Abstract
Hydrogen peroxide is a major by-product of peroxisomal metabolism and has the potential to cause critical oxidative damage. In all eukaryotes, catalase is thought to be instrumental in removing this H2O2. However, plants also contain a peroxisomal membrane–associated ascorbate-dependent electron transfer system, using ascorbate peroxidase and monodehydroascorbate reductase (MDAR). Here, I report that the conditional seedling-lethal sugar-dependent2 mutant of Arabidopsis thaliana is deficient in the peroxisomal membrane isoform of MDAR (MDAR4). Following germination, Arabidopsis seeds rely on storage oil breakdown to supply carbon skeletons and energy for early seedling growth, and massive amounts of H2O2 are generated within the peroxisome as a by-product of fatty acid β-oxidation. My data suggest that the membrane-bound MDAR4 component of the ascorbate-dependent electron transfer system is necessary to detoxify H2O2, which escapes the peroxisome. This function appears to be critical to protect oil bodies that are in close proximity to peroxisomes from incurring oxidative damage, which otherwise inactivates the triacylglycerol lipase SUGAR-DEPENDENT1 and cuts off the supply of carbon for seedling establishment.
INTRODUCTION
In eukaryotes, the oxidative metabolism that takes place in peroxisomes generates hydrogen peroxide (H2O2), which must be detoxified to prevent damage to proteins, lipids, and DNA (van den Bosch et al., 1992; Singh, 1997; Willekens et al., 1997). This important task is performed universally by catalase, which catalyzes the decomposition of H2O2 into molecular oxygen and water within the peroxisomal matrix. Defects in catalase cause peroxisome dysfunction and are detrimental in mammals, plants, and yeast (Zhang et al., 1993; Willekens et al., 1997; Sheikh et al., 1998; Horiguchi et al., 2001).
Uniquely, in addition to catalase, higher plant peroxisomes possess a membrane-bound ascorbate-dependent electron transfer system that is also able to remove H2O2 (Yamaguchi et al., 1995; Bunkelmann and Trelease, 1996; Mullen and Trelease, 1996; Karyotou and Donaldson, 2005). This system is thought to rely on the cooperative action of ascorbate peroxidase (APX) and monodehydroascorbate reductase (MDAR). APX initiates electron transfer from two molecules of ascorbate to convert H2O2 to water, and the monodehydroascorbate is then recycled to reduced ascorbate by MDAR via electron transfer from NADH. Monodehydroascorbate can also spontaneously disproportionate to ascorbate and dehydroascorbate, and the latter can be reduced back to ascorbate by glutathione-dependent dehydroascorbate reductase (Jiménez et al., 1997; del Rio et al., 1998).
Interestingly, although catalase is highly active in plant peroxisomes, it has a much lower affinity for H2O2 than APX, and this has led to the suggestion that at low concentrations, H2O2 might be preferentially scavenged by the APX/MDAR system (Bunkelmann and Trelease, 1996; Mullen and Trelease, 1996). It has been hypothesized that the membrane association of APX/MDAR allows the system to protect membrane lipids and integral proteins from oxidative damage and act as a cordon to limit the escape of H2O2 into the cytosol (Yamaguchi et al., 1995; Mullen and Trelease, 1996; Karyotou and Donaldson, 2005). MDAR may also play a role in reductant balance within the peroxisome by recycling NAD+ (Bowditch and Donaldson, 1990; Mullen and Trelease, 1996).
The physiological importance of catalase in maintaining redox balance in plant peroxisomes has been demonstrated in several studies using mutants or antisense suppression (Kendall et al., 1983; Takahashi et al., 1997; Willekens et al., 1997; Vandenabeele et al., 2004), but less attention has been paid to peroxisomal APX and MDAR. Overexpression of APX3, encoding a peroxisomal isoform from Arabidopsis thaliana, has been reported to increase protection against oxidative stress (Wang et al., 1999). However, Narendra et al. (2006) have recently shown that a null mutant in APX3 is healthy under normal growth conditions. It therefore remains unclear whether the peroxisomal APX/MDAR system is required for plant growth and development and whether it plays a different role from that of catalase.
In this study, I provide molecular genetic evidence for the function of the ascorbate-dependent electron transfer system by characterizing an Arabidopsis mutant in the peroxisomal membrane isoform of MDAR (MDAR4; Lisenbee et al., 2005). This mutant was originally designated sugar-dependent2 (sdp2; Eastmond, 2006), and it exhibits a conditional seedling-lethal phenotype because its seeds are unable to break down storage oil to provide carbon skeletons and energy for early seedling growth. In oilseeds such as Arabidopsis, massive amounts of H2O2 are generated within the peroxisome as a by-product of fatty acid β-oxidation (Graham and Eastmond, 2002). My data suggest that without MDAR4, the escaping H2O2 inactivates the lipases on the oil body surface that are responsible for catalyzing the first step in storage oil breakdown.
RESULTS
SDP2 Is Required for Storage Oil Hydrolysis in Germinating Arabidopsis Seeds
In Arabidopsis seeds, the breakdown of stored triacylglycerol (TAG) following germination is essential to drive the initial phase of seedling growth and allow photosynthetic establishment (Hayashi et al., 1998). To investigate this process, a forward genetic screen was used to isolate a collection of Arabidopsis mutants that require an alternate exogenous source of carbon (sucrose) to support postgerminative growth (Eastmond, 2006). Screening these sdp mutants for defects in TAG hydrolysis revealed that sdp1, sdp2, and sdp3 are defective in the lipase activity that is associated with the oil body membranes. I have previously characterized the patatin domain TAG lipase encoded by SDP1 (Eastmond, 2006). Here, I investigate SDP2.
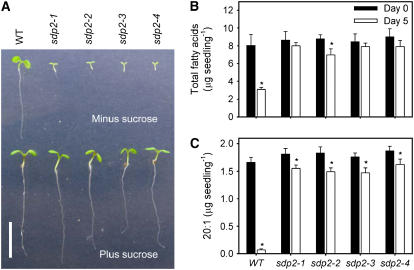
The sdp2 mutant germinates normally (see Supplemental Table 1 online), but the cotyledons fail to expand or green, and seedling growth arrests (Figure 1A). Providing sucrose rescues the growth phenotype (Figure 1A), and once sdp2 seedlings establish photosynthetic competence, they grow normally and complete their life cycle. Analysis of the fatty acid content of sdp2 seeds germinated in the presence of sucrose revealed that the mutant is severely impaired in its ability to breakdown TAG (Figures 1B and 1C). Eicosinoic acid is specifically found in TAG in Arabidopsis (Lemieux et al., 1990); therefore, it can be used as a convenient marker to monitor TAG breakdown (Eastmond et al., 2000a). The levels of both total fatty acids and eicosinoic acid (20:1) decline by <20% over the course of the first 5 d of postgerminative growth in sdp2, while in wild-type seedlings, they fall by 60 and 95%, respectively.
Figure 1.
Postgerminative Growth and Fatty Acid Breakdown in sdp2.
(A) Images of 5-d-old sdp2 and wild-type seedlings grown on medium with and without 1% (w/v) sucrose. Bar = 1 cm.
(B) and (C) Total fatty acid (B) and eicosenoic acid (20:1) (C) content of sdp2 and wild-type seeds and 5-d-old seedlings grown on medium with 1% (w/v) sucrose. Values are the mean ± se of measurements on five batches of 20 seeds or seedlings. *Significantly different from seeds (P < 0.05).
SDP2 Encodes a Component of the Peroxisomal Antioxidant System
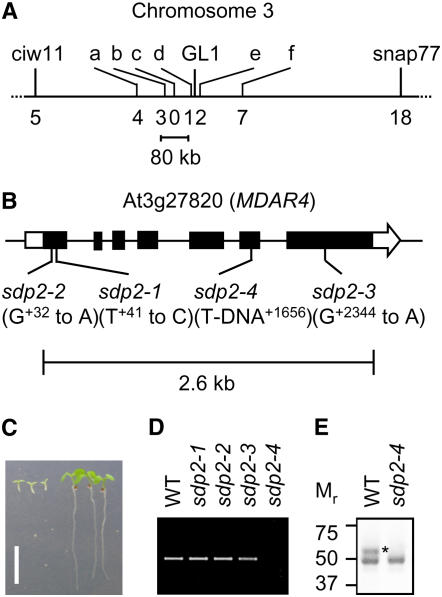
The SDP2 locus was mapped to a region on chromosome 3 near GLABRA1 and between PCR-based markers ciw11 and snap77 (Figure 2A). Further mapping reduced the interval to an ∼80-kb region, containing 22 open reading frames. Sequencing candidate genes within this region revealed that three independent ethyl methanesulfonate sdp2 alleles contained mutations in At3g27820 (Figure 2B). This gene encodes an isoform of MDAR (MDAR4) that is associated with the peroxisomal membrane (Lisenbee et al., 2005). MDAR4 forms part of an ascorbate-dependent electron transfer system on the peroxisomal membrane that has been proposed to play a role in scavenging H2O2, produced as a by-product of oxidative metabolism (Yamaguchi et al., 1995; Mullen and Trelease, 1996; Karyotou and Donaldson, 2005).
