Abstract
Viruses with separately encapsidated genomes could have their genomes introduced into different leaves of a plant, thus necessitating long-distance trafficking of the viral RNAs for successful infection. To examine this possibility, individual or combinations of genome segments from the tripartite Brome mosaic virus (BMV) were transiently expressed in leaves of Nicotiana benthamiana plants using engineered Agrobacterium tumefaciens. BMV RNA3 was found to traffic from the initial site of expression to other leaves of the plant, as detected by RNA gel blot analyses and also by the expression of an endoplasmic reticulum–targeted green fluorescent protein. When RNA3 trafficked into leaves containing the BMV replication enzymes, RNA replication, transcription, and virion production were observed. RNA3 trafficking occurred even when it did not encode the movement or capsid proteins. However, coexpression of the movement protein increased the trafficking of BMV RNAs. BMV RNA1 and RNA2 could also traffic throughout the plant, but less efficiently than RNA3. All three BMV RNAs trafficked bidirectionally to sink leaves near the apical meristem as well as to the source leaves at the bottom of the stem, suggesting that trafficking used the phloem. These results demonstrate that BMV RNAs can use a replication-independent mechanism to traffic in N. benthamiana.
INTRODUCTION
RNA plays important roles in regulating cellular metabolism and development, in innate defense, and in establishing systemic infection by viruses (Carrington et al., 1996; Wassarman, 2002; Baulcombe, 2004; Lough and Lucas, 2006). These mechanisms require proper processing and transport of the RNAs from cell to cell. In plants, RNA-based communication has the additional challenge of being trafficked through the plasmodesmata and the plant vascular system (Lucas, 1995). The transport of small silencing-associated RNAs that spread systemically through the plasmodesmata (Voinnet et al., 1998; Lucas et al., 2001; Himber et al., 2003) and the phloem has been characterized (Yoo et al., 2004). In addition, several transcription factors have been documented to bind mRNAs and traverse the plasmodesmata as protein-RNA complexes (Lucas et al., 1995; Gilbertson and Lucas, 1996; Kühn et al., 1997; Haywood et al., 2005).
Viruses have evolved their own systems to traffic their RNA and RNA replication complexes (for reviews, see Waigmann et al., 2004; Nelson, 2005; Scholthof, 2005; Lucas, 2006). Studies showing how plant viral RNAs and proteins spread through the cells can be informative as a model for RNA trafficking in plants. The virally coded movement proteins (MPs) bind viral RNA in a sequence-nonspecific manner and are believed to interact with components of the plasmodesmata to increase their size exclusion limit, thus mediating the translocation of the viral RNA-MP complexes to adjacent cells (Waigmann et al., 2004).
Viral RNA trafficking is a complex phenomenon. Among the plant viruses, three modes have been distinguished by the requirements for MP and the capsid protein (CP) (Scholthof, 2005; Lucas, 2006, and references within). Tobamovirus, dianthovirus, and umbravirus genera comprise one group that requires only the MP for cell-to-cell trafficking of their RNAs. However, systemic movement by Tobacco mosaic virus also requires the CP and a component of the replication complex (Takamatsu et al., 1987; Dawson et al., 1988; Nelson et al., 1993; Bao et al., 1996; Liu et al., 2005). A second group, represented by potyviruses, hordeiviruses, and potexviruses, requires multiple MPs and the CP for both cell-to-cell and systemic trafficking of the viral RNA. The role of the CP may be to facilitate MP activity or to protect the genome. A third group, represented by comovirus and closteroviruses, requires the MP and the CP for cell-to-cell and long-distance movement. These viruses traffic as virion particles, thus explaining the requirement for CP in movement.
Bromoviruses, the subject of this study, exhibit an interesting phenomenon, where MP is required for cell-to-cell movement and, in some cases, the CP is also required for the trafficking through the plasmodesmata (Mise and Ahlquist, 1995; Schmitz and Rao, 1996). Depending on the viral strain and the plant host, however, different requirements for BMV cell-to-cell and systemic movement have also been reported (Sasaki et al., 2005, and references within).
Brome mosaic virus (BMV), the type member of the bromoviruses, has served as a model system for studies of viral replication and gene expression (Lane, 1981; Kao and Sivakumaran, 2000; Noueiry and Ahlquist, 2003). The BMV genome is divided into three positive-strand RNAs, designated RNA1, RNA2, and RNA3 (see Supplemental Figure 1A online). RNA1 and RNA2 are monocistronic, and each encodes a replication-associated enzyme that acts together in a membrane-associated complex called the replicase. BMV RNA1 and RNA2 together can replicate in barley (Hordeum vulgare) protoplasts in the absence of RNA3 (Kiberstis et al., 1981). RNA3 encodes both the MP and the CP. The MP is translated directly from RNA3, while the CP is translated from RNA4, which is synthesized from (−)-RNA3 (see Supplemental Figure 1B online) (Miller et al., 1985; Siegel et al., 1997).
An interesting feature of BMV biology is the separate encapsidation of its RNAs. RNA1 and RNA2 are encapsidated individually, while RNA3 and RNA4 are packaged together (Lane, 1981). All three genomic RNAs are required for successful BMV replication and systemic infection of plants. The arrangement of the BMV genome raises the possibility that the three particles could initially be introduced into different cells, especially when the viral titer is low. A potential solution to this problem would be for one or more viral RNAs to traffic from cell to cell or even to travel long distances to reconstitute a complete infection. To address the issue of BMV RNA trafficking within a host plant, we used an Agrobacterium tumefaciens–mediated system to deliver viral RNAs to different Nicotiana benthamiana leaves. BMV RNAs were found to traffic long distances independent of viral replication within the host plant.
RESULTS
Trafficking of BMV RNA3 to the Site of the BMV Replicase
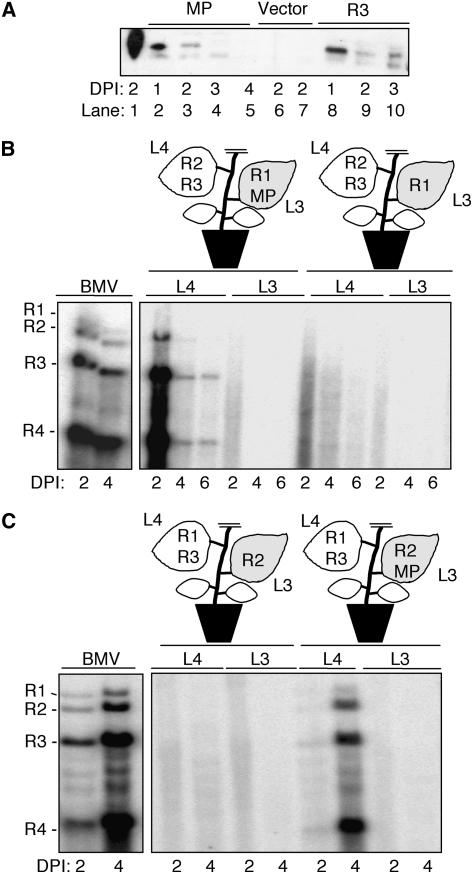
We have previously reconstituted BMV RNA replication and systemic infection in N. benthamiana by infiltrating plants with a mixture of Agrobacterium-containing recombinant T-DNA plasmids that encode the BMV cDNAs under the control of the 35S promoter of Cauliflower mosaic virus (CaMV). The replication of BMV RNAs transcribed from T-DNA mimics BMV replication in protoplasts derived from the natural host, barley (Gopinath et al., 2005). An advantage of the agroinfiltration system is that we can mix and match cultures to examine the cis- and trans-acting requirements for BMV infection in plants. A leaf agroinfiltrated to express all three BMV RNAs produces viral RNAs that are easily detectable by RNA gel blots starting at 22 h after infiltration, and the RNAs will accumulate for at least 5 d in the infiltrated leaves (Figure 1A, lanes 1 to 4).
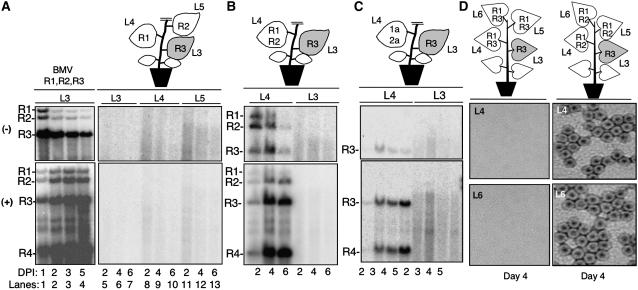
Figure 1.
