|---|
| tniCF | CGATCTCTGCGAAGAACTCG |
| 5′CS | GCCTGTTCGGTTCGTAAGCT |
| 3′CS | CGGATGTTGCGATTACTTCG |
| INDAM2638F | CGGTCTAGACTTGCTCAAGC |
| INDAM3059F | TTATCCTGTTGCGGCACTGG |
| INDAM3497F | CGCAGAGAAGGCATAGCTAC |
| INDAM4027F | TGGCGGCGGTCATCTTGAAG |
| INDAM3122R | GCCGTTGAGTGGAGCACTTC |
| INDAM3609R | GCTGGTTGGTCTTCTACGCC |
| ISPa21404F | GATATGTACAGGAGCAGCCG |
| ISPa21409R | CATATCGACCGAATGCCTCC |
FIG. 1.
Hypothetical model of class 1 integron evolution. Open reading frames are represented by open boxes, with the arrows indicating the direction of transcription. Solid black circles represent 59-base elements, and open ellipses represent the attI1 site of the integron. Inverted repeats are depicted as parallel vertical lines. Open reading frames that are identical in all three blaVIM-2-containing integrons are shaded gray.
The lack of a 3′CS is characteristic of the class 1 integron harbored by transposon Tn5090 (also called Tn402), the progenitor of the common type of class 1 integron structure that contains the 3′CS, as seen, for example, in transposon Tn21 (5) (Fig. 1). The addition of the sul1 gene cassette and its subsequent fusion to the Tn5090/Tn402 qacE gene cassette by integration and deletion events, respectively, gave rise to the common form of the class 1 integron (Fig. 1).
These three blaVIM-2-harboring integrons found in P. aeruginosa strains isolated from widely separated geographical locations probably originated from a widely dispersed Tn5090 transposon. This transposon has evolved by normal integrase-mediated acquisition and loss of gene cassettes to include the blaVIM-2 gene. The wide dispersal of this genetic structure with this particular gene array may be the reason that the blaVIM-2 MBL is reported more often than any other MBL gene (14). A hypothetical model of Tn5090/Tn402 evolution that gives rise to the blaVIM-2-harboring integrons described in this study as well as the more common form of class 1 integron found in Tn21 is depicted in Fig. 1. Notably, the majority of integrons with a 3′CS are contained within Tn5090/Tn402 transposons defective in transposition functions, often with the loss of tniC and a section of tniB (Fig. 1) (5). The Tn5090/Tn402 transposon is fully functional (8), and therefore, it may be expected that these three class 1 integron structures harboring blaVIM-2 are also present on a functional transposon, enhancing mobility. Experiments are under way to determine if this is indeed the case. Tn5090 was initially sequenced from the IncP plasmid R751, isolated from Enterobacter aerogenes (11). Plasmids were not detected in Indian P. aeruginosa strain 42 by the alkaline lysis procedure with a QIAGEN mini-prep kit (13) and could not be conjugated to P. aeruginosa PAO1 or Escherichia coli DH5α by standard methods (13).
The finding of a number of class 1 integron structures without a 3′CS draws attention to the fact that the frequency of class 1 integrons in clinically important bacterial pathogens is probably underestimated in the literature, since most studies use PCR analysis with primers designed to be specific for the 5′CSs and 3′CSs. Indeed, a recent study has also highlighted the fact that class 1 integrons are also commonly found in forest soil and lake sediments and that these integrons lack both antibiotic resistance gene cassettes and Tn402 transposon genes (10).
Nucleotide sequence accession number.
The nucleotide sequence reported in this paper has been submitted to GenBank under accession number AM296017.
Acknowledgments
Mark Toleman is funded by the EC through COBRA contract LSHM-CT-2003-503335.
Footnotes
Published ahead of print on 16 April 2007.
