Abstract
Rhizobial bacteria activate the formation of nodules on the appropriate host legume plant, and this requires the bacterial signaling molecule Nod factor. Perception of Nod factor in the plant leads to the activation of a number of rhizobial-induced genes. Putative transcriptional regulators in the GRAS family are known to function in Nod factor signaling, but these proteins have not been shown to be capable of direct DNA binding. Here, we identify an ERF transcription factor, ERF Required for Nodulation (ERN), which contains a highly conserved AP2 DNA binding domain, that is necessary for nodulation. Mutations in this gene block the initiation and development of rhizobial invasion structures, termed infection threads, and thus block nodule invasion by the bacteria. We show that ERN is necessary for Nod factor–induced gene expression and for spontaneous nodulation activated by the calcium- and calmodulin-dependent protein kinase, DMI3, which is a component of the Nod factor signaling pathway. We propose that ERN is a component of the Nod factor signal transduction pathway and functions downstream of DMI3 to activate nodulation gene expression.
INTRODUCTION
Legume/rhizobial symbiosis plays an important role in the nitrogen cycle through the fixation of atmospheric dinitrogen. Recognition of rhizobial bacteria by the appropriate legume host induces the formation of a unique structure, the nodule, on the plant root. Microaerobic conditions are maintained within the nodule to allow activity of the bacterial enzyme nitrogenase that is responsible for nitrogen fixation.
The early signaling and transcriptional events leading to nodule formation have been well studied. Rhizobia recognize flavonoids released from the host legume roots and respond with the production of lipochito-oligosaccharide signaling molecules termed Nod factors (Long, 1996). Nod factors can induce many of the early plant responses associated with nodulation: within seconds of Nod factor perception, the root hair cells exhibit ion fluxes followed by calcium spiking (Ehrhardt et al., 1996); after a few hours, early nodulation genes (ENODs) are induced (Vernoud et al., 1999; Journet et al., 2001; Chabaud et al., 2002; Mitra et al., 2004a) and root hair deformation is observed (Lerouge et al., 1990); and after 24 h, cortical cells reactivate mitotic growth to form the nodule primordium (Franssen et al., 1995).
Root hair deformation leads to the entrapment of rhizobial bacteria within an infection focus inside the curled root hair. Infection threads elongate from this infection focus, and this allows bacterial invasion into the developing nodule primordium. The infection thread is an invagination of the cell wall and plasma membrane, forming a tube-like structure that extends from the infection focus toward the cortex (Gage, 2002). Cytoplasmic changes, likened to the formation of the premitotic plate, precede the initiation of infection thread growth (Brewin, 2004). Extension of the infection thread is coupled with bacterial division at the elongating tip such that the bacteria constantly fill the growing thread (Gage, 2002). The infection thread ramifies into the dividing cortical cells where the bacteria are released, bound by plant-derived membrane, into the host cells where they differentiate into nitrogen-fixing bacteroids.
Although biochemical proof is yet to be obtained, it is widely believed that Nod factors are perceived by receptor-like kinases that contain sugar binding LysM domains. Such receptor-like kinases exist as gene families with at least three members, NFP, LYK3, and LYK4 (Ben Amor et al., 2003; Limpens et al., 2003; Arrighi et al., 2006), in Medicago truncatula and two members, NFR1 and NFR5 (Radutoiu et al., 2003), in Lotus japonicus. It has been proposed that heterodimers between different family members may act in concert to bind and perceive Nod factors and transduce the signal to the intracellular environment (Madsen et al., 2003; Parniske and Downie, 2003; Radutoiu et al., 2003; Oldroyd and Downie, 2004). Mutant alleles of several putative receptors show little or no response to Nod factors, thereby strengthening the hypothesis that they participate in Nod factor perception (Ben Amor et al., 2003; Limpens et al., 2003; Radutoiu et al., 2003).
Downstream of the hypothetical Nod factor receptors in M. truncatula are a receptor-like kinase with Leu-rich repeat motifs, DMI2 (Catoira et al., 2000; Endre et al., 2002; Stracke et al., 2002), and a gene encoding a putative cation channel, DMI1 (Catoira et al., 2000; Ane et al., 2004). In addition, NUP133 and NUP85 of L. japonicus that encode core components of the nucleoporin are necessary for Nod factor signaling (Kanamori et al., 2006; Saito et al., 2007). Mutations in these genes abolish Nod factor signal transduction, including the activation of calcium spiking, indicating a role early in the Nod factor signaling pathway. A calcium- and calmodulin-dependent protein kinase (CCaMK), encoded by DMI3 (Mitra et al., 2004b), is a likely candidate for decoding the calcium spiking signal since mutations in this gene abolish Nod factor–induced gene expression but maintain Nod factor–induced calcium spiking (Wais et al., 2000). Unlike the putative Nod factor receptor genes, DMI1, DMI2, DMI3, NUP133, and NUP85 are also required for the association with mycorrhizal fungi, indicating conservation in the early signaling steps in these two symbioses (Catoira et al., 2000; Kanamori et al., 2006; Saito et al., 2007). NSP1 and NSP2 are two GRAS family transcriptional regulators that function in Nod factor signaling downstream of CCaMK (Catoira et al., 2000; Oldroyd and Long, 2003; Kaló et al., 2005; Smit et al., 2005).
It is likely that the Nod factor–induced calcium spiking signal is transduced via CCaMK to activate gene expression. This is supported by pharmacological evidence indicating that the inhibition of calcium signaling blocks Nod factor–induced gene expression (Charron et al., 2004). It has recently been shown that gain-of-function mutations in CCaMK of both M. truncatula and L. japonicus lead to spontaneous nodulation in the absence of either rhizobial bacteria or Nod factor, indicating the central role that calcium and CCaMK play in the activation of nodulation (Gleason et al., 2006; Tirichine et al., 2006). Spontaneous nodulation induced by CCaMK is independent of NFR5, DMI1, and DMI2 but dependent on NSP1 and NSP2, suggesting that NSP1 and NSP2 transduce the signal downstream of CCaMK leading to the activation of the nodulation developmental pathway (Gleason et al., 2006; Tirichine et al., 2006). GRAS domain family proteins such as NSP1 and NSP2 appear to regulate gene expression but have not been shown to have direct DNA binding activity (Bolle, 2004). However, it was recently shown that the GRAS protein SHR associates with DNA elements in vivo (Levesque et al., 2006); while this association may be direct, it may also be indirect through other associated proteins. In addition to these GRAS domain proteins, a third putative transcription factor, NIN, is required for nodulation in L. japonicus and encodes a transmembrane transcriptional regulator with homology to Notch of Drosophila (Schauser et al., 1999). However, it has been shown that NIN is not necessary for Nod factor signaling; hence, this protein most likely plays a role in later stages of the nodulation process (Marsh et al., 2007).
Transcriptome analysis in M. truncatula of the Nod factor signaling mutants nfp, dmi1, dmi2, dmi3, nsp1, and nsp2 revealed that all six genes are absolutely required for rhizobial and Nod factor–induced gene transcription, suggesting that these six genes may function in a linear genetic pathway leading to early gene induction (Mitra et al., 2004a). Hence, the current model for Nod factor signal transduction involves Nod factor perception at the plasma membrane by the LysM receptor-like kinases, followed by calcium spiking, which causes CCaMK to activate GRAS proteins leading to gene expression changes. Because of the ambiguity of the GRAS protein mechanism, there have been to date no defined transcription factors that could function in the Nod factor signal transduction pathway to account for the assumed need for direct cis-regulation of nodulin gene promoters.
Here, we report the identification of ERN, an AP2-like transcription factor in the ERF subfamily, which is necessary for nodulation and functions in early Nod factor signaling. Like the GRAS proteins previously identified, this gene functions downstream of CCaMK. ERF transcription factors have well-defined DNA binding activity; thus, ERN is an excellent candidate for a transcription factor that could work with the GRAS proteins to activate the expression of nodulation genes.
RESULTS
Identification of a Gene Necessary for Infection Thread Development
We undertook a genetic screen in a population of fast neutron mutagenized M. truncatula plants to identify components necessary for nodulation (Starker et al., 2006). We identified a number of mutants that were unable to form nodules, classified as Nod−. For one of these Nod− mutants, bit1-1 (Table 1), the F2 of a backcrossed population segregated in a 3:1 ratio (37 Nod+:12 Nod−; χ2 = 0.007), indicating a single recessive mutation. When inoculated with mycorrhizal fungi, we saw normal colonization in this mutant, indicating that the mutated gene was not necessary for mycorrhizal infection (M. Harrison, personal communication).
Table 1.
Allelism Test of bit1 Relative to Other Nod− Mutants
| Male/Female | bit1-1 | nfp-1 | dmi1-2 | dmi2-1 | dmi3-1 | nsp1-2 | nsp2-1 | hcl | rit | lin | bit1-2 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| bit1-1 | + (8/6) | + (10/3) | + (5/3) | + (4/2) | + (1/1) | ND | ND | ND | ND | − (14/4) | |
| nfp-1 | ND | ||||||||||
| dmi1-2 | + (18/4) | ||||||||||
| dmi2-1 | + (6/1) | ||||||||||
| dmi3-1 | + (7/2) | ||||||||||
| nsp1-2 | + (7/1) | ||||||||||
| nsp2-1 | + (8/2) | ||||||||||
| hcl | + (5/1) | ||||||||||
| rit | + (5/1) | ||||||||||
| lin | + (10/3) | ||||||||||
| bit1-2 | − (12/3) |
ND, not determined.