Reconstitution of BMV RNA Replication in N. benthamiana by Agroinfiltration.
(A) RNA gel blot analysis demonstrating BMV replication for up to 6 d in N. benthamiana. The three lanes labeled “BMV” contain RNAs extracted from leaf tissue where all three BMV RNAs were expressed. The schematics of the plants illustrate the locations where BMV components were infiltrated into each plant. This schematic is intended to denote the location of and the composition of the agroinfiltration and not to denote the relative sizes of the leaves in the plants used. R1, RNA1; R2, RNA2; R3, RNA3. L1 to L6 denote the leaf number. The shaded leaf identifies the location of the RNA3 that will be the focus of the RNA trafficking. Lanes in the image of the RNA gel blots contain RNAs extracted from the leaves shown in the schematic of the plant. The identities of the BMV (+)- and (−)-strand RNAs are indicated to the left of the autoradiogram image. DPI, days after infiltration.
(B) Replication of RNA3 in leaf 4 after agroinfiltration into leaf 3. Leaf 4 was infiltrated with constructs that express RNA1 and RNA2.
(C) RNA3 expressed by agroinfiltration in one leaf replicated in neighboring leaves agroinfiltrated to transiently express the BMV replication proteins.
(D) Production of BMV virions in leaves that reconstituted BMV RNA3 replication. The schematics above the electron micrographs denote the location of BMV components expressed in the leaves relevant to this analysis. The fourth and sixth leaves were used to extract the virions shown in the electron micrographs. The electron micrographs show the presence of the BMV virions (∼28 nm) in leaves 4 and 6.
To test whether BMV RNAs initially expressed in separate N. benthamiana leaves could traffic independently to initiate systemic BMV infection, Agrobacterium cultures that singly express RNA1, RNA2, or RNA3 were infiltrated into three different leaves of the same plant (Figure 1A). No replicated viral RNAs were detected in RNA gel blots in any of the three leaves over the 6-d period (Figure 1A, lanes 5 to 13). Thus, individual BMV RNAs cannot result in BMV replication when all three RNAs are introduced in different leaves.
A likely explanation for the failure to detect BMV RNA replication is that formation of BMV replicase requires proper intermolecular interaction of the proteins expressed from RNA1 and RNA2 (Kao et al., 1992; Dinant et al., 1993; Chen et al., 2001). Another possibility is that the trafficking may be an inefficient event and the need to traffic two RNAs could decrease the efficiency to a level where we cannot easily detect the replication products. To overcome both events, an N. benthamiana plant was agroinfiltrated to express transiently both RNA1 and RNA2 in the fourth leaf, while the third leaf was agroinfiltrated with the RNA3 construct (Figure 1B). Throughout this manuscript, the leaf number is designated consecutively from the oldest, located at the base of the stem (leaf 1), to the youngest near the meristem (leaf 6). RNA3 was introduced to the third leaf by agroinfiltration to examine whether it could traffic along with the photoassimilates to the higher young sink leaves using the plasmodesmata-phloem symplastic network (Lucas et al., 1993). Two days after infiltration, all three of the negative-strand [(−)-strand] and positive-strand [(+)-strand] BMV genomic RNAs, as well as subgenomic RNA4, were detected. The detection of subgenomic RNA4 in leaf 4 was particularly informative, since it can only be expressed after successful replication of (−)-strand RNA3 (see Supplemental Figure 1B online) (Kao and Sivakumaran, 2000). Furthermore, the ratios of RNAs accumulated in the reconstitution experiment were comparable to that of a wild-type control (cf. Figures 1A and 1B). The trafficking of BMV RNA3 to the lower leaf 2 was also observed (see Supplemental Figure 2 online). Interestingly, these RNAs were detected in the leaf originally infiltrated to express RNA1 and RNA2, but not in leaf 3, which was infiltrated to express RNA3 (Figure 1B). A longer exposure of the RNA gel blot from leaf 3 revealed the presence of RNA3, but RNA4 was never detected (data not shown), suggesting that RNA3 trafficked to the location of BMV RNA1 and RNA2 but not vice versa.
To confirm that the coexpression of the BMV replicase proteins is required for RNA3 replication, we transiently expressed the replicase proteins 1a and 2a from constructs driven by the CaMV 35S promoter in leaf 4 (Gopinath et al., 2005). This produces mRNAs that encode 1a and 2a, but, unlike the experiment shown in Figure 2B, these RNAs lack the BMV 5′ and 3′ untranslated regions and cannot replicate. RNA3 was expressed by agroinfiltration in leaf 3 of the same plant. As seen in Figure 1B, leaf 4, but not leaf 3, was found to contain both (−)- and (+)-strand BMV RNA3 and also subgenomic RNA4 (Figure 1C), consistent with the results in Figure 1B. The level of (−)-strand RNA3 accumulation is lower than in leaves that contained all of the BMV RNAs, but the presence of RNA4 demonstrates that RNA3 had trafficked and replicated. Plants agroinfiltrated to express 1a and 2a in different leaves did not exhibit BMV RNA replication products (data not shown).
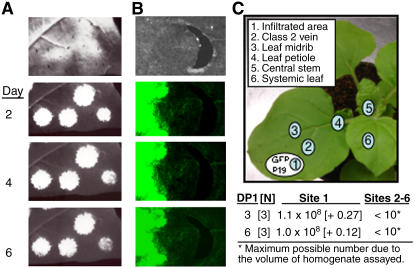
Figure 2.
Agrobacterium Does Not Freely Traffic in the Infiltrated Leaves.
(A) Macroscopic examination of N. benthamiana plants infiltrated to express GFP along with the silencing suppressor protein P19 from tomato bushy stunt virus. The top image is a part of the infiltrated leaf viewed under normal light. The needle pricks were used to demarcate the edge of the infiltrated areas. The images labeled Day 2, 4, and 6 denote the times after infiltration in which the same area of the leaf was photographed.
(B) Fluorescence microscopic examination of a spot infiltrated to express GFP and P19. The top image is of the infiltrated area that had been marked with a needle prick; the image was taken under normal light. The next three panels show microscopic images of the same infiltrated area and the lack of GFP expression between the infiltrated areas and the edge of the needle pricks.
(C) An examination of the location of Agrobacterium within an infiltrated N. benthamiana plant. The sites sampled for Agrobacterium colonies are labeled 1 to 6, where the site 1 represents the location where cells expressing both GFP and P19 were initially infiltrated. The numbers below the image of the plant are the means and 1 se of the CFUs per milliliter of plant lysates. The results were obtained from three independent experiments. Agrobacterium colonies were observed only in site 1, the site of infiltration, but the maximal possible number of CFUs is stated as <10 since we plated only 0.1 mL of leaf lysates and did not observe any colonies.
To determine whether reconstituted BMV RNA replication leads to a successful infection cycle, we purified BMV particles from leaf lysates by ultracentrifugation over a 10% sucrose cushion, which concentrates the virus at the bottom of the cushion (Rao et al., 1994). No viral particles were observed in leaf 3 (agroinfiltrated to express RNA3 only) (data not shown) or in leaves 4 and 6 of a control plant where agrobacterial constructs for RNA1 and RNA3 were infiltrated in the absence of RNA2 (Figure 1D, left two panels). However, abundant virions were detected by transmission electron microscopy in leaves 4 and 6 of plants agroinfiltrated with RNA1 and RNA2 constructs in these leaves, along with RNA3 in leaf 3 (Figure 1D, right two panels). The purified virions caused systemic infection after inoculation onto barley seedlings (data not shown), demonstrating that the virus produced was infectious. Thus, production of infectious BMV occurred as a result of trafficking of RNA3 from leaf 3 to higher leaves.
Agrobacterium Is Not Responsible for RNA3 Trafficking
Since RNA3 trafficked to the leaf containing the replicase proteins (or containing RNA1 and RNA2) but not vice versa, it seemed unlikely that the infiltrated Agrobacterium was responsible for the observed trafficking of RNA3. Nevertheless, to address this possibility directly, we agroinfiltrated leaf 3 and leaf 4 with constructs that direct the synthesis of enhanced green fluorescent protein (eGFP) (Kasschau, and Carrington, 1998) and the silencing suppressor protein P19, which increases the stability of the eGFP mRNA (Voinnet et al., 2003). After blotting excess culture from the leaf surface, pinpricks were used to mark the borders of the agroinfiltrated areas (∼4 mm in diameter), and eGFP expression was assessed by a handheld UV source over a 6-d period. If the inoculated agrobacteria were capable of movement from cell to cell, the eGFP fluorescence would spread beyond the infiltration spot. eGFP expression was readily detected by 24 h in the infiltration spots, but no significant spread of the fluorescence relative to the inoculation site was observed over a 6-d period (Figure 2A). Examination of the infiltrated cells and neighboring cells by fluorescence microscopy confirmed this observation (Figure 2B).