Figure 2.
Molecular Characterization of SDP2.
(A) Mapping of SDP2. PCR-based simple sequence polymorphism, cleaved amplified polymorphic sequence, and single nucleotide amplified polymorphism markers were used to map SDP2 to an 80-kb region on the top arm of chromosome 3 near GL1. The positions of markers a to f (listed in Supplemental Table 2 online) are denoted by bars, and the number of recombination events/total number of chromosomes (380) is listed below each.
(B) Schematic diagram of the SDP2 locus showing mutations in At3g27820 (MDAR4). Black bars are exons, and the white bar and arrow represent the 5′ and 3′ untranslated regions, respectively. Insertion/substitution positions are numbered relative to the ATG. A full-length cDNA clone of SDP2 has previously been published (GenBank accession number AY039980).
(C) Complementation of sdp2-4 by transformation with a genomic copy of MDAR4. Images of 5-d-old sdp2-4 (left) and sdp2-4 MDAR4 (right) seedlings grown on medium without sucrose. Bar = 1 cm.
(D) SDP2 transcript levels in 2-d-old wild-type and sdp2 seedlings detected using RT-PCR. The PCR primers amplify a 568-bp product from the 3′ end of the cDNA.
(E) SDP2 protein in peroxisomes purified from wild-type and sdp2-4 seedlings detected using protein gel blotting. *The larger polypeptide band corresponding to MDAR4 is 54 kD.
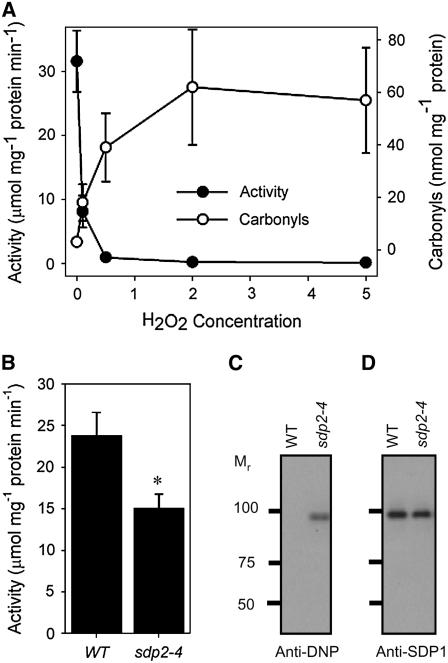
In the sdp2-1 mutant allele, a T-to-C mutation causes a Val-to-Ala amino acid substitution at position +14 in the polypeptide sequence (Figure 2B). In the sdp2-2 and sdp2-3 mutant alleles, A-to-G mutations give rise to Gly-to-Glu amino acid substitutions in the polypeptide sequence at positions +11 and +386, respectively (Figure 2B). An amino acid sequence alignment indicates that the Gly residues at positions +11 and +386 in MDAR4 are conserved among all Arabidopsis MDAR isoforms (Obara et al., 2002). The Val residue at position +14 is also conserved in MDAR1, 2, 3, and 4 but not in MDAR5 and 6 (Obara et al., 2002). Plant MDAR polypeptide sequences contain two putative nucleotide binding regions with the consensus motif V/IXGXGXXGXXXG/A (Drew et al., 2006). These motifs are likely to form Rossmann folds and interact with the adenine moieties of either the cofactor flavin adenine dinucleotide or the electron donor NADH (Drew et al., 2006). The mutations in sdp1-1 and sdp1-2 give rise to amino acid substitutions within the first putative nucleotide binding region. A T-DNA allele of MDAR4 (sdp2-4) that contains an insertion in exon 6 (Figure 2B) was also obtained from the SALK collection (Alonso et al., 2003). This line exhibits the same phenotype as the ethyl methanesulfonate alleles (Figure 1A). To confirm that the phenotype of sdp2-4 is caused by a defect in MDAR4, the mutant was complemented by transformation with a construct carrying a genomic copy of the gene (Figure 2C).
RT-PCR performed on RNA from 2-d-old seedlings showed that MDAR4 transcripts are present in the wild type, sdp2-1, sdp2-2, and sdp2-3 but are absent from sdp2-4 (Figure 2D). To investigate whether MDAR4 protein is present in the sdp2-4 allele, protein gel blots were performed on peroxisome-enriched fractions obtained by sucrose gradient centrifugation of crude seedling extracts (Eastmond et al., 2000b). A polyclonal antibody was used that was raised against a 47-kD MDAR from cucumber (Cucumis sativus). This antibody has been shown previously to recognize two polypeptides in purified Arabidopsis peroxisomes (Lisenbee et al., 2005). The 47-kD band corresponds to the matrix isoform MDAR1, and the 54-kD band corresponds to the membrane isoform MDAR4. Both bands were present in peroxisomal fractions from the wild type, but the 54-kD band was specifically absent in peroxisomal fractions from sdp2-4 (Figure 2E).
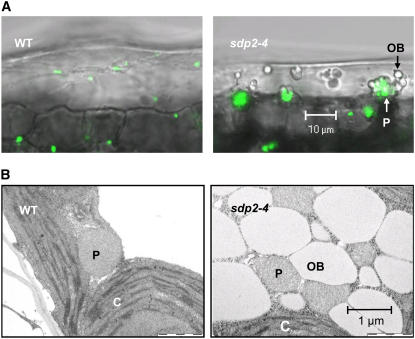
Peroxisomes and Oil Bodies Cluster Together in sdp2 Seedlings
It has been proposed that the APX/MDAR system on the peroxisomal membrane acts as a cordon to intercept H2O2 that is not detoxified by catalase and prevent it from damaging membrane lipids and integral proteins and escaping from the immediate vicinity of the peroxisome surface out into the cytosol (Yamaguchi et al., 1995; Mullen and Trelease, 1996). The lack of oil body lipase activity in germinating sdp2 seeds (Eastmond, 2006) might therefore be a symptom of oxidative damage. Confocal microscopy was used to investigate peroxisome morphology and proximity to oil bodies in living cells of sdp2 seedlings. In epidermal cells from the cotyledons of 5-d-old sdp2-4 seedlings, green fluorescent protein (GFP)–labeled peroxisomes formed large (5 to 10 μm) clusters that are associated with groups of transparent spherical undegraded oil bodies (Figure 3A). By contrast, in the wild type, essentially no oil bodies remained by this stage of development, and the individual peroxisomes were dispersed throughout the cytosol (Figure 3A). Electron microscopy images of sections through parenchyma cells from the cotyledons of 5-d-old seedlings also suggest an association between oil bodies and peroxisomes (Figure 3B). The striking peroxisome clustering phenotype in sdp2-4 was only observed in seedling tissues that contained undegraded oil bodies and was not obvious in tissues from other stages in plant development under the growth conditions used in this study (data not shown).
Figure 3.
Peroxisome Clustering and Proximity to Oil Bodies in sdp2 Seedlings.
Confocal images of epidermal cells (A) and electron microscopy images of parenchyma cells (B) of cotyledons from 5-d-old sdp2-4 and wild-type seedlings grown on medium containing 1% (w/v) sucrose. For confocal microscopy, peroxisomes were visualized using a genetic background containing PTS1-targeted GFP driven by the 35S promoter (Cutler et al., 2000). P, peroxisome; OB, oil body; C, chloroplast. Bars = 10 μm in (A) and 1 μm in (B).
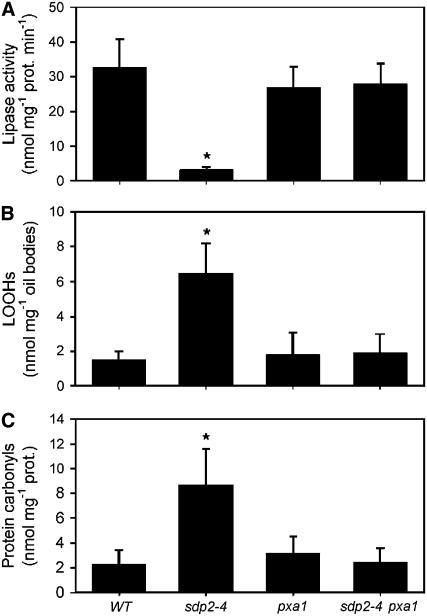
β-Oxidation Causes Oxidative Damage to Oil Bodies in sdp2 Seedlings
During the postgerminative growth of oilseeds, acyl-CoA oxidase, which is the first enzyme of peroxisomal fatty acid β-oxidation, is likely to be the major source of H2O2 production in the cell (Graham and Eastmond, 2002). To determine whether fatty acid β-oxidation inhibits oil body lipase activity in sdp2 seedlings, the sdp2-4 mutant was crossed into the fatty acid catabolism–deficient mutant pxa1. The PXA1/CTS/PED3 protein is an ABC transporter, which is required to import substrate (fatty acids or acyl-CoAs) into the peroxisome for β-oxidation (Zolman et al., 2001). TAG lipase activity was almost undetectable in oil body membranes prepared from 2-d-old sdp2-4 seedlings (Figure 4A). However, lipase activity was recovered to wild-type levels in the sdp2-4 pxa1 double mutant (Figure 4A). To investigate whether oil bodies from sdp2 incur oxidative damage, lipid peroxidation was estimated using the ferrous oxidation xylenol orange assay (Griffiths et al., 2000), and protein oxidation was monitored by detecting protein carbonyls (Levine et al., 1990). Both lipid hydroperoxides (LOOHs) and protein carbonyl levels were significantly elevated in oil bodies purified from 2-d-old sdp2-4 seedlings verses the wild type, and in both cases, these increased levels were suppressed in the pxa1 background (Figures 4B and 4C).