Upon further characterization, we found that bit1-1 could form small bumps when in the presence of M. truncatula's rhizobial partner Sinorhizobium meliloti for in excess of 2 months (Figure 1F), indicating the formation of underdeveloped nodule primordia. At the equivalent time point, large and numerous nodules had formed on the wild-type plants (Figure 1C). Infection with S. meliloti carrying the hemA-LacZ fusion allowed us to assess the level of bacterial infection in this mutant. We found that the formation of infection foci in the bit1-1 mutant was similar to that of the wild type (bit1-1, 28 ± 6; wild type, 32 ± 8 infection events per root), but in most cases, there was no further development of infection structures beyond the infection focus in bit1-1 (Figure 1D). However, a limited number of infection threads did develop in this mutant, but these generally terminated within the root hair cell (Figures 1G and 1H). In contrast with the wild type, which had small infection foci and unbranched infection treads (Figure 1A), the infection foci of bit1-1 were often distended (Figure 1D), and infection threads commonly showed complex branched structures within the root hair cell (Figures 1G and 1H); hence, we chose to call this mutant allele branching infection threads1-1 (bit1-1). To determine whether infection threads could progress successfully in this mutant, we assessed the infection events associated with the small bumps forming on bit1-1 after long periods of time in the presence of S. meliloti. When we stained these bumps, it was possible to visualize the ramification of infection threads into the cortex (Figure 1E), but again these infection threads terminated prematurely with little expansion into the nodule primordium as seen in wild-type plants (Figure 1B).
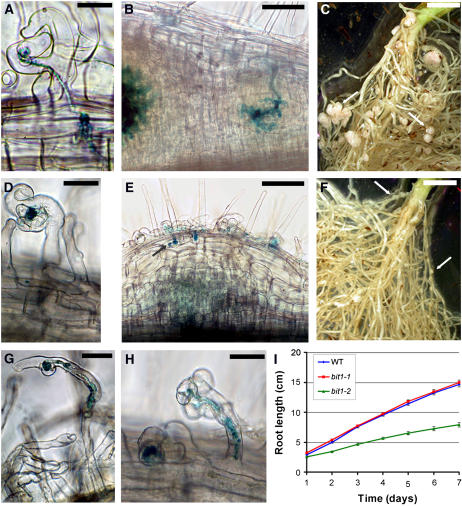
Figure 1.
bit1-1 Shows Defects in Infection Thread Initiation and Maintenance.
(A) to (F) S. meliloti infection in wild-type ([A] to [C]) and bit1-1 ([D] to [F]) plants.
(A) Five days after inoculation with S. meliloti, the infection threads on wild-type plants have exited the root hair cell and begin to divide toward the cortex.
(B) One week after inoculation, the wild-type infection threads have ramified into the dividing cells of the nodule primordium.
(C) After 2 months in the soil with S. meliloti, large pink nodules can be seen on wild-type plants.
(D) By contrast, bit1-1 mutants show only infection foci development within the first week of infection with S. meliloti.
(E) Three weeks after infection, bit1-1 mutants show some cortical cell division, and infection threads may invade the first cortical cell layer.
(F) After 2 months in the soil with S. meliloti, bit1-1 plants show small undeveloped nodules, indicated with arrows.
(G) and (H) Infection threads that do initiate on bit1-1 are often branched.
(I) The root growth of bit1-2 (green) is much slower than either bit1-1 (red) or the wild type (blue). Bars represent se.
Bacteria were visualized using X-galactosidase staining ([A], [B], [D], [E], [G], and [H]). Bars = 20 μm in (A), (D), (G), and (H), 100 μm in (B) and (E), and 5 mm in (C) and (F).
The bit1-1 mutant phenotype is similar to mutants previously described with defects in infection thread development and shows particular similarity to lin (Kuppusamy et al., 2004). Allelism tests with lin revealed that these two mutants represent two separate complementation groups (Table 1). From these data, we concluded that we had identified a gene that is necessary for initiation and maintenance of infection thread growth.
In parallel to analysis of fast neutron populations, phenotypic screens for Nod− mutants were conducted in ethyl methanesulfonate–mutagenized populations (Penmetsa and Cook, 2000). Among several mutants identified, one displayed early arrest of infection thread growth, a phenotype reminiscent of bit1-1. This mutant segregated as a single Mendelian locus (231 Nod+:67 Nod−; χ2 = 0.30). Genetic crosses between this ethyl methanesulfonate mutant and bit1-1 yielded nod− F1 progeny (Table 1), indicating that this mutant is a second allele of bit1 (bit1-2).
Despite the general similarity of infection arrest phenotypes, bit1-2 is distinguished by abnormal root development (Figures 1I and 2). Root hairs develop in alternating regions of dense and sparse hairs, giving the root a “poodle's tail” appearance (Figure 2A); hence, the previous reference to this allele as poodle. Here, we refer to this allele as bit1-2. Similarly to bit1-1 infection threads initiated in bit1-2, however, they were never observed to exit the epidermal root hair cells, and as in bit1-1, the formation of infection foci was comparable to the wild type (bit1-2, 94 ± 18; wild type, 116 ± 20 infection events per root) (Figure 2B). Growth conditions for quantification of infection events for bit1-1 and bit1-2 differ (see Methods), and this explains the discrepancies in the levels of infection observed. Semithin longitudinal sections through wild-type and bit1-2 roots revealed that the patches of dense root hair development were accompanied by a disorganized subtending root cortex (Figures 2C and 2E). Such patches were characterized by reduced longitudinal cell expansion, increased radial cell expansion, the presence of occasional intracellular spaces between cortical cells, and adjacent clustering of root hair cells. Wild-type cells of the inner cortex have rectangular length and width dimensions (162 ± 117 and 23 ± 13 μm, respectively), whereas the homozygous bit1-2 plants have regions where inner cortical cell dimensions approximate to that of a square (72 ± 19 μm length × 69 ± 18 μm width). In contrast with bit1-1, which had a wild-type root system in the absence of S. meliloti, the roots of bit1-2 had a much reduced root growth rate (Figure 1I). Scoring for nodule structures and bacterial infection 21 d after inoculation revealed fully developed functional nodules in the wild type (Figure 2F) in contrast with bit1-2, which showed arrested infections and no noticeable inner cortical cell division (Figure 2D). Cosegregation of the Nod− and poodle-like phenotypes in all 485 F2 progeny from backcrossed plants suggests that these phenotypes derive from the same single gene mutation.
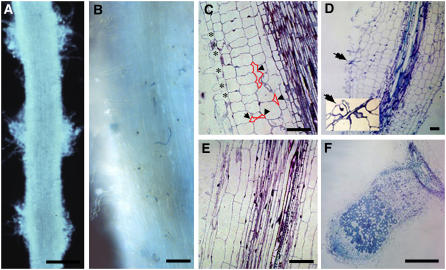
Figure 2.
Cytological Analysis Reveals bit1-2 Phenotypes Affecting Both Nodulation and Root Development.
Light micrographs of roots of bit1-2 ([A] to [D]) and the wild type ([E] and [F]). (A), (C), and (E) are noninoculated roots. (B) is 3 d after inoculation, and (D) and (F) are 21 d after inoculation. In (B), numerous infections can be observed at 3 d after inoculation, although these infections uniformly arrest within the epidermis. The inset in (D) shows a higher magnification of an arrested infection (arrow) at 21 d after inoculation. Note that in addition to altered cortical cell dimensions, bit1-2 is characterized by close spacing of root hairs ([C]; stars) and the presence of intercellular spaces between cortical cells ([C]; arrowheads with selected intracellular spaces outlined in red for clarity). Radial root swelling shown for bit1-2 at 21 d after inoculation was correlated with altered cortical cell morphology but not S. meliloti–induced cell division. Bars = 500 μm in (A) and (F) and 100 μm in (B) to (E).
BIT1 Is Necessary for Nod Factor Signal Transduction
The CCaMK encoded by DMI3 is essential for Nod factor signal transduction (Levy et al., 2004; Mitra et al., 2004b). For the kinase to function, and thereby relay the Nod factor–derived signal, an autoinhibitory domain must be inactivated through the binding of calmodulin (Patil et al., 1995). Gleason et al. (2006) have shown that by removing the autoinhibitory domain, the kinase is constitutively active. When plants are transformed with this construct, spontaneous nodules form in the absence of rhizobia (Gleason et al., 2006; Figure 3A). As such, this construct can be used for testing the epistatic relationship of genes within the Nod factor signaling cascade.
Figure 3.
bit1 Is Defective in Nod Factor Signaling.
(A) and (B) Spontaneous nodules form in the absence of S. meliloti in plants transformed with the constitutively active kinase of DMI3, DMI31-311 (A), but not in bit1-1 plants transformed with DMI31-311 (B), indicating that BIT1 is necessary for DMI31-311-induced spontaneous nodulation. To ensure that bit1 plants were transformed with DMI31-311, we assessed for the presence of the kanamycin gene in transformed root systems ([B], top panel of inset): (a) bit1-1, (b) bit1-2, and (c) wild type; the bottom panel shows amplification of a positive control. Bars = 5 mm.
(C) and (D) Treatment with 1 nM Nod factor leads to the induction of ENOD11:GUS in wild-type (C) but not bit1-1 (D) plants.
(E) and (F) Nonsymbiotic ENOD11:GUS activity is present in the cotyledons of both wild-type (E) and bit1-1 (F) plants. Bars = 10 mm in (C) to (F).
