Abstract
The SOS (for Salt Overly Sensitive) pathway plays essential roles in conferring salt tolerance in Arabidopsis thaliana. Under salt stress, the calcium sensor SOS3 activates the kinase SOS2 that positively regulates SOS1, a plasma membrane sodium/proton antiporter. We show that SOS3 acts primarily in roots under salt stress. By contrast, the SOS3 homolog SOS3-LIKE CALCIUM BINDING PROTEIN8 (SCABP8)/CALCINEURIN B-LIKE10 functions mainly in the shoot response to salt toxicity. While root growth is reduced in sos3 mutants in the presence of NaCl, the salt sensitivity of scabp8 is more prominent in shoot tissues. SCABP8 is further shown to bind calcium, interact with SOS2 both in vitro and in vivo, recruit SOS2 to the plasma membrane, enhance SOS2 activity in a calcium-dependent manner, and activate SOS1 in yeast. In addition, sos3 scabp8 and sos2 scabp8 display a phenotype similar to sos2, which is more sensitive to salt than either sos3 or scabp8 alone. Overexpression of SCABP8 in sos3 partially rescues the sos3 salt-sensitive phenotype. However, overexpression of SOS3 fails to complement scabp8. These results suggest that SCABP8 and SOS3 are only partially redundant in their function, and each plays additional and unique roles in the plant salt stress response.
INTRODUCTION
Soil salinity is a major abiotic stress that reduces plant growth and ranks among the leading factors limiting agricultural productivity. The sessile nature of plants has favored the evolution of mechanisms to cope with varying salt concentrations in the environment. Na+ is not essential for plant growth, and under salt stress, it hinders uptake of the important mineral nutrient K+ and competes for its enzyme binding sites. Maintaining a high K+ and low Na+ homeostasis in the cytoplasm is thus essential for plant salt tolerance. In Arabidopsis thaliana, an ionic homeostasis regulatory pathway activated by salt stress has been identified through molecular and genetic characterization of several salt overly sensitive (sos) mutants that are defective in K+/Na+ homeostasis (Liu and Zhu, 1998; Liu et al., 2000; Shi et al., 2000).
The SOS pathway is known to be defined by three protein components, SOS1, SOS2, and SOS3. SOS3 encodes an EF hand–type calcium sensor with an N-myristoylation signal peptide (Liu and Zhu, 1998). Both calcium binding activity and N-myristoylation are essential for SOS3 function (Ishitani et al., 2000). In vitro, SOS3 physically interacts with and activates a Ser/Thr protein kinase, SOS2. Calcium is not required for the SOS2 and SOS3 interaction but was observed to enhance the phosphorylation of synthesized substrate p3 by the SOS2-SOS3 complex (Halfter et al., 2000). One of the downstream targets of the SOS3-SOS2 complex is SOS1, a plasma membrane–localized sodium/proton antiporter (Shi et al., 2000; Qiu et al., 2002; Quintero et al., 2002).
Genetic evidence indicates that SOS3 and SOS2 function by activating SOS1. Mutations in SOS1, SOS2, or SOS3 all reduce the Na+/H+ exchange activity, and a constitutively active SOS2 enhances Na+/H+ exchange activity in a SOS1-dependent and SOS3-independent manner (Qiu et al., 2002). In yeast, SOS3 recruits SOS2 to the plasma membrane, and SOS1 increases NaCl tolerance in yeast mutants lacking Na+ transporters. Only when both SOS2 and SOS3 exist together does the system function to further increase SOS1-dependent NaCl resistance due to the phosphorylation of SOS1 by the SOS2-SOS3 complex (Quintero et al., 2002). However, evidence is lacking for an interaction of SOS2 and SOS3 in planta.
The growth inhibition of sos1 and sos2 mutants is more severe than that of sos3 under mild salt stress (Zhu et al., 1998). SOS1 and SOS2 are expressed in both root and shoot tissues, suggesting that they function in diverse tissue types (Liu et al., 2000; Shi et al., 2002). However, our examination of public microarray results (http://jsp.weigelworld.org/expviz/expviz.jsp) indicated that SOS3 is mainly expressed in the root tissue. It thus seems plausible that an additional calcium sensor(s) may exist to regulate SOS2 in shoot tissues as well. Here, we report that SOS3-LIKE CALCIUM BINDING PROTEIN8 (SCABP8) interacts with and recruits SOS2 to the plasma membrane in vivo and is required for salt tolerance in shoots.
RESULTS
Expression of SOS2, SOS3, and SCABP8
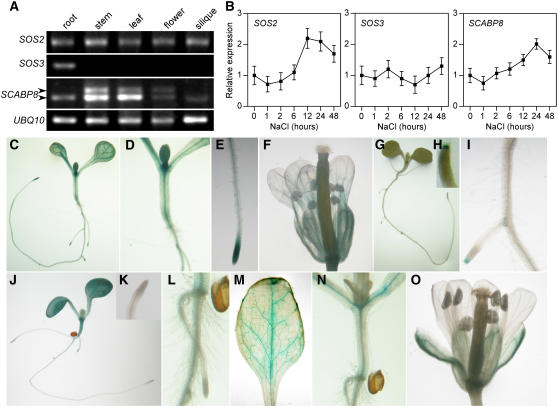
Liu et al. (2000) and Shi et al. (2002) reported that SOS1 and SOS2 were expressed in both root and shoot tissues in Arabidopsis. By contrast, an examination of the microarray results in the public domain (http://jsp.weigelworld.org/expviz/expviz.jsp) indicated that SOS3 was strongly expressed in roots but only at background level in shoots, suggesting that SOS3 mainly functions in root. We therefore speculated that an SOS3 homolog or SOS2-regulatory protein may function in shoots to regulate SOS2. Interestingly, we noticed that of the other nine SOS3-like calcium binding proteins, SCABP8 is the only one expressed strongly in shoots but very weakly in roots in this microarray data set. To confirm these results, RT-PCR analysis was performed. The transcripts of SOS2 could be amplified from roots, stems, leaves, flowers, and siliques, whereas SOS3 was only detected in roots. SCABP8 was expressed at a high level in leaves and stems but at a very low level in roots, flowers, and siliques (Figure 1A), consistent with microarray data in the public domain.
Figure 1.
Expression of SOS2, SOS3, and SCABP8 in Arabidopsis.
(A) Expression of SOS2, SOS3, and SCABP8 in root, stem, leaf, flower, and silique determined by 30 cycles of RT-PCR with gene-specific primers. UBQ10 was a loading control.
(B) Expression of SOS2, SOS3, and SCABP8 in Arabidopsis seedlings in response to salt stress. Error bars represent sd (n = 3).
(C) to (F) SOS2 promoter-GUS expression in 10-d-old wild-type seedling (C), stem (D), root tip (E), and flower (F).
(G) to (I) SOS3 promoter-GUS expression in 10-d-old wild-type seedling (G), root tip (H), and root (I).
(J) to (O) SCABP8 promoter-GUS expression in 10-d-old wild-type seedling (J), root tip (K), root (L), leaf (M), stem (N), and flower (O).
To examine whether expression of SOS2, SOS3, and SCABP8 is induced by NaCl treatment, total RNA was extracted from wild-type plants treated with 100 mM NaCl for 0, 1, 2, 6, 12, 24, and 48 h. Consistent with previous results (Liu et al., 2000), the expression of SOS2, but not SOS3, was induced by NaCl treatment (Figure 1B). The transcript of SCABP8 was slightly induced 6 h after treatment and decreased after 24 h (Figure 1B).
The SCABP8 gene contains nine exons and eight introns and encodes a 246–amino acid protein with a predicted molecular mass of 28.3 kD and isoelectric point of 4.3. Two SCABP8 cDNAs were amplified and sequenced. Two bands pertaining to SCABP8 were amplified from aerial parts, whereas only the lower band yielded in roots (Figure 1A). The shorter sequence is identical to At4g33000.2 (http://www.arabidopsis.org), while the longer retains its seventh intron, which contains a premature stop codon that results in a truncated protein 194 amino acids long. Overexpression of this mis-spliced SCABP8 cDNA could not complement the scabp8 salt sensitivity phenotype (data not shown).
To further determine the tissue specificity of these genes, promoter regions of SOS2 (1.6 kb), SOS3 (1.4 kb), and SCABP8 (1.7 kb) were fused to a β-glucuronidase (GUS) reporter gene. The resulting constructs were transferred into an Arabidopsis Columbia (Col-0) background. For each construct, 12 independent F2 transgenic lines were analyzed by a GUS staining assay. Though the intensity of the GUS staining from each of the independent lines was variable, the tissue-specific localization was the same. As shown in Figures 1C to 1F, GUS driven by the SOS2 promoter was expressed in the entire young seedling, leaf, stem, and flower and was particularly strong in root tips. Similar expression has been detected in SOS1 promoter-GUS transgenic plants (Shi et al., 2002). Consistent with microarray and RT-PCR results, GUS expression driven by the SOS3 promoter was only detected in root and very strongly in the root tip (Figures 1G to 1I) but not other tissues. However, SCABP8-GUS was expressed only in the stem, leaf, flower, and weakly in root but not in the root tip (Figures 1J to 1O). The combined pattern of SCABP8 and SOS3 expression overlaps with that of SOS2 and suggests that SCABP8 and SOS3 may assume similar roles in different tissue types to activate SOS2 in response to salt stress.
scabp8 Is Hypersensitive to Salt in Shoot Tissues
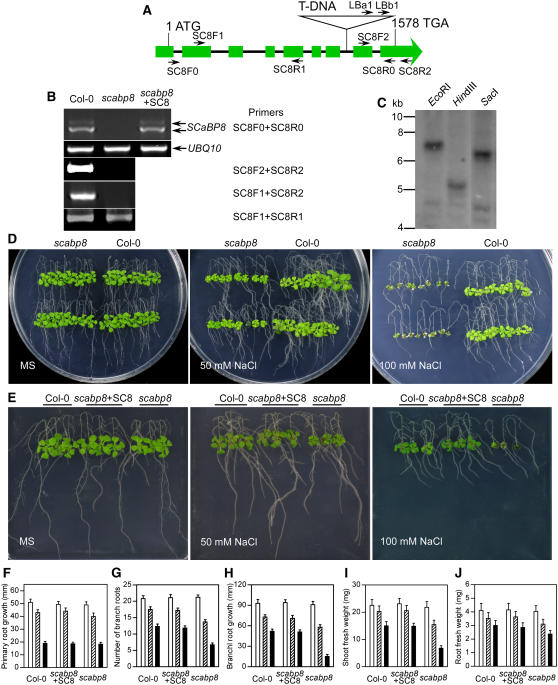
To test the hypothesis that SCABP8 is a tissue-specific activator of SOS2, the Arabidopsis mutant line SALK_056042 with a T-DNA insertion in the SCABP8 gene was obtained from the ABRC (Alonso et al., 2003). The T-DNA insertion is located in the seventh intron of the SCABP8 gene and was confirmed by PCR using a SCABP8-specific primer and T-DNA left border primers (data not shown; Figure 2A). The insertion was further confirmed by a DNA gel blot probed with a neomycin phosphotransferase coding sequence (nptII). Genomic DNA was extracted from scabp8, digested by three restriction enzymes, EcoRI, HindIII, and SacI, which are the enzymes with a single digesting site in the pBIN-pROK2 vector, and subjected to DNA gel blot analysis. Only one strongly hybridizing band from each digestion was observed with different molecular sizes (Figure 2C). Two weaker bands of similar sizes were detected in all three digestions, suggesting they were nonspecific. These data suggest that there was a single T-DNA insertion in the scabp8 mutant. To precisely map the border sequences flanking the T-DNA insertion, PCR reactions were performed with combinations of the primers annealing upstream or downstream of the T-DNA insertion into the SCABP8 gene and primers specific to the left border or right border of the T-DNA. The PCR products were sequenced, and the results indicated that the T-DNA insertion had generated an 18-bp deletion from 1264 to 1281 bp downstream of the SCABP8 translation start codon, in the seventh intron. This insertion point matches the sequence shown on the SALK website. The absence of a full-length SCABP8 transcript in the scabp8 mutant was confirmed by RT-PCR analysis (Figure 2B). However, a truncated SCABP8 transcript corresponding to a fragment before the T-DNA insertion, but not after, still existed in the scabp8 mutant (Figure 2B). Overexpression of this truncated SCABP8 transcript did not rescue the scabp8 salt phenotype (data not shown).