Figure 4.
β-Oxidation–Dependent Oxidative Damage to Oil Bodies in sdp2 Seedlings.
Rate of [14C]triolein hydrolysis (A), LOOH levels (B), and protein carbonyl levels (C) in oil bodies purified from 2-d-old sdp2-4, pxa1, sdp2-4 pxa1, and wild-type seedlings. The concentration of triolein was equivalent to 10 mM. Values are the mean ± se of measurements on five separate preparations. *Significantly different from the wild type (P < 0.05).
SDP1 Is a Target of Oxidative Damage in sdp2 Seedlings
A large proportion of the TAG lipase activity that is associated with oil body membranes is attributable to SDP1 (Eastmond, 2006), suggesting that this is a likely target of oxidative damage. In vitro incubation of purified recombinant SDP1 with H2O2 showed that the protein can be inactivated by concentrations >0.5 mM (Figure 5A). The loss of catalytic activity caused by H2O2 was accompanied by oxidation of the protein, as indicated by a 20-fold increase in the level of carbonylation (Figure 5A). Enzyme assays showed that recombinant SDP1 is still able to hydrolyze oil bodies purified from 2-d-old sdp2 seedlings but at a significantly lower rate than from the wild type (Figure 5B). This suggests that peroxidation of substrate and/or oxidation of additional oil body proteins also forms a partial barrier to oil hydrolysis by SDP1.
Figure 5.
Carbonylation and Inactivation of SDP1.
(A) Effect of H2O2 concentration on recombinant SDP1 activity and level of carbonylation. Purified recombinant SDP1 was preincubated with H2O2 for 2 h at 22°C before the protein was assayed for either lipase activity or carbonyl levels. Lipase assays were performed using 10 mM [14C]triolein as a substrate. Prior to the addition of the substrate, H2O2 was removed from the samples by adding 10 units mL−1 of catalase and incubating for 10 min at 22°C. Catalase assays confirmed that essentially all the H2O2 is removed by this treatment. Values are the mean ± se of measurements from four separate incubations.
(B) Activity of recombinant SDP1 on oil bodies purified from 2-d-old sdp2-4 and wild-type seedlings. Values are the mean ± se of measurements from four separate incubations. *Activity significantly different from the wild type (P < 0.05).
(C) Detection using anti-DNP antibodies of carbonyl groups in SDP1 immunoprecipitated from oil body membranes of 2-d-old sdp2-4 and wild-type seedlings.
(D) Detection of SDP1 protein in purified oil body membranes from 2-d-old sdp2-4 and wild-type seedlings using anti-SDP1 antibodies.
To investigate whether SDP1 suffers oxidative damage in sdp2-4 seedlings, the protein was purified from oil body membranes by immunoprecipitation using affinity-purified polyclonal antibodies raised against an SDP1 peptide. The purified protein was then reacted with 2,4-dintrophenylhydrazine (DNP) to derivatize carbonyl groups, and the carbonyl groups were detected with anti-DNP antibodies. Carbonyls could only be detected in SDP1 purified from sdp2-4 oil body proteins and not from the wild type (Figure 5C). The polyclonal antibodies raised against the SDP1 peptide detected similar amounts of SDP1 in oil body protein samples from sdp2-4 and the wild type (Figure 5D). Together, these data suggest that SDP1 is transcribed, translated, and targeted to oil bodies in sdp2 mutant seedlings but that it is then inactivated by oxidative damage.
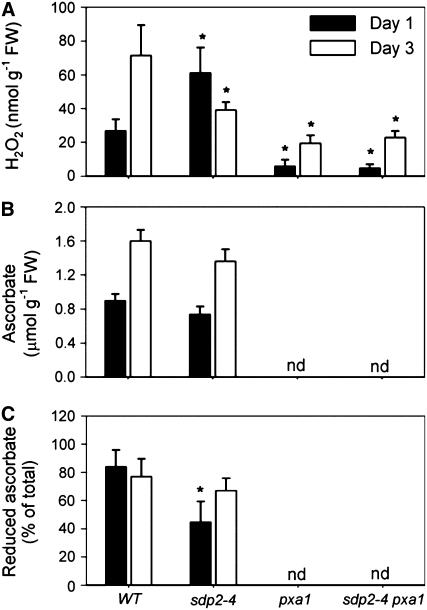
An Initial Increase in H2O2 Levels in sdp2 Is Likely to Result from a Failure to Recycle Ascorbate
The sdp2 mutant is defective in the second component of the APX/MDAR system (MDAR4). This mutation might be expected to reduce the capacity of seedlings to recycle ascorbate immediately following germination and lead to a shortage of substrate for APX and consequently cause a rise in H2O2 levels. To test this hypothesis, H2O2, the total ascorbate pool (ascorbate + dehydroascorbate), and the percentage of the ascorbate pool that is reduced were measured in 1- and 3-d-old sdp2-4 seedlings grown on medium with sucrose (Huang et al., 2005). At day 1, the total ascorbate pool in sdp2-4 was similar to the wild type, but the percentage of reduced ascorbate was significantly lower, and the level of H2O2 was increased (Figure 6). By contrast, at day 3, H2O2 levels in sdp2-4 were lower than those of the wild type (Figure 6A). The production of H2O2 in sdp2-4 pxa1 and pxa1 seedlings was strongly suppressed relative to sdp2-4 and the wild type, respectively, at both days 1 and 3 (Figure 6A). These data are consistent with a model in which immediately following sdp2 germination, the production of H2O2 by fatty acid β-oxidation leads to a negative feedback loop whereby the H2O2 inactivates SDP1 and progressively cuts off the supply of substrate for β-oxidation, which in turn reduces H2O2 production.
Figure 6.
Metabolite Levels in Germinating sdp2 Seedlings.
H2O2 (A), total ascorbate (ascorbate + dehydroascorbate) (B), and reduced ascorbate (C) as a percentage of the total in 1- and 3-d-old sdp2-4, pxa1, sdp2-4 pxa1, and wild-type seedlings germinated on medium containing 1% (w/v) sucrose. nd, not determined. Values are the mean ± se of measurements on five separate extracts. *Significantly different from the wild type (P < 0.05).
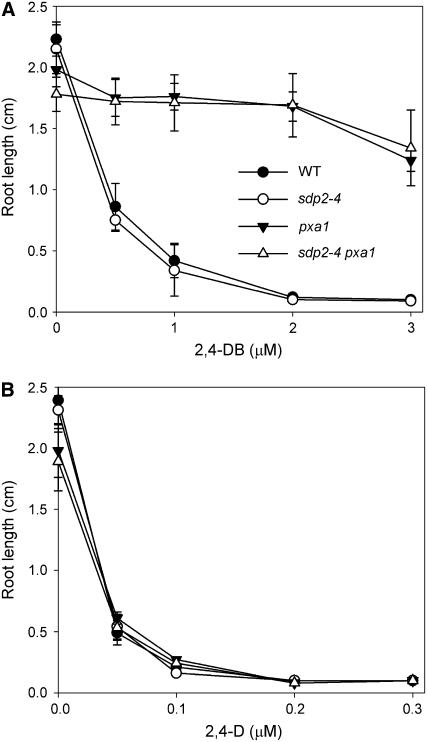
Seedlings of sdp2 Retain the Capacity to β-Oxidize 2,4-Dichlorophenoxybutyric Acid
By detoxifying H2O2 that escapes the peroxisome, the ascorbate-dependent electron transfer system has the potential to protect components within the peroxisomal matrix and membrane. MDAR4 may also play a specific role in fatty acid catabolism by recycling NAD+ within the peroxisome to support the l-3-hydroxyacyl-CoA dehydrogenase reaction of β-oxidation (Bowditch and Donaldson, 1990; Mullen and Trelease, 1996). To investigate whether either of these factors is likely to play a major role in the phenotype of sdp2, the mutant was grown on medium containing 2,4-dichlorophenoxybutyric acid (2,4-DB). This compound is converted to the herbicide 2,4-D by a single cycle of β-oxidation (Hayashi et al., 1998). Arabidopsis mutants in many genes that are either directly required for fatty acid β-oxidation or for general peroxisome function exhibit a 2,4-DB–resistant phenotype (Baker et al., 2006). To date, this list of mutants includes pxa1/cts/ped3, acx1, acx3, acx4, aim1, ped1/kat2, cys2 cys3, chy1, ped2/pex14, pmdh1 pmdh2, pex4, pex5, and pex6. However, unlike these mutants, root growth in sdp2-4 seedlings does not exhibit increased resistance to 2,4-DB (Figure 7), indicating that many peroxisomal membrane and matrix proteins remain at least partially functional. In vitro assays performed on extracts of 2-d-old sdp2-4 seedlings also show that the activities of peroxisomal marker enzymes, acyl-CoA oxidase, enoyl-CoA hydratase, l-3-hydroxyacyl-CoA dehydrogenase, 3-ketoacyl-CoA thiolase, isocitrate lyase, malate synthase, and catalase, are all similar to the wild type (Table 1).