(G) Forty-six tentative consensus sequences (TC) previously identified as being induced/repressed by Nod factor and S. meliloti within 24 h of treatment were assessed in bit1-1 using Affymetrix GeneChips. The induction/repression of these tentative consensus sequences is shown in the wild type, dmi3-1, bit1-1, and hcl (the data for dmi3 and hcl have been reported previously in Mitra et al., 2004a). A pooled log2 fold change was calculated from three replicates of buffer-treated versus S. meliloti 1021–treated plants. The brightest green represents a log2 fold change of −3, and the brightest magenta a log2 fold change of +3. Analysis was performed using SAM2.11; for q-values of data, see Supplemental Table 1 online.
When 98 bit1-1 and 61 bit1-2 plants were transformed with autoactivated CCaMK (DMI31-311), spontaneous nodules failed to form (Figure 3B). By contrast, spontaneous nodules did form on seven out of 17 dmi3-1 plants transformed with DMI31-311. To ensure that the bit1 roots were transformed with this CCaMK construct, 10 transformed root systems were pooled and the presence of the cotransformed kanamycin gene was shown to be present (Figure 3B, inset). This result indicates that DMI3 is dependent on the downstream action of BIT1 for the production of spontaneous nodules, showing that BIT1 functions downstream of CCaMK in the activation of nodule organogenesis. These results suggest that BIT1 plays a role not only in infection thread growth but also in nodule formation.
The necessity for BIT1 function in CCaMK induction of spontaneous nodulation was surprising because the mutant phenotype indicated a role for BIT1 in the initiation and maintenance of infection threads. However, the results of the autoactivated CCaMK experiments suggested that BIT1 may function in the Nod factor signal transduction pathway. To further test this hypothesis, we analyzed the ability of the Nod factor to activate the Nod factor reporter ENOD11:β-glucuronidase (GUS) in the bit1-1 mutant background. Within 6 h of Nod factor application, wild-type plants showed strong induction of ENOD11:GUS (Figure 3C), while bit1-1 mutants showed no or severely diminished ENOD11:GUS induction (Figure 3D). Although widely reported (Journet et al., 2001), we found the nonsymbiotic root cap expression of ENOD11:GUS to be highly variable both in mutant and wild-type lines. To ensure plants were transformed with the ENOD11:GUS construct, GUS activity was assessed at the seedling stage where GUS activity is reliably observed in the cotyledons (Figures 3E and 3F). This result indicates that BIT1 is required for Nod factor induction of ENOD11. Together with the CCaMK spontaneous nodulation experiments, these data strongly implicate a role for BIT1 in the Nod factor signal transduction pathway.
To further explore the role of BIT1 in the induction of early nodulation gene expression, we assessed rhizobial-induced genes that have previously been shown to require Nod factor signaling components. Mitra et al. (2004a) identified a set of 46 transcripts that were induced or repressed by Nod factor and S. meliloti within 24 h of treatment. None of these genes were induced/repressed in the nfp, dmi1, dmi2, dmi3, nsp1, or nsp2 mutants following S. meliloti inoculation, while a subset of 25 of these genes were induced or repressed by S. meliloti in the hcl mutant (Mitra et al., 2004a). We assessed rhizobial induction and repression of these 46 genes in bit1-1 using an Affymetrix GeneChip containing probe sets for 9935 M. truncatula genic regions. While some of the 46 rhizobial induced or repressed genes behave similarly in both bit1-1 and wild-type plants, it is clear that bit1-1 plants are less responsive to S. meliloti with respect to the induction of gene expression (Figure 3G). However, it is equally clear that unlike dmi3-1 (and other members of the Nod factor signaling pathway) bit1-1 is not insensitive to the S. meliloti stimulus. Of the 46 S. meliloti–induced genes, 29 are not significantly induced or repressed by S. meliloti by at least twofold in bit1-1 plants compared with 21 that are not induced or repressed by S. meliloti in hcl mutants (see Supplemental Table 1 online). This expression profile suggests that BIT1 is necessary for early rhizobial-induced gene expression changes, but the bit1-1 defect is less severe than that of plants mutated in the previously defined components of the Nod factor signaling pathway.
Transcript-Based Gene Cloning Identified Downregulated Genes in bit1-1
To further understand the function of BIT1 in Nod factor signaling, we undertook the isolation of the gene. It has been shown that mutations that alter transcript production or stability can be identified by comparing transcript abundance of mutant plants to wild-type plants (Mitra et al., 2004b). Since bit1-1 was generated by fast neutron bombardment, a treatment that is known to induce deletions, we hypothesized that this mutant would be a good candidate for such transcript-based gene cloning. To identify downregulated genes, we used the Affymetrix GeneChip to compare the RNA profile of bit1-1 with that of wild-type plants in four different treatments: 1 or 4 d after inoculation with either buffered nodulation media (BNM) buffer or S. meliloti 1021. In all four treatments, three transcripts were consistently downregulated in bit1-1 by >2.5-fold, though some of the data points are not highly significant (Table 2; see Supplemental Table 2 online). Importantly, these transcripts were downregulated in the absence of S. meliloti treatment and thus were considered good candidates genes for BIT1.
Table 2.
Downregulated Genes in bit1-1 Plants
| Log2 (Fold Change) Wild Type versus bit1
|
|||||
|---|---|---|---|---|---|
| TC Number | Sequence Homology | 1 DAI S. meliloti | 4 DAI S. meliloti | 1 DAI Buffer | 4 DAI Buffer |
| TC103682 | α-Tubulin | −3.8* | −4.2 | −4.4 | −4.3* |
| TC99463 | ERF TF | −5.7* | −4.9 | −3.9 | −2.5* |
| TC102418 | AG-peptide | −3.6 | −4.9 | −3.7 | −4.7 |
Not significant at the 95% confidence level; q-values are presented in Supplemental Table 2 online. TC, tentative consensus sequence; DAI, days after inoculation; TF, transcription factor.
All three of these transcripts occur within the sequenced BAC mte1.32m6 located on LG7, suggesting that a single deletion might be causal of the reduced transcript levels. We found that BIT1 mapped to LG7 and cosegregated with marker 002E05, which resides within the same BAC contig as mte1.32m6, suggesting that the deletion we had identified may contain BIT1. PCR analysis with primers specific for one of the candidate transcripts (an arabinogalactan-peptide gene) confirmed the presence of a DNA deletion that cosegregated with the bit1-1 mutant phenotype in a backcrossed population (Figure 4C). The extent of the deletion was estimated by progressive amplification of genomic DNA fragments flanking the AGP transcript. Using this method, a region of ∼40 kb is estimated to be deleted in bit1-1. The centromeric deletion boundary was only tentatively identified, since the PCR product was contained within a multicopy retrotransposon, thus confounding the interpretation of the PCR amplification. The PCR results confirm that at least five genes are affected by this deletion: an arabinogalactan peptide, an unknown gene, an α-tubulin, an ERF transcription factor, and a karyopherin. Neither the unknown gene nor the karyopherin was detected by the transcript-based cloning because the unknown gene was not on the Affymetirx GeneChip and the expression of the karyopherin was very low in the wild type, such that its absence in bit1-1 could not be detected.
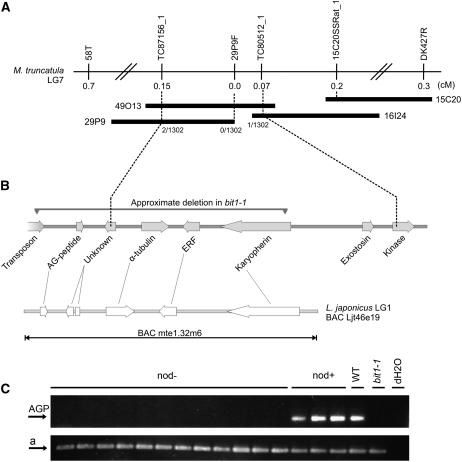
Figure 4.
BIT1 Identification by Positional and Transcript-Based Cloning.
(A) and (B) The bit1-2 mutation was found to be located between the genetic markers TC87156_1 and TC80512_1 on linkage group 7 (A). The physical region between these markers is spanned by the three BACs 29P9, 49O13, and 16I24 and contains four genes (B). The deletion that causes the bit1-1 mutation covers a similar region as indicated. A region on L. japonicus linkage group 1 shows absolute colinearity. As shown, the bit1-1 deletion affects at least five genes.
(C) Amplification of a 200-bp fragment of TC102418 encoding the AGP contained within the bit1-1 deletion in a segregating F2 backcrossed population of bit1-1 reveals cosegregation of the deletion with the nod− phenotype. Bottom panel (a) shows amplification of a positive control.
