Abstract
LasA protease is a 20-kDa elastolytic and staphylolytic enzyme secreted by Pseudomonas aeruginosa. LasA is synthesized as a preproenzyme that undergoes proteolysis to remove a 22-kDa amino-terminal propeptide. Like the propeptides of other bacterial proteases, the LasA propeptide may act as an intramolecular chaperone that correctly folds the mature domain into an active protease. To locate regions of functional importance within proLasA, linker-scanning insertional mutagenesis was employed using a plasmid containing lasA as the target. Among the 5 missense insertions found in the mature domain of proLasA, all abolished enzymatic activity but not secretion. In general, the propeptide domain was more tolerant to insertions. However, insertions within a 9-amino-acid region in the propeptide caused dramatic reductions in LasA enzymatic activity. All mutant proLasA proteins were still secreted, but extracellular stability was low due to clustered insertions within the propeptide. The codons of 16 residues within and surrounding the identified 9-amino-acid region were subjected to site-directed mutagenesis. Among the alanine substitutions in the propeptide that had a major effect on extracellular LasA activity, two (L92A and W95A) resulted in highly unstable proteins that were susceptible to proteolytic degradation and three (H94A, I101A, and N102A) were moderately unstable and allowed the production of a LasA protein with low enzymatic activity. These data suggest that these clustered residues in the propeptide may play an important role in promoting the correct protein conformation of the mature LasA protease domain.
Pseudomonas aeruginosa is a gram-negative opportunistic pathogen that primarily infects immunocompromised patients and causes chronic pneumonia in those with cystic fibrosis. This bacterium secretes elastolytic proteases as virulence factors that include LasA protease and LasB elastase. LasA has been shown to greatly enhance the activity of LasB in the degradation of elastin (10, 19), an important component of connective tissue, blood vessels, and lung tissue. LasA promotes the shedding of syndecan-1, the predominant cell-surface heparin sulfate proteoglycan of epithelial cells, which can enhance microbial virulence (17, 18). LasA is also known as a potent staphylolytic enzyme, an activity resulting from its ability to cleave peptide bonds within the pentaglycine cross-bridges in the peptidoglycan of Staphylococcus aureus (9, 12). It is therefore also given the descriptive name staphylolysin, which is used as a synonym for this protease (9). LasA staphylolytic activity may confer an important advantage to P. aeruginosa over S. aureus during colonization of the cystic fibrosis lung. The potential of LasA as an antistaphylolytic therapeutic agent has recently been confirmed in vivo in an experimental model of S. aureus keratitis (2).
LasA protease belongs to the M23 family of β-lytic Zn-metallo-endopeptidases. It shares high homology with the mature domains of the β-lytic proteases of Achromobacter lyticus and Lysobacter enzymogenese (9, 12), although a crystal structure for these has not yet been solved. LasA is synthesized as an inactive precursor that is composed of a signal peptide (3 kDa), a propeptide (22 kDa), and a mature catalytic domain (20 kDa) (6, 21). The signal peptide is cleaved as the protein crosses the inner membrane, and the propeptide remains covalently attached until the protein is secreted from the cell (11). LasA differs from most propeptide-containing proteases in that processing of its propeptide is not autocatalytic (6). Instead, the LasA propeptide is processed by other endopeptidases secreted by P. aeruginosa, primarily LasB and LysC, and a lysine-specific protease (also known as protease IV or PrpL) (5, 26), which are thought to initiate cleavage at or near Lys168 (11). The maturation process in which the 42-kDa proLasA is converted to the 20-kDa mature LasA includes a 28-kDa intermediate, possibly due to cleavage(s) near Lys168 (11). A less-efficient maturation pathway by alkaline protease cleavage can also occur which does not show a 28-kDa intermediate (11).
A number of bacterial proteases have now been shown to require the assistance of a covalently attached N-terminal propeptide to fold them into an active conformation. A term that has been used to describe such a propeptide is intramolecular chaperone (IMC) (24). This propeptide region generally functions as an inhibitor and a facilitator of folding, stability, and even secretion of the protein. In most cases, the propeptide domain is cleaved from the enzymatic domain autocatalytically to release an active protease. The propeptide domains of many proteases have also been demonstrated to initiate folding, stability, and secretion of their cognate protease even when produced as separate proteins in trans (1, 16, 23, 25). An example of a well-characterized bacterial IMC is α-lytic protease, a chymotrypsin-like serine protease secreted by the soil bacterium Lysobacter enzymogenes. It has a 166-residue amino-terminal propeptide that facilitates folding of the 198-residue protease to its stable native state (8). The propeptide promotes a critical step in the folding reaction, and the more stable the proregion, the faster the folding reaction can occur (13). The crystal structure of α-lytic protease in a complex with its propeptide has been solved, which shows a C-shaped propeptide folding around a β-barrel domain of the protease, forming specific contact points with the enzyme (20).
In P. aeruginosa, the propeptide of LasB-elastase, a thermolysin-like zinc-metalloprotease, has also been shown to be a classic IMC (3, 16). Processing of proLasB is autocatalytic, and secretion of active LasB occurs even if the propeptide is expressed as a separate protein (3, 16). LasB is secreted as a complex of propeptide and mature domain through a type II secretion apparatus (11). Some of the critical residues in the LasB propeptide that are required for proper folding were recently identified (15).