Figure 2.
scabp8 Is a Salt-Sensitive Mutant.
(A) Structure of the SCABP8 gene. Filled green boxes indicate the exons, and the lines between boxes indicate introns. The primers used to identify T-DNA insertion and transcription of SCABP8 are marked with arrows.
(B) Expression of SCABP8 in the wild type, the scabp8 mutant, and the complementary line. RT-PCR products were amplified from scabp8 with 30 cycles. The primer positions were identified in (A). UBQ10 was used as a loading control.
(C) DNA gel blotting analysis of scabp8 genomic DNA probed with NPTII.
(D) Five-day-old wild-type and scabp8 seedlings were transferred onto MS medium containing 0, 50, and 100 mM NaCl, respectively. The pictures were taken after 10 d of treatment.
(E) Complementation of scabp8 by the wild-type SCABP8 gene. Five-day-old wild-type (left), transgenic scabp8 plants containing the wild-type SCABP8 gene (center), and scabp8 (right) seedlings were grown on MS medium with or without NaCl.
(F) to (J) Primary root elongation (F), number of lateral roots (G), lateral root elongation (H), shoot fresh weight (I), and root fresh weight (J) were measured at day 10 after transfer. Open bars, without NaCl treatment; hatched bars, 50 mM NaCl treatment; closed bars, 100 mM NaCl treatment. Error bars represent sd (n > 15).
To test the salt sensitivity of the scabp8 mutant, 5-d-old mutant and wild-type plants were transferred to Murashige and Skoog (MS) medium with or without NaCl. Indeed, scabp8 was hypersensitive to NaCl, mainly in the shoot tissues, but developed normally when grown on MS medium lacking NaCl (Figure 2D). On MS medium containing 50 mM NaCl, the primary root growth of scabp8 was indistinguishable from the wild type (Figures 2D and 2F). The number and length of secondary roots were reduced relative to the wild type, but the overall root fresh weight was reduced only ∼12% (Figures 2D, 2G, 2H, and 2J). However, shoot growth was more severely inhibited (30% fresh weight reduction) and leaves were smaller and darker in color compared with the wild type (Figures 2D, 2E, and 2I). On 100 mM NaCl medium, growth inhibition was more pronounced. The shoot fresh weight of scabp8 was less than half of the wild type. The lateral root growth was also further restricted, as evidenced by reduced fresh weight and shorter and lesser lateral roots. However, the primary root growth was significantly less affected (Figures 2E and 2F).
To complement the scabp8 salt-sensitive phenotype, a 3783-bp genomic DNA fragment corresponding to the sequence from 1736 bp upstream of the SCABP8 translation start codon to 469 bp downstream of the stop codon was excised from the BAC clone F26P21 with XbaI and PstI. The fragment was then cloned into the pCAMBIA1200 vector, and the resulting construct was transferred into the scabp8 mutant. All 12 independent T2 transgenic lines tested under salt treatment showed growth patterns similar to wild-type plants (Figures 2E to 2J), demonstrating that the NaCl sensitivity phenotype of scabp8 is due to the loss of function of SCABP8. The transcription level of SCABP8 in these transgenic lines was similar to the wild type (Figure 2B).
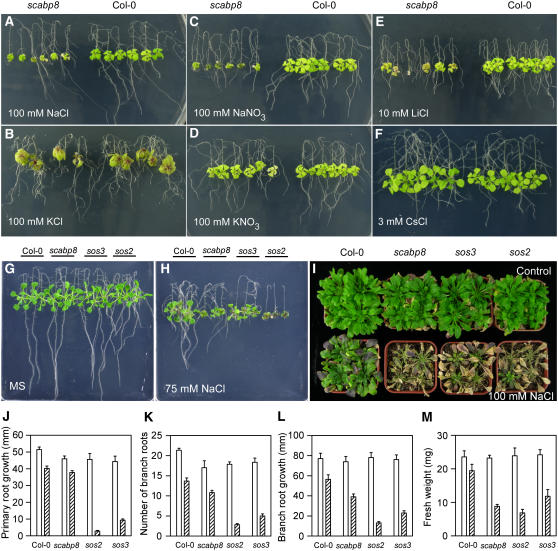
To determine whether the growth defects of the scabp8 mutant are specific to the sodium response, wild-type and scabp8 seedlings were treated with various salts, polyethylene glycol, mannitol, and plant hormones. The results are shown in Figure 3. Though scabp8 seedlings were very sensitive to 100 mM NaCl and NaNO3 (Figures 3A and 3C), they failed to show significant growth differences with the wild type on the medium containing 100 mM KCl or KNO3 (Figures 3B and 3D). The scabp8 seedlings were also as tolerant as the wild type to 15 and 20% polyethylene glycol, 150 and 200 mM mannitol, and plant hormones abscisic acid, indole-3-acetic acid, and gibberellic acid (data not shown). Individual sos1, sos2, and sos3 mutants are hypersensitive to Na+ and Li+ but not to Cs+ (Zhu et al., 1998). Our findings indicate that the scabp8 mutant behaves similarly (Figures 3E and 3F). Though more sensitive to 10 mM LiCl than the wild type (Figure 3E), the scabp8 mutant did not show obvious differences in growth on MS medium with 3 mM CsCl, which inhibited wild-type growth by half (Figure 3F). The similar behaviors of sos and scabp8 mutants under ionic stresses suggest that scabp8 may perform a critical function in the SOS pathway.
Figure 3.
Na+ and Li+ Are the Toxic Ions to scabp8.
(A) to (F) Five-day-old seedlings of the wild type and scabp8 were transferred onto MS medium with 100 mM NaCl (A), 100 mM KCl (B), 100 mM NaNO3 (C), 100 mM KNO3 (D), 10 mM LiCl (E), and 3 mM CsCl (F). The pictures were taken at 10 d after transfer.
(G) to (I) Salt sensitivity of sos2, sos3, and scabp8 mutants. Five-day-old seedlings of the wild type, sos2, sos3, and scabp8 were transferred onto MS medium without (G) and with (H) 75 mM NaCl or transferred into soil (I). After 1 month of growth in soil, the plants were then treated with 100 mM NaCl for 2 weeks and photographed.
(J) to (M) Primary root elongation (J), lateral root number (K), lateral root elongation (L), and fresh weight (M) regarding to (G) and (H) were measured at day 10 after transfer. Open bars, without NaCl treatment; hatched bars, 75 mM NaCl treatment. Error bars represent sd (n > 15).
Compared with sos3, salt sensitivity of scabp8 is more prominent in shoot tissues. On a MS medium, sos2, sos3, scabp8, and wild-type plants showed similar shoot growth and root bending patterns (Figure 3G). When grown on MS medium containing 75 mM NaCl, both root bending and shoot growth of sos2 were dramatically reduced (Figure 3H). Root growth was completely arrested and fresh weight was less than one-third that of the wild type (Figure 3M). Both primary and lateral root growth of sos3 were also reduced relative to the wild-type level, though less severely than in sos2. The fresh weight of seedlings, which mainly represents shoot growth, was the highest in sos3 among the three mutants (Figure 3M), suggesting that SOS3 function was less critical to mediate salt response in shoots. However, scabp8 showed severely reduced shoot growth when compared with the wild type or sos3. The fresh weight of scabp8 was reduced to less than half of that of the wild type and was also less than that of sos3 (Figures 3H and 3M). By contrast, both the primary root length and lateral root length and number of scabp8 under this treatment were reduced much less than in sos3 (Figures 3J to 3L). When 4-week-old soil-grown plants were treated with 100 mM NaCl for 2 weeks, the growth of wild-type plants was slightly inhibited and old leaves accumulated anthocyanin. The older leaves from all three mutants were bleached. A few young leaves of sos3 and sos2 survived but none from scabp8 (Figure 3I).
We further tested whether expression of stress-responsive genes is affected by scabp8 mutation in response to NaCl in an RNA gel blot assay. These genes included RD29A, Cor15A, Cor47, and Kin1. No significant difference was detected between the mutant and the wild type (data not shown).
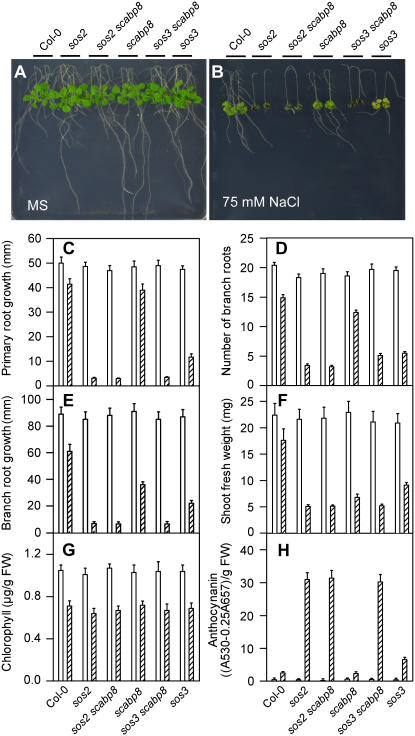
Genetic Interaction between SCABP8 and SOS2
The sos2 sos3 double mutant shows a salt-sensitive phenotype similar to sos2 and greater than sos3, suggesting that SOS2 and SOS3 genetically function in the same pathway and that SOS2 has additional functions in salt tolerance relative to SOS3 (Halfter et al., 2000). To determine whether SCABP8 and SOS2 interact genetically, the scabp8 T-DNA insertion line was crossed with sos2 and sos3 mutants. From ∼100 F2 plants of each cross, five sos2 scabp8 and seven sos3 scabp8 double mutants were identified. The double mutants were then crossed into their parental backgrounds to confirm the genotypes (the sos2-2 allele bears an untagged 2-bp deletion that prevented PCR-based genotyping). We transferred 5-d-old wild-type, mutant, and double mutant seedlings onto MS medium with or without 75 mM NaCl. In the absence of NaCl treatment, none of the mutants showed any significant growth differences compared with the wild type (Figure 4A). As in the sos2 sos3 double mutant (Halfter et al., 2000), sos2 scabp8 seedlings also displayed salt-sensitive features similar to sos2, shown in shoot and root growth, and chlorophyll and anthocynanin accumulation (Figures 4B to 4H). No additional phenotypic difference was observed between sos2 and sos2 scabp8, suggesting that SOS2 and SCABP8 function in the same pathway. Under 75 mM NaCl treatment, both root and shoot growth of the sos3 scabp8 double mutant plants were more severely reduced compared with the sos3 and scabp8 single mutants. The phenotype of sos3 scabp8 showed slightly higher lateral root number than sos2 but otherwise resembled sos2 (Figures 4B to 4H). These results further support a model in which SCABP8 and SOS3 function together to modulate the activity of SOS2.
Figure 4.
Genetic Interaction of scabp8 with sos2 or sos3.
Five-day-old seedlings of the wild type, sos2, sos2 scabp8, scabp8, scabp8 sos3, and sos3 were transferred onto MS medium without (A) and with (B) 75 mM NaCl. The pictures were taken at day 10. Primary root elongation (C), lateral root number (D), lateral root elongation (E), fresh shoot weight (FW) (F), chlorophyll (G), and anthocynanin contents (H) were measured at day 10 after transfer. Open bars, without NaCl treatment; hatched bars, 75 mM NaCl treatment. Error bars represent sd (n > 15).