Positional Cloning Identifies bit1-2
In parallel to deletion analysis, we undertook the positional cloning of the bit1-2 allele. bit1-2 was located on contig 995 (http://medicago.org/genome) between markers TC87156_1 and TC80512_1 on LG7 and cosegregated with marker 29P9F that was generated from the end sequence of BAC 29P9 (Figure 4A). This genetic region was contained within a single BAC clone (Figure 4A), and the sequencing of this region revealed four genes between markers TC87156_1 and TC80512_1: an α -tubulin, an ERF transcription factor, a karyopherin, and an exostosin-like protein. Sequencing these four genes in the bit1-2 mutant revealed a single mutation within the ERF transcription factor and wild-type alleles of the other three candidate genes. The mutation causes an amino acid change of Gly to Asp at a residue that is completely conserved within the DNA binding domain of all 147 Arabidopsis AP2-like genes. The affected ERF in bit1-2 is the same gene as the ERF transcription factor contained within the deleted region of bit1-1 (Figure 4B). Importantly, in mapping bit1-2, both the nonsymbiotic and symbiotic phenotypes were used, and all recombinants were mutant for both symbiotic and nonsymbiotic phenotypes. As all genes between the cosegregating markers were sequenced and only the mutation in the ERF was found, we can attribute both phenotypes to this gene.
bit1-1 Is Complemented by Agrobacterium rhizogenes–Mediated Transformation Using the ERF Transcription Factor
As both bit1 mutant alleles contained an alteration to the ERF transcription factor, we tested if this gene could complement bit1-1 plants. Genomic DNA encoding the ERF transcription factor under the control of its own promoter (promoter of ERF Required for Nodulation [pERN]) or the 35S promoter was introduced into bit1-1 roots using A. rhizogenes. This resulted in chimeric plants with untransformed shoots and transformed hairy roots (Boisson-Dernier et al., 2001). The roots of bit1-1 plants transformed with this ERF transcription factor developed nodules in the presence of S. meliloti (Figures 5A and 5B). Four out of 11 plants and 10 out of 34 plants showed complementation of bit1-1 when transformed with 35S:ERN and pERN:ERN, respectively, while six out of 23 dmi3 plants showed complementation when transformed with pDMI3:DMI3. By contrast, no nodules were observed on bit1-1 plants transformed with either the empty vector (15 plants tested) or with the AGP gene (40 plants tested) that is also contained within the bit1-1 deletion (Figure 5C). A t test analysis showed no significant differences between the 35S:ERN and pERN:ERN complementation of bit1-1 and the pDMI3:DMI3 complementation of dmi3. The nodules on bit1-1 complemented plants were elongated, pink, and developed within 2 weeks of inoculation, a period where no nodule-like structures are normally observed in bit1-1 plants. In addition, unlike the infection threads observed on bit1-1 plants, the complemented plants showed extensive infection thread growth and ramification throughout the developing nodule (Figure 5A). The combination of two independent mutations in the same gene coupled with the complementation of bit1-1 indicates that BIT1 encodes a protein with homology to ERF transcription factors. We have chosen to call this protein ERF Required for Nodulation (ERN).
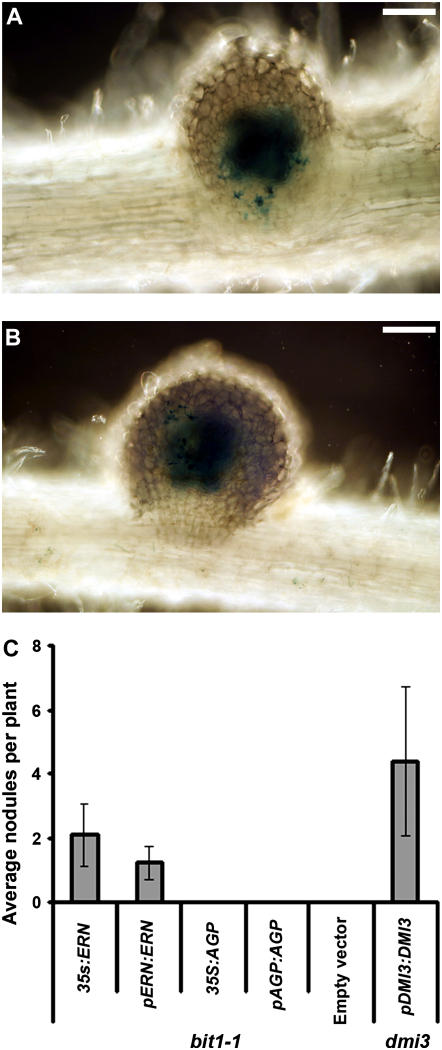
Figure 5.
Complementation of the bit1-1 Phenotype with ERN.
(A) and (B) The transformation of bit1-1 plants with pERN:ERN (ERN driven by its native promoter) leads to complementation of the mutant allowing nodulation after S. meliloti infection. X-galactosidase staining reveals S. meliloti contained within infection threads that ramify throughout the nodule primordium. Bars = 50 μm.
(C) Average number of nodules on all transformed plants, including those showing no nodulation, 20 d after inoculation with S. meliloti. bit1-1 plants were transformed with five constructs; the ERN gene regulated by the 35S promoter or its native promoter and the AGP that is also contained within the bit1-1 deletion regulated by the 35S promoter or its native promoter and an empty vector. As a positive control, dmi3 plants were complemented with DMI3 driven by its native promoter. Error bars represent se. There is no significant difference (P = 0.1) among 35S:ERN, pERN:ERN, and pDMI3:DMI3.
ERN Is a Member of a Previously Uncharacterized Group of ERF Transcription Factors
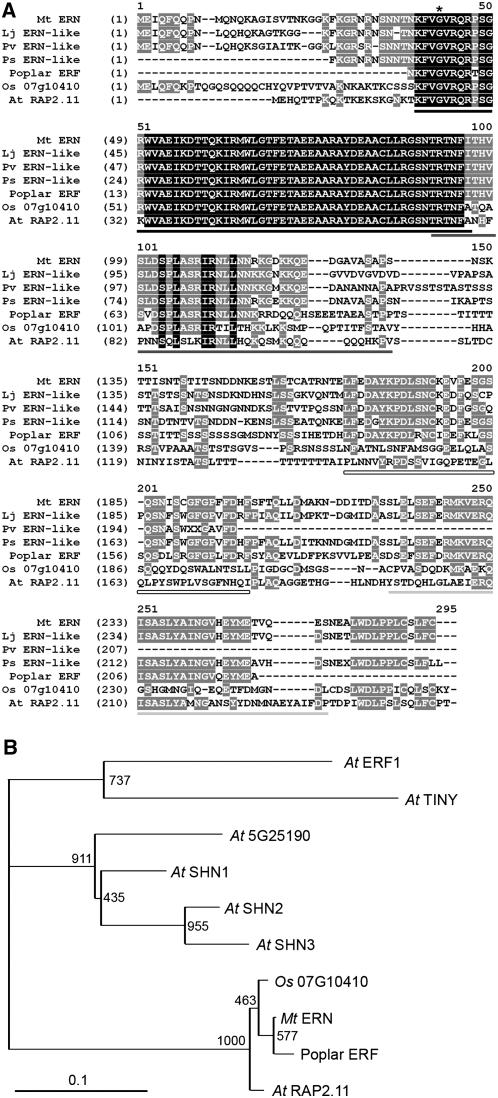
Among the AP2 transcription factors of Arabidopsis thaliana, ERN shows closest similarity to RAP2.11, a member of the ERF subfamily of AP2 transcription factors, with 41.3% overall identity and 96.6% identity within the AP2 DNA binding domain (Figure 6A). Close homologs were identified in L. japonicus, Pisum sativum, Phaseolus vulgaris, Glycine max, and Populus alba (poplar). The L. japonicus homolog is contained within a genomic region that is highly syntenic to that of ERN in M. truncatula (Figure 4B). Within the AP2 DNA binding domain, there is 100% amino acid sequence identity between the legume proteins and near complete conservation among all plants (Figure 6A). Two additional domains, CMV3 and CMV4 (conserved motif of Group V), have been bioinformatically identified in this class of ERF transcription factors in Arabidopsis and rice (Oryza sativa) (Nakano et al., 2006). These domains show moderate conservation between the legume proteins and other plant homologs of ERN (Figure 6A). However, there are significant differences in both composition and size of these domains in the legume and poplar proteins compared with Arabidopsis and rice. Yet within the legumes and poplar there is considerable similarity. In addition, a fourth domain of homology is apparent in the legume and poplar proteins between the CMV3 and CMV4 domains (Figure 6A). Arabidopsis RAP2.11 has previously been classed in Group V of the ERF group of AP2 transcription factors together with a number of other Arabidopsis ERF transcription factors, including the SHN1 gene (Aharoni et al., 2004; Nakano et al., 2006). Using the AP2 domain, we assessed the phylogenetic relationship of the Group V class of ERFs to the closest homologs of ERN in legumes and nonlegumes (Figure 6B). ERN clusters with RAP2.11 along with the poplar and rice homologs, but this grouping appears to be distinct from the other Group V members in Arabidopsis.
Figure 6.
M. truncatula ERN Is an ERF Transcription Factor.
(A) Alignments of M. truncatula ERN with homologs form L. japonicus, P. sativum, P. vulgaris, poplar, rice, and Arabidopsis. The AP2 domain is underlined in black, CMV-3 is underlined in dark gray, and CMV-4 is underlined in light gray. A novel conserved domain that is specific to plants in the Rosid 1 clade is also apparent and underlined by a hollow bar. The asterisk denotes the site of the bit1-2 mutation.
(B) A phylogenetic tree of the AP2 domains of M. truncatula ERN and its homologs. Since the AP2 domains of the legume homologs are identical, only M. truncatula ERN is shown along with the AP2 domains of rice, poplar, the group V Arabidopsis ERFs, and the Arabidopsis ERFs TINY and ERF1 as outgroups.