REFERENCES
- 1.Cardoso, O., R. Leitao, A. Figueiredo, J. C. Sousa, A. Duarte, and L. V. Peixe. 2002. Metallo-β-lactamase VIM-2 in clinical isolates of Pseudomonas aeruginosa from Portugal. Microb. Drug Resist. 893-97. [DOI] [PubMed] [Google Scholar]
- 2.Lee, K., J. H. Yum, D. Yong, H. M. Lee, H. D. Kim, J. D. Docquier, G. M. Rossolini, and Y. Chong. 2005. Novel acquired metallo-β-lactamase gene, blaSIM-1, in a class 1 integron from Acinetobacter baumannii clinical isolates from Korea. Antimicrob. Agents Chemother. 494485-4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levings, R. S., S. R. Partridge, D. Lightfoot, R. M. Hall, and S. P. Djordjevic. 2005. New integron-associated gene cassette encoding a 3-N-aminoglycoside acetyltransferase. Antimicrob. Agents Chemother. 491238-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levings, R. S., D. Lightfoot, L. D. H Elbourne, S. P. Djordjevic, and R. M. Hall. 2006. New integron gene cassette encoding a trimethoprim-resistant DfrB-type dihydrofolate reductase. Antimicrob. Agents Chemother. 502863-2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liebert, C. A., R. M. Hall, and A. O. Summers. 1999. Transposon Tn21, flagship of the floating genome. Microbiol. Mol. Biol. Rev. 63507-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lolans, K., A. M. Queenan, K. Bush, A. Sahud, and J. P. Quinn. 2005. First nosocomial outbreak of Pseudomonas aeruginosa producing an integron-borne metallo-β-lactamase (VIM-2) in the United States. Antimicrob. Agents Chemother. 493538-3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Radstrom, P., O. Skold, G. Swedberg, J. Flensberg, P. H Roy, and L. Sundstrom. 1994. Transposon Tn5090 of plasmid R751, which carries an integron, is related to Tn7, mu, and the retroelements. J. Bacteriol. 1763257-3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shapiro, J. A., and P. Sporn. 1977. Tn402: a new transposable element determining trimethoprim resistance that inserts in bacteriophage lambda. J. Bacteriol. 1291632-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shaw, K. J., C. A. Cramer, M. Rizzo, R. Mierzwa, K. Gewain, G. H. Miller, and R. S. Hare. 1989. Isolation, Characterisation, and DNA Sequence Analysis of an aac(″)-II gene from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 332052-2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stokes, H. W., C. L. Nesbo, M. Holley, M. I. Bahl, M. R. Gillings, and Y. Boucher. 2006. Class 1 integrons potentially predating the association with Tn402-like transposition genes are present in a sediment microbial community. J. Bacteriol. 1885722-5730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thorsted, P. B., D. P. Macartney, P. Akhtar, A. S. Haines, N. Ali, P. Davidson, T. Stafford, M. J. Pocklington, and C. M. Thomas. 1998. Complete sequence of the IncPbeta plasmid R751: implications for evolution and organization of the IncP backbone. J. Mol. Biol. 282969-990. [DOI] [PubMed] [Google Scholar]
- 12.Toleman, M. A., A. M. Simm, T. A. Murphy, A. C. Gales, D. J. Biedenbach, R. N. Jones, and T. R. Walsh. 2002. Molecular characterization of SPM-1, a novel metallo-β-lactamase isolated in Latin America: report from the SENTRY Antimicrobial Surveillance Programme. J. Antimicrob. Chemother. 50673-679. [DOI] [PubMed] [Google Scholar]
- 13.Toleman, M. A., D. Biedenbach, D. M. Bennett, R. N. Jones, and T. R. Walsh. 2005. Italian metallo-beta-lactamases: a national problem? Report from the SENTRY Antimicrobial Surveillance Programme. J. Antimicrob. Chemother. 5561-70. [DOI] [PubMed] [Google Scholar]
- 14.Walsh, T. R., M. A. Toleman, L. Poirel, and P. Nordmann. 2005. Metallo-β-lactamases: the quiet before the storm? Clin. Microbiol. Rev. 18306-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang, C., J. Wang, and Z. Mi. 2006. Pseudomonas aeruginosa producing VIM-2 metallo-β-lactamases and carrying two aminoglycoside modifying enzymes in China. J. Hosp. Infect. 62522-524. [DOI] [PubMed] [Google Scholar]
- 16.Yong, D., K. Lee., J. H. Yum., H. B. Shin., G. M. Rossolini, and Y. Chong. 2002. Imipenem-EDTA disk method for differentiation of metallo-β-lactamase-producing clinical isolates of Pseudomonas spp. and Acinetobacter spp. J. Clin. Microbiol. 403798-3801. [DOI] [PMC free article] [PubMed] [Google Scholar]