The propeptide of LasA protease may also be an IMC that properly folds the mature LasA protease domain. However, the LasA propeptide is unusual for an IMC in that processing is not autocatalytic (6) and secretion of LasA does not occur if the propeptide is expressed as a separate protein (J. Gustin and D. Ohman, unpublished data). The propeptide sequence of LasA shows little identity to those of other β-lytic proteases, thus making it difficult to predict regions of functional importance. Also, the secretion signals directing proLasA into the type II secretion pathway are unknown and could be in the propeptide. The primary goal of this study was to identify functional domains within proLasA and understand the potential role of the propeptide domain in the folding, secretion, and activation of the mature enzyme.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Bacteria were cultured in L broth (1% tryptone, 0.5% yeast extract, 0.5% NaCl). Escherichia coli strain DH10B was used for the construction and maintenance of plasmids and was grown in L broth with ampicillin (100 μg/ml). P. aeruginosa strain PDO801 was a lasA::aacCI (gentamicin resistance cartridge) mutant of PAO1 constructed by allelic exchange and used for complementation by plasmid-borne lasA alleles. The P. aeruginosa lasA gene with its promoter was cloned on a 2.1-kb SalI-SphI fragment into the E. coli/P. aeruginosa shuttle vector pUCP18 (22) to form pKKG08 for mutagenesis of lasA. Restriction endonucleases and T4 DNA ligase were purchased from New England Biolabs.
Linker insertion mutagenesis.
Linker insertion mutagenesis was performed according to the GPS-LS linker-scanning system from New England Biolabs to generate random transposon insertions, referred to as transprimers (Tp), conferring kanamycin resistance (Kanr) in lasA using pKKG08 as the target. Briefly, a mixture of donor plasmid (pGPS5), target plasmid (pKKG08), TnsABC transposase and start solution (magnesium acetate) were incubated for 1 h at 37°C, and then the reaction was inactivated at 75°C for 10 min. The mutagenized plasmids were electroporated into E. coli DH10B with selection for Kanr on L agar containing kanamycin (30 μg/ml), and the colonies were pooled for isolation of plasmids using the QIAprep Spin miniprep kit (QIAGEN). The Tp-containing XbaI-SphI fragments from pools of targeted plasmids were cloned back into pKKG08, so that the region of transposon mutagenesis was only within the fragment containing lasA. Five hundred individual E. coli transformants were screened by restriction analysis to identify a wide variety of Tp insertion sites. The PmeI sites present in the Tp and an SphI site in pKKG08 were used for mapping the sites of insertion by agarose gel electrophoresis and measuring the sizes of fragments produced. Selected pKKG08::Tp plasmids were digested with PmeI, gel extracted from a 1% agarose gel using the QIAprep Spin miniprep kit from QIAGEN, and religated using T4 DNA ligase at 16°C overnight. Upon religation, all but the 15-bp insertion remained at a unique PmeI site. The plasmids were transformed by electroporation into E. coli DH10B with selection for Ampr and were screened for Kans by patching onto L agar containing kanamycin (30 μg/ml). Representative linker insertion mutants were sequenced in the ∼500-bp area surrounding the linker insertions using primers designed from the published sequence of lasA (6), accession no. U68175. Selected pKKG08 lasA::In plasmids were electroporated into PDO801 with selection on Pseudomonas isolation agar (Difco) containing carbenicillin (150 μg/ml).
Growth conditions for LasA production.
P. aeruginosa cultures were grown under standardized conditions in which overnight cultures were diluted 1:100 in L broth, grown to an optical density at 600 nm (OD600) of 0.6, subcultured at a 1:100 dilution in L broth containing carbenicillin (150 μg/ml), and then grown for 18 h at 37°C with rapid aeration. Culture supernatants containing LasA were obtained from cultures by centrifugation to remove bacteria. A washed-cell culture method was also used to detect transient protease precursors and unstable mutant proteins secreted into the supernatant of P. aeruginosa cultures as previously described (11). Briefly, bacteria from 18 h cultures were collected by centrifugation (10,000 × g for 10 min), washed with saline, and grown in fresh L broth for 0.5 h. Some unstable proteins were examined over time from washed-cell cultures by taking samples at 1, 10, 20, 30, and 60 min. Then the supernatants were obtained by centrifugation for Western blot analysis.
Assay of LasA protease activity.
The effect of linker insertions in lasA on the activity of the resulting LasA::In proteins was determined by measuring the cleavage of pentaglycine in the peptidoglycan of S. aureus, which is also known as staphylolytic activity (12). A sample (50 μl) of standard P. aeruginosa 18-h culture supernatant was added to a 1.0-ml cell suspension of S. aureus strain ATCC 25904, previously heat-killed by boiling an overnight culture, washing the cells, and resuspending in 20 mM Tris-HCl (pH 8.0) at an OD595 of ∼0.8. The rate of cell lysis was determined spectrophotometrically over ∼5 min, and the specific activity was defined as the decline in OD595/min/mg protein over the linear portion of the curve as previously described (12). The values shown represent the averages of results from three experiments ± standard deviations.
Western blot analysis of LasA protein.
LasA-related proteins were demonstrated by Western blot analysis using rabbit polyclonal anti-LasA antibodies as previously described (6). Supernatants were obtained from 18-h standard cultures or washed-cell cultures. Bovine serum albumin (1 μg/ml) was added to culture supernatants as a carrier protein to improve protein precipitation by 15% trichloroacetic acid (TCA) on ice for 30 min followed by centrifugation. Intracellular proLasA was examined in P. aeruginosa grown under standard conditions for 18 h. Cells from a 1-ml culture were collected by centrifugation and washed in saline to remove extracellular proteins (e.g., proteases), and 15% TCA was added on ice for 30 min. All of the samples were then washed twice with 1 ml cold acetone, dried under a vacuum, and resuspended in 50 μl Laemmli sample buffer. For Western blot analyses, samples were heated to 100°C for 5 min. Protein concentrations were determined using a commercial Bradford assay (Bio-Rad). In all cases, 1.5 μg total protein was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis using 3% stacking and 12% resolving gels. Proteins were transferred to polyvinylidene difluoride membranes at 12 V overnight in ethanolamine buffer (25 mM ethanolamine, 38 mM glycine, 20% methanol, and 0.01% sodium dodecyl sulfate) (12). Membranes were blocked in Tris-buffered saline-Tween with 1% bovine serum albumin, and the primary antibody, polyclonal anti-LasA antibody raised in rabbits (6), was added at a 1:2,000 or higher dilution. A peroxidase-conjugated goat anti-rabbit secondary antibody (Sigma) was used at a 1:10,000 dilution. LasA-related protein bands were detected using chemiluminescence with 1.25 mM luminol (Sigma), 0.2 mM p-coumeric acid (Sigma), 0.01% hydrogen peroxide, and 50 mM Tris-HCl (pH 8.5). For film exposure, clear blue X-ray film (CL-X Posure film; Pierce) was used.