This experiment does not rule out the possibility that a few Agrobacterium cells could escape the initially inoculated area. Therefore, we infiltrated a mixture of cultures expressing GFP and P19 near the tip of the third and fourth leaves and then collected discs from different parts of the leaves and other parts of the plant after 3 and 6 d (Figure 2C). The leaf discs were homogenized in 500 μL of infiltration buffer, and 100 μL of the homogenate was plated onto media containing kanamycin (a marker for the binary T-DNA plasmid). An average of 1.0 × 108 colony-forming units (CFUs) per milliliter were obtained in the agroinfiltrated areas at both 3 and 6 d, but no Agrobacterium colonies were detected in other parts of the same leaf, the petiole, the stem, or in higher systemic leaf samples (Figure 2C). These results were reproduced in three independent experiments and were consistent between leaves 3 and 4. Together with the examination of eGFP (Figures 2A and 2B), these results show that Agrobacterium trafficking is not responsible for the establishment of RNA3 replication. Thus, the only viable explanation remaining is that BMV RNA3 is capable of trafficking long distances.
Roles of the RNA3-Encoded Proteins for Viral RNA3 Trafficking
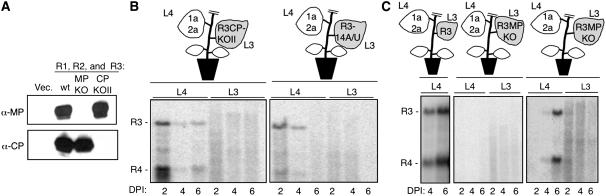
We wanted to determine whether RNA3-encoded proteins were required to traffic RNA3. The coat protein was unlikely to play an essential role because it is translated only after the replication of (−)-strand RNA3 synthesis. Even so, we tested a mutant version of RNA3, R3CPKOII, in which the first two CP initiation codons are mutated to CUG. It has been previously shown that these mutations abolish BMV CP translation (Rao, 1997). To confirm that CP expression was indeed affected by this mutation, extracts from N. benthamiana leaves agroinfiltrated to express RNA1, RNA2, and R3CPKOII were analyzed by protein gel blot analysis using CP antibody. Lysates expressing wild-type BMV RNAs had an abundance of CP, but none was detected in leaves infiltrated with R3CPKOII (Figure 3A). R3CPKOII was examined for its ability to traffic by expressing it in leaf 3 of a plant while the replicase proteins 1a and 2a were expressed in leaf 4. RNA gel blot analysis revealed the presence of R3CPKOII and subgenomic RNA4 in leaf 4 after 2 d, indicating that CP is not required for RNA3 trafficking (Figure 3B). The accumulation of R3CPKOII was lower than that of wild-type RNA3, but this is to be expected since the CP contributes to increased accumulation of (+)-strand BMV RNAs (Annamalai and Rao, 2005; Gopinath et al., 2005).
Figure 3.
CP Is Not Required for RNA3 Trafficking.
(A) Protein gel blot for CP and MP in leaves infiltrated to express the RNAs labeled above the gel image. “Vec.” denotes a control plant infiltrated with the Agrobacterium vector only. The other three samples were infiltrated to express RNA1, RNA2, and the mutant version of RNA3 noted above the lanes. The antisera used to probe the blots are indicated on the left.
(B) Results of RNA replication with two versions of BMV RNA3 defective for CP expression. The schematic of the plant is intended to denote the BMV components infiltrated into the leaf, not to denote the relative sizes of the leaves in the plants used. The RNA gel blots contain (+)-strand RNAs extracted from leaf 2 (L3) or leaf 4 (L4). DPI denotes the days after infiltration at which the RNA samples were harvested.
(C) MP contributes to RNA trafficking but is not absolutely required. The schematics of the plants depict the design of the experiments, and the leaves are labeled to show the infiltration of the BMV components. The RNA gel blots detect the synthesis of (+)-strand RNAs in two independent experiments where MP translation was prevented by a mutation in the initiation codon.
To confirm that the CP is not required for RNA3 trafficking, we tested a construct that will express an RNA named R3-14A/U, which expresses an RNA3 with a substitution in the subgenomic core promoter that debilitates RNA4 production (Sivakumaran et al., 2004; Gopinath et al., 2005). The agroconstruct of this mutant was expressed in leaf 3 of a plant, while 1a and 2a were transiently expressed by agroinfiltration in leaf 4. RNA gel blot analysis revealed the presence of RNA3 in leaf 4 in which only 1a and 2a had been infiltrated. As expected, RNA4 levels were much decreased with R3-14A/U. These results support our contention that the CP is not required to traffic RNA3 between leaves (Figure 3B).
To determine whether the MP is required for RNA3 trafficking, we used an RNA named R3MPKO, which has the MP initiation codon mutated to CUG (Gopinath et al., 2005). In protein gel blots of lysates made from leaves agroinfiltrated to express RNA1, RNA2, and R3MPKO, MP was not detected, although abundant CP was (Figure 3A). To analyze whether R3MPKO could traffic between leaves, it was expressed by agroinfiltration in leaf 3 of an N. benthamiana plant, while leaf 4 of the same plant was agroinfiltrated to express RNA1 and RNA2. Of the six independently infiltrated plants, two did not accumulate RNA3 and RNA4 in leaf 4 even after 6 d, but four accumulated some level of RNA3 and RNA4. A representative of each class is shown in Figure 3C. In the four plants where there was trafficking, RNA3 and RNA4 accumulation was reliably observed at day 4 but not at day 2, indicating a delay in replication compared with the wild-type control.
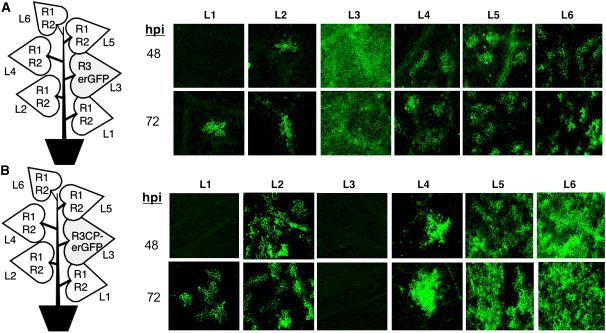
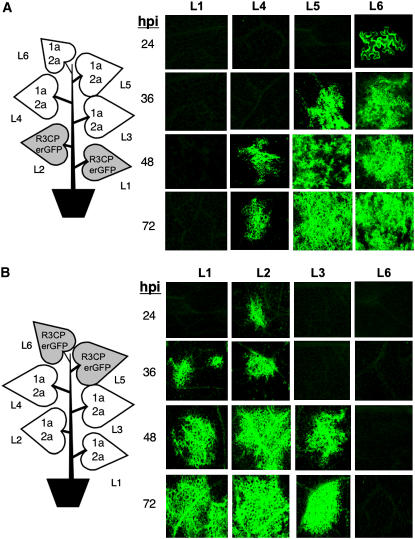
BMV cell-to-cell movement in N. benthamiana was previously reported to require MP (Mise and Ahlquist, 1995). Our results suggest that a low level of RNA trafficking could take place in the absence of MP. To confirm that MP is not absolutely required for RNA trafficking, we replaced the MP coding sequence in RNA3 with an endoplasmic reticulum–targeted green fluorescent protein (ERGFP) (Haseloff et al., 1997). ERGFP is restricted to the endoplasmic reticulum and does not significantly diffuse from cell to cell in the mature source or young sink leaves (Oparka et al., 1999; Roberts et al., 2001). This construct, R3erGFP, was infiltrated into leaf 3 of an N. benthamiana plant while all other leaves were agroinfiltrated to express BMV RNA1 and RNA2. In this construct, ERGFP can be translated from R3erGFP without RNA replication because it replaces the MP. Within 24 h after infiltration, ERGFP fluorescence was observed in every cell in the infiltrated area of leaf 3 (data not shown). The fluorescence level was maximal 2 d after agroinfiltration but decreased by 72 h (Figure 4A). There was no detectable fluorescence in leaf 1, while fluorescent single cells and occasionally clusters of cells were observed in the higher leaves (Figure 4A). At day 2 post infiltration, there was no detectable fluorescence in leaf 1, although single cells were observed on day 3. The upper leaves (leaves 4, 5, and 6) had more fluorescent cells than the lower leaves, and occasionally clusters of cells were observed above the leaves infiltrated with R3erGFP (Figure 4A). The abundance of ERGFP expressed in epidermal cells in leaves 4, 5, and 6 (Figure 4A) also increased from 48 to 72 h. Within the higher leaves, the fluorescent epidermal cells were primarily observed in the proximity of the class 3 veins or in the spaces between class 4 and 5 veins (Roberts et al., 2001), where the images were taken in Figure 4A. These results further confirm the idea that the MP is not absolutely required for the trafficking of RNA3 and eliminates the possibility that a reversion event could have restored MP levels.