ERN Is Rapidly Induced in Response to S. meliloti Inoculation
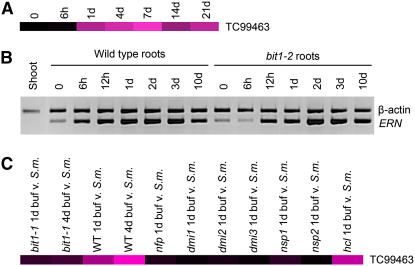
It was previously shown that TC102418, encoding ERN, is upregulated in response to S. meliloti inoculation based on data from Affymetrix arrays (Starker et al., 2006). The 1.5-fold induction 1 d after rhizobial inoculation is maintained for at least 21 d after inoculation (Figure 7A; see Supplemental Table 3 online). Semiquantitative RT-PCR confirmed that ERN is rapidly induced (Figure 7B). However, in the RT-PCR work, induction was observed at 6 h after inoculation with S. meliloti and this induction is maintained. Despite slight differences between the Affymetrix and RT-PCR data, both implicate the induction of ERN by S. meliloti. This pattern is also observed in the roots of bit1-2 plants, although the onset of expression induction appears to be delayed (Figure 7B). ERN is expressed in M. truncatula roots prior to S. meliloti infection but is not expressed in the shoot.
Figure 7.
ERN Is Induced upon Rhizobial Inoculation, and This Requires the Nod Factor Signaling Pathway.
(A) Affymetrix GeneChip analysis reveals that TC99463, which encodes ERN, is induced following S. meliloti inoculation, with maximal induction at 7 d after infection. Expression levels were calculated by comparing three replicates of infected plants with three replicates of uninfected plants. Black represents no induction and the brightest magenta a 2.0 log2 fold change. For q-values, see Supplemental Table 3 online.
(B) RT-PCR of ERN reveals induction following S. meliloti inoculation, with maximal induction at 72 h in the time points assessed in both wild-type and bit1-2 roots.
(C) Affymetrix analysis of the induction of ERN (TC99463) at 24 h after rhizobial infection requires components of the Nod factor signaling pathway but not HCL. Black represents no induction and the brightest magenta a 1.7 log2 fold change compared with uninfected plants. For q-values, see Supplemental Table 3 online. Some data from (A) and (C) have been published previously (Mitra et al., 2004a; Starker et al., 2006).
To assess whether ERN induction by rhizobia required components of the Nod factor signaling pathway, we looked at the rhizobial induction of ERN on Affymetrix gene profiles in the Nod factor signaling mutants. ERN was not induced in nfp, dmi1, dmi2, dmi3, nsp1, or nsp2 plants 24 h after S. meliloti inoculation but was induced in hcl at an equivalent level to the wild type (Figure 7C). This shows that the S. meliloti induction of ERN is dependent on the Nod factor signaling components but is independent of HCL.
DISCUSSION
In the last few years, several components of the Nod factor signaling pathway have been defined from the model legumes M. truncatula and L. japonicus (Oldroyd and Downie, 2004, 2006). Nod factor perception at the plasma membrane involves multiple receptor-like kinases that are necessary to activate oscillatory calcium changes associated with the nucleus (Ben Amor et al., 2003; Limpens et al., 2003; Radutoiu et al., 2003). Perception of the calcium signal is likely to be facilitated through CCaMK, the activation of which is sufficient to induce nodule organogenesis (Levy et al., 2004; Mitra et al., 2004b; Gleason et al., 2006; Tirichine et al., 2006). Downstream of CCaMK are two GRAS family transcriptional regulators (Kaló et al., 2005; Smit et al., 2005). Such GRAS proteins have been proposed to act as transcriptional activators, though the mechanism remains elusive (Bolle, 2004). Here, we show that ERN, a protein with homology to ERF transcription factors and with a highly conserved AP2 DNA binding domain, is necessary for Nod factor signaling. We show that ERN functions in the Nod factor signaling pathway downstream of CCaMK and, based on the mutant phenotype, probably downstream of the GRAS family proteins. ERN is a strong candidate to provide specific binding to the nodulin promoters for Nod factor–dependent activation of gene expression.
ERN is a member of Group V of the ERF subfamily of AP2 transcription factors (Nakano et al., 2006). There are five Arabidopsis genes within this family, and ERN shows highest similarity to RAP2.11, a gene of unknown function (Okamuro et al., 1997). Of the Arabidopsis ERF Group V genes, the SHN genes have been shown to regulate cuticular waxes and thus modify drought tolerance (Aharoni et al., 2004). However, our own phylogenetic analysis with a number of ERN homologs suggests that RAP2.11 clusters with ERN and its homologs in other plants and indicates that ERF Group V may be split into two separate subgroups. The DNA binding AP2 domain is highly conserved among ERN, RAP2.11, and other homologs in legumes, poplar, and rice. Domains CMV-3 and CMV-4 defined by Nakano et al. (2006) are also conserved in the legume and poplar ERN homologs. In addition, a third domain, located between CMV3 and CMV4, is conserved in the legume and poplar genes but shows only limited conservation in rice and Arabidopsis.
As ERN encodes a transcription factor, genes downregulated in the mutant may give clues as to the function of ERN. Of the 46 genes known to be up- or downregulated by twofold in response to rhizobial inoculation, 29 of these genes were not induced or repressed in the bit1-1 mutant. While predefined nodulins and other transcription factors were among these genes (see Supplemental Table 1 online), it is difficult to draw conclusions regarding the regulation of nodulation by ERN from these regulated genes.
Three lines of evidence strongly support the statement that ERN functions in Nod factor signal transduction. First, nodulin reporter gene studies show that Nod factor–dependent ENOD11 expression is absent or greatly reduced in bit1-1 compared with the wild type. Second, the spontaneous nodulation induced by a modified CCaMK requires ERN activity. Finally, Nod factor–induced genes show altered expression in bit1-1 plants. However, unlike nfp, dmi1, dmi2, dmi3, nsp1, and nsp2, the rhizobial induction of these genes in not completely blocked in bit1-1. A function for ERN in Nod factor signaling would implicate this protein in regulating processes activated by the bacterial signal, namely, nodulin gene expression, infection thread development, and induction of the nodule meristem. Consistent with this hypothesis, we see defects in all of these processes in the bit1 mutants. ERF transcription factors are known to function in hormone signaling, most notably that of ethylene (Ohme-Takagi and Shinshi, 1995), and hormones are also known to regulate nodulation. This correlation may indicate a mechanism of action for ERN. However, we believe that caution is required when ascertaining function from related transcription factors, and preliminary analyses indicate that bit1-1 and bit1-2 nodulation defects cannot be rescued through the regulation of ethylene levels (data not shown).
Despite the requirement for ERN in Nod factor signaling, both bit1 mutants have phenotypes less severe than plants mutated in the previously defined components of the Nod factor signaling pathway. Specifically, in contrast with earlier mutants, rhizobial-induced gene expression still occurs in bit1-1 despite a reduced number of up- or downregulated genes showing significant expression changes in bit1-1 compared with wild-type plants. Additionally, bit1-1 and bit1-2 are infected by S. meliloti though show only limited infection thread development. Because bit1-1 is a complete deletion of ERN, the capacity of bit1-1 plants to partially transduce the Nod factor signal suggests that ERN is not as essential for Nod factor signaling as the other transcriptional regulators in the pathway, NSP1 and NSP2. It is perhaps important to note from the Affymetrix analysis that it is apparent that the expression of DMI2, DMI3, and NSP2 are unaffected in the bit1-1 mutant (data not shown). The limited Nod factor signaling activity remaining in the absence of ERN is sufficient to activate a subset of early responses but not adequate to complete the rhizobial infection process. This is consistent with previous observations showing greater stringency for Nod factor perception during infection stages compared with the induction of earlier responses, such as gene expression (Ardourel et al., 1994; Limpens et al., 2003, 2005; Capoen et al., 2005).
The bit1-2 phenotype shows distinct differences to bit1-1. In particular, bit1-2 has altered root cortical and epidermal cell development in the absence of rhizobia or the Nod factor. It is possible that the inability of the roots of bit1-2 plants to nodulate stems not just from abnormal Nod factor signaling but an inability for either the clustered root hairs or aberrant cortical cells to support, respectively, root hair–mediated infection or rhizobia-induced cortical cell division. However, the absence of any discernible aberrant root hair or cortical cell phenotype in the null allele bit1-1 suggests that the bit1-2 mutant protein (BIT1-2) has a novel function that interferes with normal root development. The fact that bit1-2 results from a single amino acid change in the AP2 DNA binding motif is consistent with the possibility that BIT1-2 has altered DNA binding properties, perhaps binding nontarget promoter sequences or altered affinity to cognate promoter sequences. Regardless of functional changes to BIT1-2, we note that ERN and other components of the Nod factor signaling pathway are expressed in nonsymbiotic tissue (e.g., all of the DMI genes are expressed prior to Nod factor application), consistent with the idea that presymbiotic cells contain receptors and transduction pathways ready to initiate Nod factor signaling. Mutations that change the function of such pathway components may display phenotypes in the absence of rhizobial signals, such as has been demonstrated for particular C-terminal truncations of DMI3 (Gleason et al., 2006). The combined symbiotic and presymbiotic phenotypes of bit1-2 are also consistent with this interpretation. Thus, we propose that ERN and other Nod factor signal transduction components are present in the absence of Nod factor, establishing a presymbiotic state that is primed for Nod factor signaling.