Figure 7.
β-Oxidation of 2,4-DB by sdp2 Seedlings.
Effect of 2,4-DB concentration (A) and 2,4-D concentration (B) on root length of 5-d-old sdp2-4, pxa1, sdp2-4 pxa1, and wild-type seedlings grown on medium containing 1% (w/v) sucrose. Values are the mean ± se of measurements on five batches of 20 seedlings.
Table 1.
Peroxisomal and Oil Body Membrane Enzyme Activities in Wild-Type, sdp2, and CAT2HP2 Seedlings
| Activity (nmol mg−1 Protein min−1)
|
||||
|---|---|---|---|---|
| Enzyme | Wild Type (Col-0) | sdp2-4 | Wild Type (Col4) | CAT2HP2 |
| Peroxisome | ||||
| ACX (16:0) | 4.1 ± 0.7 | 3.6 ± 0.5 | 4.6 ± 0.3 | 4.0 ± 0.6 |
| ACX (10:0) | 10.7 ± 1.7 | 8.9 ± 0.9 | 11.9 ± 2.0 | 7.9 ± 3.3 |
| ACX (4:0) | 42.9 ± 3.8 | 40.1 ± 4.2 | 36.7 ± 2.9 | 34.8 ± 4.8 |
| ECH (4:0) | 46.3 ± 3.7 | 47.5 ± 4.9 | 48.6 ± 5.2 | 43.1 ± 5.0 |
| HAD (4:0) | 36.5 ± 3.8 | 39.0 ± 2.7 | 41.7 ± 4.4 | 37.6 ± 3.6 |
| KAT (4:0) | 54.9 ± 7.2 | 58.4 ± 9.4 | 57.9 ± 7.0 | 44.1 ± 11 |
| ICL | 79.6 ± 6.6 | 81.7 ± 5.2 | 87.3 ± 7.9 | 16.5 ± 2.0* |
| MLS | 47.2 ± 2.0 | 52.6 ± 3.7 | 50.0 ± 5.3 | 20.5 ± 1.9* |
| CAT | 2302 ± 245 | 2459 ± 319 | 2511 ± 304 | 211 ± 29* |
| Oil body | ||||
| Lipase | 21.9 ± 3.9 | 0.4 ± 0.1* | 23.9 ± 4.0 | 17.8 ± 3.5 |
| Cytosol | ||||
| GAPDH | 179 ± 24 | 166 ± 31 | nd | nd |
Seeds were germinated on medium containing 1% (w/v) sucrose. Peroxisomal enzyme activities were measured in whole extracts from 2-d-old seedlings. Acyl-CoA oxidase (ACX) activity was assayed using palmitoyl-CoA, decanoyl-CoA, and butyryl-CoA. Enoyl-CoA hydratase (ECH) was assayed using crotonyl-CoA. l-hydroxyacyl-CoA dehydrogenase (HAD) and 3-ketoacyl-CoA thiolase (KAT) activities were assayed using acetoacetyl-CoA. Lipase activity was measured in purified oil body membranes using 10 mM [14C]triolein as a substrate. Values are the mean ± se of measurements on three separate extracts. *Activity significantly different from the wild type (P < 0.001). ICL, isocitrate lyase; MLS, malate synthase; CAT, catalase; nd, not determined.
The activity of the cytosolic marker enzyme NADH-dependent glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was also measured in 2-d-old seedlings of sdp2-4 grown on medium containing sucrose (Table 1). This enzyme is an indicator of oxidative damage caused by H2O2 in Arabidopsis (Hancock et al., 2005; Job et al., 2005). The activity of GAPDH was not affected, suggesting that the deficiency in MDAR4 is unlikely to have caused a general increase in oxidative damage to cytosolic proteins.
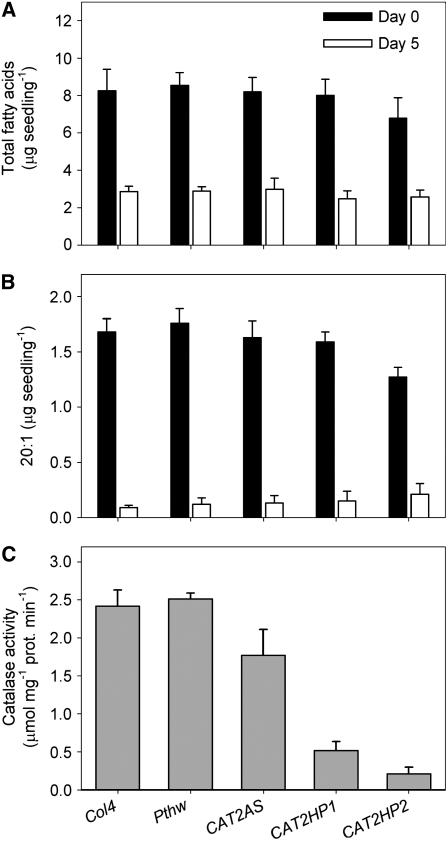
Catalase Deficiency Has a Comparatively Small Effect on Storage Oil Breakdown
To compare the effect of MDAR4 deficiency with that of catalase, a series of Arabidopsis CATALASE2 antisense lines (Vandenabeele et al., 2004) were screened for their ability to breakdown storage oil following germination. Surprisingly, unlike sdp2, none of these lines were strongly impaired in total fatty acid or eicosinoic acid breakdown when grown on medium containing sucrose (Figure 8). Enzyme assays confirmed that the lines had reduced catalase activity following germination, with CAT2HP2 exhibiting the lowest level (only 10% of the wild type; Figure 8C). Further analysis of CAT2HP2 seedlings revealed that the activities of specific peroxisomal enzymes (isocitrate lyase and malate synthase) are significantly reduced in comparison with the wild type, while oil body membrane lipase activity is not affected (Table 1). This indicates that a 90% reduction in catalase activity does impact negatively on peroxisome function, albeit insufficiently to prevent fatty acid β-oxidation from occurring when seedlings are provided with sucrose.
Figure 8.
Effect of Catalase Deficiency on Fatty Acid Breakdown during Postgerminative Growth.
(A) and (B) Total fatty acid (A) and eicosenoic acid (20:1) (B) content of seeds and 5-d-old seedlings grown on medium with 1% (w/v) sucrose. Col4, the wild type; Pthw, empty vector control; CAT2AS, CAT2HP1, and CAT2HP2, catalase-deficient lines (Vandenabeele et al., 2004). Values are the mean ± se of measurements on five batches of 20 seeds or seedlings.
(C) Catalase activities in extracts from 2-d-old seedlings of Col4, Pthw, CAT2AS, CAT2HP1, and CAT2HP2. Values are the mean ± se of measurements on three separate extracts.
DISCUSSION
Plant peroxisomes contain two major enzymatic antioxidant mechanisms capable of removing the highly toxic H2O2 that is generated as a by-product of oxidative metabolism (Mullen and Trelease, 1996). The first mechanism exploits catalase in the matrix and the second uses a membrane-bound ascorbate-dependent electron transfer system, requiring the cooperative action of APX and MDAR. Although the importance of catalase is well established, the physiological requirement for a component of the APX/MDAR system has yet to be demonstrated. In this study, I provide genetic evidence to show that a component of this system plays an essential role during the life cycle of Arabidopsis.
The seeds of many plants contain oil that serves as an essential source of carbon to drive postgerminative growth and allow photosynthetic establishment (Hayashi et al., 1998). The breakdown of this oil is accompanied by the generation of massive amounts of H2O2 within the peroxisome because H2O2 is formed as a by-product of the acyl-CoA oxidase step of fatty acid β-oxidation (Graham and Eastmond, 2002). To identify genes that are required for storage oil breakdown in Arabidopsis, I previously used a genetic screen in which mutants that require sucrose to support postgerminative growth were isolated (Eastmond, 2006). Three of these sdp mutants (sdp1, sdp2, and sdp3) were found to lack the lipase activity that is associated with their oil body membranes, explaining why they are unable to breakdown storage oil. SDP1 encodes a TAG lipase (Eastmond, 2006). Here, I show that SDP2 is the peroxisomal membrane isoform of MDAR (MDAR4), which constitutes one of the components of the ascorbate-dependent electron transfer system (Lisenbee et al., 2005).