Figure 4.
Visualization of ERGFP Expressed from R3erGFP and R3CPerGFP in N. benthamiana.
(A) Detection of ERGFP expressed from R3erGFP. The schematic of a plant illustrates where agroinfiltration was used to express the components shown. The shaded leaf 3 denotes the source of the RNA that could express ERGFP. The fluorescence micrographs are representative images of ERGFP from each leaf of the infiltrated plant. hpi denotes the hours after infiltration at which the samples were photographed. All of the images were taken at ×10 magnification.
(B) Microscopic examination of ERGFP expression in N. benthamiana leaves from R3CPerGFP. Except for L5 and L6 (×5), all of the other images were taken at ×10 magnification.
We next examined whether replacement of the CP with ERGFP would affect movement from leaf to leaf (Figure 4B). The CP coding sequence from codon eight through the termination codon was replaced with the sequence coding for the ERGFP resulting in the construct R3CPerGFP. In this case, ERGPF was expressed from RNA4 and could not be synthesized unless RNA3 was replicated to produce (−)-strand RNA3 (see Supplemental Figure 1B online). N. benthamiana plants were agroinfiltrated with R3CPerGFP in leaf 3, while the other leaves were infiltrated with constructs to express RNA1 and RNA2. RNA gel blots to probe for the GFP-coding sequence detected R3CPerGFP in all of the leaves above and below leaf 3 (see Supplemental Figure 3 online). Notably, all leaves replicated R3CPerGFP and also the subgenomic RNA4 except leaf 3, demonstrating that the regulatory sequences in this RNA remained functional. ERGFP was detected in single epidermal cells starting around 24 h in leaf 6, but not in other leaves at this time (data not shown). By 48 h, several clusters of ∼10 cells each were found in the second leaf, and the fluorescence signals were stronger in the 4th, 5th, and 6th leaves. At 72 h, the ERGFP signals were observed in several islands of epidermal cells in the 1st and 2nd leaves and spread progressively in the higher leaves (Figure 4B).
The results obtained with R3erGFP and R3CPerGFP differ, likely because the latter produces functional MP, which alters the size exclusion limit of the plasmodesmata to increase the rate of trafficking. While the kinetics of spread were different depending on whether CP or MP was present, the results in Figures 4A and 4B demonstrate that neither CP nor MP was absolutely required for RNA trafficking. Furthermore, R3CPerGFP could traffic from leaf 3 throughout the plant and in both the basal and apical directions. Additional examination of the directionality of BMV RNA trafficking will be presented later.
Trafficking of RNA1 and RNA2
Thus far we have not presented evidence for the trafficking of BMV RNA1 and RNA2. Since the presence of MP increased RNA3 trafficking, we examined whether RNA1 and RNA2 could traffic when the MP was expressed in trans (Gopinath et al., 2005). An N. benthamiana plant was agroinfiltrated to express the MP along with RNA1 in the third leaf and RNA2 and RNA3 in the fourth leaf (Figure 5B). Immunoblot analysis showed that the amount of MP expression in trans was comparable to the amount expressed from BMV RNA3 at 24 h after agroinfiltration (Figure 5A). Within 2 d after infiltration, RNA3 replication and RNA4 transcription were observed in leaf 4, and some persisted through day 6. As controls, plants agroinfiltrated to express RNA1 and an empty vector did not produce replication products in either leaf 3 or 4 (Figure 5B). These results indicate that RNA1 must have moved from its site of infiltration in leaf 3 to the replication site in leaf 4 and that this migration required MP. The lack of trafficking of RNA1 and RNA3 was reproducible in six independently tested plants (data not shown). Note, however, that the relative ratios of the four BMV RNAs were different than a normal wild-type BMV infection, suggesting that RNA1 movement was not as efficient as that of RNA3 when both were expressed from leaf 3 of comparable plants.
Figure 5.
Examination of the Effects of BMV MP on RNA1 and RNA2 Trafficking.
(A) Protein gel blot demonstrating that the MP expressed transiently from a CaMV 35S promoter is comparable to the levels expressed from wild-type RNA3.
(B) Trans-expression MP supports RNA1 trafficking. Locations of the BMV components expressed by agroinfiltration are noted in the schematic of the plants. The shaded leaf 3 denotes agroinfiltration of RNA2 and the MP. The RNA gel blot image contains (+)-strand viral RNAs extracted from the leaves as diagrammed in the schematic.
(C) Trans-expression of MP supports trafficking of RNA2. The shaded leaf 3 denotes the site of agroinfiltration with cultures that express RNA2 and the MP.
BMV RNA replication was also reconstituted in plants agroinfiltrated to express the MP and RNA2 in leaf 3 and RNA3 and RNA1 in leaf 4 (Figure 5C). Together, these results demonstrate that the MP can function in trans to mediate long-distance trafficking of both RNA1 and RNA2. The presence of RNA3 did not noticeably aid in the trafficking of RNA2 (Figure 5B) or RNA1 (Figure 5C), perhaps due to RNA3 titrating away some of the MP.
Leaf and Plant Development and RNA Trafficking
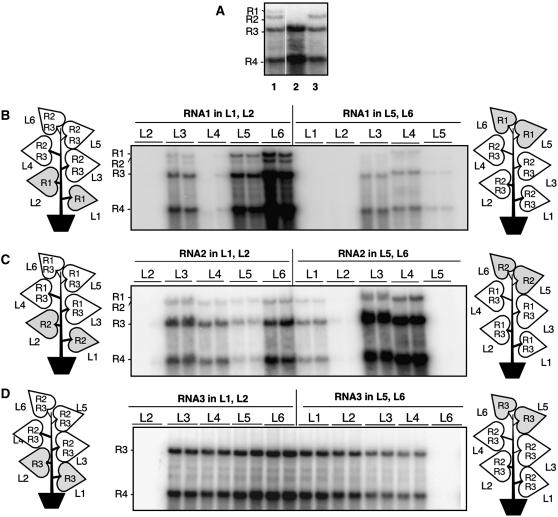
It is known that the developmental stage of a plant can lead to alterations of plasmodesmatal permeability during the sink/source transition from simple to branched plasmodesmata (Itaya et al., 1998; Oparka et al., 1999; Roberts et al., 2001). We wanted to determine whether the position of the leaf would influence the efficiency of RNA trafficking. Since RNA1 or RNA2 trafficked inefficiently from leaf 3 in comparison to RNA3, we first infiltrated a construct that can express RNA1 from the bottom two mature source leaves or the top two young sink leaves. The other leaves were agroinfiltrated to express BMV RNA2 and RNA3. All of the leaves were harvested 4 d after infiltration, and two samples from each leaf were analyzed for BMV RNAs in RNA gel blots, including blots performed with different combinations of RNAs that would help to identify each (Figure 6A). When RNA1 was initially expressed in the bottom two leaves, we detected BMV RNA replication products in three of the five leaves assayed (Figure 6B). The level of replicated RNAs in each leaf differed significantly. Interestingly, expressing RNA1 from the top two leaves also resulted in some replication products in several, but not all, of the leaves. Similar results were obtained when construct expressing RNA2 was initially infiltrated in the lower two or the higher two leaves and the other leaves were made to express RNA1 and RNA3 (Figure 6C). Results from these experiments differed from those wherein RNA1 or RNA2 was initially expressed in the third leaves of plants (Figure 5). Therefore, we conclude that the leaf position could significantly influence the trafficking of the BMV RNAs and that all three BMV RNAs could traffic, albeit with differing efficiencies.
Figure 6.