Site-directed mutagenesis.
A sequential PCR method was used to change selected codons to encode alanine using a two-step PCR previously described (7). The first step involved the amplification of the region of interest using a mutagenic forward primer that encoded an alanine substitution at the site of mutagenesis and a downstream reverse primer. A second primary PCR was also performed using an upstream forward primer and a mutagenic reverse primer. Both of these primary PCRs were gel extracted and subjected to a secondary PCR using the wild-type forward and reverse primers used above. This final product was gel extracted, digested with XbaI and SphI, and cloned into pKKG08. All mutations were confirmed by sequencing of approximately 500 bp of the surrounding area.
RESULTS
Random insertional mutagenesis of lasA.
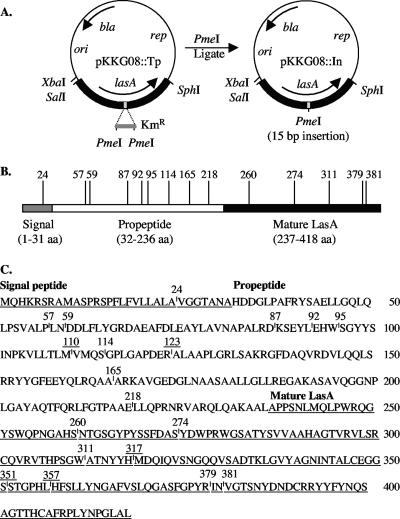
Linker scanning is a method used for the genetic study of protein structure where domains are potentially disrupted by the insertion of a small number of amino acids. Here we wanted to identify functional domains within proLasA, so we tested the effects of random 5-amino-acid insertions on the formation of stable and enzymatically active LasA protease. The target was pKKG08 (Fig. 1A) containing lasA on a 2.1-kb fragment in an E. coli/P. aeruginosa shuttle vector. An in vitro mutagenesis system (GPS-LS linker-scanning; NEB) was employed that generated random insertions in lasA of a Tn7-based transposon, which is called a Tp. The sites of ∼500 insertions in pKKG08::Tp plasmids were mapped by restriction analysis, and 19 of these clearly showed unique insertion sites in lasA. These were subjected to PmeI digestion and religation, which removed all but 15 bp from within the Tp at the site of insertion (Fig. 1A). If in-frame, this 15-bp insertion resulted in a 5-amino-acid insertion in preproLasA. The unique pKKG08::In plasmids were sequenced to determine the precise location of the 15-bp linker insertion (Fig. 1B). The specific 5 amino acids inserted were also revealed by the sequence analysis (Table 1), which varied because the transposition reaction resulted in a 5-bp duplication of the variable insertion site in addition to the insertion of 10 bp from the transposon. Five of the insertions contained a nonsense codon, leading to a truncated proLasA (Table 1). Thus, 14 unique insertion sites each resulted in the addition of 5 amino acids, with 1 insertion in the signal sequence, 8 in the propeptide, and 5 in the mature (i.e., proteolytic) domain of preproLasA (Fig. 1C).
FIG. 1.
Locations of Tp insertions within preproLasA. (A) Target pKKG08 (6,682 bp) contained the lasA gene and its promoter on a 2.1-kb SalI-SphI fragment cloned into pUCP18, an E. coli/P. aeruginosa shuttle vector. The GPS-LS linker-scanning system (New England Biolabs) was used to generate random Tp insertions conferring Kanr, which were mapped relative to the SphI site. Digestion of the plasmid with PmeI and religation left a 15-bp insertion to potentially encode a 5-amino-acid insertion in the encoded proLasA, which was shown by sequence analysis. (B) The domain structure of preproLasA (product of lasA) shows a signal peptide (3.3 kDa), a propeptide (22.3 kDa), and the mature LasA protease (20.0 kDa) (6). Vertical lines on the domain map mark the sites of Tp insertions in lasA that produced in-frame insertions and are marked with the number of the preceding amino acid in preproLasA. (C) The amino acid sequence of preproLasA is shown with the signal and mature domains underlined. The sites of selected linker insertions found within preproLasA are indicated, and the position numbers are underlined for stop codons (causing truncations).
TABLE 1.
Amino acids inserted within preproLasA by linker-scanning mutagenesisa
| Mutant no. | Linker sequence inserted | Location |
|---|---|---|
| 24 | VVFKH | Signal sequence |
| 57 | LPKQP | Propeptide |
| 59 | CLNMN | Propeptide |
| 87 | ACLNN | Propeptide |
| 92 | CLNNL | Propeptide |
| 95 | MFKHW | Propeptide |
| 110 | VV.XM | Propeptide stop |
| 114 | CLNKS | Propeptide |
| 123 | APV.T | Propeptide stop |
| 165 | ALFKH | Propeptide |
| 218 | LFKQE | Propeptide |
| 260 | CLNNS | Mature |
| 274 | RCLNR | Mature |
| 311 | CLNSW | Mature |
| 317 | IV.TH | Mature stop |
| 351 | SV.TS | Mature stop |
| 357 | HFV.T | Mature stop |
| 379 | INCLN | Mature |
| 381 | CLNIN | Mature |
The mutant number indicates the last amino acid number (relative to the initiating methionine of preproLasA) preceding the site of insertion. A dot in the linker sequence inserted indicates a stop codon.