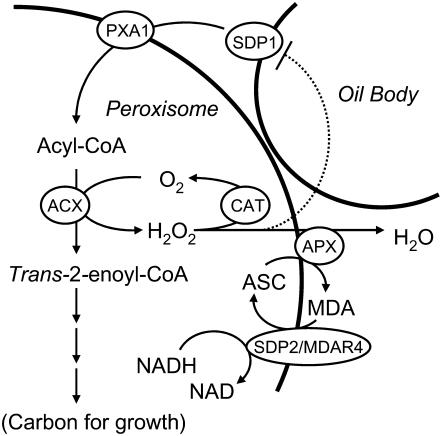
It has previously been reported that during periods of rapid β-oxidation, mammalian peroxisomes can leak H2O2 (Mueller et al., 2002). The most plausible explanation for the phenotype of sdp2 is that without the APX/MDAR system some of the H2O2 produced by acyl-CoA oxidase following seed germination escapes from the peroxisome and causes oxidative damage to oil bodies, inactivating SDP1 (Figure 9). Substantial data were obtained to support this hypothesis. Analysis of sdp2 seedlings immediately following germination confirmed that they have elevated levels of H2O2 and that their oil body proteins and lipids become oxidized. Furthermore, when sdp2 was introduced into a pxa1 background, which cannot β-oxidize fatty acids (Zolman et al., 2001), H2O2 levels, oxidative damage to oil bodies, and loss of lipase activity were all suppressed. The activity of SDP1 can be inhibited by H2O2 in vitro. Finally, oxidized SDP1 can be detected in oil bodies from sdp2 seedlings but not from the wild type. Inactivation of SDP1 is sufficient to account for much of the sugar-dependent phenotype of sdp2 (Eastmond, 2006). However, it cannot be discounted that additional proteins, which are necessary for oil hydrolysis, might also be damaged. Proteomic analysis by Job et al. (2005) has established that in wild-type Arabidopsis seeds and seedlings, proteins from many subcellular compartments exhibit a significant level of oxidation. However, no oxidized oil body proteins were identified in their study.
Figure 9.
Diagram Illustrating the Proposed Role of MDAR4 in Storage Oil Breakdown in Germinating Arabidopsis Seeds.
ASC, ascorbate; MDA, monodehydroascorbate; SDP1, lipase; PXA1, ABC transporter; ACX, acyl-CoA oxidase; CAT, catalase.
Unlike oil bodies, peroxisomes do not appear to depend so greatly on the APX/MDAR system for protection against H2O2. Seedlings of sdp2 are able to β-oxidize 2,4-DB. This capability implies that numerous proteins in the peroxisomal membrane and matrix retain at least part of their normal capacity (Baker et al., 2006). The activities of seven peroxisomal marker enzymes are also not adversely affected in sdp2. It is possible that a defect in the APX/MDAR system is not severely detrimental to peroxisomes because catalase remains active within the matrix. However, I cannot exclude the possibility that peroxisomes in sdp2 are damaged. Analysis of catalase antisense lines (Vandenabeele et al., 2004) showed that following Arabidopsis seed germination, certain peroxisomal enzyme activities are reduced, while lipase activity on oil body membranes is unaffected. In particular, the activity of the key glyoxylate cycle enzyme isocitrate lyase (Eastmond et al., 2000a) is reduced by 80% in the strongest catalase antisense line. This observation is consistent with recent in vitro data, which show that isocitrate lyase from castor bean (Ricinus communis) endosperm is readily inactivated by H2O2 and that this enzyme physically associates with catalase in the peroxisome (Nguyen and Donaldson, 2005; Yanik and Donaldson, 2005).
A number of studies have previously presented electron micrographic evidence to suggest that associations exist in oilseed cells between oil bodies and other subcellular compartments, principally peroxisomes (e.g., Wanner and Theimer, 1978; Hayashi et al., 2001). Interestingly, Chapman and Trelease (1991) have also provided biochemical evidence that in cotton (Gossypium hirsutum) seedlings, neutral lipids are transferred directly from the oil body into the peroxisomal membrane. It is therefore possible that physical contact plays a role in storage oil breakdown in oilseeds by facilitating the transfer of fatty acids from oil bodies to peroxisomes so that they can be β-oxidized. Using confocal microscopy, I was able to show that peroxisomes and oil bodies do cluster together in the cotyledon cells of living sdp2 seedlings and that this association persists as long as the oil bodies remain undegraded. It is not known whether peroxisome/oil body associations are influenced by H2O2 production, but it is noteworthy that H2O2 and UV-B light treatments have previously been reported to cause peroxisomes to cluster together with one another in animal and plant cells (Lingard and Trelease, 2006).
An association between oil bodies and peroxisomes would help explain why the deficiency in MDAR4 is so detrimental to storage oil hydrolysis since it would result in a close proximity between SDP1 and acyl-CoA oxidases, which generate H2O2. In yeast, a close association between oil bodies and peroxisomes has also recently been reported, and evidence has been provided that proteins involved in β-oxidation (e.g., acyl-CoA oxidase) are localized close to the inner surface of the peroxisomal membrane at sites of contact with the oil body (Binns et al., 2006). The concentration of H2O2 required to inhibit SDP1 in vitro (∼0.5 mM) is 16-fold higher than the estimated cytosolic concentration in 1-d-old sdp2 seedlings (∼0.03 mM). This concentration was calculated using the data from Figure 6A, assuming that the cytosol constitutes ∼20% of the seedling volume. However, H2O2 concentrations are unlikely to be uniform and could be heightened at, or near, the peroxisomal membrane, particularly if acyl-CoA oxidases are situated there.
The sdp2 mutant is defective in the second enzyme in the APX/MDAR system (MDAR4). Metabolite measurements suggest that in sdp2, the monodehydroascorbate produced at the peroxisomal membrane cannot be recycled efficiently, causing the availability of ascorbate to diminish and the APX/MDAR system to collapse. There are several alternative ways in which ascorbate could still be replenished for APX in the absence of MDAR4. In addition to MDAR4, Arabidopsis peroxisomes also contain the soluble matrix isoform MDAR1 (Lisenbee et al., 2005). Monodehydroascorbate can also disproportionate to ascorbate and dehydroascorbate, and biochemical studies have suggested that glutathione-dependent dehydroascorbate reductase activity is present in plant peroxisomes and therefore could convert dehydroascorbate back to ascorbate (Jiménez et al., 1997; del Rio et al., 1998). Finally, it has been argued that ascorbate and monodehydroascorbate are likely to be shuttled across the peroxisomal membrane since the catalytic site of MDAR4 is situated on the matrix side of the peroxisomal membrane, while the catalytic site of APX3 is on the cytosolic side (Lisenbee et al., 2005). Despite these possibilities, the phenotype of sdp2 suggests that none can complement the function of MDAR4 in protecting oil bodies against β-oxidation–dependent oxidative damage.
The predominant APX isoform from the peroxisomal membranes of Arabidopsis is APX3 (At4g35000). Surprisingly, Narendra et al. (2006) have recently reported that the APX3 gene is dispensable for growth and development. Therefore, a deficiency in APX activity might not give rise to the same phenotype as sdp2. Redundancy cannot be dismissed as an explanation for the apparent disparity, considering that there are nine APX genes in the Arabidopsis genome (Lisenbee et al., 2003). Specifically, there is a homolog of APX3 (APX5; At4g35970) with sequence similarity throughout the polypeptide sequence, including the C-terminal transmembrane domain and targeting motif. However, the level of expression of APX5 is very low relative to APX3 (Narendra et al., 2006).
My data do not preclude the possibility that MDAR4 functions independently of APX. Sattler et al. (2004) have recently shown that a deficiency in the lipid-soluble antioxidant tocopherols (vitamin E) also leads to lipid peroxidation after the germination of Arabidopsis seeds and that this adversely effects storage oil breakdown and seedling growth. Furthermore, sucrose can partially relieve the postgerminative growth defect. Tocopherols can scavenge lipid peroxy radicals, yielding a tocopheroxyl radical that could be recycled by reacting with ascorbate to produce monodehydroascorbate (Liebler, 1993). It is therefore possible that MDAR4 functions in a one-electron redox cycle that regenerates tocopherol from the tocopheroxyl radical at the peroxisomal membrane. However, it is unlikely that MDAR4 can operate solely by this mechanism since the sdp2 mutant has a more severe postgerminative growth arrest phenotype than the tocopherol-deficient vte2 mutant under normal growth conditions (Sattler et al., 2004). It is also possible that ascorbate could scavenge reactive oxygen species directly or that MDAR4 might be capable of recycling the oxidation products of other powerful antioxidants, such as phenolics (Sakihama et al., 2000). Indeed MDAR is unique in that it is the only enzyme known to use organic radicals as substrates (Hossain et al., 1984). Apart from its enzyme activity, MDAR4 might also act as an anchor for proteins associated with the peroxisomal inner membrane that lack a transmembrane domain, such as the long-chain acyl-CoA synthases LACS6 and LACS7 (Fulda et al., 2004).