The Leaf Position Where BMV RNAs Were Initially Expressed Affected RNA Trafficking.
(A) Three samples of replicated BMV RNAs to aid in the identification of the RNAs.
(B) Trafficking of BMV RNA1 when initially expressed in the bottom two leaves (L1 and L2) or the top two leaves (L5 and L6) of the plants. The schematics of N. benthamiana plants denote the locations where BMV constructs were infiltrated. The leaves in gray denote the location where the RNAs of interest were infiltrated. The leaves from which the RNA samples were isolated are identified above the image of the RNA gel blot probed to detect (+)-strand BMV RNAs. The identities of the bands are shown to the left of the autoradiogram images.
(C) Trafficking of BMV RNA2 when initially expressed in the bottom two leaves (L1 and L2) or the top two leaves (L5 and L6) of the plants. The layout of this panel is identical to that of (B).
(D) Trafficking of BMV RNA3 when initially expressed in the bottom two (L1 and L2) or the top two leaves (L5 and L6) of the plants.
A similar analysis of construct expressing BMV RNA3 was infiltrated in the top two or the bottom two leaves, while the others leaves were made to express transiently the 1a and 2a proteins. As would be expected, RNA3 trafficked from these leaves throughout the plant and notably more efficiently toward the apical meristem, except where RNA3 was initially expressed in the absence of replicase (Figure 6D).
To confirm the bidirectional trafficking of BMV RNA3 and to better characterize the kinetics of RNA trafficking, R3CPerGFP was agroinfiltrated either into the two most mature source leaves at the bottom of the plant or into the two youngest sink leaves near the apical meristem. The young leaves are typically small (<2 to 3 cm in diameter) and accommodated only ∼0.4 mL of agrobacterial culture compared with the mature leaves 3 or 4, where we can infiltrate up to 3 mL of culture into each leaf. The youngest and older leaves are thought to differ in the structures of their plasmodesmata (Oparka et al., 1999; Roberts et al., 2001). In both case, all of the other leaves were agroinfiltrated to transiently express the BMV replicase proteins 1a and 2a. No ERGFP signals were detected in the leaves infiltrated to express R3CPerGFP at the top or the bottom of the plant (Figure 7, panels L1 and L6). ERGFP fluorescence was observed at 24 h in the epidermal cells of the 5th and 6th leaves but not in leaves that are closer to the source of R3CPerGFP (Figure 7A). The ERGFP signal increased and spread into the leaf lamina by 72 h. The presence of ERGFP in the other leaves, such as leaf 4, was detected starting at 48 h, indicating efficient trafficking toward the apical meristem where the young leaves are sinks for importing photoassimilates. In plants where R3CPerGFP was infiltrated in the top two leaves, ERGFP fluorescence was first observed at the end of class 3 veins in leaf 2 at 24 h and leaf 1 at 36 h (Figure 7B). The ERGFP signal was not observed in the leaves closer to the source of R3CPerGFP until after 36 h (Figure 7B), confirming that RNA can traffic through the phloem toward the apical meristem and the roots. These results also demonstrate the feasibility of trafficking engineered BMV RNA molecules throughout the plant.
Figure 7.
Directional Trafficking of R3CPerGFP.
(A) Trafficking of R3CPerGFP infiltrated in the bottom two most mature leaves of the plant. The schematic of the plant indicates the locations where the BMV components were expressed by agroinfiltration. The shaded leaves denote the source of the RNA that expresses ERGFP. The fluorescent micrographs were taken at the hour after infiltration (hpi) indicated. All the images were taken at ×5 magnification, except L6, which was at ×40 magnification.
(B) Trafficking of R3CPerGFP expressed by agroinfiltration in the top two leaves of the plant.
DISCUSSION
Several virus families, including the Bromoviridae, Chrysoviridae, Furoviridae, Pomoviridae, and Artiviridae, have genomes that are encapsidated into multiple viral particles, raising the possibility that the viral genomes have evolved the ability to traffic their RNAs from cell to cell to reconstitute an infectious genome. We have successfully demonstrated that all three BMV RNAs could traffic long distances in N. benthamiana in the presence of the MP. RNA3 was the most efficient in trafficking, and it did so in a manner independent of the MP. Furthermore, BMV RNA trafficking does not require a prior round of RNA replication or encapsidation by the CP. These results rule out an essential role for viral proteins in BMV RNA3 trafficking. Furthermore, the trafficked RNA3 resulted in the accumulation of both viral RNA replication products and the production of viral particles. The movement of the RNAs and their subsequent amplification could take place bidirectionally in the plant, with somewhat more efficient trafficking toward the meristem. This result suggests that RNA is transported through the phloem in the net direction of photoassimilate flow (Jorgensen et al., 1998; Golecki et al., 1999; Ruiz-Medrano et al., 2001). Lastly, BMV RNA could also mediate long-distance trafficking of recombinant ERGFPs expressed from BMV RNAs, suggesting that RNA3 may have value as a targeted gene delivery system in planta.
The best-studied RNA trafficking systems reported thus far are viroids, which are the smallest single-stranded and circular RNAs (275 to 400 nucleotides) that infect plants. Viroids do not encode proteins and are not encapsidated, indicating that the information within the RNA is responsible for trafficking, likely through the aid of cellular factors (Ding et al., 2005; Ding and Itaya, 2007). The other emerging system is the umbraviruses (Taliansky and Robinson, 2003), which do not encode a functional CP and do not form virion particles. Nevertheless, umbraviruses can traffic long distances in plants in the presence of the ORF3 protein (Taliansky et al., 2003). The existing view regarding long-distance transport is that once a protein or RNA molecule enters the sieve tube in the phloem, it moves from source to sink organs by simple diffusion following mass flow (Sjölund, 1997). However, studies from Potato spindle tuber viroid in tomato (Solanum lycopersicum), N. benthamiana, and Nicotiana tabacum suggest that it doesn't simply follow the mass flow from source to sink organs, rather it traffics into selective sink organs but fails to move into source leaves (Zhu et al., 2001, 2002).
The presence of the MP expressed along with the viral RNA increased RNA trafficking, suggesting that the MP either enhances the activities of the cellular proteins and/or traffics the RNAs by a more direct mechanism, perhaps by altering the plasmodesmatal size exclusion limit. This activity of the MP is consistent with the large body of literature on the properties of viral MPs (Lucas, 2006), including the trafficking of an animal virus RNA through a plant when the MP is expressed from transgenic plants (Dasgupta et al., 2001). MPs characterized thus far exhibit sequence-nonspecific RNA binding in vitro (Jansen et al., 1998; Carvalho et al., 2004; Waigmann et al., 2004). Increased efficiency of trafficking in cis may reflect better access to the RNA by the MP.
Several features of BMV RNA3 trafficking that we observed can be compared with published results. With regard to the requirement for the MP, not only is it required to traffic RNA1 and RNA2, we observed that the ERGFP expressed in place of the MP in R3erGFP was mostly confined to single epidermal cells or at most to clusters of four to five cells even after 6 d (Figure 4A). In the absence of the MP, R3erGFP may not be able to traffic efficiently through the plasmodesmata to the neighboring cells. Also in support of a role for the MP is the observation that the ERGFP generated from R3CPerGFP could spread throughout the leaf within 48 h, indicating that plasmodesmatal gating was more efficient in the presence of functional MP (Figure 4B). These observations are consistent with published reports (Mise and Ahlquist, 1995; Schmitz and Rao, 1996; Rao, 1997; Sasaki et al., 2003, 2005). Given that both R3erGFP and R3CPerGFP could traffic within the plant, we would propose that the RNAs are not trafficking solely from the epidermal cells. Consistent with this, we have noticed substantial ERGFP signals in the cortical mesophyll cells underneath the fluorescent epidermal cell clusters (data not shown).
Our finding that BMV RNAs can traffic to the site of the BMV replicase but not vice versa suggests that replication is not a prerequisite for viral RNA3 trafficking. This claim is supported by the absence of the subgenomic RNA in the leaves infiltrated with only RNA3 (Figure 1B) and is in contrast with the trafficking of small RNAs. Himber et al. (2003) and Voinnet et al. (2003) have reported that long-distance trafficking of small RNAs requires the amplification of the RNAs with the replication-associated proteins SDE1 and SDE3. Kawakami et al. (2004) have reported that fragmented membranes containing the replicase of Tobacco mosaic virus could traffic through the plasmodesmata. If this occurs in our system, it may be at a level that is insufficient to significantly amplify RNA. More likely, movement of the replicase occurs only after the cell has undergone robust viral infection with the fragmentation of the cellular membranes that can accompany viral infection (Schwartz et al., 2004).