The AP2 domain of Arabidopsis ERF1 has been resolved and shown to bind to DNA (Allen et al., 1998). To our knowledge, no other functional domains have been assigned to proteins containing AP2 domains. ERF transcription factors have been shown to be involved in the regulation of developmental processes as well as playing roles in many other diverse processes, including responses to pathogens (Feng et al., 2005). NSP1 and NSP2 encode GRAS domain proteins that, although thought to regulate transcription, have yet to be shown to have direct DNA binding activity (Bolle, 2004). When fused to a DNA binding domain, the GRAS protein LISCL functioned as a transactivator in yeast and plants (Morohashi et al., 2003). This suggests that when GRAS proteins are brought to promoter elements they can activate the RNA polymerase complex. It is tempting to speculate that ERN may be one protein that can interact with NSP1 and NSP2 to allow DNA binding of a protein complex that includes the GRAS proteins, allowing them to induce transcription. We are currently assessing interactions between ERN and NSP1 and NSP2 as well as assessing the binding of ERN to early nodulin promoter elements. To our knowledge, ERN represents the first Nod factor signaling protein with a highly conserved DNA binding domain. The further characterization of ERN and its interactions with Nod factor signaling components will help elucidate the regulation of nodulation gene expression.
METHODS
Plant Growth and Bacterial Strains
Medicago truncatula cv Jemalong A17 was used as the wild type. Backcrosses of bit1-1 and bit1-2 were to A17 and mapping crosses to A20. ENOD11 promoter GUS fusions were introduced into bit1-1 plants by crossing with a wild-type ENOD11:GUS line and selection through a series of generations to produce plants homozygous for both bit1-1 and ENOD11:GUS. For plate assays, seeds were scarified with sandpaper and sterilized in bleach for 3 min. Seeds were imbibed for 2 h in water and geminated inverted on damp filter paper for 2 d at 4°C, followed by 1 d at room temperature. Seedlings were then transferred to BNM medium supplemented with 0.1 μM aminoethoxyvinylglycine. Plates were infected with Sinorhizobium meliloti by flooding and subsequent draining of the plate with 1 mL of overnight culture, spun down and resuspended in BNM. For infection thread analyses and visualization, S. meliloti 1021 carrying pXLGD4, expressing LacZ under a hemA promoter, was used to inoculate plants. Agrobacterium rhizogenes ARqua was used as the transformation vector in all hairy root transformations.
Phenotypic Characterization
Roots of plants infected with S. meliloti 1021 pXLGD4 were fixed for 1 h in 1.25% gluteraldehyde, rinsed three times with phosphate buffer, pH 7.0, stained overnight with 5 mM of both potassium ferrocyanide and potassium ferricyanide and 0.06% X-gal in phosphate buffer at room temperature, and rinsed three times in phosphate buffer and twice in sterile distilled water. For ENOD11:GUS expression analysis, plants were germinated and grown on plates for 3 d before submersion in 1 nM Nod factor in liquid BNM. Plants were then stained for GUS activity with 2 mM X-gluc and 5 mM EDTA overnight at 37°C.
For infection analysis of bit1-1, plants were grown on BNM supplemented with 0.1 μM aminoethoxyvinylglycine. By contrast, bit1-2 infection was assessed in aeroponic culture. In both treatments, plants were inoculated with S. meliloti 1021 carrying pXLGD4 and stained 3 d after inoculation. The number of infection events was assessed.
Affymetirx GeneChip construction, RNA preparation, and chip hybridizations were performed on bit1-1 plants at 1 or 4 d after inoculation with either S. meliloti 1021 or BNM as described by Mitra et al. (2004a). The statistical analysis of the microarray data is described below in the expression analysis section. The grouping of genes with similar expression patterns was facilitated through hierarchical clustering based on Euclidean distance and average linkage clustering using The Institute for Genomic Research (TIGR) MultiExperiment Viewer.
For A. rhizogenes–mediated hairy root transformation of the DMI31-311 construct (Gleason et al., 2006), plants were germinated as described above. The tip of the radicle was removed with a scalpel and the cut end dipped into an A. rhizogenes ARqua overnight culture before plating onto modified Farhreus media. Plants were transferred to growth pouches 1 month after transformation (Boisson-Dernier et al., 2001). Spontaneous nodulation was scored for 2 to 4 weeks thereafter. Roots that did not show spontaneous nodulation were tested for transformation using primers specific to the cotransformed kanamycin gene: KanF 5′-CGCGACGTCAATGATTGAACAAGATGGATTGCA-3′, KanR 5′-CAATCTTAAGAAACTTTTCAGAAGAACTCGTCAAGAAGG-3′. As a positive DNA control, a genomic DNA fragment was amplified from the BAC mte1.32m6: mte1 F, 5′-AAACACAAGCGGAATCAACC-3′; mte1 R, 5′-GGCCAGACTTGCAGGATAAA-3′.
Gene Isolation
The transcript-based cloning of the bit1-1 allele made use of the previously described M. truncatula/S. meliloti Affymetrix GeneChip analyses (Mitra et al., 2004b). cRNA of mutant and wild-type plants was hybridized to the chip with three biological replicates for both genotypes. Model-based analysis and pairwise comparisons were performed as described by Mitra et al. (2004b) to identify transcripts with lowered abundance in bit1-1. The cosegregation of the deleted AGP was confirmed in a bit1-1 backcrossed F2 population using primers specific to AGP: AGP F, 5′-GACAATGGAGGCATTGAAGATGAAC-3′; AGP R, 5′-CCAAATGCAAGTGCTATGAGAGAAGC-3′. As a positive DNA control, a genomic DNA fragment was amplified from the BAC mte1.32m6 as described above. Fifteen nod− plants and three nod+ plants were analyzed. Twelve nod− plants of F2 mapping population individuals from a cross between bit1-2 and M. truncatula A20 plants were used to show complete cosegregation of the bit1-1 phenotype with the marker 002E05. Primers were as follows: 002E05 F, 5′-ATGGAAGGTGGAACCTATCT-3′; 002E05 R, 5′-GGTGTCGACTGATCCTAGC-3′.
Using an F2 mapping population of 1302 individuals from a cross between bit1-2 and M. truncatula A20 plants, the nod− phenotype and the cosegregating bit1-2 root phenotype were located between cleaved-amplified polymorphic sequence markers 58T and DK427R at a genetic distance of 0.7 and 0.3 centimorgans, respectively, on linkage group 7. Marker DK427R was selected for chromosome walking and used to identify the sequenced BAC clone Mth2_15C20 (GenBank accession number AC126009). Sequence information from this clone was used to generate a new genetic marker, 15c20SSRat_1, and identify BAC clone Mth2_16I24 that had been survey sequenced. Sequence information of Mth2_16I24 allowed the generation of a new genetic marker TC80512_1 by which BAC clone Mth2_49o13 was identified and then survey sequenced. Based on this, the marker TC87156_1 was generated, and the forward end sequence of BAC Mth2_29P9 (GenBank accession number CG929807) was identified from which 29P9F-specific primer pairs were generated. The sequenced region between the two bit1-2 recombinant markers was submitted to GenBank (accession number DQ984942). Primers are as follows: 58T forward primer, 5′-AAGGGCTTTTAATTTCTGTCTG-3′; 58T reverse primer, 5′-CCAAACCAATCTAAAATAATAAAC-3′; DK427R forward primer, 5′-CCAAACAAGGAAAAGTGTTGGTGTCA-3′; DK427R reverse primer, 5′-ATGAGAAACTTTTGAAATTTAGGATACGATAG-3′; 15C20SSRat_1 forward primer, 5′-AATAGGGACATTAAGATG-3′; 15C20SSRat_1 reverse primer, 5′-TTGTGAAATAGTCGTGAT-3′; TC80512_1 forward primer, 5′-CATAACAAAAACCCAAAGTGAGA-3′; TC80512_1 reverse primer, 5′-AGTTATAATATGTCGGTGCCAATC-3′; TC87156_1 forward primer, 5′-TGGAGCTATATGTTCTTTTGGTGT-3′; TC87156_1 reverse primer, 5′-GTGGTGGCCTGGGTTTGA-3′; 29P9F 1 forward primer, 5′-TATCACAACATCTTTTCCAC-3′; 29P9F 1 reverse primer, 5′-GCTCTAGCTGCTTCCTCA-3′.
The genomic DNA from M. truncatula plants was isolated using the ZenoGene40 plant DNA purification kit (Zenon Bio). BAC DNA purifications were performed according to a slightly modified method described previously (Nam et al., 1999). For survey sequencing, BAC clones were subcloned into pUC19 following either restriction enzyme digestion or random fragmentation with sonication. DNA sequencing was performed on an ABI-PRISM 3100 automatic sequencer using Hi-Di Formamide and POP6 polymer.
A. rhizogenes–Mediated Complementation of bit1-1
Complementation constructs were generated using a Gateway-based system. PCR products were cloned into the entry vector pDONR201 (Invitrogen) and recombined into either pK7WG2D, for 35S promoter experiments, or pKGW, for native promoter experiments. Seedlings were transformed as described above. After transfer to pouches, plants were inoculated with S. meliloti 1021 pXLGD4. Nodulation was assessed 2 to 4 weeks after inoculation in at least three biological replicates. Roots were stained for LacZ activity as described. Primers are as follows: for pAGP:AGP, forward primer, 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTCCCCCAAACCTACGTCAGACCA-3′, and reverse primer, 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCCTACGAATTGAACTCTAACCCCTTG-3′; for p35S:AGP, forward primer, 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTCCGTCTCAAAATTTTCTCTCTC-3′, and reverse primer, 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCCTAGATCAAGAGACTTAAATTTC-3′; for pERN:ERN, forward primer, 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTAGGGGAAGAAAGGGGACAAG-3′, and reverse primer, 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCATTTAACTGCGATTCTCTGTA-3′; for p35S:ERN, forward primer, 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTATGGAAATTCAATTTCAGCAAC-3′, and reverse primer, 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTTTAACAGAACAAGGAGCACAAG-3′.