Peroxisomal MDAR has also been implicated in fatty acid catabolism through the provision of NAD+ cofactor for l-3-hydroxyacyl-CoA dehydrogenase and malate dehydrogenase (Bowditch and Donaldson, 1990; Mullen and Trelease, 1996). These enzymes are required for β-oxidation and glyoxylate cycle function, respectively; theoretically, if H2O2 was detoxified entirely by APX/MDAR, the system could recycle sufficient NAD+ for both pathways (Mullen and Trelease, 1996). However, Mettler and Beevers (1980) have proposed an alternative scheme in which peroxisomal malate dehydrogenase operates in reverse to supply NAD+ for l-3-hydroxyacyl-CoA dehydrogenase via a metabolite shuttle, coupled to the mitochondrial electron transport chain. Malate dehydrogenase has since been shown to perform this role in yeast (van Roermund et al., 1995). Analysis of sdp2 suggests that MDAR4 is not essential for providing NAD+ in Arabidopsis peroxisomes since the mutant can β-oxidize 2,4-DB. By contrast, Pracharoenwattana et al. (2007) have shown that the glyoxylate cycle can still function in Arabidopsis pmdh1 pmdh2 mutant seedlings, which lack peroxisomal malate dehydrogenase activity, but that this double mutant is impaired in both the β-oxidation of 2,4-DB and oil breakdown. These data provide evidence to suggest that the metabolite shuttle proposed by Mettler and Beevers (1980) is necessary to supply NAD+ for fatty acid β-oxidation (Pracharoenwattana et al., 2007). Within the peroxisome, MDAR4 (and MDAR1) activity must regenerate some NAD+. However, it is likely that a proportion of the H2O2 generated by acyl-CoA oxidase is detoxified by catalase, and in this event, the regeneration of NAD+ by the APX/MDAR system alone could not sustain both l-3-hydroxyacyl-CoA dehydrogenase and forward malate dehydrogenase activities. The result would be a negative feedback loop, which could stall β-oxidation. The reversible activity of peroxisomal malate dehydrogenase might be essential for β-oxidation, not because it supplies all the NAD+ but because it can balance the shortfall.
In addition to fatty acid β-oxidation, the photorespiratory pathway also generates large quantities of H2O2 in plant peroxisomes as a result of the activity of glycolate oxidase (Willekens et al., 1997). Catalase has been shown to play a major role in detoxifying this H2O2. Antisense suppression of catalase results in oxidative damage and triggers cell death in tobacco (Nicotiana tabacum) and Arabidopsis plants that are subjected to high light treatment (Willekens et al., 1997; Vandenabeele et al., 2004). Although public microarray data show that MDAR4 is expressed in leaves, sdp2 plants do not exhibit more obvious phenotypic symptoms of oxidative damage than the wild type when they are subjected to high light (data not shown). Further studies will be required to determine if MDAR4 plays a role associated with photorespiration. Reactive oxygen species also play important signaling roles in plants (Hancock et al., 2005), and it is conceivable that inhibition of SDP1 activity might be a significant mechanism in the regulation of oil hydrolysis during postgerminative growth.
In conclusion, catalase and the APX/MDAR system are both important parts of the peroxisomal antioxidant machinery during the postgerminative growth of Arabidopsis seedlings. Our data suggest that their roles are physiologically different and that neither can fully compensate for the loss of the other. Catalase protects constituents of the peroxisomal matrix from oxidative damage, while the main role of MDAR4 appears to be to prevent H2O2 from escaping beyond the outer surface of the peroxisomal membrane. The consequences of H2O2 escape appear to be fatal primarily because inactivation of TAG hydrolysis on closely associated oil bodies prevents the seedling from releasing the carbon skeletons and energy that it needs for initial postgerminative growth.
METHODS
Plant Material and Growth Conditions
Wild-type Arabidopsis thaliana (ecotypes Columbia-0 and Landsberg erecta) were obtained from the Nottingham Arabidopsis Stock Centre (University of Nottingham, UK). The sdp2-4 mutant (SALK_068667) was obtained from T-DNA express (Alonso et al., 2003). The PTS1-targeted GFP line A5 (Cutler et al., 2000), the pxa1 mutant (Zolman et al., 2001), and catalase-deficient lines (Vandenabeele et al., 2004) were kindly provided by Chris Somerville (Carnegie Institution, Stanford University, CA), Bonnie Bartel (Rice University, Houston, TX), and Frank Van Breusegem (Ghent University, Flanders Interuniversity Institute for Biotechnology, Belgium), respectively. Seeds were surface-sterilized, applied to agar plates containing half-strength Murashige and Skoog salts (Sigma-Aldrich), and imbibed in the dark for 4 d at 4°C. The plates were then transferred to a growth chamber set to 21°C (16 h light/8 h dark; PPFD = 150 μmol m−2 s−1). In some experiments, 1% (w/v) sucrose, 2,4-DB, or 2,4-D were added to the agar medium.
Metabolite Analysis
The fatty acid composition of seed and seedling lipids was measured by gas chromatography analysis after combined digestion and fatty acid methyl ester formation from frozen tissue using the method of Browse et al. (1986). H2O2 and ascorbate (ascorbate and dehydroascorbate) were extracted from 0.1 g fresh weight of seedlings and measured spectophotometrically using the methods described by Huang et al. (2005).
Mapping
The sdp2-1 mutant was outcrossed to the wild-type ecotype Landsberg erecta. F1 plants were allowed to self-fertilize, and the F2 progeny were screened for the sugar-dependent phenotype. Genomic DNA was isolated from 190 F2 sdp2-1 lines using the Extract-N-Amp Plant PCR kit (Sigma-Aldrich). Mapping was performed using simple sequence polymorphisms (Bell and Ecker, 1994), cleaved amplified polymorphic sequences (Konieczny and Ausubel, 1993), and single nucleotide amplified polymorphisms (Drenkard et al., 2000) using information from the Monsanto Arabidopsis polymorphism collection (Jander et al., 2002). New markers used in this study are listed in Supplemental Table 2 online. Candidate genes within the mapping interval were amplified from sdp2-1, sdp2-2, and sdp2-3 genomic DNA by PCR and sequenced to identify mutations.
Transcript and Protein Analysis
DNase-treated total RNA was isolated from Arabidopsis seedlings using the RNeasy kit from Qiagen. The synthesis of single-stranded cDNA was performed using SuperScript II RNase H− reverse transcriptase from Invitrogen. SDP2 transcripts were detected by PCR using primers SDP2F (5′-CAAAGACGGGAGCCACTTAC-3′) and SDP2R (5′-CTGCTGACTCACAACCGTGT-3′). Measurement of total protein levels, SDS-PAGE, and protein gel blot analysis were performed as described previously (Eastmond, 2004). Rabbit anti-cucumber MDAR antiserum, anti-DNP antibody (Millipore), and anti-SDP1 antibody were used as primary antibodies at dilutions of 1 in 1000, 1 in 30, and 1 in 500, respectively. The SDP1 antibody was raised against the SDP1-specific peptide SSEDSGLQEPVSGSVC by Eurogentec and affinity purified.
Microscopy
Transmission electron microscopy was performed as described previously (Eastmond, 2006). Five-day-old Arabidopsis seedlings were fixed for 2 h in 2.5% (v/v) glutaraldehyde and 4% (v/v) formaldehyde in 100 mM phosphate buffer, pH 7.0, with a secondary fixation of 1% (w/v) osmium tetroxide in phosphate buffer for 1 h. The tissue was embedded in Spurr's resin, sectioned, and stained with uranyl acetate and Reynolds lead citrate. Confocal microscopy was performed using a Zeiss LSM 510 Meta on an Axioplan 2M, fitted with a ×63 PlanApo lens (numerical aperture of 1.4). The sample was excited with a 488-nm argon laser and GFP emission collected via a 505- to 530-nm band-pass filter. Bright-field images were captured simultaneously with the transmission detector.
Organelle Purification, Protein Purification, and Measurement of Oxidative Damage
Oil bodies and oil body membranes were purified from 2-d-old Arabidopsis seedlings, and recombinant N-terminal His6-tagged SDP1 was expressed and purified using protocols that were described previously (Eastmond, 2006). Peroxisomal fractions were obtained from homogenates of 5-d-old etiolated Arabidopsis seedlings using sucrose density gradient centrifugation as described previously (Eastmond et al., 2000b). The levels of LOOHs in purified oil body lipids were estimated using the ferrous oxidation xylenol orange assay following the protocol described by Sattler et al. (2004). The only modification to the method was that the lipids were extracted from oil bodies in 1.5-mL tubes without homogenization. The levels of protein carbonyls in oil body membranes and purified recombinant SDP1 were determined using the spectrophotometric quantification method described by Nguyen and Donaldson (2005). SDP1 was immunoprecipitated from oil body membranes using the IP50 Protein G Immunoprecipitation kit (Sigma-Aldrich), and carbonyl groups were detected using the OxyBlot protein oxidation detection kit (Millipore) as described by Nguyen and Donaldson (2005).