Effects of insertion mutations in the mature domain of LasA.
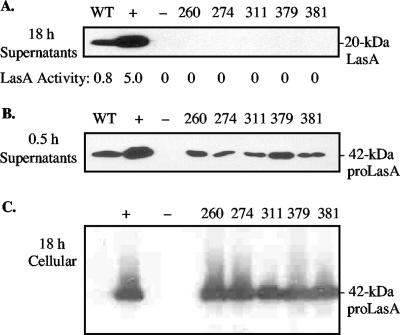
The 19 defined pKKG608 lasA::In derivatives were then transferred to PDO801, a lasA mutant of wild-type PAO1, to characterize the effects of the insertions when expressed in trans. The 18-h culture supernatant of the lasA+ positive control strain, PDO801(pKKG08), contained high levels of LasA staphylolytic activity (5.0 units), and LasA protein was readily detected by Western blot analysis (Fig. 2A). PDO801(pKKG08) produced ∼5 times more LasA than wild-type PAO1 due to having lasA in multicopy. The insertions obtained in the mature domain of LasA (excluding nonsense codons) followed residues 260, 274, 311, 379, and 381 (Table 1). Under the same conditions, none of the 18-h culture supernatants from PDO801 strains carrying plasmids with mature domain mutations contained detectable LasA protein or any enzymatic activity (Fig. 2A). However, P. aeruginosa is known to secrete LasA in the 42-kDa proLasA form, which then undergoes extracellular processing by other P. aeruginosa proteases to form the stable and protease-resistant mature LasA (11). To determine if these mature domain mutations adversely affected the secretion of proLasA, bacteria from 18-h cultures were washed to remove other exoproteases produced by P. aeruginosa that would degrade an unstable proLasA protein and then were cultured in fresh medium for 0.5 h. All of these washed-cell culture supernatants clearly showed the secretion of proLasA, although the amount observed according to Western blotting was less than that seen in the PDO801(pKKG08) lasA+ control (Fig. 2B). To determine if the reduced extracellular level of proLasA could be due to an intracellular build-up, such as from a secretion defect, we examined cell lysates by Western blot analysis. However, no increased accumulation of 42-kDa proLasA protein was observed (Fig. 2C). Thus, the mutant proLasA proteins appeared to be secreted normally but were unstable once outside the cell in the protease-rich culture supernatant of P. aeruginosa.
FIG. 2.
Effect of linker insertion mutations within the mature domain of lasA. (A) A Western blot analysis is shown of 18-h culture supernatants probed with an anti-LasA antibody. Tops of lanes are marked with strains examined (WT, PAO1 LasA+ wild-type strain; +, LasA+ control PDO801(pKKG08); −, LasA− control PDO801) and numbers that indicate residues preceding a linker insertion. Note that stable LasA was rapidly processed in the P. aeruginosa culture supernatants to 20 kDa, but unstable LasA was degraded. Activity of LasA protease in culture supernatants was determined as the rate of lysis of Staphylococcus aureus cells (OD595/min/mg protein). (B) Western blot analysis of washed-cell culture supernatants after 0.5 h of incubation that were used to detect short-lived LasA protease precursors. Note that all of the linker mutants were secreted, even though they were unstable and thus degraded by 18 h (as shown in panel A). (C) Western blot analysis of intracellular LasA to detect any abnormal accumulation of proLasA, which was not observed.
The linker insertions at positions 317, 351, and 357 within the mature catalytic domain resulted in the generation of termination codons. These permitted a preliminary deletion analysis of 42-kDa proLasA with respect to secretion. The predicted sizes of these proLasA(stop) proteins were approximately 32, 36, and 37 kDa, which corresponded to mature domains with truncations of 81, 115, and 121 amino acids, respectively. None of the three nonsense mutants showed enzymatic activity in P. aeruginosa culture supernatants. Also, no LasA protein was detected by Western analysis in 18-h cultures or in 0.5-h washed-culture supernatants (data not shown). Whole-cell lysates were also examined for the presence of truncated LasA-related proteins, but none could be detected (data not shown). Thus, a deletion of 81 amino acids or more from the C terminus of proLasA caused major instability, which presumably led to its degradation by intracellular proteases before secretion could take place.
Effects of insertion mutations in the propeptide of LasA.
An insertion in the signal peptide following amino acid 24 had little effect on LasA secretion (Fig. 3A), probably because 3 of the 5 residues inserted were hydrophobic (Table 1) and so did not interfere with membrane insertion. Eight unique lasA insertion mutations (encoding 5 amino acid residues) were identified in the propeptide domain of lasA, which followed amino acids 57, 59, 87, 92, 95, 114, 165, and 218 (Fig. 1). Unlike the insertions in the mature domain of LasA described above, the propeptide domain was much more tolerant to such mutations. Although LasA enzymatic activity was reduced in all 18-h culture supernatants of the mutants, insertions that caused the most dramatic reduction in LasA enzymatic activity followed amino acids 87, 92, and 95, thus placing them within a 9-amino-acid region of the central area of the propeptide (Fig. 3A). In fact, there appeared to be a stepwise decrease in activity as the linker insertions approached this region from either side (Fig. 3A). The enzymatic activities in these mutant culture supernatants showed good correlation with the amount of LasA protein in 18-h standard culture supernatants as shown by Western analysis (Fig. 3B). Only the insertion at position 92 nearly prevented detection of LasA in the 18-h standard culture supernatant. However, all propeptide mutant proteins were shown to be secreted in the washed-cell culture assay and detectable through Western analysis (Fig. 3C). This indicated that the domains potentially recognized by the type II secretion apparatus were unlikely to have been affected. However, all of these insertion mutants appeared to have less proLasA protein in the 0.5-h washed-cell culture supernatant compared to the PDO801(pKKG08) positive control. This may be related to varying degrees of protein stability and, thus, susceptibility to degradation by other Pseudomonas proteases also produced during the 0.5-h incubation period.