We report here that long-distance BMV RNA trafficking does not require the CP, in contrast with previous reports of several groups (Mise and Ahlquist, 1995; Schmitz and Rao, 1996; Okinaka et al., 2001). With respect to RNA3, we obtained trafficking in three independent constructs that affected CP expression, including one that had all CP coding sequence replaced with ERGFP except for the first eight codons. The trafficking of BMV RNA1 and RNA2 also supports our contention that the CP is not required for their trafficking. We also note that there are strains of BMV that do not require the CP for cell-to-cell movement (Takeda et al., 2005) and that some changes in the MP can convert a BMV strain that requires the CP for trafficking to one that does not (Sasaki et al., 2005). These results indicate that the requirement for the CP in BMV trafficking may not be stringent.
There may be several reasons for our observations, perhaps acting in varying combinations, depending on the developmental stage of the plant. First, it is possible that the level of expression we achieved by agroinfiltration overcame restrictions observed with other inoculation methods. BMV has been selected as a model system because it replicates to very high levels in its monocot hosts, such as barley. The levels of BMV RNAs we produced in N. benthamiana are similar to those seen in barley plants (Gopinath et al., 2005). Second, we are amplified the trafficked RNAs by expressing the BMV replicase in other leaves. We could, however, observe ERGFP signals in cells expressed from R3erGFP in leaves that lacked the replicase, suggesting that the replicase is merely amplifying the RNAs that have successfully trafficked. Third, all of our results are obtained in the model plant N. benthamiana, which is a susceptible host for a wide range of viruses. Furthermore, it has been shown to be partially defective in RNA-dependent RNA polymerase 1, which participates in the RNA silencing pathway (Yang et al., 2004). It is possible that other plant species will differ in the ability to support RNA trafficking. However, this remains to be determined, and having a model plant and RNA virus should prove to be instructive for an in-depth understanding of viral RNA trafficking.
An area that is ripe for future analysis is the interaction of cellular proteins with BMV RNA. Several proteins that specifically traffic RNAs in the phloem have been identified and characterized (Lucas et al., 1995; Gilbertson and Lucas, 1996) and could be examined for effects on BMV RNA trafficking. In addition, from a proteome-wide screen, we have identified a yeast protein, Actin patch protein 1 (App1), which can recognize BMV RNAs and, when overexpressed in N. benthamiana, prevented systemic infection by BMV (Zhu et al., 2007). App1 likely interacts with the cellular actin network (Samanta and Liang, 2003) and may affect RNA trafficking.
In terms of identifying the cis-acting sequences required to interact with the cellular proteins, BMV RNA3 is an obvious choice. It is the best adapted among the three BMV RNAs to take advantage of the MP-independent trafficking mechanism, perhaps because it needs to get to the site of RNA replication while the other two RNAs replicate in cis to the replication proteins they encode. RNA sequences involved in trafficking have been identified in the 5′ untranslated region of Potato virus X (Lough et al., 2006). Using Potato spindle tuber viroid as a model system, Qi et al. (2004) identified functional motifs that are required for efficient trafficking from bundle sheath into mesophyll cells to establish systemic infection in N. tabacum. This motif, however, is not necessary for the trafficking in the reverse direction that is from mesophyll to bundle sheath cells. Retroviral RNAs have evolved sequences that can regulate the trafficking of the spliced and unspliced RNAs from the nucleus to the cytoplasm (Tang et al., 1997; Strebel, 2003). In the absence of a functional role for the BMV-encoded proteins MP and CP in the trafficking, 5′ and 3′ noncoding regions (NCRs) and the intercistronic region in RNA3 are candidate regions to contain functional motifs involved in the interaction with the phloem proteins. A comparison of the motifs required for this activity should yield clues regarding the requirements for protein–RNA interaction that contribute to the long-distance trafficking of viral RNAs.
METHODS
Materials and Chemicals
All chemicals were purchased from Sigma-Aldrich, unless stated otherwise, and were of the purity certified by the ACS.
Nicotiana benthamiana Growth Conditions
N. benthamiana plants were grown in a growth chamber with 26/22°C day/night temperature cycles and 16-h-light/8-h-dark cycles. For each set of experiments, we used 6- to 8-week-old plants that were typically at the six leaf stage, and each experiment was repeated three times unless otherwise stated. A representative set has been presented for each experiment.
Molecular Manipulations
The M1 (Russian) strain was used to generate Agrobacterium tumefaciens constructs to express BMV RNAs and proteins (Ahlquist et al., 1984; Dreher et al., 1989). A number of the expression constructs were described in Gopinath et al. (2005). All mutations were engineered by site-directed mutagenesis using the QuikChange kit (Invitrogen). Pfu polymerase was used to amplify PCR products. Each PCR product was cloned into the pGEM-T Easy vector (Promega), and the sequences of all constructs were validated by DNA sequencing using the Big Dye sequencing kit (US Biochemicals).
Two PCR fragments were generated in an effort to replace the MP open reading frame (ORF) in RNA3 with the ERGFP ORF. The first was generated with the cDNA to BMV RNA3, pBR3, as a template and primers 1 and 2 (see Supplemental Table 1 online), which add flanking BglII and NcoI sites, respectively, to the CaMV 35S promoter and the 5′ NCR of RNA3. The second PCR product was generated using pBR3 and primers 3 and 4 that add flanking XbaI and AseI sites, respectively, to the intercistronic region. The two DNA fragments were released from pGEM-T Easy by digestion with BglII and AseI. DNA encoding for ERGFP ORF was amplified from pBIN-mGFP5-ER using primers 5 and 6 (see Supplemental Table 1 online). mGFP5-ER is an endoplasmic reticulum–localized form of the plant optimized GFP with an N-terminal Arabidopsis thaliana basic chitinase signal sequence and a C-terminal KDEL endoplasmic reticulum retention signal (Haseloff et al., 1997). Primers 5 and 6 add flanking NcoI and XbaI sites, respectively, and the ERGFP sequence along with the other two PCR products was cloned into pBR3 at the BglII and AseI restriction sites to generate plasmid R3erGFP.
Two PCR products were used to express ERGFP in place of the CP. The first was generated using pBIN-mGFP5-ER as a template and primers 7 and 6 (see Supplemental Table 1 online), which add an AseI site and the sequence for the first eight codons of the CP ORF to the 5′ end of ERGFP and an XbaI restriction site at the 3′ end, respectively. The second PCR product was generated using pBR3 as a template and primers 8 and 9 (see Supplemental Table 1 online) that add an XbaI site at the start of the RNA3 3′ NCR and an XmaI restriction site at the 3′ end of the BMV RNA3 cDNA. The resultant plasmid that should express ERGFP is named R3CPerGFP.
Agrobacterium Cultures and Plant Infiltrations
Plasmids were transformed into Agrobacterium strain C58C1 by electroporation (Gopinath et al., 2005). Agroinfiltration into N. benthamiana was performed as described by Llave et al. (2000). Briefly, cultures harboring each plasmid were grown at 30°C from single colonies in Luria-Bertani broth containing kanamycin (50 μg/mL), 10 mM MES, pH 5.9, and 50 μM acetosyringone. The cultures were centrifuged at 6000 rpm for 15 min, and the pellets were suspended in the infiltration medium (10 mM MgCl2, 10 mM MES, pH 5.9, and 150 μM acetosyringone) and incubated at room temperature for a minimum of 3 h. Agrobacterium cultures (at 0.5 OD600) were mixed at equal proportions prior to infiltration to result in the same bacterial concentrations for all infiltrated samples. N. benthamiana plants containing up to six leaves were infiltrated by pressing the end of a 3-mL syringe loaded with the appropriate culture to the lower side of a leaf and exerting gentle pressure to flood the interstitial areas of the leaf.
Analysis of BMV RNAs
Total nucleic acids were extracted from ∼50 mg of leaf tissue after macerating the tissue with a disposable pestle that fit into a microcentrifuge tube in the presence of a lysis buffer (0.1 M glycine, pH 9.2, 40 mM EDTA, 100 mM NaCl, 2% SDS, and 0.05% bentonite) and then extracted with an equal volume of phenol and chloroform (1:1 mix) and precipitated with 1 volume of isopropanol. RNA gel blots were performed with 5 μg of glyoxylated total RNAs and were probed with strand-specific 32P-labeled riboprobes generated from the 200 nucleotides of BMV RNA3, as described by Choi et al. (2004) and Hema and Kao (2004).