Enzyme Assays
TAG lipase activity was measured in purified oil body membranes and recombinant SDP1 using an emulsion of [14C]triolein as described previously (Eastmond, 2006). Purified oil bodies were also used as a substrate for recombinant SDP1 using the assay procedure described by Eastmond (2006). For assays of peroxisomal enzymes, 2-d-old seedlings grown on medium containing 1% (w/v) sucrose were ground in a pestle and mortar with 1 mL of buffer (150 mM Tris/HCl, pH 7.5, 10 mM KCl, 1 mM EDTA, 10 mM flavin adenine dinucleotide, and 10% [v/v] glycerol). The extract was clarified by centrifugation at 15,000g for 30 min at 4°C, and the supernatant was desalted using a Sephadex G-50 spin column. The extract was assayed spectrophotometrically for acyl-CoA oxidase, enoyl-CoA hydratase, l-3-hydroxyacyl-CoA dehydrogenase, 3-ketoacyl-CoA thiolase, isocitrate lyase, malate synthase, catalase, and NADH-dependent glyceraldehyde-3-phosphate dehydrogenase activitities according to methods described previously (Takahashi et al., 1997; Eastmond et al., 2000a; Hancock et al., 2005; Rylott et al., 2006).
Complementation of sdp2-4
A region of genomic DNA containing MDAR4 was amplified from Arabidopsis using primers 5′-ATCTCATGATTGAGTGGGTGATTGGTTG-3′ and 5′-AGCTTCTTCGAGGGTTAGGGATGAGATT-3′, and the product was cloned into the pCR2.1-TOPO vector from Invitrogen. Using standard molecular biology techniques, MDAR4 was excised and cloned into the pGREENII vector (Hellens et al., 2000). The MDAR4 construct was transformed into Agrobacterium tumefaciens strain GV3101 containing the pSOUP vector (Hellens et al., 2000) by electroporation and into Arabidopsis sdp2-4 ecotype Columbia by the floral dip method (Clough and Bent, 1998). Transformants containing the T-DNA were selected by screening for loss of a sugar-dependent phenotype, and the presence of wild-type MDAR4 transcripts was confirmed by RT-PCR.
Accession Number
Sequence data for SDP2 (MDAR4) cDNA can be found in the GenBank/EMBL data libraries under accession number AY039980.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table 1. Frequency of sdp2 Germination.
Supplemental Table 2. New SSLP and CAPS Markers Used to Map SDP2.
Supplementary Material
Acknowledgments
I thank Lynda Sainty (University of York, UK) for her invaluable technical assistance. I also thank Karen Chance and Meg Stark from the University of York, Biology Department Technology Facility for their assistance with sample preparation and analysis using confocal and electron microscopy. Satoshi Sano (Kyoto Prefectural University, Japan) generously provided the cucumber MDAR antiserum used in this study. Frank Van Breusegem (Ghent University, Flanders Interuniversity Institute for Biotechnology, Belgium) kindly provided the catalase antisense lines. I also appreciate the helpful suggestions made by the coeditor and reviewers. The work was funded by the Biotechnology and Biological Sciences Research Council via a David Phillips Research Fellowship (87/JF/16985) awarded to P.J.E.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Peter J. Eastmond (p.j.eastmond@warwick.ac.uk).
Online version contains Web-only data.
References
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Baker, A., Graham, I.A., Holdsworth, M., Smith, S.M., and Theodoulou, F.L. (2006). Chewing the fat. New roles for β-oxidation in plant signalling and development. Trends Plant Sci. 11 124–132. [DOI] [PubMed] [Google Scholar]
- Bell, C.J., and Ecker, J.R. (1994). Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19 137–144. [DOI] [PubMed] [Google Scholar]
- Binns, D., Januszewski, T., Chen, Y., Hill, J., Markin, V.S., Zhao, Y., Gilpin, C., Chapman, K.D., Anderson, R.G., and Goodman, J.M. (2006). An intimate collaboration between peroxisomes and lipid bodies. J. Cell Biol. 173 719–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowditch, M.L., and Donaldson, R.P. (1990). Ascorbate free-radical reduction by glyoxysomal membranes. Plant Physiol. 94 531–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browse, J., Mc Court, P.J., and Somerville, C.R. (1986). Fatty acid composition of leaf lipids determined after combined digestion and fatty acid methyl ester formation from fresh tissue. Anal. Biochem. 152 141–145. [DOI] [PubMed] [Google Scholar]
- Bunkelmann, J.R., and Trelease, R.N. (1996). Ascorbate peroxidase: A promenent membrane protein in oilseed glyoxysomes. Plant Physiol. 110 589–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman, K.D., and Trelease, R.N. (1991). Acquisition of membrane lipids by differentiating glyoxysomes: Role of lipid bodies. J. Cell Biol. 115 995–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Cutler, S.R., Ehrhardt, D.W., Griffitts, J.S., and Somerville, C.R. (2000). Random GFP∷cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc. Natl. Acad. Sci. USA 97 3718–3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Rio, L.A., Sandalio, L.M., Corpas, F.J., López-Huertas, E., Palma, J.M., and Pastori, G.M. (1998). Activated oxygen-mediated metabolic functions of leaf peroxisomes. Physiol. Plant 104 673–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drenkard, E., Richter, B.G., Rozen, S., Stutius, L.M., Angell, N.A., Mindrinos, M., Cho, R.J., Oefner, P.J., Davis, R.W., and Ausubel, F.M. (2000). A simple procedure for the analysis of single nucleotide polymorphisms facilitates map-based cloning in Arabidopsis. Plant Physiol. 124 1483–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew, D.P., Lunde, C., Lahnstein, J., and Fincher, G.B. (2006). Heterologous expression of cDNAs encoding monodehydroascorbate reductases from the moss Physcomitrella patens and characterization of the expressed enzymes. Planta 225 945–954. [DOI] [PubMed] [Google Scholar]
- Eastmond, P.J. (2004). Cloning and characterization of the acid lipase from castor beans. J. Biol. Chem. 279 45540–45545. [DOI] [PubMed] [Google Scholar]
- Eastmond, P.J. (2006). SUGAR-DEPENDENT1 encodes a patatin domain triacylglycerol lipase that initiates storage oil breakdown in germinating Arabidopsis seeds. Plant Cell 18 665–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastmond, P.J., Germain, V., Lange, P.R., Bryce, J.H., Smith, S.M., and Graham, I.A. (2000. a). Postgerminative growth and lipid catabolism in oilseeds lacking the glyoxylate cycle. Proc. Natl. Acad. Sci. USA 97 5669–5674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastmond, P.J., Hooks, M.A., Williams, D., Lange, P., Bechtold, N., Sarrobert, C., Nussaume, L., and Graham, I.A. (2000. b). Promoter trapping of a novel medium-chain acyl-CoA oxidase, which is induced transcriptionally during Arabidopsis seed germination. J. Biol. Chem. 275 34375–34381. [DOI] [PubMed] [Google Scholar]
- Fulda, M., Schnurr, J., Abbadi, A., Heinz, E., and Browse, J. (2004). Peroxisomal Acyl-CoA synthetase activity is essential for seedling development in Arabidopsis thaliana. Plant Cell 16 394–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham, I.A., and Eastmond, P.J. (2002). Pathways of straight and branched chain fatty acid catabolism in higher plants. Prog. Lipid Res. 41 156–181. [DOI] [PubMed] [Google Scholar]
- Griffiths, G., Leverentz, M., Silkowski, H., Gill, N., and Sanchez-Serrano, J.J. (2000). Lipid hydroperoxide levels in plant tissues. J. Exp. Bot. 51 1363–1370. [PubMed] [Google Scholar]
- Hayashi, Y., Hayashi, M., Hayashi, H., Hara-Nishimura, I., and Nishimura, M. (2001). Direct interaction between glyoxysomes and lipid bodies in cotyledons of the Arabidopsis thaliana ped1 mutant. Protoplasma 218 83–94. [DOI] [PubMed] [Google Scholar]
- Hayashi, H., Toriyama, K., Kondo, M., and Nishimura, M. (1998). 2,4-Dichlorophenoxybutyric acid-resistant mutants of Arabidopsis have defects in glyoxysomal fatty acid beta-oxidation. Plant Cell 10 183–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock, J.T., Henson, D., Nyirenda, M., Desikan, R., Harrison, J., Lewis, M., Hughes, J., and Neill, S.J. (2005). Proteomic identification of glyceraldehyde 3-phosphate dehydrogenase as an inhibitory target of hydrogen peroxide in Arabidopsis. Plant Physiol. Biochem. 43 828–835. [DOI] [PubMed] [Google Scholar]
- Hellens, R.P., Edwards, E.A., Leyland, N.R., Bean, S., and Mullineaux, P.M. (2000). pGreen: A versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 42 819–832. [DOI] [PubMed] [Google Scholar]