FIG. 3.
Effects of linker insertions within the propeptide domain of lasA. (A) LasA proteolytic activity in 18-h culture supernatants determined as the rate of staphylolysis (OD595/min/mg protein). The strains examined were as follows: WT, wild-type PAO1 (LasA+ strain); +, LasA+ control PDO801(pKKG08); −, LasA− control PDO801(pUCP18). Numbers indicate the residues preceding a linker insertion in strain PDO801(pKKG08). (B) Western blot analysis of mature LasA (20 kDa) in 18-h culture supernatants. Note that propeptide linker mutants 87, 92, and 95 were especially reduced in both activity and protein production at 18 h. (C) Western blot analysis of 0.5-h washed-cell culture supernatants to detect secretion of proLasA (42 kDa), which showed that all propeptide mutant proteins were secreted. Note that propeptide mutant 165 displayed a double band at the position predicted for the 28-kDa intermediate form of LasA. (D) Western blot analysis of intracellular proLasA in propeptide mutants 87, 92, and 95 grown for 18 h. The proLasA was detected but did not appear to be accumulating inside the cell.
Extracellular processing of wild-type proLasA often includes a 28-kDa intermediate, containing part of the propeptide, prior to formation of the final 20-kDa mature form (11). We also observed this in the 0.5-h culture supernatant with the lasA+ control strain PDO801(pKKG08) and in mutants pKKG08-24 (insertion in signal sequence), pKKG08-114, and pKKG08-165 (Fig. 3C). The predicted site of processing to 28 kDa is at Lys-168 by P. aeruginosa lysine-specific protease or at one of the surrounding alanine residues by LasB-elastase (11). Interestingly, mutant pKKG08-165 has its insertion (ALFKH) at this site (…AÂARKA…), and it produced a double band at 28 kDa (Fig. 3C). Thus, the cleavage site was apparently altered due to this insertion.
We also examined intracellular proLasA within insertion mutants pKKG08-87, -92, and -95 because they showed the most dramatic reduction in extracellular LasA at 18 h, which might be due to a secretion defect. However, when whole-cell lysates of these mutants from 18-h cultures were examined by Western blot analysis, no unusual intracellular accumulation of proLasA was observed (Fig. 3D). Thus, these propeptide mutants yielded LasA proteins that were secreted but apparently unstable in the protease-rich 18-h culture supernatants of P. aeruginosa. This was consistent with the hypothesis that the propeptide provides chaperone activity for its cognate domain, mature LasA.
Effects of propeptide mutations on the stability of secreted LasA protein over time.
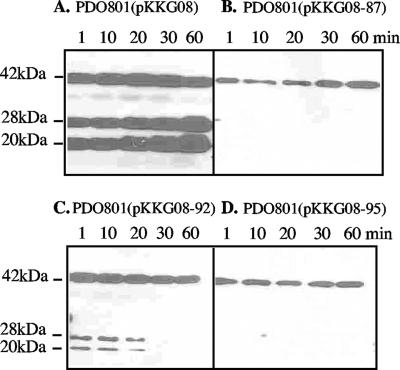
To monitor the fate of LasA from its proform to mature form and evaluate the stability of the mature form over time, we again used the washed-cell culture method. This reduced the general proteolytic environment of P. aeruginosa and allowed us to observe changes in the extracellular LasA-related proteins after secretion. Culture supernatants of cells in fresh medium were sampled at 1, 10, 20, 30, and 60 min of incubation, and the proteins were rapidly stabilized by TCA on ice. In the LasA+ positive control strain, PDO801(pKKG08), proLasA was readily detected outside the cell. It was already processed to the 28-kDa form and to the 20-kDa mature LasA at the earliest time point (Fig. 4A). By 60 min, mature LasA had accumulated in excess of the 42-kDa proLasA. Under the same conditions, proLasA was secreted by mutants PDO801(pKKG08-87, -92, and -95) and detected by 1 min of incubation (Fig. 4B, C, and D). However, the amount of extracellular 42-kDa proLasA accumulating in the supernatants of these strains was much lower than in the positive control (Fig. 4A), and no mature LasA accumulated over time (Fig. 4B, C, and D). This suggested that the insertions in the propeptide prevented folding to a protease-resistant stable form. It was interesting that mature LasA could be visualized from pKKG08-92 at early time points (Fig. 4C), even though 18-h cultures showed no mature LasA as described above (Fig. 3B). Although under these conditions pKKG08-87 and pKKG08-95 did not show conversion of proLasA to mature LasA during the first 60 min (Fig. 4B and D), the 18 h cultures did contain low levels of mature LasA (Fig. 3B). Thus, pKKG08-87, -92, and -95 produced proLasA that could be processed to the mature form, so the insertions of 5 amino acids apparently caused defects in the ultimate stability of the protein. To rule out the possibility of artifacts due to having lasA alleles in multicopy, proLasA secretion and processing over time were examined in the parent strain with lasA in a single chromosomal copy. Like PDO801(pKKG08), proLasA from PAO1 was detected at the earliest time point and accumulated in the supernatant; mature LasA was not observed until 20 to 30 min, probably due to the reduced amount of protein resulting from lasA in single copy (data not shown). As expected, the LasA− strain PDO801(pUCP18) produced no LasA under these conditions (data not shown).
FIG. 4.
Early time course of proLasA processing and stability in propeptide linker insertion mutants. Washed-cell cultures were periodically sampled following 1 to 60 min of incubation and subjected to a Western analysis to detect secretion of LasA-related proteins. (A) LasA+ control strain, PDO801(pKKG08); (B) PDO801(pKKG08-87); (C) PDO801(pKKG08-92), shown at a slightly longer exposure to reveal mature LasA at 1, 10, and 20 min; (D)/ PDO801(pKKG08-95).