Interaction of SOS2 and SCABP8
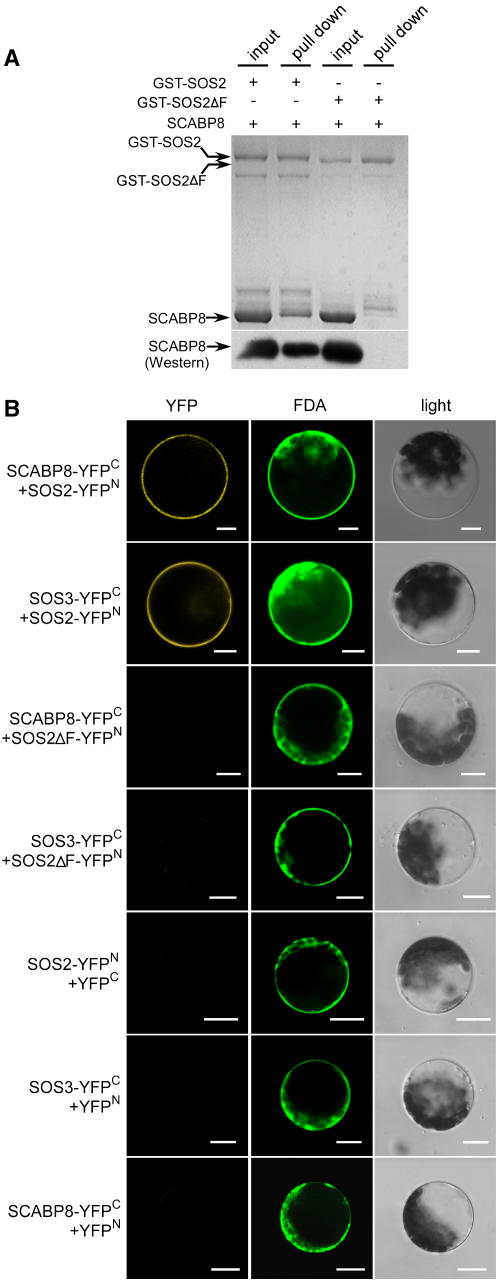
If SCABP8 has partial functional redundancy with SOS3 and regulates SOS2 in the tissues that do not express SOS3, SCABP8 also would be expected to interact with and activate SOS2. This was tested by cloning the SCABP8 cDNA into the pGEX-6P-1 vector containing a glutathione S-transferase (GST) tag, with similar constructs made for SOS2 and SOS3. The FISL motif in the SOS2 regulatory domain is necessary and sufficient for SOS3 interaction (Guo et al., 2001). As a negative control, an SOS2ΔFISL fragment (SOS2 with deletion of the FISL motif) was also generated by PCR mutagenesis and cloned into the pGEX-6P-1 vector. The GST-SCABP8, -SOS3, -SOS2, and -SOS2ΔFISL fusion proteins were purified from Escherichia coli strain BL21(DE3) using glutathione affinity chromatography. SOS3 and SCABP8 were digested from their GST fusion protein by PreScission protease and subjected to in vitro pull-down assays. As expected, SOS3 was pulled down by GST-SOS2 but not by GST-SOS2ΔFISL (data not shown). Similarly, the FISL motif was also required for the interaction of SOS2 with SCABP8. SCABP8 was pulled down by GST-SOS2 but not by GST-SOS2ΔFISL (Figure 5A). To confirm whether or not the pulled-down band was SCABP8, 1% of the input and pull-down samples were subjected to protein gel blot analysis with anti-SCABP8 antibodies. The result showed that SCABP8 protein was only detected in GST-SOS2 samples but not in GST-SOS2ΔFISL (Figure 5A). To further test whether the interaction of SOS2 with SOS3 or SCABP8 is calcium dependent, 250 μM calcium was included in the buffer in the presence or absence of 500 μM EGTA. The interaction of SOS2 and SCABP8 was not affected by the presence of free calcium (data not shown). This result is consistent with the previous findings that the SOS3 and SOS2 interaction in vitro does not require calcium (Halfter et al., 2000).
Figure 5.
Interaction of SOS2 with SOS3 and SCABP8.
(A) SOS2 interacts with SCABP8. Lane 1, input of 1 μg of GST-SOS2 and 5 μg of SCABP8; lane 3, input of 1 μg of GST-SOS2ΔFISL and 5 μg of SCABP8; lanes 2 and lane 4, the pull-down products of lines 1 and 3 after washing four times. Protein gel blot analysis of 1% of the input proteins and pull-down proteins with anti-SCABP8 antibodies (bottom).
(B) SOS2 interacts with SOS3 and SCABP8 in vivo. Different combinations of the N-YFP and C-YFP fusion constructs as indicated were cotransferred into Arabidopsis protoplasts. The images were collected under the Zeiss confocal microscope. Left, YFP signal; right, the light microscope pictures; middle, the protoplast stained with FDA (green). Bars = 10 μm.
To determine the interaction of SOS2 and SCABP8 in vivo, we cloned SOS2 or SOS2ΔFISL cDNA into the split–yellow fluorescence protein (YFP) vector pUC-SPYNE (Walter et al., 2004) and cloned SCABP8 or SOS3 into the pUC-SPYCE vector. The resulting plasmids and empty vectors were purified by CsCl gradient centrifugation and used for transient transfection assays with protoplasts. Different combinations of SOS2/SOS2ΔFISL/pUC-SPYNE and SOS3/SCABP8/pUC-SPYCE were cotransfected into the Col-0 leaf protoplasts. The protoplasts were analyzed under a confocal microscope to detect YFP bimolecular complementation and then stained with 5 μg/mL fluorescein diacetate (FDA) and 1 μg/mL propidium iodide to check protoplast viability. The YFP signal was detected in SOS3-YFPC/SOS2-YFPN and SCABP8-YFPC /SOS2-YFPN cotransfection assays in protoplasts stained normally with FDA (Figure 5B). However, we were not able to observe interaction between SOS2ΔFISL and SOS3 or SCABP8, although >90% of protoplasts were stained with FDA after transfection. The YFP signal was not detected with any single construct individually or with the combinations of SOS2 and pUC-SPYCE or SOS3/SCABP8 and pUC-SPYNE. Together, these results indicate that SOS2 interacts with SOS3 and SCABP8 in vivo and that the FISL motif is required for these interactions (Figure 5B; data not shown). Images obtained by bimolecular complementation were consistent with the localization of protein complexes near or at the plasma membrane, although we could not resolve whether these interactions between SOS2 and SOS3 or SCABP8 occurred in other cell compartments also.
SCABP8 Recruits SOS2 to the Plasma Membrane
Quintero et al. (2002) showed that SOS3 recruits SOS2 to the plasma membrane in a yeast system. To determine if SCABP8 recruited SOS2 to the plasma membrane in planta, YFP-SOS2 and YFP-SCABP8 fusion constructs were transferred to Col-0, scabp8, and sos2 plants. YFP-SOS2 and YFP-SCABP8 rescued the sos2 and scabp8 salt-sensitive phenotypes, respectively, and did not cause additional phenotypes, suggesting that both YFP fusion proteins are functional in plants (data not shown). The YFP-SOS2 and YFP-SCABP8 localization was imaged in the T2 generation using a confocal microscope (see Supplemental Figure 1 online). Twelve independent T2 transgenic lines expressing the individual constructs were analyzed in different genetic backgrounds. In Col-0, both YFP-SOS2 and YFP-SCABP8 were localized to the cell periphery, and a low level of YFP signal also could be detected at the cytoplasm (see Supplemental Figures 1A and 1C online). To exclude the possibility of cell wall association of SOS2 and SCABP8, the transgenic plants were plasmolyzed by mannitol treatment. Both YFP-SOS2 and YFP–SCABP8 signals were detached from the cell wall (see Supplemental Figures 1B and 1D online). Transgenic plants expressing YFP alone (see Supplemental Figures 1G and 1H online), the cytoplasmic acidic ribosomal green fluorescent protein (GFP) (see Supplemental Figures 1I to 1J) (Cutler et al., 2000), and the plasma membrane water channel protein (PIP2a)-GFP (see Supplemental Figures 1K and 1L online) (Cutler et al., 2000) were used as controls. When YFP-SOS2 was expressed in the scabp8 mutant, YFP-SOS2 was detected more abundantly in the cytoplasm in comparison to the wild-type background (see Supplemental Figure 1E online). After mannitol treatment, YFP-SOS2 was clearly aggregated in the cytosol (see Supplemental Figure 1F online). These determinations were conducted in the upper part of seedling roots where the analysis of promoter-GUS fusions demonstrated a significant expression of SOS2 and SCABP8 genes, whereas that of SOS3 was undetectable (Figure 1). These results indicate that although the localization of SOS2 and SCABP8 was not exclusively at the plasma membrane, SCABP8 enhanced the fraction of SOS2 bound to the plasma membrane.
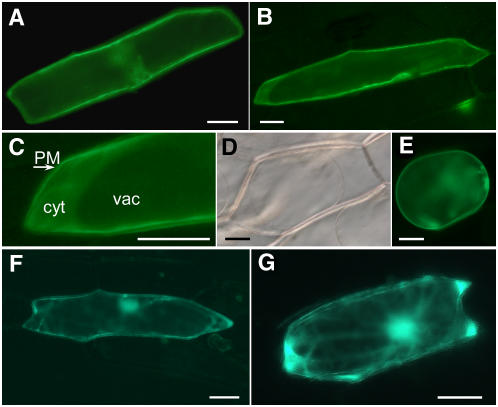
To further test the localization of SCABP8 in plant cells, a translational fusion was constructed in which GFP was added to the C terminus of SCABP8. The chimeric construct was delivered into onion epidermal cells by particle bombardment, and transient expression of SCABP8-GFP was monitored by epifluorescence. As shown in Figure 6A, SCABP8-GFP was predominantly localized to the plasma membrane at 20 h after DNA delivery. However, at longer times (40 h), significant labeling of the cytoplasm was also observed (Figures 6B and 6C). The large vacuole of epidermal cells confined most of the cytoplasm to a thin layer, except in places where the tonoplast was separated from the cell periphery by nuclei and pockets of cytosol. Plasmolization with 0.5 M sorbitol permitted a clear visualization of the detached plasma membrane labeled with SCABP8-GFP (Figures 6D and 6E). Removal of a hydrophobic domain near the N terminus of SCABP8 (see below) resulted in a much more prominent labeling of cytosol and nuclei (Figures 6F and 6G) similar to GFP alone (data not shown). These results indicate that SCABP8 is partially associated with the plasma membrane and that this interaction requires the hydrophobic N terminus of the protein.
Figure 6.
Transient Expression of GFP-Tagged SCABP8 in Onion Epidermal Cells Visualized by Epifluorescence Microscopy.
(A) Epifluorescence after 20 h of transient expression of SCABP8-GFP.
(B) Epifluorescence after 40 h of transient expression of SCABP8-GFP.
(C) Magnification of the cell shown in (B). PM, plasma membrane; cyt, cytosol; vac, vacuole.
(D) Clear field image of cells plasmolized with 0.5 M sorbitol for 10 min.
(E) Epifluorescence of the same cell shown in (D).
(F) Fluorescence pattern of truncated SCABP8ΔN fused to GFP.
(G) Fluorescence pattern of SCABP8ΔN-GFP.
Bars = 50 μm.