Sequence Analysis
Sequences of ERN homologs were aligned using VectorNTI. A bootstrap neighbor-joining tree, from 1000 trials, was constructed based on ClustalX alignments using TreeView. Both At ERF1 and At TINY were used as outgroups.
Expression Analysis
The expression profile of the ERN gene was analyzed by semiquantitative RT-PCR in shoots and roots of wild-type M. truncatula and bit1-2 plants following inoculation with S. meliloti ABS7. Roots from four plants at each time point were collected, and total RNA was purified with the RNeasy Plant Mini Kit (Qiagen) and treated with DNaseI (Invitrogen) to eliminate genomic DNA contamination. The amount of total RNA was normalized by measuring the RNA concentration at 260 nm, and 10 ng of total RNA was added to individual tubes to synthesize first-strand cDNA using gene-specific primers. The reverse transcription and the amplification of the cDNA templates were performed sequentially in the same tubes using the One-Step RT-PCR kit (Qiagen) according to the supplier's instructions. The amplification of bit1-2 and β-ACTIN genes was performed in the same tube at each time point, and samples without reverse transcriptase were used as a control to prove the absence of genomic DNA. The bit1-2 transcripts were amplified using the forward 5′-TTTCAGCAACCAAACATGCAGA-3′ and reverse 5′-GAGCAGAAGCAACAGCACCATC-3′ primers (0.64 μM). The forward 5′-GCAGATGCTGAGGATATTAACCCC-3′ and reverse 5′-CGACCACTTGCATAGAGGGAGAGG-3′ primers (0.64 μM) were used to amplify the β-ACTIN transcript as an internal constitutive control. PCR reactions in 12.5 μL final volume were started with an initial activation step of 15 min at 95°C and performed in 30 cycles of 40 s at 94°C, 50 s at 58°C, and 1 min at 72°C. The reactions were terminated at 72°C for 10 min. The amplified products were separated in 1% agarose gels and visualized by ethidium bromide staining. The experiment was repeated four times to confirm the expression profile of ERN.
Affymetrix-based expression analysis of the ERN transcript with time has been previously described (Starker et al., 2006). In all Affymetrix analyses, three independent biological replicates of each treatment were performed. Data are displayed using the TIGR MultiExperiment Viewer.
For microarray analysis, Affymetrix MAS 5.5 software was used to extract pixel values from scan files, and the .cel files were analyzed using DCIP 1.3 (www.dchip.org).Values were normalized across independent chips using invariant set normalization. To assess the significance of expression changes, Significance Analysis of Microarrays (SAM2.11) was used (Tusher et al., 2001), with the expectation of fewer than one false-positive genes and a greater than twofold change in expression. Such analysis provides an error measure called false discovery rate, which is a valid alternative for type I family-wise error corrections (Storey, 2002) and outputs q-values that are provided for all data in Supplemental Tables 1 to 3 online.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: sequenced region between bit1-2 recombinant markers, DQ984942; SHN1, At1g15360; SHN2, At5g11190; SHN3, At5g25390; RAP2.11, At5g19790; At5g25190; LjT26E19, AP006677; Pv ERN-like, CV535404; Ps ERN-like, EF396329; poplar ERF, LG_XIII:4164146-4164813; Os 07g10410, BAC79791; Mt ERN TC99463, EF396330.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table 1. Gene Induction by S. meliloti in the Wild Type, dmi3, bit1, and hcl.
Supplemental Table 2. Q-Values of Downregulated Genes in bit1-1.
Supplemental Table 3. Q-Values Associated with ERN Expression.
Supplementary Material
Acknowledgments
We thank A. Downie, S. Hirsch, G. Deák, G. Endre, and E. Kiss for their scientific help and advice as well as K. Karchesz and I. Szívós for skillful technical assistance. This study was supported by the Biotechnology and Biological Sciences Research Council, the Royal Society, European Grain Legumes for Food and Feed (Grant FOOD-CD-2004-506223); Hungarian national grants NKFP Medicago Genomics (Grant 4/023/2001, Ministry of Education); Medicago Biotechnology (Grant 4/031/2004); Biotechnology 2001 (Grant OMFB-00229/2002); OTKA (Hungarian Research Scientific Fund) Grants OTKA T046645 and OTKA T046819; the GVOP-3.1.1-2004-05-0101/3.0 and the U.S. Department of Energy Biosciences program.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Giles E.D. Oldroyd (giles.oldroyd@bbsrc.ac.uk).
Online version contains Web-only data.
References
- Aharoni, A., Dixit, S., Jetter, R., Thoenes, E., van Arkel, G., and Pereira, A. (2004). The SHINE clade of AP2 domain transcription factors activates wax biosynthesis, alters cuticle properties, and confers drought tolerance when overexpressed in Arabidopsis. Plant Cell 16 2463–2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen, M.D., Yamasaki, K., Ohme-Takagi, M., Tateno, M., and Suzuki, M. (1998). A novel mode of DNA recognition by a beta-sheet revealed by the solution structure of the GCC-box binding domain in complex with DNA. EMBO J. 17 5484–5496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ane, J.M., et al. (2004). Medicago truncatula DMI1 required for bacterial and fungal symbioses in legumes. Science 303 1364–1367. [DOI] [PubMed] [Google Scholar]
- Ardourel, M., Demont, N., Debelle, F.D., Maillet, F., Debilly, F., Prome, J.C., Denarie, J., and Truchet, G. (1994). Rhizobium meliloti lipooligosaccharide nodulation factors: Different structural requirements for bacterial entry into target root hair cells and induction of plant symbiotic developmental responses. Plant Cell 6 1357–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrighi, J.F., et al. (2006). The Medicago truncatula lysine motif-receptor-like kinase gene family includes NFP and new nodule-expressed genes. Plant Physiol. 142 265–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Amor, B., Shaw, S.L., Oldroyd, G.E.D., Maillet, F., Penmetsa, R.V., Cook, D., Long, S.R., Denarie, J., and Gough, C. (2003). The NFP locus of Medicago truncatula controls an early step of Nod factor signal transduction upstream of a rapid calcium flux and root hair deformation. Plant J. 34 495–506. [DOI] [PubMed] [Google Scholar]
- Boisson-Dernier, A., Chabaud, M., Garcia, F., Becard, G., Rosenberg, C., and Barker, D.G. (2001). Agrobacterium rhizogenes-transformed roots of Medicago truncatula for the study of nitrogen-fixing and endomycorrhizal symbiotic associations. Mol. Plant Microbe Interact. 14 695–700. [DOI] [PubMed] [Google Scholar]
- Bolle, C. (2004). The role of GRAS proteins in plant signal transduction and development. Planta 218 683–692. [DOI] [PubMed] [Google Scholar]
- Brewin, N.J. (2004). Plant cell wall remodelling in the rhizobium-legume symbiosis. CRC Crit. Rev. Plant Sci. 23 293–316. [Google Scholar]
- Capoen, W., Goormachtig, S., De Rycke, R., Schroeyers, K., and Holsters, M. (2005). SrSymRK, a plant receptor essential for symbiosome formation. Proc. Natl. Acad. Sci. USA 102 10369–10374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catoira, R., Galera, C., de Billy, F., Penmetsa, R.V., Journet, E.P., Maillet, F., Rosenberg, C., Cook, D., Gough, C., and Denarie, J. (2000). Four genes of Medicago truncatula controlling components of a nod factor transduction pathway. Plant Cell 12 1647–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabaud, M., Venard, C., Defaux-Petras, A., Becard, G., and Barker, D.G. (2002). Targeted inoculation of Medicago truncatula in vitro root cultures reveals MtENOD11 expression during early stages of infection by arbuscular mycorrhizal fungi. New Phytol. 156 265–273. [DOI] [PubMed] [Google Scholar]
- Charron, D., Pingret, J.L., Chabaud, M., Journet, E.P., and Barker, D.G. (2004). Pharmacological evidence that multiple phospholipid signaling pathways link rhizobium nodulation factor perception in Medicago truncatula root hairs to intracellular responses, including Ca2+ spiking and specific ENOD gene expression. Plant Physiol. 136 3582–3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrhardt, D.W., Wais, R., and Long, S.R. (1996). Calcium spiking in plant root hairs responding to Rhizobium nodulation signals. Cell 85 673–681. [DOI] [PubMed] [Google Scholar]
- Endre, G., Kereszt, A., Kevei, Z., Mihacea, S., Kalo, P., and Kiss, G.B. (2002). A receptor kinase gene regulating symbiotic nodule development. Nature 417 962–966. [DOI] [PubMed] [Google Scholar]
- Feng, J.X., Liu, D., Pan, Y., Gong, W., Ma, L.G., Luo, J.C., Deng, X.W., and Zhu, Y.X. (2005). An annotation update via cDNA sequence analysis and comprehensive profiling of developmental, hormonal or environmental responsiveness of the Arabidopsis AP2/EREBP transcription factor gene family. Plant Mol. Biol. 59 853–868. [DOI] [PubMed] [Google Scholar]
- Franssen, H., Mylona, P., Pawlowski, K., Vandesande, K., Heidstra, R., Geurts, R., Kozik, A., Matvienko, M., Yang, W.C., Hadri, A.E., Martinezabarca, F., and Bisseling, T. (1995). Plant genes involved in root nodule development on legumes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 350 101–107. [Google Scholar]