Site-directed mutagenesis of the propeptide.
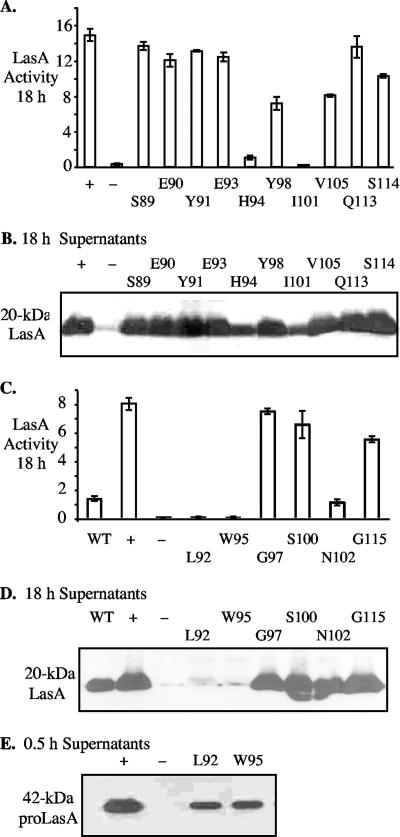
The linker insertions near residues 87 and 95 in the propeptide had produced dramatic effects on LasA protein stability. This provided an opportunity to identify specific residues in this region that were critical for its potential chaperone function. Ten amino acids between positions 89 and 114 were selected for alanine substitution, and the mutations were confirmed by nucleotide sequence analysis. DNA fragments containing the mutations were cloned into pKKG08, transferred to PDO801, and tested for their effects on LasA enzymatic activity and stability as reflected by LasA protein accumulation. Five of the 10 mutations (S89A, E90A, Y91A, E93A, and Q113A) showed no obvious effect on LasA enzymatic activity (Fig. 5A) or accumulation of stable LasA protein in 18-h culture supernatants (Fig. 5B). ProLasA-Y98A, -V105A, and -S114A mutants were reduced in enzymatic activity, which was consistent with the somewhat lower levels of mature LasA protein in the respective culture supernatants (Fig. 5A and B). However, proLasA mutants H94A and I101A were drastically affected in both enzymatic activity and yield of mature LasA in 18-h culture supernatants (Fig. 5A and B). Thus, propeptide residues His-94 and Ile-101 were apparently critical for stable LasA formation.
FIG. 5.
Effects of alanine substitutions in the propeptide domain of LasA. (A, C) Staphylolytic activity resulting from mutant lasA alleles. (B, D) Western blot detection of mature LasA (20 kDa) in 18-h culture supernatants. (E) Western blot detection of proLasA (42 kDa) in 0.5-h washed-cell culture supernatants to detect secretion. Strains examined include the following: WT, wild-type PAO1 (LasA+ strain); +, LasA+ control PDO801(pKKG08); −, LasA− control PDO801(pUCP18). Numbers indicate amino acid residues converted to alanine.
We then targeted 6 additional amino acids in the same region of the propeptide for alanine replacement mutagenesis: L92A, W95A, G97A, S100A, N102A, and G115A. These mutants were also tested for LasA enzymatic activity, protein stability, and secretion as described above. Mutant proLasA-G115A was slightly reduced for activity and proLasA-N102 was dramatically reduced for enzymatic activity (Fig. 5C), but both produced stable LasA (Fig. 5D). In contrast, proLasA-L92A and proLasA-W95A were negative for LasA enzyme activity (Fig. 5C), and almost no stable LasA was detected in 18-h supernatants of these mutants (Fig. 5D). However, both proLasA-L92A and proLasA-W95A were still secreted, as shown by the washed-cell culture assay (Fig. 5E). Overall, this study showed that the clustered residues L92, H94, W95, I101, and N102 were critical for the ability of the propeptide to promote the folding of mature LasA into a stable and/or active enzyme.
DISCUSSION
Little is known about the role of the propeptide of LasA, an M23A family protease, and whether the propeptide promotes folding and/or contains signals for recognition by the type II secretion apparatus (which is referred to as Xcp in P. aeruginosa). In this study, random linker scanning mutagenesis was used to generate 5-amino-acid insertions to identify regions of functional importance within the mature and propeptide domains of proLasA. The lasA alleles were expressed from a plasmid, and the mutant proLasA::In proteins were screened for altered phenotypes. Each mutant was examined for LasA protease activity, secretion, stability, and intracellular accumulation.
All of the linker insertions in the mature domain resulted in a secreted proLasA that was highly unstable, causing it to be completely degraded by 18 h of culture incubation. A limited deletion analysis showed that removal of 57 amino acids or more from the C terminus resulted in a highly unstable protein. In contrast, the propeptide was more tolerant to random 5-amino-acid insertions, and mature LasA could readily be detected in the supernatants of all 18-h cultures except for a mutant with an insertion at position 92. This mutant showed the most drastic defect on LasA accumulation in the supernatant and thus revealed a critical region in the propeptide. The linker insertions nearest to Leu-92 also showed clear defects in forming a stable LasA. These mutant proteins were all found to be secreted and did not accumulate within the cell, suggesting that no secretion signal was altered by any of the linker insertions. A time course study of mutant proLasA-92 secretion, into a low-protease environment, showed some conversion to mature-sized (20-kDa) LasA, but it was completely degraded within 30 min, presumably by other accumulating proteases produced by P. aeruginosa. This suggested that the region of the propeptide near Leu-92 was important for propeptide-mediated folding of LasA and, thus, for its stability in the protease-rich extracellular environment typically generated by P. aeruginosa.