We were not able to detect YFP-SOS2 signal in shoot tissues of transgenic plants. To test the subcellular localization of SOS2 in leaf cells, a YFP-SOS2 plasmid construct was transferred into leaf protoplasts derived from wild-type and scabp8 plants. The YFP-SOS2 signal was observed at both the cell periphery and the cytoplasm in wild-type protoplasts. However, in the scabp8 protoplast, YFP-SOS2 was more dispersed throughout the cytoplasm (see Supplemental Figures 2B and 2C online). When YFP-SCABP8, YFP-AtVAM3 (Uemura et al., 2002), and CFP-PIP2a were expressed in wild-type leaf protoplasts, YFP-AtVAM3 signal was detected in the vacuolar membrane, CFP-PIP2a was in the plasma membrane, and YFP-SCABP8 showed a pattern similar to that of YFP-SOS2 in scabp8 (see Supplemental Figures 2A to 2E online). Coexpression of CFP-PIP2a and YFP-SCABP8 indicated that part of the YFP and CFP signals overlapped in the plasma membrane, although YFP-SCABP8 was also detected in the cytoplasm (see Supplemental Figure 2F online). Similar results were obtained from the coexpression of CFP-PIP2a and YFP-SOS2 in wild-type protoplasts (see Supplemental Figure 2G online).
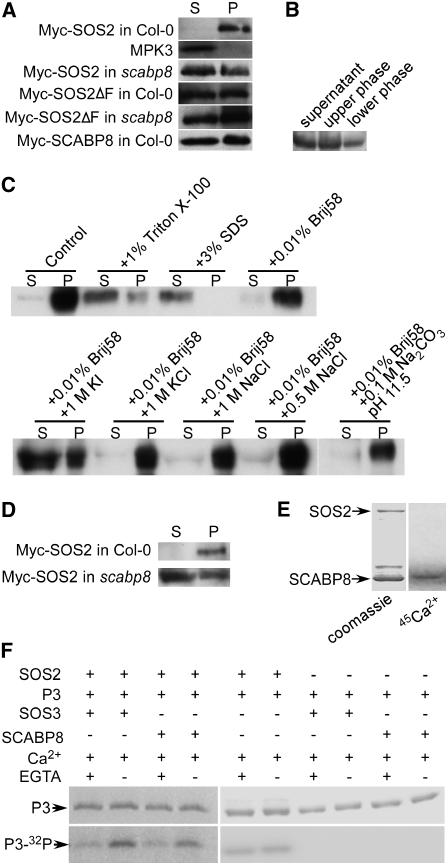
To test further the recruitment of SOS2 by SCABP8 to the plasma membrane, a myc tag in a cluster of six tandem repeats was fused to the N terminus of SOS2, SOS2ΔFISL, and SCABP8. The tagged proteins were expressed in leaf protoplasts of the wild type and scabp8 mutant. Pellets of microsomal membrane fraction and soluble fraction were prepared from the protoplasts and subjected to protein gel blot analysis with anti-myc antibody. The results are shown in Figure 7A. The tagged SOS2 protein was detected almost exclusively in the membranous fraction of wild-type plants. Using the same protein samples, MPK3 protein kinase was found in the soluble fraction. However, in the scabp8 mutant, myc-SOS2 was found in both soluble and microsomal fractions, consistent with the YFP-SOS2 results (see Supplemental Figure 1 online), showing that SCABP8 enhances the association of SOS2 with the membranes. When the FISL motif was removed from SOS2, myc-SOS2ΔFISL was also detected in both pellet and soluble fractions and independently of the presence or no presence of SCABP8, suggesting that the FISL motif is required for the full association of SOS2 with the membrane, possibly through its interaction with SCABP8. However, these results also indicate that SOS2 can associate with membranes by a mechanism independent of SCABP proteins and the FISL interaction domain.
Figure 7.
SCABP8 Binds to Calcium, Increases SOS2 Activity, and Is Required for Association of SOS2 with the Plasma Membrane.
(A) Myc-SOS2, -SOS2ΔF, and -SCABP8 plasmids were transferred into wild-type and scabp8 leaf protoplasts. The membrane fraction (P) and soluble fraction (S) were extracted from the protoplasts and probed with anti-myc or anti-MPK3 antibodies.
(B) Vesicles from Arabidopsis overexpressing 35S:myc-SCABP8 were prepared by two-phase partitioning. Five micrograms of proteins from the soluble fraction, upper phase, and lower phase were subjected to protein gel blot analysis with anti-Myc antibody.
(C) Plasma membrane–enriched vesicles were treated with 0.01% Brij58, 1% Triton X-100, 3% SDS, 1 M KI, 1 M KCl, 1 M NaCl, 0.5 M NaCl, and 0.1 M Na2CO3, pH 11.5, for 30 min. After centrifugation at 150,000g at 4°C for 60 min, equal aliquots of supernatants (S) and pellets (P) were loaded and detected with anti-Myc antibody.
(D) Plasma membrane–enriched vesicles were purified by a two-phase method from protoplasts transformed with 35S:myc-SOS2, and 5 μg of soluble proteins (S) and 1 μg of plasma membrane–enriched proteins (P) were loaded and detected with anti-Myc.
(E) SCABP8 binds calcium. GST-SOS2 and SCABP8 proteins were separated by SDS-PAGE and stained with Coomassie blue (left) or electroblotted onto a cellulose membrane, incubated with 45Ca2+, and autoradiographed (right).
(F) Phosphorylation of GST-P3 by SOS2 kinase. Coomassie blue–stained GST-P3 in an SDS-PAGE gel (top); GST-P3 phosphorylation activity (bottom).
We also generated transgenic plants expressing the tagged myc-SCABP8 protein to be able to purify large quantities of purified plasma membrane fractions. The plasma membrane vesicles were isolated by the two-phase method (Qiu et al., 2002), and membrane purity was determined by measuring the inhibition of H+-ATPase activity with different H+-ATPase inhibitors (see Supplemental Table 1 online). The results showed that >88% ATPase activity in the vesicles obtained from the upper phase were blocked by 1 mM Na3VO4, an inhibitor of plasma membrane H+-ATPase. Five micrograms of protein from upper- and lower-phase fractions was subjected to a protein gel blot assay with anti-myc antibody. Tagged myc-SCABP8 was detected in the plasma membrane–enriched fraction (upper phase), the lower phase enriched in other cell membranes, and the soluble protein fraction (Figure 7B). However, myc-SCABP8 appeared to be moderately more abundant in the plasma membrane–enriched upper phase than in the lower phase. This result and presence of myc-SCABP8 in the soluble protein fraction are consistent with the microscopy localization of SCABP8.
Because plasma membrane vesicles in the upper phase are mainly in right-side-out form, they could embrace some cytoplasmic proteins. We used 0.01% (w/v) Brij58 to treat the upper-phase vesicles to turn right-side-out vesicles to inside-out for releasing the embraced cytoplasmic proteins, and vesicles and soluble proteins were subjected to a protein gel blot assay with anti-myc antibody. Myc-SCABP8 protein was mainly present in the pellet fraction (Figure 7C). To further test how tightly SCABP8 associates with the plasma membrane, the upper-phase vesicles were treated with 0.01% Brij58 and with either 0.5 M NaCl, 1 M NaCl, 1 M KCl, 1 M KI, 0.1 M Na2CO3 at pH 11.5, 1% Triton X-100, or 3% SDS, respectively. Salt treatments with NaCl or KCl and alkaline treatment with Na2CO3 at pH 11.5 were not able to disassociate SCABP8 from the vesicles; the nonionic detergent Triton X-100 and a chaotropic reagent KI could partially disassociate SCABP8, and only the anionic detergent SDS completely removed SCABP8 from the vesicles (Figure 7C). These data demonstrate that SCABP8 is tightly associated with cellular membranes.
We transfected a myc-SOS2 construct into a large amount of leaf protoplasts of both the wild type and scabp8 mutant. Vesicles enriched in plasma membrane were purified by the two-phase method. The membrane purity was again determined by measuring the inhibition of ATPase activity with different H+-ATPase inhibitors (see Supplemental Table 2 online). One microgram of upper-phase protein and 5 μg of soluble proteins were subjected to a protein gel blot assay with anti-myc antibody. The results showed that myc-SOS2 was detected in both plasma membrane–enriched and soluble fractions in the scabp8 mutant but mainly in the vesicle fraction of wild-type protoplasts (Figure 7D). These results indicate that SOS2 is bound to the plasma membrane of leaf protoplasts and that it is partially released in the absence of SCABP8.
Recruitment of SOS2 to the Plasma Membrane and Activation of SOS1 in Yeast
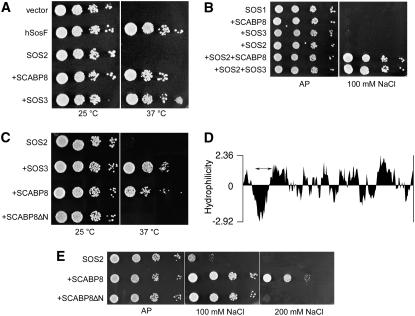
Results in planta indicate that SCABP8 enhances the recruitment of SOS2 to the plasma membrane, albeit other mechanisms for membrane targeting of SOS2 acting independently of SCABP8 are also in place. Using a reconstituted SOS system in yeast, Quintero et al. (2002) showed that SOS3 activates and recruits SOS2 to the plasma membrane to facilitate the phosphorylation and stimulation of the Na+ efflux protein SOS1. To test whether SCABP8 achieves similar functions, we used the Sos Recruitment System (SRS) to monitor the recruitment of SOS2 to the plasma membrane of yeast in the absence of other ancillary elements specific to plants. A full-length SOS2 protein was translationally fused to the human hSos protein, a functional homolog of the yeast Ras guanyl nucleotide exchange factor CDC25. When protein–protein interaction occurs at the plasma membrane, recruitment of the human hSos protein restores Ras activation, allowing the growth at 37°C of the thermosensitive yeast mutant cdc25-2. As shown in Figure 8A, expression of the hSos-SOS2 reporter alone did not restore growth at the restrictive temperature, indicating a cytosolic localization, but coexpression of either SCABP8 or SOS3 resulted in the recruitment of SOS2 to the plasma membrane. Moreover, SCABP8 was fully competent for the activation of SOS1 when coexpressed with SOS2 (Figure 8B), as demonstrated by growth in plates containing 100 mM NaCl. Similarly to SOS3, SCABP8 alone or in combination with SOS2 did not affect the salt tolerance of yeast in the absence of SOS1 (data not shown). Together, these results indicate that SCABP8 and SOS3 perform similar roles in the activation of SOS1 by SOS2 and are in agreement with the sodium-sensitive phenotypes of sos2, sos3, and scabp8 mutant plants.
Figure 8.
SCABP8 Recruits SOS2 to the Plasma Membrane and Activates SOS1 in Yeast.
(A) Recruitment of SOS2 to the plasma membrane. A yeast cdc25-2 mutant was transformed with an empty pADNs plasmid (vector), a full-length human hSos protein as positive control (hSosF), and the hSos:SOS2 translational fusion (SOS2) in the same vector pADNs. Cells expressing hSos:SOS2 were cotransformed with plasmids directing the expression of SCABP8 (+SCABP8) or SOS3 (+SOS3). Five microliters of serial decimal dilutions were spotted onto YPD plates and incubated at 25 or 37°C for 3 d.
(B) Activation of SOS1. Yeast AXT3K cells expressing SOS1 and transformed with the indicated combinations of Arabidopsis genes were grown overnight in liquid AP medium with 1 mM KCl. Five microliters of serial decimal dilutions were spotted onto plates of the same medium (AP) or supplemented with 100 mM NaCl. Plates were incubated at 28°C for 4 d.
(C) Truncated SCABP8 does not recruit SOS2 to the plasma membrane. Mutant cdc25-2 expressing the hSos:SOS2 translational fusion (SOS2) was cotransformed with SOS3 (+SOS3), full-length SCABP8 (+SCABP8), or a truncated form of SCABP8 (+SCABP8ΔN) and processed as in (A).