Protein Analysis
Approximately 50 mg of leaf sample were suspended in 0.5 mL of a TB buffer containing 50 mM Tris acetate, pH 7.4, 10 mM potassium acetate, 1 mM EDTA, 5 mM DTT, and 0.5 mM PMSF (Gopinath et al., 2000). The homogenate was centrifuged at 3000g for 10 min at 4°C to remove insoluble materials, and the supernatant was subjected to 30,000g for 20 min at 4°C. The green pellet was considered the membrane-associated fraction and the supernatant the soluble fraction. When used, the membrane pellet was suspended in a volume equivalent to the soluble fraction. All samples were suspended in Laemmli's sample buffer, and the proteins were separated on 4 to 12% gradient gels. For protein gel blots, the proteins were transferred onto polyvinylidene difluoride membranes. The primary antisera were added to the membranes preblocked with 5% (w/v) nonfat milk powder in phosphate-buffered saline. Polyclonal antiserum to the MP was a kind gift of K. Mise, and the antiserum to the CP was purchased from the American Type Culture Collection. The secondary antibody was goat anti-rabbit conjugated to horseradish peroxidase. The antigens were visualized with the ECL Plus detection system (Amersham Biosciences).
Fluorescence Microscopy
GFP and ERGFP fluorescence from leaf samples were visualized by fluorescence microscopy using a Zeiss Axio 2 microscope with a GFP-optimized fluorescein isothiocyanate filter set at 543-nm excitation and 505- to 530-nm emission parameters. All series from one treatment were at the same magnification. Virus particles were negatively stained with 1% uranyl acetate, and images were taken with a JEOL 1200EX electron microscope set at 100 kV and a magnification of ×39,000. Virus particles were purified from N. benthamiana leaves by the protocol of Rao et al. (1994).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table 1. Oligonucleotides Used to Construct BMV RNA3 to Express Endoplasmic Reticulum–Targeted GFP.
Supplemental Figure 1. Background Information on BMV Biology.
Supplemental Figure 2. The BMV RNA3 Expressed by Agroinfiltration in Leaf 3 Can Traffic to Neighboring Leaves That Are Higher or Lower Than Leaf 3.
Supplemental Figure 3. RNA Gel Blot Analysis of R3CPerGFP and the Subgenomic RNA Derived from RNA3 Showing Movement to the Noninfiltrated Leaves.
Supplementary Material
Acknowledgments
We thank the Texas A&M University Cereal Killers for helpful discussions, A. Murali for the electron microscopic images of BMV, Kazuyuki Mise for the polyclonal antibodies to the MP, Jim Haseloff for the plasmid of ERGFP, and Laura Kao and Linda Guarino for editorial suggestions. The Kao lab is supported by the National Science Foundation MCB Grant 0332259.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: C. Cheng Kao (ckao@tamu.edu).
Online version contains Web-only data.
References
- Ahlquist, P., French, R., Janda, M., and Loesch-Fries, L.S. (1984). Multicomponent RNA plant virus infection derived from cloned viral cDNA. Proc. Natl. Acad. Sci. USA 81 7066–7070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annamalai, P., and Rao, A.L.N. (2005). Replication-independent expression of genome components and capsid protein of brome mosaic virus in planta: A functional role for viral replicase in RNA packaging. Virology 338 96–111. [DOI] [PubMed] [Google Scholar]
- Bao, Y., Carter, S.A., and Nelson, R.S. (1996). The 126- and 183-kilodalton proteins of tobacco mosaic virus, and not their common nucleotide sequence, control mosaic symptom formation in tobacco. J. Virol. 70 6378–6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baulcombe, D. (2004). RNA silencing in plants. Nature 431 356–363. [DOI] [PubMed] [Google Scholar]
- Carrington, J.C., Kasschau, K.D., Mahajan, S.K., and Schaad, M.C. (1996). Cell-to-cell and long-distance transport of viruses in plants. Plant Cell 8 1669–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho, C.M., Pouwels, J., van Lent, J.W., Bisseling, T., Goldbach, R.W., and Wellink, J. (2004). The movement protein of cowpea mosaic virus binds GTP and single-stranded nucleic acid in vitro. J. Virol. 78 1591–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J., Noueiry, A., and Ahlquist, P. (2001). Brome mosaic virus protein 1a recruits viral RNA2 to RNA replication through a 5′ proximal RNA2 signal. J. Virol. 75 3207–3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, S.K., Hema, M., Gopinath, K., Santos, J., and Kao, C. (2004). Replicase binding sites on plus- and minus-strand BMV RNAs and identification of their roles in RNA replication in plant cells. J. Virol. 78 13420–13429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta, R., Garcia II, B.H., and Goodman, R.M. (2001). Systemic spread of an RNA insect virus in plants expressing plant viral movement protein genes. Proc. Natl. Acad. Sci. USA 98 4910–4915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson, W.O., Bubrick, P., and Grantham, G.L. (1988). Modifications of the tobacco mosaic virus coat protein gene affecting replication, movement and symptomatology. Phytopathology 78 783–789. [Google Scholar]
- Dinant, S., Janda, M., Kroner, P.A., and Ahlquist, P. (1993). Bromovirus RNA replication and transcription require compatibility between the polymerase and helicase-like viral RNA synthesis proteins. J. Virol. 67 7181–7189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, B., and Itaya, A. (2007). Viroid: A useful model for studying the basic principles of infection and RNA biology. Mol. Plant Microbe Interact. 20 7–20. [DOI] [PubMed] [Google Scholar]
- Ding, B., Itaya, A., and Zhong, X. (2005). Viroid trafficking: A small RNA makes a big move. Curr. Opin. Plant Biol. 8 606–612. [DOI] [PubMed] [Google Scholar]
- Dreher, T.W., Rao, A.L.N., and Hall, T.C. (1989). Replication in vivo of mutant brome mosaic virus RNAs defective in aminoacylation. J. Mol. Biol. 206 425–438. [DOI] [PubMed] [Google Scholar]
- Golecki, B., Schulz, A., and Thompson, G.A. (1999). Translocation of structural P proteins in the phloem. Plant Cell 11 127–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopinath, K., Dragnea, B., and Kao, C. (2005). Interaction between Brome mosaic virus proteins and RNAs: Effects on RNA replication, protein expression, and RNA stability. J. Virol. 79 14222–14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopinath, K., Wellink, J., Porta, C., Taylor, K.M., Lomonossoff, G.P., and Van Kammen, A. (2000). Engineering cowpea mosaic virus RNA-2 into a vector to express heterologous proteins in plants. Virology 267 159–173. [DOI] [PubMed] [Google Scholar]
- Gilbertson, R.L., and Lucas, W.J. (1996). How do viruses traffic on the “vascular highway”? Trends Plant Sci. 1 260–268. [Google Scholar]
- Haseloff, J., Siemering, K.R., Prasher, D.C., and Hodge, S. (1997). Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc. Natl. Acad. Sci. USA 94 2122–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haywood, V., Yu, T.S., Huang, N.C., and Lucas, W.J. (2005). Phloem long-distance trafficking of gibberellic acid-insensitive RNA regulates leaf development. Plant J. 42 49–68. [DOI] [PubMed] [Google Scholar]
- Hema, M., and Kao, C. (2004). Template sequence near the initiation nucleotide can modulate brome mosaic virus RNA accumulation in plant protoplasts. J. Virol. 78 1169–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himber, C., Dunoyer, P., Moissiard, G., Ritzenthaler, C., and Voinnet, O. (2003). Transitivity-dependent and -independent cell-to-cell movement of RNA silencing. EMBO J. 22 4523–4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itaya, A., Woo, Y.M., Masuta, C., Bao, Y., Nelson, R.S., and Ding, B. (1998). Developmental regulation of intercellular protein trafficking through plasmodesmata in tobacco leaf epidermis. Plant Physiol. 118 373–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen, K.A., Wolfs, C.J., Lohuis, H., Goldbach, R.W., and Verduin, B.J. (1998). Characterization of the brome mosaic virus movement protein expressed in E. coli. Virology 242 387–394. [DOI] [PubMed] [Google Scholar]
- Jorgensen, R.A., Atkinson, R.G., Forster, R.L., and Lucas, W.J. (1998). An RNA-based information superhighway in plants. Science 279 1486–1487. [DOI] [PubMed] [Google Scholar]
- Kao, C., and Sivakumaran, K. (2000). Brome mosaic virus, good for an RNA virologist's basic needs. Mol. Plant Pathol. 1 91–97. [DOI] [PubMed] [Google Scholar]
- Kao, C.C., Quadt, R., Hershberger, R.P., and Ahlquist, P. (1992). Brome mosaic virus RNA replication proteins 1a and 2a from a complex in vitro. J. Virol. 66 6322–6329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasschau, K.D., and Carrington, J.C. (1998). A counter-defensive strategy of plant viruses: Suppression of posttranscriptional gene silencing. Cell 95 461–470. [DOI] [PubMed] [Google Scholar]
- Kawakami, S., Watanabe, Y., and Beachy, R.N. (2004). Tobacco mosaic virus infection spreads cell to cell as intact replication complexes. Proc. Natl. Acad. Sci. USA 101 6291–6296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiberstis, P.A., Fries, L., and Hall, T.C. (1981). Viral protein synthesis in barley protoplasts inoculated with native and fractionated brome mosaic virus RNA. Virology 112 804–808. [DOI] [PubMed] [Google Scholar]
- Kühn, C., Franceschi, V.R., Schulz, A., Lemoine, R., and Frommer, W.B. (1997). Macromolecular trafficking indicated by localization and turnover of sucrose transporters in enucleate sieve elements. Science 275 1298–1300. [DOI] [PubMed] [Google Scholar]
- Lane, L.C. (1981). Bromoviruses. In Handbook of Plant Virus Infections and Comparative Diagnosis, E. Kurstak, ed (Amsterdam: Elsevier/North Holland Biomedical Press), pp. 333–375.