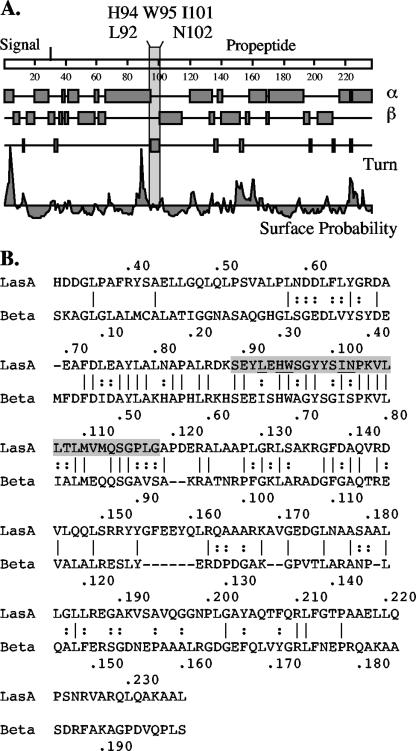
If the propeptide acts as a chaperone, then it was possible that the spacing of residues in the Leu-92 region was important for critical amino acids to interact with certain residues within the mature domain, such that insertion of 5 amino acids here altered the conformation that favored this interaction. Alternatively, an insertion of 5 amino acids within this region displaced important residues, causing proLasA folding defects that could render it susceptible to extracellular proteases. Thus, to better understand this potentially important region of the propeptide for chaperone activity, site-directed mutagenesis was employed in an effort to identify critical residues. In all, 15 residues within the region were changed to Ala to evaluate their importance. Indeed, residues Leu-92, His-94, Trp-95, Ile-101, and Asn-102 were clearly shown to be critical for the activity and/or stability of secreted LasA and thus may play a direct role in the propeptide's chaperone function. The variants with Ala substitutions at H94, I101, and N102 in the propeptide are interesting in that a relatively stable LasA protein is found in culture supernatants with low enzyme activity. The role of these residues in the formation of an enzymatically active LasA may become more apparent once the crystal structures of LasA and proLasA are solved. Computer-generated secondary structure of the propeptide predicts that these critical amino acids are in a loop between a surface-exposed alpha-helix and a beta-turn region located in the N-terminal half of the propeptide (Fig. 6A). In general, loops in proteins can participate in forming binding sites or enzyme active sites. This may be a signature conformation for regions important for stability or for important contact points with the mature domain. Interestingly, this region is within a zone of homology between the propeptides of LasA and β-lytic protease of Achromobacter lyticus (Fig. 6B).
FIG. 6.
Secondary structure prediction for the propeptide of LasA. (A) Secondary structure prediction for the propeptide of LasA suggests that the critical amino acids Leu-92, His-94, Trp-98, and I-101 are located (shown by a shaded rectangle) near a hydrophilic region at the site of a predicted turn between an alpha helix (α) and beta sheet (β). (B) Sequence alignment of the propeptides of P. aeruginosa LasA and Achromobacter lyticus β-lytic protease (Beta). The region targeted for alanine-scanning mutagenesis is shaded, which is also a region of highest homology. Critical amino acids for LasA activity and protein stability (Leu92, His94, Trp98, I101, and N102) are underlined in the shaded region. Identical residues are indicated by vertical lines; similar residues are indicated by two stacked dots.
Studies of propeptide-mediated folding of the α-lytic protease provide a model for future LasA-folding studies. Point mutations within the conserved residues of the amino-terminal domain of the α-lytic protease proregion have been demonstrated to remove hydrogen bonds and consequently disturb binding to the protease domain (4). Also, a functional linkage between the active site of α-lytic protease and a distant surface loop has been demonstrated through alanine-scanning mutagenesis (14). Specific interactions between the propeptide amino-terminal domain and α-lytic protease are required for efficient folding of the protease (4). The propeptide amino-terminal domain is thought to fold against the propeptide C-domain to facilitate the packing of the α-lytic protease core (4). There may be a comparable mechanism in the folding of proLasA. Future studies will examine the potential for interactions between residues in the critical domain of the LasA propeptide and its catalytic mature domain.
Acknowledgments
We thank Althea Bolden for her excellent technical assistance and the Virginia Commonwealth University Nucleic Acid Core Laboratory for accurate DNA sequence analyses.
This work was supported by Public Health Service Grant AI-26187 from the National Institute of Allergy and Infectious Diseases (to D.E.O.), by an NRSA training grant (to D.E.O.), and in part by Veterans Administration Funds (to D.E.O.).
Footnotes
Published ahead of print on 9 March 2007.