(D) Hydrophilicity plot of SCABP8. The internal deletion from residues Val-13 to Val-35 spanning the hydrophobic domain is indicated by the double arrow.
(E) Truncated SCABP8 interacts with SOS2 to activate SOS1. Yeast AXT3K cells coexpressing SOS1 and SOS2 (SOS2) were transformed with wild-type SCABP8 (+SCABP8) or truncated SCABP8 (+SCABP8ΔN) and plated in AP medium with the indicated NaCl concentrations as in (B).
N-terminal myristoylation of SOS3 is essential for its function in vivo (Ishitani et al., 2000). Three other SCABP/CALCINEURIN B-LIKE (CBL) proteins, CBL1, CBL5, and CBL9, also appear to become myristoylated, and this lipid modification is thought to be important for protein–membrane interaction (Kolukisaoglu et al., 2004). SCABP8 does not contain any consensus sequence for acylation or prenylation, but it has a conspicuous hydrophobic domain between amino acid residues Val-13 and Val-35, which is long enough to span across a membrane once (Figure 8D). To test whether this hydrophobic domain mediates the tight association of SCABP8 with membranes (Figure 7), an internal deletion removing amino acids between Val-13 and Val-35 was generated in SCABP8 by mutagenic PCR. The truncated protein (SCABP8ΔN) became unable to recruit the hSos-SOS2 reporter protein to the plasma membrane as evidenced by the SRS assay (Figure 8C). This failure could not be attributed to lack of interaction with SOS2 because the truncated protein retained the ability to activate SOS1 via SOS2 (Figure 8E). A SCABP8ΔN-hSos translational fusion failed to show targeting to the plasma membrane in contrast with wild-type SCABP8 (data not shown). Thus, the differential salt tolerance conveyed by SCABP8 and SCABP8ΔN likely results from the lack of plasma membrane targeting of the later and reduced activation of SOS1 by a cytoplasmic SOS2 protein kinase (Quintero et al., 2002).
SCABP8 Is a Calcium Binding Protein, and Calcium Enhances SOS2 Kinase Activity
The data suggest that SCABP8 regulates SOS2 activity. The cDNA sequence of SCABP8 corresponds to The Arabidopsis Information Resource accession At4g33000.2 and protein sequence 1009053152, which has three predicted EF hand domains. To test whether SCABP8 could bind calcium, SOS2 and SCABP8 protein were mixed and resolved on a 10% SDS-PAGE gel, and the proteins were transferred to a cellulose membrane. The membrane was then used to perform a calcium binding assay. SCABP8 was able to bind calcium, but SOS2 did not (Figure 7E). The interaction of SOS3 with SOS2 does not require calcium, but activation of SOS2 by SOS3 is calcium dependent (Halfter et al., 2000). To quantitate the activation of SOS2, a P3 peptide (Halfter et al., 2000) was fused to GST in the pGEX-6P-1 vector. GST-SOS2, GST-SCABP8, GST-SOS3, and GST-P3 proteins were purified, and kinase activity assays were performed. The results are shown in Figure 7F. All reactions contained 250 μM calcium. The SOS2 kinase activity was significantly higher in the absence of EGTA than with supplemental 500 μM EGTA, demonstrating that calcium enhances SOS2 activity in the presence of SOS3 or SCABP8. Neither SCABP8 nor SOS3 showed kinase activity.
Overexpression of SCABP8 Partially Suppresses sos3 Salt Sensitivity
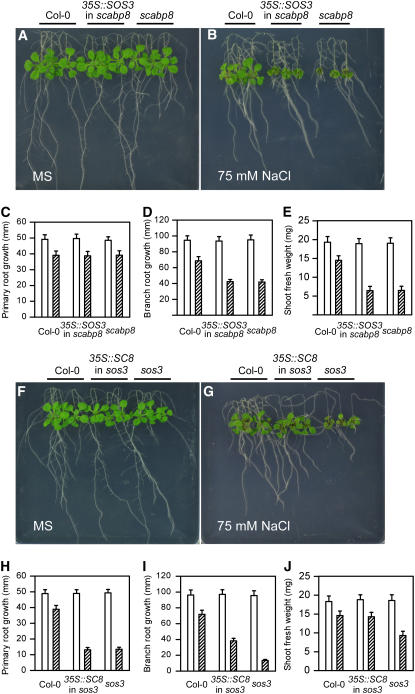
SCABP8 was observed to differ from SOS3 only in its expression pattern. If SCABP8 and SOS3 have identical functions but are expressed in different tissues due to their tissue-specific promoters, ectopic overexpression of either gene in the sos3 or scabp8 mutant background should rescue their salt-sensitive phenotype. To test this hypothesis, SCABP8 and SOS3 cDNA were expressed under the control of the cauliflower mosaic virus 35S promoter in both sos3 and scabp8 mutants. Overexpression of the transgenes was confirmed by RT-PCR (data not shown), and the resulting transgenic plants were tested for salt sensitivity. Consistent with previous studies (data not shown; Ishitani et al., 2000), overexpression of SOS3 in the sos3 mutant complemented its salt-sensitive phenotype. However, overexpression of SOS3 in the scabp8 mutant did not suppress the salt sensitivity of this mutant, including the growth of shoot, primary root, and branch root (Figures 9A to 9E), suggesting that SCABP8 performs an activity required for salt tolerance in shoots that cannot be fulfilled by SOS3. As expected, overexpression of SCABP8 in the scabp8 mutant complemented its salt-sensitive phenotype (data not shown). However, overexpression of SCABP8 in the sos3 mutant only suppressed salt sensitivity in the shoots and partially in the lateral root but not in the primary root (Figures 9F to 9J). These results suggest that SOS3 and SCABP8 have partially overlapping and unique biochemical functions in the salt-tolerant response of Arabidopsis.
Figure 9.
SCABP8 Partially Suppresses the sos3 Phenotype.
(A) to (E) Five-old-day seedlings of the wild type (left), overexpression of SOS3 in scabp8 (center), and scabp8 (right) were transferred onto MS medium without (A) or with (B) 75 mM NaCl. Primary root elongation (C), lateral root elongation (D), and shoot fresh weight (E) were measured.
(F) to (J) Five-old-day seedlings of the wild type (left), overexpression of SCABP8 in sos3 (center), and sos3 (right) were transferred onto MS medium without (F) or with (G) 75 mM NaCl. Primary root elongation (H), lateral root elongation (I), and shoot fresh weight (J) were measured. The pictures were taken at day 15. Open bars, without NaCl treatment; hatched bars, 75 mM NaCl treatment. Error bars represent sd (n > 15).
DISCUSSION
Although functionally equivalent genes often display spatial and temporal differences in transcriptional regulation (Yamada et al., 1995; Kirik et al., 2005), how the unique functions of those complementary genes are achieved is not well understood. Gene duplication and functional diversification during evolution endow plants with manifold protection mechanisms and precision to cope with various environmental changes. In the plant salt response, root-expressed genes are thought to function mainly in the control of ion uptake and transport Na+ to the shoots, whereas shoot-expressed genes are involved in preventing Na+ toxicity and maintaining leaf turgor (Munns, 2005). In this study, we report that two similar calcium binding proteins, SCABP8 and SOS3, protect complementary spectra of tissues or organs (such as shoot and root tissues, respectively) from salt stress by activating and recruiting the SOS2 kinase. Our data also suggest that SOS3 and SCABP8 differ in additional regulatory mechanisms that are specific to each protein.
SCABP8 and SOS3 Together Protect Shoots and Roots from Salt Stress
On growth medium supplemented with 100 mM NaCl, sos3 and scabp8 mutants showed severe growth inhibition in both shoot and root tissues, but nearly complete growth arrest occurred only in the root of sos3 and the shoot of scabp8 plants (Figure 2; Zhu et al., 1998). At a lower NaCl concentration (50 mM), the effect of sos3 mutation on salt tolerance was primarily limited to root growth (Zhu et al., 1998), while the effect of the scabp8 mutation was more prominent in shoot growth (Figures 2 and 3). These results indicate that salt tolerance in planta under extreme situations requires the functional integration of all tissues but also that SOS3 and SCABP8 proteins are needed for salt tolerance in a tissue-specific manner. These results are in agreement with their respective expression patterns as determined by promoter-GUS fusions (Figure 1). SOS3 was only expressed in root tissues, particularly in root tips, whereas SCABP8 was preferentially expressed in shoot tissues. Notably, the combined expression patterns of these genes resembled that of SOS2 and SOS1 (Figure 1; Shi et al., 2002), and the salt sensitivity of the sos3 scabp8 double mutant closely matched that of the sos2 mutant (Figure 4). Moreover, the combination of either sos3 or scabp8 with the sos2 mutation did not affect the overall salt sensitivity associated with the later (Figure 4; Zhu et al., 1998). Thus, the shoot- and root-specific salt sensitivities of the scabp8 and sos3 mutants, respectively, at least in part arise from the different expression patterns of the SCABP8 and SOS3 genes. Genetic interactions also indicate that SCABP8 and SOS3 regulate SOS2 coordinately.
SCABP8 Partially Associates to Plasma Membranes and Recruits SOS2
Our results demonstrate that SCABP8 shares most of the biochemical properties of SOS3. Like SOS3, SCABP8 encodes a protein with EF hand domains that bind calcium in vitro, is capable of binding to SOS2, and activates SOS2 in a calcium-dependent manner (Figures 5A and 7). Two-phase partitioning of microsomal membranes indicated that SCABP8 was associated with the plasma membrane and cytosol. This bipartite localization was corroborated by microscopy inspection of root cells and leaf protoplasts expressing SCABP8 fused to YFP and its colocalization with other cytological markers (see Supplemental Figures 1 and 2 online) and by transient expression of SCABP8 fused to GFP in onion epidermal cells (Figure 6). Based on biochemical evidence, a fraction of the SCABP8 protein pool was tightly associated with cell membranes because it could only be released by treating the membrane vesicles with detergents and the chaotropic salt KI (Figure 7).
Although SOS3 is known to recruit SOS2 to the plasma membrane in yeast (Quintero et al., 2002), this function had not yet been demonstrated in plants. Bimolecular fluorescence complementation of split-YFP modules fused to SCABP8 and SOS2 indicated that these two proteins primarily interact at the plasma membrane (Figure 5B). Consistent with a recruitment role for SCABP8, SOS2 was also detected at the plasma membrane, and this localization was partially dependent on the presence of SCABP8 (see Supplemental Figure 1 online; Figure 7). In the upper part of seedling roots, where the expression of SOS3 is below detection (Figure 1), the relative amount of cytosolic YFP-SOS2 was greater in the scabp8 mutant than in the wild type (see Supplemental Figure 1 online). In protoplasts prepared from leaf, where SOS3 promoter expression was also undetectable, tagged myc-SOS2 fractionated with the microsomal pellet in the wild type but became partly solubilized in the scabp8 mutant (Figure 7). CBL1 is expressed in leaf tissue, interacts with SOS2, and localizes to the plasma membrane (Cheong et al., 2003; Batistic and Kudla, 2004; D'Angelo et al., 2006), which could account for the SOS2 fraction that remained attached to microsomal membranes in the absence of SCABP8. The FISL motif is required for the interaction of SOS2 with SOS3 and SCABP8 (Figure 5). Deletion of the FISL motif in SOS2 results in the reduction of SOS2 plasma membrane localization (Figure 7A) and disrupts SOS2 function in planta (Guo et al., 2004), demonstrating that recruitment of SOS2 to the plasma membrane is essential for salt tolerance. Notably, even without the FISL motif, some of the SOS2ΔF protein was still bound to the microsomal fraction in leaf protoplasts of the wild type and scabp8 mutant (Figure 5A), suggesting that SOS2 can be recruited to cell membranes independently of SCABP proteins. SOS2 interacts with the vacuolar Ca2+/H+ antiporter CAX1 independently of SOS3 (Cheng et al., 2004). Presumably, this protein interaction occurs without involvement of the FISL domain. In summary, recruitment of SOS2 to cell membranes seems to involve multiple proteins and mechanisms, both dependent and independent of SCABP proteins, and our data clearly suggest that SCABP8 is instrumental in this process.