- Horiguchi, H., Yurimoto, H., Toh-kheng, G., Nakagawa, T., Kato, N., and Sakai, Y. (2001). Peroxisomal catalase in the methylotrophic yeast Candida boidinni: Transport efficiency and metabolic significance. J. Bacteriol. 183 6372–6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain, M.A., Nakano, Y., and Asada, K. (1984). Monodehydroascorbate reductase in spinach chloroplasts and its participation in regeneration of ascorbate for scavenging hydrogen peroxide. Plant Cell Physiol. 25 385–395. [Google Scholar]
- Huang, C., He, W., Guo, J., Chang, X., Su, P., and Zhang, L. (2005). Increased sensitivity to salt stress in an ascorbate-deficient Arabidopsis mutant. J. Exp. Bot. 56 3041–3049. [DOI] [PubMed] [Google Scholar]
- Jander, G., Norris, S.R., Rounsley, S.D., Bush, D.F., Levin, I.M., and Last, R.L. (2002). Arabidopsis map-based cloning in the post-genome era. Plant Physiol. 129 440–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez, A., Hernández, J.A., del Rio, L.A., and Sevilla, F. (1997). Evidence for the presence of the ascorbate-glutathione cycle in mitochondria and peroxisomes of pea leaves. Plant Physiol. 114 275–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Job, C., Rajjou, L., Lovigny, Y., Belghazi, M., and Job, D. (2005). Patterns of protein oxidation in Arabidopsis seeds and during germination. Plant Physiol. 138 790–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karyotou, K., and Donaldson, R.P. (2005). Ascorbate peroxidase, a scavenger of hydrogen peroxide in glyoxysomal membranes. Arch. Biochem. Biophys. 434 248–257. [DOI] [PubMed] [Google Scholar]
- Kendall, A.C., Keys, A.J., Turner, J.C., Lea, P.J., and Miflin, B.J. (1983). The isolation and characterization of a catalase-deficient mutant of barley (Hordeum vulgare). Planta 159 505–511. [DOI] [PubMed] [Google Scholar]
- Konieczny, A., and Ausubel, F.M. (1993). A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 4 403–410. [DOI] [PubMed] [Google Scholar]
- Lemieux, B., Miquel, M., Somerville, C., and Browse, J. (1990). Mutants of Arabidopsis with alterations in seed lipid fatty-acid composition. Theor. Appl. Genet. 80 234–240. [DOI] [PubMed] [Google Scholar]
- Levine, R.L., Garland, D., Oliver, C.N., Amici, A., Climent, I., Lenz, A.G., Ahn, B.W., Shaltiel, S., and Stadtman, E.R. (1990). Determination of carbonyl content in oxidatively modified proteins. Methods Enzymol. 186 464–478. [DOI] [PubMed] [Google Scholar]
- Liebler, D.C. (1993). The role of metabolism in the antioxidant function of vitamin E. Crit. Rev. Toxicol. 23 147–169. [DOI] [PubMed] [Google Scholar]
- Lingard, M.J., and Trelease, R.N. (2006). Five Arabidopsis peroxin 11 homologs individually promote peroxisome elongation, duplication or aggregation. J. Cell Sci. 119 1961–1972. [DOI] [PubMed] [Google Scholar]
- Lisenbee, C.S., Heinze, M., and Trelease, R.N. (2003). Peroxisomal ascorbate peroxidase resides within a subdomain of rough endoplasmic reticulum in wild-type Arabidopsis cells. Plant Physiol. 132 870–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisenbee, C.S., Lingard, M.J., and Trelease, R.N. (2005). Arabidopsis peroxisomes possess functionally redundant membrane and matrix isoforms of monodehydroascorbate reductase. Plant J. 43 900–914. [DOI] [PubMed] [Google Scholar]
- Mettler, I.J., and Beevers, H. (1980). Oxidation of NADH in glyoxysomes by a malate-aspartate shuttle. Plant Physiol. 66 555–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller, S., Weber, A., Fritz, R., Mutze, S., Rost, D., Walczak, H., Volkl, A., and Stremmel, W. (2002). Sensitive and real-time determination of H2O2 release from intact peroxisomes. Biochem. J. 363 483–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen, R.T., and Trelease, R.N. (1996). Biogenesis and membrane properties of peroxisomes: Does the boundary membrane serve and protect? Trends Plant Sci. 1 389–394. [Google Scholar]
- Narendra, S., Venkataramani, S., Shen, G., Wang, J., Pasapula, V., Lin, Y., Kornyeyev, D., Holaday, A.S., and Zhang, H. (2006). The Arabidopsis ascorbate peroxidase 3 is a peroxisomal membrane-bound antioxidant enzyme and is dispensable for Arabidopsis growth and development. J. Exp. Bot. 57 3033–3042. [DOI] [PubMed] [Google Scholar]
- Nguyen, A.T., and Donaldson, R.P. (2005). Metal-catalyzed oxidation induces carbonylation of peroxisomal proteins and loss of enzymatic activities. Arch. Biochem. Biophys. 439 25–31. [DOI] [PubMed] [Google Scholar]
- Obara, K., Sumi, K., and Fukuda, H. (2002). The use of multiple transcription starts causes the dual targeting of Arabidopsis putative monodehydroascorbate reductase to both mitochondria and chloroplasts. Plant Cell Physiol. 43 697–705. [DOI] [PubMed] [Google Scholar]
- Pracharoenwattana, I., Cornah, J.E., and Smith, S.M. (March 21, 2007). Arabidopsis peroxisomal malate dehydrogenase functions in β-oxidation but not in the glyoxylate cycle. Plant J. http://dx.doi.org/10.1111/j.1365–313X.2007.03055.x. [DOI] [PubMed]
- Rylott, E.L., Eastmond, P.J., Gilday, A.D., Slocombe, S.P., Larson, T.R., Baker, A., and Graham, I.A. (2006). The Arabidopsis thaliana multifunctional protein gene (MFP2) of peroxisomal β-oxidation is essential for seedling establishment. Plant J. 45 930–941. [DOI] [PubMed] [Google Scholar]
- Sakihama, Y., Mano, J., Sano, S., Asada, K., and Yamasaki, H. (2000). Reduction of phenoxyl radicals mediated by monodehydroascorbate reductase. Biochem. Biophys. Res. Commun. 279 949–954. [DOI] [PubMed] [Google Scholar]
- Sattler, S.E., Gilliland, L.U., Magallanes-Lundback, M., Pollard, M., and DellaPenna, D. (2004). Vitamin E is essential for seed longevity and for preventing lipid peroxidation during germination. Plant Cell 16 1419–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheikh, F.G., Pahan, K., Khan, M., Barbosa, E., and Singh, I. (1998). Abnormality in catalase import into peroxisomes leads to severe neurological disorder. Proc. Natl. Acad. Sci. USA 95 2961–2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, I. (1997). Biochemistry of peroxisomes in health and disease. Mol. Cell. Biochem. 167 1–29. [DOI] [PubMed] [Google Scholar]
- Takahashi, H., Chen, Z., Du, H., Liu, Y., and Klessig, D.F. (1997). Development of necrosis and activation of disease resistance in transgenic tobacco plants with severely reduced catalase levels. Plant J. 11 993–1005. [DOI] [PubMed] [Google Scholar]
- Vandenabeele, S., Vanderauwera, S., Vuylsteke, M., Rombauts, S., Langebartels, C., Seidlitz, H.K., Zabeau, M., Van Montagu, M., Inze, D., and Van Breusegem, F. (2004). Catalase deficiency drastically affects gene expression induced by high light in Arabidopsis thaliana. Plant J. 39 45–58. [DOI] [PubMed] [Google Scholar]
- van den Bosch, H., Schutgens, R.B., Wanders, R.J., and Tager, J.M. (1992). Biochemistry of peroxisomes. Annu. Rev. Biochem. 61 157–197. [DOI] [PubMed] [Google Scholar]
- van Roermund, C.W., Elgersma, Y., Singh, N., Wanders, R.J., and Tabak, H.F. (1995). The membrane of peroxisomes in Saccharomyces cerevisiae is impermeable to NAD(H) and acetyl-CoA under in vivo conditions. EMBO J. 14 3480–3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J., Zhang, H., and Allen, R.D. (1999). Overexpression of an Arabidopsis peroxisomal ascorbate peroxidase gene in tobacco increases protection against oxidative stress. Plant Cell Physiol. 40 725–732. [DOI] [PubMed] [Google Scholar]
- Wanner, G., and Theimer, R.R. (1978). Membranous appendices of spherosomes (oleosomes). Possible role in fat utilization in germinating oil seeds. Planta 140 163–169. [DOI] [PubMed] [Google Scholar]
- Willekens, H., Chamnongpol, S., Davey, M., Schraudner, M., Langebartels, C., Van Montagu, M., Inzé, D., and Van Camp, W. (1997). Catalase is a sink for H2O2 and is indispensable for stress defense in C3 plants. EMBO J. 16 4806–4816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi, K., Mori, H., and Nishimura, M. (1995). A novel isoenzyme of ascorbate peroxidase localized on glyoxysomal and leaf peroxisomal membranes in pumpkin. Plant Cell Physiol. 36 1157–1162. [DOI] [PubMed] [Google Scholar]
- Yanik, T., and Donaldson, R.P. (2005). A protective association between catalase and isocitrate lyase in peroxisomes. Arch. Biochem. Biophys. 435 243–252. [DOI] [PubMed] [Google Scholar]
- Zhang, J.W., Han, Y., and Lazarow, P.B. (1993). Novel peroxisome clustering mutants and peroxisome biogenesis mutants of Saccharomyces cerevisiae. J. Cell Biol. 123 1133–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman, B.K., Silva, I.D., and Bartel, B. (2001). The Arabidopsis pxa1 mutant is defective in an ATP-binding cassette transporter-like protein required for peroxisomal fatty acid beta-oxidation. Plant Physiol. 127 1266–1278. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.