REFERENCES
- 1.Anderson, D. E., R. J. Peters, B. Wilk, and D. A. Agard. 1999. Alpha-lytic protease precursor: characterization of a structured folding intermediate. Biochemistry 38:4728-4735. [DOI] [PubMed] [Google Scholar]
- 2.Barequet, I. S., G. J. Ben Simon, M. Safrin, D. E. Ohman, and E. Kessler. 2004. Pseudomonas aeruginosa LasA protease in treatment of experimental staphylococcal keratitis. Antimicrob. Agents Chemother. 48:1681-1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Braun, P., J. Tommassen, and A. Filloux. 1996. Role of the propeptide in folding and secretion of elastase of Pseudomonas aeruginosa. Mol. Microbiol. 19:297-306. [DOI] [PubMed] [Google Scholar]
- 4.Cunningham, E. L., T. Mau, S. M. Truhlar, and D. A. Agard. 2002. The pro region N-terminal domain provides specific interactions required for catalysis of alpha-lytic protease folding. Biochemistry 41:8860-8867. [DOI] [PubMed] [Google Scholar]
- 5.Engel, L. S., J. M. Hill, A. R. Caballero, L. C. Green, and R. J. O'Callaghan. 1998. Protease IV, a unique extracellular protease and virulence factor from Pseudomonas aeruginosa. J. Biol. Chem. 273:16792-16797. [DOI] [PubMed] [Google Scholar]
- 6.Gustin, J. K., E. Kessler, and D. E. Ohman. 1996. A substitution at His-120 in the LasA protease of Pseudomonas aeruginosa blocks enzymatic activity without affecting propeptide processing or extracellular secretion. J. Bacteriol. 178:6608-6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horton, R. M., H. D. Hunt, S. N. Ho, J. K. Pullen, and L. R. Pease. 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77:61-68. [DOI] [PubMed] [Google Scholar]
- 8.Jaswal, S. S., J. L. Sohl, J. H. Davis, and D. A. Agard. 2002. Energetic landscape of alpha-lytic protease optimizes longevity through kinetic stability. Nature 415:343-346. [DOI] [PubMed] [Google Scholar]
- 9.Kessler, E., and D. E. Ohman. 2004. Staphylolysin (LasA endopeptidase), p. 1001-1003. In A. Barrett, N. Rawlings, and J. Woessner (ed.), The handbook of proteolytic enzymes, 2nd ed., vol. 1. Elsevier Academic Press, Amsterdam, The Netherlands. [Google Scholar]
- 10.Kessler, E., M. Safrin, W. R. Abrams, J. Rosenbloom, and D. E. Ohman. 1997. Inhibitors and specificity of Pseudomonas aeruginosa LasA. J. Biol. Chem. 272:9884-9889. [DOI] [PubMed] [Google Scholar]
- 11.Kessler, E., M. Safrin, J. K. Gustin, and D. E. Ohman. 1998. Elastase and LasA protease of Pseudomonas aeruginosa are secreted with their propeptides. J. Biol. Chem. 273:30225-30231. [DOI] [PubMed] [Google Scholar]
- 12.Kessler, E., M. Safrin, J. C. Olson, and D. E. Ohman. 1993. Secreted LasA of Pseudomonas aeruginosa is a staphylolytic protease. J. Biol. Chem. 268:7503-7508. [PubMed] [Google Scholar]
- 13.Ma, B., C. J. Tsai, and R. Nussinov. 2000. Binding and folding: in search of intramolecular chaperone-like building block fragments. Protein Eng. 13:617-627. [DOI] [PubMed] [Google Scholar]
- 14.Mace, J. E., B. J. Wilk, and D. A. Agard. 1995. Functional linkage between the active site of alpha-lytic protease and distant regions of structure: scanning alanine mutagenesis of a surface loop affects activity and substrate specificity. J. Mol. Biol. 251:116-134. [DOI] [PubMed] [Google Scholar]
- 15.McIver, K. S., E. Kessler, and D. E. Ohman. 2004. Identification of residues in the Pseudomonas aeruginosa elastase propeptide required for chaperone and secretion activities. Microbiology 150:3969-3977. [DOI] [PubMed] [Google Scholar]
- 16.McIver, K. S., E. Kessler, J. C. Olson, and D. E. Ohman. 1995. The elastase propeptide functions as an intramolecular chaperone required for elastase activity and secretion in Pseudomonas aeruginosa. Mol. Microbiol. 18:877-889. [DOI] [PubMed] [Google Scholar]
- 17.Park, P. W., G. B. Pier, M. J. Preston, O. Goldberger, M. L. Fitzgerald, and M. Bernfield. 2000. Syndecan-1 shedding is enhanced by LasA, a secreted virulence factor of Pseudomonas aeruginosa. J. Biol. Chem. 275:3057-3064. [DOI] [PubMed] [Google Scholar]
- 18.Park, S.-H., K. T. O'Neil, and H. Roder. 1997. An early intermediate in the folding reaction of the B1 domain of protein G contains a native-like core. Biochemistry 36:14277-14283. [DOI] [PubMed] [Google Scholar]
- 19.Peters, J. E., and D. R. Galloway. 1990. Purification and characterization of an active fragment of the LasA protein from Pseudomonas aeruginosa: enhancement of elastase activity. J. Bacteriol. 172:2236-2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sauter, N. K., T. Mau, S. D. Rader, and D. A. Agard. 1998. Structure of alpha-lytic protease complexed with its pro region. Nat. Struct. Biol. 5:945-950. [DOI] [PubMed] [Google Scholar]
- 21.Schad, P. A., and B. H. Iglewski. 1988. Nucleotide sequence and expression in Escherichia coli of the Pseudomonas aeruginosa lasA gene. J. Bacteriol. 170:2784-2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schweizer, H. P. 1991. Escherichia-Pseudomonas shuttle vectors derived from pUC18/19. Gene 97:109-112. [DOI] [PubMed] [Google Scholar]
- 23.Shinde, U., S. Chatterjee, and M. Inouye. 1993. Folding pathway mediated by an intramolecular chaperone. Proc. Natl. Acad. Sci. USA 90:6924-6928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shinde, U., and M. Inouye. 2000. Intramolecular chaperones: polypeptide extensions that modulate protein folding. Semin. Cell Dev. Biol. 11:35-44. [DOI] [PubMed] [Google Scholar]
- 25.Takagi, H., M. Koga, S. Katsurada, Y. Yabuta, U. Shinde, M. Inouye, and S. Nakamori. 2001. Functional analysis of the propeptides of subtilisin E and aqualysin I as intramolecular chaperones. FEBS Lett. 508:210-214. [DOI] [PubMed] [Google Scholar]
- 26.Wilderman, P. J., A. I. Vasil, Z. Johnson, M. J. Wilson, H. E. Cunliffe, I. L. Lamont, and M. L. Vasil. 2001. Characterization of an endoprotease (PrpL) encoded by a PvdS-regulated gene in Pseudomonas aeruginosa. Infect. Immun. 69:5385-5394. [DOI] [PMC free article] [PubMed] [Google Scholar]