The mechanism by which SCABP8 attaches itself to membranes is novel among this class of proteins. SOS3 has an N-terminal myristoylation signature sequence that is essential for SOS3 function in salt tolerance, although it remains to be shown whether this posttranslational modification is required for the plasma membrane localization of SOS3 or binding to SOS2 in plant cells (Ishitani et al., 2000; Quintero et al., 2002). CBL1 and CBL9 also have N-myristoylation consensus sequences and localize to the plasma membrane (D'Angelo et al., 2006). Interestingly, SCABP8 does not have such a signature sequence for N-myristoylation or any other recognizable acylation, but it is nevertheless localized to the plasma membrane in plant and yeast cells. Removal of an N-terminal hydrophobic domain in SCABP8 preserved the capacity for activation of SOS2 and SOS1 but abrogated recruitment of SOS2 to the plasma membrane of yeast (Figure 8). A truncated SCABP8ΔN protein lacking the N-terminal hydrophobic domain fused to GFP showed a fluorescence pattern similar to soluble GFP, indicating a predominant cytosolic localization (Figure 6). These results, together with the tight association of SCABP8 to the plasma membrane (Figure 7), suggest that the N-terminal part of this protein becomes immersed in the plasma membrane. However, it still remains to be determined whether a regulated process is involved in SCABP8 targeting to the plasma membrane, how the SCABP8-SOS2 complex is formed, and whether this complex includes the target substrate for this protein kinase in a ternary conjugate.
Common and Differential Regulatory Mechanisms of SCABP8 and SOS3
Although capable of performing similar functions in biochemical and cellular tests, SCABP8 and SOS3 must fulfill distinct regulatory functions in the salt stress response of Arabidopsis plants. SOS3 is expressed in root tissues only, particularly in the root tips. SOS1, SOS2, and SOS3 are all strongly expressed in this tissue, suggesting that the SOS cascade plays an important role in root elongation in response to salt stress. On the other hand, SCABP8 is mainly expressed in shoot tissues (Figure 1), and expression of SCABP8 but not SOS3 is slightly induced by salt stress, indicating that SOS3 and SCABP8 differ in their transcriptional regulation in response to salinity. The Na+/H+ exchanger SOS1 is preferentially expressed in the xylem parenchyma of shoots and roots, where it is thought to control sodium loading in the xylem sap (Shi et al., 2002). Activation of SOS2 and SOS1 in shoot tissues requires components other than SOS3, and our data suggest that SCABP8 is one of them. Functional analyses in yeast, together with the specific sodium sensitivity of sos1, sos2, sos3, and scabp8 mutants, suggest that SOS3 and SCABP8 have an overlapping function in the activation of SOS2 and its downstream target SOS1. The shoot- and root-specific salt sensitivities of the scabp8 and sos3 mutants may arise, at least in part, from the different spatial expression patterns of the SCABP8 and SOS3 genes (Figure 1; Shi et al., 2002).
Like the sos2 and sos3 mutants, scabp8 is hypersensitive to Na+ and Li+ but not to Cs+ (Figure 2; Zhu et al., 1998). The sos2 scabp8, sos2 sos3, and sos3 scabp8 double mutants displayed a sos2-like phenotype, suggesting that SCABP8 and SOS3 perform similar functions in regulating SOS2 (Figure 4; Halfter et al., 2000). One critical difference, however, is that SCABP8 mainly protects shoot tissues, whereas SOS3 protects root tissues from salt stress. Guo et al. (2004) have shown that expression of an active form of SOS2, T/DSOS2, which has a single amino acid Thr-168-to-Asp substitution in the kinase activation loop, suppressed the salt sensitivity of sos3 mutants only in the shoots, suggesting that both SOS2 kinase activity and precise localization facilitated by SCABP8 are important for salt tolerance in shoot tissues. Accordingly, deletion of the FISL domain necessary for interaction with SOS3 abrogated suppression by T/DSOS2, indicating that activated SOS2 could bypass the requirement for SOS3 if it retained the ability to bind to alternative SCABP proteins. Our data indicate that SCABP8 could possibly fulfill this role. Binding of SOS3 by SOS2 is necessary for both protein kinase activation and recruitment to the plasma membrane (Quintero et al., 2002; Guo et al., 2004), and we have shown that SCABP8 can replace SOS3 in the reconstituted yeast system (Figure 8), interacts with SOS2 primarily in the plasma membrane of Arabidopsis protoplasts (Figure 5), and significantly enhances the fraction of membrane-bound SOS2 (Figure 7).
The question then remains why SOS3 and SCABP8 could not replace each other in genetic complementation tests (Figure 9). Several SCABP proteins show preferential interaction with only a subset of PKS/CIPK partners (Guo et al., 2001; Albrecht et al., 2003; Kolukisaoglu et al., 2004). SOS3 interacts very specifically, both biochemically and functionally, with SOS2 (Guo et al., 2001; Albrecht et al., 2003), but SOS2 also binds SCABP8 (this work), CBL1, and CBL9 (Kolukisaoglu et al., 2004). Mutants deficient in SOS3, SCABP8, and CBL1, but not CBL9, are affected in salt tolerance to various degrees. On the other hand, CBL1 deficiency affects plant responses to drought, salt, and cold stresses through its interaction with CIPK1, SOS2, and other unidentified protein kinases, respectively (Albrecht et al., 2003; Batistic and Kudla, 2004). Thus, different combinations of protein kinase complexes may generate temporal and spatial specificity signaling, connecting various stimuli to defined cellular responses (Guo et al., 2001). Presumably, these alternative complexes also exhibit different specificities toward their targets. Indeed, SOS2 has several known targets that are determinants of salt tolerance that might involve diverse mechanisms of regulation and recruitment of SOS2 to various subcellular compartments. Together with SOS3, SOS2 activates SOS1 at the plasma membrane (Qiu et al., 2002; Quintero et al., 2002). SOS2 also regulates the activity of vacuolar cation/proton antiporters of the NHX family (Qiu et al., 2004) and V-ATPase (J.-K. Zhu, personal communication); the SCABP subunit presumably involved in recruitment of SOS2 to the tonoplast is presently unknown. Moreover, SOS2 regulates the vacuolar Ca2+/H+ exchanger CAX1 by direct interaction, and no phosphorylation or ancillary SCABP subunit seem to be required (Cheng et al., 2004). Finally, SOS2 interacts with the protein phosphatase ABI2, a master regulation of dehydration stress in plants (Ohta et al., 2003). Any of these processes in which SOS2 is involved could have an effect on the salt tolerance imparted by this protein kinase, and these diverse functions could be regulated differently by SCABP8 and SOS3. Hence, it is not entirely unexpected that alternative protein kinase complexes SOS2-SOS3 and SOS2-SCABP8 may share common target substrates, like SOS1, while affecting differentially distinct targets and processes related to salt stress. Detailed investigation of such tissue- and process-specific regulatory mechanisms involved in the salt response will be an important area for future research.
METHODS
Plant Materials
The homozygous scabp8 mutant was identified from a SALK line (Alonso et al., 2003) harboring a T-DNA insertion in the seventh intron of At4g33000 (seed stock no. SALK_056042) using the gene-specific primers 5′-CAGGAGAAGACCGATTGTAAGG-3′ and 5′-GCGTCGACTCAGTCTTCAACCTCAGTG-3′ and the T-DNA left border primers Lba1 5′-TGGTTCACGTAGTGGGCCATCG-3′ and LBb1 5′-GCGTGGACCGCTTGCTGCAACT-3′. Both of the sos2 and sos3 mutants were in the gl1/gl1 Col-0 background. The mutation gl1 did not affect the salt tolerance phenotype. The sos2 scabp8 and sos3 scabp8 double mutants were obtained by crossing scabp8 to sos2-2 and sos3, respectively. From F2 plants, scabp8 homozygotes were identified as described above. SOS3 gene-specific primers 5′-TCTCATGAATTTGCAGTTGC-3′ and 5′-AAACTGTTTAATCTGGAGGG-3 (Halfter et al., 2000) were used to amplify a 112-bp DNA fragment from sos3 and a 121-bp fragment from the wild type. For sos2-2, primers 5′-CTATACGAGCAAGGGAAGAAG-3′ and 5′-AATGAAGGCGAAGCACAAAC-3′ were used for PCR amplification. The products were 86 bp from sos2-2 and 88 bp from the wild type. All of the PCR products were separated on a 4% agarose gel.
Salt Sensitivity
Seeds of the mutants and wild-type Col-0 were sterilized in the solution containing 20% sodium hypochlorite and 0.1% Triton X-100 for 10 min, washed five times with sterilized water, and sown on MS medium with 0.6% Phytagel (Sigma-Aldrich). The plates were placed at 4°C for 2 d, and then the seeds were germinated vertically at 23°C under continual illumination. Four-day-old seedlings with a root length ∼1.5 cm were transferred onto MS medium with salt added as described.
scabp8 Phenotype Complementation
For scabp8 mutant complementation, a 3783-bp genomic DNA fragment containing 1736 bp upstream of the translation start site and 469 bp downstream of the stop codon of SCABP8 was obtained by the digestion of BAC plasmid F26P21 (ABRC) with XbaI and PstI. The DNA fragment corresponded to the sequence from 54,144 to 57,927 bp of the BAC clone. This fragment was then cloned into the XbaI and PstI sites of pCAMBIA1200 vector. The resultant plasmid 1200-SCABP8CP was introduced into Agrobacterium tumefaciens GV3101 by electroporation and transformed to scabp8 mutants by the flower dip method.
DNA Gel Blot Analysis
Genomic DNA was extracted from 6-week-old scabp8 plants using the Wizard Genomic DNA extraction kit (Promega). For DNA gel blot analysis, 10 μg of genomic DNA was digested with EcoRI, HindIII, and SacI and fractionated on agarose gel (0.8% [w/v]). The separated DNA was transferred to Hybond N+ nucleic acid transfer membranes (Amersham). The 795-bp neomycin phosphotransferase coding sequence (NPTII CDS) probe was gel purified and radiolabeled with 32P by a random primer labeling kit (Takara). DNA gel blot hybridization was performed at 42°C for 16 h using hybridization solution (200 mM sodium phosphate buffer, pH 7.2, 1 mM EDTA, pH 8.0, 50% formamide, 10% BSA, and 7% SDS) with 32P-labeled NPTII CDS probes, followed by washing at 65°C in 2× SSC and 0.5% SDS, 1× SSC and 0.5% SDS, and 0.1× SSC and 0.5% SDS for 15 min sequentially. The 32P-labeled membranes were exposed to a phosphor screen (Amersham). After 12 h of exposure, signals were captured with a Typhoon 9410 phosphor imager (Amersham).
Promoter GUS Analysis
The plasmid 1200-SCABP8CP was digested with EcoRI and AccIII. The resulting fragment from 35 to 1736 bp upstream of the translation start site of SCABP8 was cloned into the EcoRI and XmaI sites of the pCAMBIA1381Z vector. SOS2 promoter DNA fragment was amplified with primers 5′-CCCAAGCTTCATTAGGGTTCATGGGTTGAG-3′ and 5′-CGGGATCCTCTTTACAAACTTTTATCTG-3′ and cloned into the HindIII and BamHI sites of pBI101 vector. SOS3 promoter fragment was amplified with primers 5′-CCGCTCGAGAAATCATGTTGGGTCTGATTGG-3′ and 5′-CGGGATCCACAAACACACCCTTCTCTCAAC-3′ and cloned into the SalI and BamHI sites of pBI101 vector. The plasmids were introduced into Arabidopsis thaliana by A. tumefaciens–mediated transformation. GUS assays using transgenic T2 lines at different developmental stages were performed as described by Jefferson et al. (1987).