- Gage, D.J. (2002). Analysis of infection thread development using Gfp- and DsRed-expressing Sinorhizobium meliloti. J. Bacteriol. 184 7042–7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleason, C., Chaudhuri, S., Yang, T.B., Munoz, A., Poovaiah, B.W., and Oldroyd, G.E.D. (2006). Nodulation independent of rhizobia induced by a calcium-activated kinase lacking autoinhibition. Nature 441 1149–1152. [DOI] [PubMed] [Google Scholar]
- Journet, E.P., El-Gachtouli, N., Vernoud, V., de Billy, F., Pichon, M., Dedieu, A., Arnould, C., Morandi, D., Barker, D.G., and Gianinazzi-Pearson, V. (2001). Medicago truncatula ENOD11: A novel RPRP-encoding early nodulin gene expressed during mycorrhization in arbuscule-containing cells. Mol. Plant Microbe Interact. 14 737–748. [DOI] [PubMed] [Google Scholar]
- Kaló, P., et al. (2005). Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science 308 1786–1789. [DOI] [PubMed] [Google Scholar]
- Kanamori, N., et al. (2006). A nucleoporin is required for induction of Ca2+ spiking in legume nodule development and essential for rhizobial and fungal symbiosis. Proc. Natl. Acad. Sci. USA 103 359–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuppusamy, K.T., Endre, G., Prabhu, R., Penmetsa, R.V., Veereshlingam, H., Cook, D.R., Dickstein, R., and Vandenbosch, K.A. (2004). LIN, a Medicago truncatula gene required for nodule differentiation and persistence of rhizobial infections. Plant Physiol. 136 3682–3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerouge, P., Roche, P., Faucher, C., Maillet, F., Truchet, G., Prome, J.C., and Denarie, J. (1990). Symbiotic host-specificity of Rhizobium meliloti is determined by a sulphated and acylated glucosamine oligosaccharide signal. Nature 19 781–784. [DOI] [PubMed] [Google Scholar]
- Levesque, M.P., Vernoux, T., Busch, W., Cui, H., Wang, J.Y., Blilou, I., Hassan, H., Nakajima, K., Matsumoto, N., Lohmann, J.U., Scheres, B., and Benfey, P.N. (2006). Whole-genome analysis of the SHORT-ROOT developmental pathway in Arabidopsis. PLoS Biol. 4 e143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy, J., et al. (2004). A putative Ca2+ and calmodulin-dependent protein kinase required for bacterial and fungal symbioses. Science 303 1361–1364. [DOI] [PubMed] [Google Scholar]
- Limpens, E., Franken, C., Smit, P., Willemse, J., Bisseling, T., and Geurts, R. (2003). LysM domain receptor kinases regulating rhizobial Nod factor-induced infection. Science 302 630–633. [DOI] [PubMed] [Google Scholar]
- Limpens, E., Mirabella, R., Fedorova, E., Franken, C., Franssen, H., Bisseling, T., and Geurts, R. (2005). Formation of organelle-like N2-fixing symbiosomes in legume root nodules is controlled by DMI2. Proc. Natl. Acad. Sci. USA 102 10375–10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, S.R. (1996). Rhizobium symbiosis: Nod factors in perspective. Plant Cell 8 1885–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen, E.B., Madsen, L.H., Radutoiu, S., Olbryt, M., Rakwalska, M., Szczyglowski, K., Sato, S., Kaneko, T., Tabata, S., Sandal, N., and Stougaard, J. (2003). A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 425 637–640. [DOI] [PubMed] [Google Scholar]
- Marsh, J.F., Rakocevic, A., Mitra, R.M., Brocard, L., Sun, J., Eschstruth, A., Long, S.R., Schultze, M., Ratet, P., and Oldroyd, G.E.D. (2007). Medicago truncatula NIN is essential for nodule organogenesis in the absence of rhizobial infection. Plant Physiol., in press. [DOI] [PMC free article] [PubMed]
- Mitra, R.M., Gleason, C.A., Edwards, A., Hadfield, J., Downie, J.A., Oldroyd, G.E.D., and Long, S.R. (2004. b). A Ca2+/calmodulin-dependent protein kinase required for symbiotic nodule development: Gene identification by transcript-based cloning. Proc. Natl. Acad. Sci. USA 101 4701–4705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra, R.M., Shaw, S.L., and Long, S.R. (2004. a). Six nonnodulating plant mutants defective for Nod factor-induced transcriptional changes associated with the legume-rhizobia symbiosis. Proc. Natl. Acad. Sci. USA 101 10217–10222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morohashi, K., Minami, M., Takase, H., Hotta, Y., and Hiratsuka, K. (2003). Isolation and characterization of a novel GRAS gene that regulates meiosis-associated gene expression. J. Biol. Chem. 278 20865–20873. [DOI] [PubMed] [Google Scholar]
- Nakano, T., Suzuki, K., Fujimura, T., and Shinshi, H. (2006). Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 140 411–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam, Y.-W., Penmetsa, R.V., Endre, G., Uribe, P., Kim, D., and Cook, D.R. (1999). Construction of a bacterial artificial chromosome library of Medicago truncatula and identification of clones containing ethylene-response genes. Theor. Appl. Genet. 98 638–646. [Google Scholar]
- Ohme-Takagi, M., and Shinshi, H. (1995). Ethylene inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell 7 173–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamuro, J.K., Caster, B., Villarroel, R., VanMontagu, M., and Jofuku, K.D. (1997). The AP2 domain of APETALA2 defines a large new family of DNA binding proteins in Arabidopsis. Proc. Natl. Acad. Sci. USA 94 7076–7081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldroyd, G.E.D., and Downie, J.A. (2004). Calcium, kinases and nodulation signalling in legumes. Nat. Rev. Mol. Cell Biol. 5 566–576. [DOI] [PubMed] [Google Scholar]
- Oldroyd, G.E.D., and Downie, J.A. (2006). Nuclear calcium changes at the core of symbiosis signalling. Curr. Opin. Plant Biol. 9 351–357. [DOI] [PubMed] [Google Scholar]
- Oldroyd, G.E.D., and Long, S.R. (2003). Identification and characterization of nodulation-signaling pathway 2, a gene of Medicago truncatula involved in Nod factor signaling. Plant Physiol. 131 1027–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parniske, M., and Downie, J.A. (2003). Plant biology - Locks, keys and symbioses. Nature 425 569–570. [DOI] [PubMed] [Google Scholar]
- Patil, S., Takezawa, D., and Poovaiah, B.W. (1995). Chimeric plant calcium/calmodulin-dependent protein kinase gene with a neural visinin-like calcium binding domain. Proc. Natl. Acad. Sci. USA 92 4897–4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penmetsa, R.V., and Cook, D.R. (2000). Production and characterization of diverse developmental mutants of Medicago truncatula. Plant Physiol. 123 1387–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radutoiu, S., Madsen, L.H., Madsen, E.B., Felle, H.H., Umehara, Y., Gronlund, M., Sato, S., Nakamura, Y., Tabata, S., Sandal, N., and Stougaard, J. (2003). Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425 585–592. [DOI] [PubMed] [Google Scholar]
- Saito, K., et al. (2007). NUCLEOPORIN85 is required for calcium spiking, fungal and bacterial symbioses, and seed production in Lotus japonicus. Plant Cell 19 610–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauser, L., Roussis, A., Stiller, J., and Stougaard, J. (1999). A plant regulator controlling development of symbiotic root nodules. Nature 402 191–195. [DOI] [PubMed] [Google Scholar]
- Smit, P., Raedts, J., Portyanko, V., Debelle, F., Gough, C., Bisseling, T., and Geurts, R. (2005). NSP1 of the GRAS protein family is essential for rhizobial Nod factor-induced transcription. Science 308 1789–1791. [DOI] [PubMed] [Google Scholar]
- Starker, C.G., Parra-Colmenares, A.L., Smith, L., Mitra, R.M., and Long, S.R. (2006). Nitrogen fixation mutants of Medicago truncatula fail to support plant and bacterial symbiotic gene expression. Plant Physiol. 140 671–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey, J.D. (2002). A direct approach to false discovery rates. J. R. Stat. Soc. Ser. B Stat. Methodol. 64 479–498. [Google Scholar]
- Stracke, S., Kistner, C., Yoshida, S., Mulder, L., Sato, S., Kaneko, T., Tabata, S., Sandal, N., Stougaard, J., Szczyglowski, K., and Parniske, M. (2002). A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature 417 959–962. [DOI] [PubMed] [Google Scholar]
- Tirichine, L., et al. (2006). Deregulation of a Ca2+/calmodulin-dependent kinase leads to spontaneous nodule development. Nature 441 1153–1156. [DOI] [PubMed] [Google Scholar]
- Tusher, V.G., Tibshirani, R., and Chu, G. (2001). Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98 5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernoud, V., Journet, E.P., and Barker, D.G. (1999). MtENOD20, a Nod factor-inducible molecular marker for root cortical cell activation. Mol. Plant Microbe Interact. 12 604–614. [Google Scholar]
- Wais, R.J., Galera, C., Oldroyd, G., Catoira, R., Penmetsa, R.V., Cook, D., Gough, C., Denarie, J., and Long, S.R. (2000). Genetic analysis of calcium spiking responses in nodulation mutants of Medicago truncatula. Proc. Natl. Acad. Sci. USA 97 13407–13412. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.