RT-PCR Analysis
Total RNA from 10-d-old seedlings and from different organs of plants at the six-week growth stage was extracted by the TRIzol method. Reverse transcription was performed using 1 μg of total RNA and MLV reverse transcriptase (Promega). The cDNA was then used for PCR amplification with the following primers. For SCABP8, the primers SC8F0 5′-CGGGATCCATGGAACAAGTTTCCTCTAG-3′ and SC8R0 5′-GCGTCGACTCAGTCTTCAACCTCAGTG-3′ were used. For SOS2, the primers 5′-CGGGATCCATGACAAAGAAAATGAGAAG-3′ and 5′-GTTCCACATGTGGTACGCAGAAGTTC-3′ were used. For SOS3, the primers 5′-CGGAATTCATGGGCTGCTCTGTATCG-3′ and 5′-CCGCTCGAGTTAGGAAGATACGTTTTGC-3′ were used. For UBQ10, the primers 5′-ACCGGAAAGACTATCACTTTG-3′ and 5′- AGAGATAACAGGAACGGAAAC-3′ were used. To identify the level of SCABP8 transcription in the SCABP8 mutant and the complementary transgenic plants by RT-PCR, the primers used were SC8F1 5′-GCGCCGTCTTTATACCTTTC-3′, SC8R1 5′-ATCGGTCTTCTCCTGGATTG-3′, SC8F2 5′-GGATGAATGGAATGTCTATG-3′, and SC8R2 5′-AACAAAGCAAAGTGCTTGAC-3′.
Real-time PCR was performed with the ABI 7500 real-time PCR system using SYBR to monitor double-stranded DNA products. For SCABP8, the primers were 5′-AACAAATGGTTTCTGCTATTCTC-3′ and 5′-ATCTGCATCAGCAAATGTTTTATC-3′. For SOS2, the primers were 5′-CGAAACTTCAAGACAAGGCTC-3′ and 5′-GTGCCACCTCGTAAATCTCTATC-3′. For SOS3, the primers were 5′-GTAATGGTGGATAAGGCTTTCG-3′ and 5′-TGAGCGATGGATTCAAGGATAC-3′. For ACT2, used as an internal control, the primers were 5′-GTCGTACAACCGGTATTGTG-3′ and 5′-GAGCTGGTCTTTGAGGTTTC-3′. For UBQ10, another internal control, the primers were 5′-CCCTGATGAATAAGTGTTCTAC-3′ and 5′-ACGAAGCGATGATAAAGAAG-3′. The relative gene expression after NaCl treatment was calculated by comparison with that of the seedlings without NaCl treatment that was defined as 1. SDS software version 1.2 (Applied Biosystems) was used for analysis. Standard deviations were calculated from three independent biological replicates.
Pigment Determination
Leaf chlorophyll was extracted with 80% acetone, the absorbance at 647 and 664 nm was determined by a DU 800 UV/visible spectrophotometer (Beckman Coulter), and total chlorophyll was calculated (Inskeep and Bloom, 1985). Anthocyanin was extracted in acid (1% HCl [w/v]) methanol, the absorbance at 530 and 657 nm was determined, and the concentration of anthocynanin was calculated by A530-0.25A657 (Rabino and Mancinelli, 1986).
Pull-Down, Kinase, and Calcium Binding Assays
The coding region of SCABP8 cDNA was amplified from Col-0 using the primers 5′-CGGGATCCATGGAACAAGTTTCCTCTAG-3′ and 5′-GCGTCGACTCAGTCTTCAACCTCAGTG-3′, containing BamHI and SalI sites, respectively. Two PCR bands were separated after electrophoresis. Both PCR products were purified and digested with BamHI and SalI and cloned into the pGEX-6P-1 vector. Sequence analysis revealed that the sequence of the smaller fragment was identical to CBL10 (Luan et al., 2002; GenBank accession no. AF513507.2). The larger fragment was the same as AF513507 except that the seventh intron of SCABP8 was not spliced out. SOS3 was amplified using primers 5′-CGGAATTCATGGGCTGCTCTGTATCG-3′ and 5′-CCGCTCGAGTTAGGAAGATACGTTTTGC-3′ and cloned into the EcoRI and XhoI sites of pGEX-6P-1 vector. SOS2 was amplified using primers 5′-CGGGATCCA TGACAAAGAAAATGAGAAG-3′ and 5′-GTTCCACATGTGGTACGCAGAAGTTC-3′. To produce GST-SOS2ΔF, two SOS2 cDNA fragments were amplified by the following primer pairs: 5′-CGGGATCCATGACAAAGAAAATGAGAAG-3′ and 5′-CAAAATCCTGTCGCAGGGGCCCTTCATCATTTCTC-3′; 5′-GAAGGGCCCCTGCGACAGGATTTTGTTAAAAG-3′ and 5′-GGAATTCTCAAAACGTGATTGTTCTGAG-3′. The resulting PCR products were used in the second round of PCR with primers 5′-CGGGATCCATGACAAAGAAAATGAGAAG-3′ and 5′-GGAATTCTCAAAACGTGATTGTTCTGAG-3′ to produce the mutant SOS2 cDNA containing a deletion of 66 bp from 924 to 990 bp (SOS2ΔF). The SOS2ΔF was cloned into the pGEX-6P-1 vector at the BamHI and EcoRI sites.
The GST fusion constructs were transformed into Escherichia coli strain BL21 (DE3). The recombinant proteins were purified with glutathione sepharose (Amersham Pharmacia) according to the manufacturer's protocol.
For the pull-down assay, 5 μg of SCABP8 or SOS3 was incubated with 1 μg each of GST-SOS2 or GST-SOS2ΔF on glutathione sepharose beads for 30 min at room temperature in 100 μL of binding buffer (20 mM Tris-HCl, pH 7.2, 10 mM MgCl2, and 2 mM DTT). After four washes with binding buffer, the beads were resuspended in 100 μL of loading buffer, 20 μL were run on a 10% SDS-PAGE gel and stained with Commassie Brilliant Blue R 250, and 1 μL was analyzed by protein gel blotting against anti-SCABP8. Kinase reaction and calcium binding assays were described by Halfter et al. (2000).
Interaction of SCABP8 and SOS2 in Plants (Split-YFP)
SCABP8 was amplified with primers 5′-CGGGATCCATGGAACAAGTTTCCTCTAG-3′ and 5′-GCGTCGACGTCTTCAACCTCAGTGTTG-3′ and cloned into the BamHI and SalI sites of a pUC-SPYCE vector (Walter et al., 2004). SOS2 and SOS2ΔFISL were amplified with primers 5′-CGGGATCCATGACAAAGAAAATGAGAAG-3′ and 5′-GCGTCGACAAACGTGATTGTTCTGAGAATC-3′ using GST-SOS2 and GST-SOS2ΔFISL plasmids as templates, respectively, and then cloned into the BamHI and SalI sites of a pUC-SPYNE vector. SOS3 was obtained by PCR using primers 5′-GCTCTAGAATGGGCTGCTCTGTATCG-3′ and 5′-CCGCTCGAGGGAAGATACGTTTTGCAATTC-3′ and cloned into the XbaI and XhoI sites of pUC-SPYCE vector. These plasmids were purified by CsCl gradient centri-fugation and transformed into Arabidopsis mesophyll protoplasts (Sheen, 2001). The YFP fluorescence of protoplasts was assayed 10 to 12 h after transformation under a Zeiss LSM 510 META confocal microscope.
Subcellular Localization of SCABP8 and SOS2
YFP-SCABP8 was constructed by excising the coding region of SCABP8 cDNA with BamHI and SalI from GST-SCABP8 and cloning it into the binary vector pCAMBIA1205-YFP downstream of YFP. SOS2 was digested with BamHI and EcoRI from GST-SOS2 and cloned into the pCAMBIA1205-YFP vector. The resulting constructs were introduced into A. tumefaciens GV3101 and transformed into Arabidopsis.
The coding sequences of SOS2, SOS2ΔF, and SCABP8 were translationally fused downstream of the c-myc tag. The plasmids were purified by CsCl gradient centrifugation and transformed into Arabidopsis mesophyll protoplasts. After overnight incubation, the protoplasts were harvested at room temperature for 2 min at 100g, resuspended in 200 μL of cold lysis buffer containing 20 mM Hepes-KOH, pH 7.1, 13.5% (w/v) sucrose, 10 mM potassium acetate, 1 mM DTT, and 1× protease inhibitor mixture (Sigma-Aldrich), and sonicated on ice in short bursts. The total crude homogenate was further centrifuged for 20 min at 1000g at 4°C, and the supernatant was centrifuged for 60 min at 140,000g at 4°C to generate a total membrane pellet and a soluble protein fraction. The resulting samples were then analyzed by SDS-PAGE and blotted onto a polyvinylidene difluoride membrane (Millipore). The blots were probed with primary antibody anti-cMyc (Sigma-Aldrich; M4439).
Plasma membrane–enriched vesicles were purified by the two-phase method, and the purity of each fraction was determined as described by Qiu et al. (2002). Seedlings overexpressing 35SP:myc-SCABP8 and protoplasts transfected with 35SP:myc-SOS2 were used for two-phase partitioning. Plasma membrane vesicles from seedlings overexpressing 35S:myc-SCABP8 were treated with 0.01% Brij58, 1% Triton X-100, 3% SDS, 1 M KI, 1 M KCl, 1 M NaCl, 0.5 M NaCl, and 0.1 M Na2CO3, pH 11.5 (Overvoorde and Grimes, 1994). After centrifugation at 150,000g at 4°C for 60 min, aliquots of supernatants and pellets were loaded onto an SDS-PAGE gel, and myc-SCABP8 and myc-SOS2 were detected with anti-myc antibody.
The intracellular localization of SCABP8 was also determined by monitoring the transient expression of a SCABP8-GFP translational fusion in onion epidermal cells after DNA particle bombardment, essentially as described by Yokoi et al. (2002). The coding regions of SCABP8 and SCABP8ΔN in which the N-terminal hydrophobic domain had been removed were amplified with primers 5′-ACGTACCATGGAACAAGTTTCCTCTAGAT-3′ and 5′-ATAGCCCATGGCGTCTTCAACCTCAGTGTTGAA-3′ that included NcoI restriction sites for subcloning into plasmid pGFP-JS in frame with GFP (Sheen et al., 1995).
Overexpression of SCABP8 and Its Splicing Variant and Overexpression of SOS3 in scabp8 and sos3
The SCABP8 open reading frame was digested from its GST fusion vector with BamHI and SalI. To produce the truncated mis-spliced SCABP8 variant, the fragment was obtained by PCR with primers 5′-CGGGATCCTGGAACAAGTTTCCTCTAG-3′ and 5′-GCGTCGACACTAGTTCAAGAACTCACTTTATCAATAATC-3′ harboring BamHI or SalI sites. Both of the fragments were inserted into the pCAMBIA1205 vector under the cauliflower mosaic virus 35S promoter, respectively. GST-SOS3 plasmid DNA was digested with XhoI and then partially digested with BamHI and cloned into the pCAMBIA1205 vector. These plasmids were introduced into A. tumefaciens GV3101 and transformed into Arabidopsis.
Yeast Methods
Saccharomyces cerevisiae strains AXT3K (ena1∷HIS3∷ena4, nha1∷LEU2, and nhx1∷KanMX4) and JP890 (ena1∷HIS3∷ena4, nha1∷LEU2, nhx1∷KanMX, and CYC1∷[PGK1:SOS1:CYC1]) have been described previously (Quintero et al., 2002; Guo et al., 2004). Sodium tolerance tests were performed in the alkali cation-free medium AP (8 mM phosphoric acid, 10 mM l-Arg, 2 mM MgSO4, 0.2 mM CaCl2, 2% glucose, plus vitamins and trace elements, brought to pH 6.5 with Arg). Aliquots (5 μL) were spotted onto AP plates supplemented with 1 mM KCl, with or without 100 mM NaCl, and grown for 3 to 4 d at 28°C. Plasmids for the expression of SOS2 and SOS3 in yeast have been described elsewhere (Quintero et al., 2002; Guo et al., 2004). Plasmid pSCABP8 was created by subcloning a 0.7-kb BamHI-SalI fragment spanning the entire open reading frame of SCABP8 into the BamHI-SalI sites of pYPGE15. An internal deletion spanning from residues Val-13 to Val-35, both included, was constructed in SCABP8 to remove an N-terminal hydrophobic domain. First, the region upstream of the deleted fragment was amplified by PCR with primers 5′-CCTGGTATCTTTATAGTCCTG-3′, annealing to the vector plasmid, and 5′- ATCGAAGCATTGTCCAGTGAGAGAGCTTGATCTAG-3′. The downstream fragment was also amplified with primers 5′-TCAAGCTCTCTCACTGGACAATGCTTCGATTGCCG-3′ and 5′-TCGAGGTCGACTCAGTCTTC-3′, both annealing to SCABP8. Both PCR products were combined and reamplified with the first and fourth primers to render a read-through fragment bearing the Val-13–Val-35 deletion in SCABP8. The resulting coding region, SCABP8ΔN, was cloned in pYPGE15 as described above to produce plasmid pSCABP8ΔN. To perform the SRS analysis, the S. cerevisiae strain cdc25-2 (MATalfa, cdc25-2, ade2, his3, leu2, lys2, trp1, ura3) was transformed with plasmids pSRS2-1 (hSos:SOS2), pSCABP8, pSCABP8ΔN, and pSOS3-2 (Quintero et al., 2002; Guo et al., 2004). As controls, the empty vector pADNS and pADNS-5′SosF encoding a full-length human Sos protein were used (Aronheim et al., 1997). Mutation cdc25-2 creates a conditional lethal phenotype at 37°C unless the reporter human protein hSos reaches the plasma membrane. Cell viability at 37°C was determined in YPD plates (1% yeast extract, 2% peptone, and 2% glucose).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: ACT2, At3g18780; SCABP8, At4g33000; SOS1, At2g01980; SOS2, At5g35410; SOS3, At5g24270; and UBQ10, At4g05320.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Enhanced Plasma Membrane Localization of SOS2 by SCABP8.
Supplemental Figure 2. Transient Expression of Fluorescent Protein–Tagged SCABP8, SOS2, PIP2a, and AtVAM3 Genes in Protoplasts.
Supplemental Table 1. Determining the Purity of Vesicles from Leaves.
Supplemental Table 2. Determination the Purity of Vesicles from Protoplasts.
Supplementary Material
Acknowledgments
We thank Xing Wang Deng, Jianmin Zhou, and Valerie Karplus for critical reading of the manuscript and stimulating discussions, Liqin Fu, Cheng Zhan, and Jun Zhang for excellent technical assistance, Jijie Chai for providing the GST-P3 plasmid, Jie Le for PIP2a-GFP and acidic ribosomal protein-GFP transgenic seeds, and the ABRC (Ohio State University) for the T-DNA insertion lines. This work was supported by National Basic Research Program of China Grant 2006CB100100 and National High Technology Research and Development Program of China 863 Grant 2003AA210100 to Y.G. and S.C. and by Spanish Ministry of Education and Science Grant BFU2006-06968 to J.M.P.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Yan Guo (guoyan@nibs.ac.cn).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Albrecht, V., Weinl, S., Blazevic, D., D'Angelo, C., Batistic, O., Kolukisaoglu, U., Bock, R., Harter, K., and Kudla, J. (2003). The calcium sensor CBL1 integrates plant responses to abiotic stresses. Plant J. 36 457–470. [DOI] [PubMed] [Google Scholar]
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Aronheim, A., Zandi, E., Hennemann, H., Elledge, S.J., and Karin, M. (1997). Isolation of an AP-1 repressor by a novel method for detecting protein-protein interactions. Mol. Cell. Biol. 17 3094–3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batistic, O., and Kudla, J. (2004). Integration and channeling of calcium signaling through the CBL calcium sensor/CIPK protein kinase network. Planta 219 915–924. [DOI] [PubMed] [Google Scholar]
- Cheng, N.-H., Pittman, J.K., Zhu, J.-K., and Hirschi, K.D. (2004). The protein kinase SOS2 activates the Arabidopsis H+/Ca2+ antiporter CAX1 to integrate calcium transport and salt tolerance. J. Biol. Chem. 279 2922–2926. [DOI] [PubMed] [Google Scholar]
- Cheong, Y.H., Kim, K.N., Pandey, G.K., Gupta, R., Grant, J.J., and Luan, S. (2003). CBL1, a calcium sensor that differentially regulates salt, drought, and cold responses in Arabidopsis. Plant Cell 15 1833–1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler, S.R., Ehrhardt, D.W., Griffitts, J.S., and Somerville, C.R. (2000). Random GFP∷cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc. Natl. Acad. Sci. USA 97 3718–3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Angelo, C., et al. (2006). Alternative complex formation of the Ca2+-regulated protein kinase CIPK1 controls abscisic acid-dependent and independent stress responses in Arabidopsis. Plant J. 48 857–872. [DOI] [PubMed] [Google Scholar]
- Guo, Y., Halfter, U., Ishitani, M., and Zhu, J.-K. (2001). Molecular characterization of functional domains in the protein kinase SOS2 that is required for plant salt tolerance. Plant Cell 13 1383–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, Y., Qiu, Q., Quintero, F.J., Pardo, J.M., Ohta, M., Zhang, C., Schumaker, K.S., and Zhu, J.-K. (2004). Transgenic evaluation of activated mutant alleles of SOS2 reveals a critical requirement for its kinase activity and C-terminal regulatory domain for salt tolerance in Arabidopsis thaliana. Plant Cell 16 435–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfter, U., Ishitani, M., and Zhu, J.-K. (2000). The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proc. Natl. Acad. Sci. USA 97 3735–3740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inskeep, W.P., and Bloom, P.R. (1985). Extinction coefficients of chlorophyll a and b in N,N-dimethylformamide and 80% acetone. Plant Physiol. 77 483–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishitani, M., Liu, J., Halfter, U., Kim, C.-S., Wei, M., and Zhu, J.-K. (2000). SOS3 function in plant salt tolerance requires myristoylation and calcium binding. Plant Cell 12 1667–1677.11006339 [Google Scholar]
- Jefferson, R.A., Kavanagh, T.A., and Bevan, M.W. (1987). GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirik, V., Lee, M.M., Wester, K., Herrmann, U., Zheng, Z., Oppenheimer, D., Schiefelbein, J., and Hulskamp, M. (2005). Functional diversification of MYB23 and GL1 genes in trichome morphogenesis and initiation. Development 132 1477–1485. [DOI] [PubMed] [Google Scholar]
- Kolukisaoglu, U., Weinl, S., Blazevic, D., Batistic, O., and Kudla, J. (2004). Calcium sensors and their interacting protein kinases: Genomics of the Arabidopsis and rice CBL-CIPK signaling networks. Plant Physiol. 134 43–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J., Ishitani, M., Halfter, U., Kim, C.-S., and Zhu, J.-K. (2000). The Arabidopsis thaliana SOS2 gene encodes a protein kinase that is required for salt tolerance. Proc. Natl. Acad. Sci. USA 97 3730–3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J., and Zhu, J.-K. (1998). A calcium sensor homolog required for plant salt tolerance. Science 280 1943–1945. [DOI] [PubMed] [Google Scholar]
- Luan, S., Kudla, J., Rodriguez-Concepcion, M., Yalovsky, S., and Gruissem, W. (2002). Calmodulins and calcineurin B-like proteins: Calcium sensors for specific signal response coupling in plants. Plant Cell 14 S389–S400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munns, R. (2005). Genes and salt tolerance: Bringing them together. New Phytol. 167 645–663. [DOI] [PubMed] [Google Scholar]
- Ohta, M., Guo, Y., Halfter, U., and Zhu, J.-K. (2003). A novel domain in the protein kinase SOS2 mediates interaction with the protein phosphatase 2C ABI2. Proc. Natl. Acad. Sci. USA 100 11771–11776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overvoorde, P.J., and Grimes, H.D. (1994). Topographical analysis of the plasma membrane-associated sucrose binding protein from soybean. J. Biol. Chem. 269 15154–15161. [PubMed] [Google Scholar]
- Qiu, Q.S., Guo, Y., Dietrich, M.A., Schumaker, K.S., and Zhu, J.-K. (2002). Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc. Natl. Acad. Sci. USA 99 8436–8441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu, Q.S., Guo, Y., Quintero, F.J., Pardo, J.M., Schumaker, K.S., and Zhu, J.K. (2004). Regulation of vacuolar Na+/H+ exchange in Arabidopsis thaliana by the SOS pathway. J. Biol. Chem. 279 207–215. [DOI] [PubMed] [Google Scholar]
- Quintero, F.J., Ohta, M., Shi, H., Zhu, J.-K., and Pardo, J.M. (2002). Reconstitution in yeast of the Arabidopsis SOS signaling pathway for Na+ homeostasis. Proc. Natl. Acad. Sci. USA 99 9061–9066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabino, I., and Mancinelli, A.L. (1986). Light, temperature, and anthocyanin production. Plant Physiol. 81 922–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen, J. (2001). Signal transduction in maize and Arabidopsis mesophyll protoplasts. Plant Physiol. 127 1466–1475. [PMC free article] [PubMed] [Google Scholar]
- Sheen, J., Hwang, S., Niwa, Y., Kobayashi, H., and Galbraith, D.W. (1995). Green-fluorescent protein as a new vital marker in plant cells. Plant J. 8 777–784. [DOI] [PubMed] [Google Scholar]
- Shi, H., Ishitani, M., Kim, C.-S., and Zhu, J.-K. (2000). The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc. Natl. Acad. Sci. USA 97 6896–6901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, H., Quintero, F.J., Pardo, J.M., and Zhu, J.-K. (2002). The putative plasma membrane Na+/H+ antiporter SOS1 controls long-distance Na+ transport in plants. Plant Cell 14 465–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura, T., Yoshimura, S.H., Takeyasu, K., and Sato, M.H. (2002). Vacuolar membrane dynamics revealed by GFP-AtVam3 fusion protein. Genes Cells 7 743–753. [DOI] [PubMed] [Google Scholar]
- Walter, M., Chaban, C., Schutze, K., Batistic, O., Weckermann, K., Nake, C., Blazevic, D., Grefen, C., Schumacher, K., Oecking, C., Harter, K., and Kudla, J. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40 428–438. [DOI] [PubMed] [Google Scholar]
- Yamada, S., Katsuhara, M., Kelly, W.B., Michalowski, C.B., and Bohnert, H.J. (1995). A family of transcripts encoding water channel proteins: Tissue-specific expression in the common ice plant. Plant Cell 7 1129–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoi, S., Quintero, F.J., Cubero, B., Ruiz, M.T., Bressan, R.A., Hasegawa, P.M., and Pardo, J.M. (2002). Differential expression and function of Arabidopsis thaliana NHX Na+/H+ antiporters in the salt stress response. Plant J. 30 529–539. [DOI] [PubMed] [Google Scholar]
- Zhu, J.-K., Liu, J., and Xiong, L. (1998). Genetic analysis of salt tolerance in Arabidopsis: Evidence for a critical role of potassium nutrition. Plant Cell 10 1181–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.