- Liu, J.Z., Blancaflor, E.B., and Nelson, R.S. (2005). The Tobacco mosaic virus 126-kilodalton protein, a constituent of the virus replication complex, alone or within the complex aligns with and traffics along microfilaments. Plant Physiol. 138 1853–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llave, C., Kasschau, K.D., and Carrington, J.C. (2000). Virus-encoded suppressor of posttranscriptional gene silencing targets a maintenance step in the silencing pathway. Proc. Natl. Acad. Sci. USA 97 13401–13406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lough, T.J., Lee, R.H., Emerson, S.J., Forster, R.L., and Lucas, W.J. (2006). Functional analysis of the 5′ untranslated region of potexvirus RNA reveals a role in viral replication and cell-to-cell movement. Virology 351 455–465. [DOI] [PubMed] [Google Scholar]
- Lough, T.J., and Lucas, W.J. (2006). Integrative plant biology: Role of phloem long-distance macromolecular trafficking. Annu. Rev. Plant Biol. 57 203–232. [DOI] [PubMed] [Google Scholar]
- Lucas, W.J. (1995). Plasmodesmata-intercellular channels for macromolecular transport in plants. Curr. Opin. Cell Biol. 7 673–680. [DOI] [PubMed] [Google Scholar]
- Lucas, W.J. (2006). Plant viral movement proteins: Agents for cell-to-cell trafficking of viral genomes. Virology 344 169–184. [DOI] [PubMed] [Google Scholar]
- Lucas, W.J., Bouche-Pillon, S., Jackson, D.P., Nguyen, L., Baker, L., Ding, B., and Hake, S. (1995). Selective trafficking of KNOTTED1 homeodomain protein and its messenger RNA through plasmodesmata. Science 270 1980–1983. [DOI] [PubMed] [Google Scholar]
- Lucas, W.J., Ding, B., and Van der Schoot, C. (1993). Plasmodesmata and the supracellular nature of plants. New Phytol. 125 435–476. [DOI] [PubMed] [Google Scholar]
- Lucas, W.J., Yoo, B.C., and Kragler, F. (2001). RNA as a long-distance information macromolecule in plants. Nat. Rev. Mol. Cell Biol. 2 849–857. [DOI] [PubMed] [Google Scholar]
- Miller, W.A., Dreher, T.W., and Hall, T.C. (1985). Synthesis of brome mosaic virus subgenomic RNA in vitro by internal initiation on (-)-sense genomic RNA. Nature 313 68–70. [DOI] [PubMed] [Google Scholar]
- Mise, K., and Ahlquist, P. (1995). Host-specificity restriction by bromovirus cell-to-cell movement protein occurs after initial cell-to-cell spread of infection in nonhost plants. Virology 206 276–286. [DOI] [PubMed] [Google Scholar]
- Nelson, R.S. (2005). Movement of viruses to and through plasmodesmata. In Plasmodesmata, K. Oparka, ed (Oxford, UK: Blackwell Publishing), pp. 188–211.
- Nelson, R.S., Li, G., Hodgson, R.A., Beachy, R.N., and Shintaku, M.H. (1993). Impeded phloem-dependent accumulation of the masked strain of tobacco mosaic virus. Mol. Plant Microbe Interact. 6 45–54. [DOI] [PubMed] [Google Scholar]
- Noueiry, A.O., and Ahlquist, P. (2003). Brome mosaic virus RNA replication: Revealing the role of the host in RNA virus replication. Annu. Rev. Phytopathol. 41 77–98. [DOI] [PubMed] [Google Scholar]
- Okinaka, Y., Mise, K., Suzuki, E., Okuno, T., and Furusawa, I. (2001). The C terminus of brome mosaic virus coat protein controls viral cell-to-cell and long-distance movement. J. Virol. 75 5385–5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oparka, K.J., Roberts, A.G., Boevink, P., Santa Cruz, S., Roberts, I., Pradel, K.S., Imlau, A., Kotlizky, G., Sauer, N., and Epel, B. (1999). Simple, but not branched, plasmodesmata allow the nonspecific trafficking of proteins in developing tobacco leaves. Cell 97 743–754. [DOI] [PubMed] [Google Scholar]
- Qi, Y., Pelissier, T., Itaya, A., Hunt, E., Wassenegger, M., and Ding, B. (2004). Direct role of a viroid RNA motif in mediating directional RNA trafficking across a specific cellular boundary. Plant Cell 16 1741–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao, A.L.N. (1997). Molecular studies on bromovirus capsid protein. III. Analysis of cell-to-cell movement competence of coat protein defective variants of cowpea chlorotic mottle virus. Virology 232 385–395. [DOI] [PubMed] [Google Scholar]
- Rao, A.L.N., Duggal, R., Lahser, F.C., and Hall, T.C. (1994). Analysis of RNA replication in plant viruses. Methods Mol. Genet. 4 216–236. [Google Scholar]
- Roberts, I.M., Boevink, P., Roberts, A.G., Sauer, N., Reichel, C., and Oparka, K.J. (2001). Dynamic changes in the frequency and architecture of plasmodesmata during the sink-source transition in tobacco leaves. Protoplasma 218 31–44. [DOI] [PubMed] [Google Scholar]
- Ruiz-Medrano, R., Xoconostle-Cazares, B., and Lucas, W.J. (2001). The phloem as a conduit for inter-organ communication. Curr. Opin. Plant Biol. 4 202–209. [DOI] [PubMed] [Google Scholar]
- Samanta, M.P., and Liang, S. (2003). Predicting protein functions from redundancies in large-scale protein interaction networks. Proc. Natl. Acad. Sci. USA 100 12579–12583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki, N., Arimoto, M., Nagano, H., Mori, M., Kaido, M., Mise, K., and Okuno, T. (2003). The movement protein gene is involved in the virus-specific requirement of the coat protein in cell-to-cell movement of bromoviruses. Arch. Virol. 148 803–812. [DOI] [PubMed] [Google Scholar]
- Sasaki, N., Kaido, M., Okuno, T., and Mise, K. (2005). Coat protein independent cell-to-cell movement of bromoviruses expressing brome mosaic virus movement protein with an adaptation-related amino acid change in the central region. Arch. Virol. 150 1231–1240. [DOI] [PubMed] [Google Scholar]
- Schmitz, I., and Rao, A.L.N. (1996). Molecular studies on bromovirus capsid protein. I. Characterization of cell-to-cell movement defective RNA3 variants of brome mosaic virus. Virology 226 281–293. [DOI] [PubMed] [Google Scholar]
- Scholthof, H.B. (2005). Plant virus transport: Motions of functional equivalence. Trends Plant Sci. 10 378–382. [DOI] [PubMed] [Google Scholar]
- Schwartz, M., Chen, J., Lee, W.M., Janda, M., and Ahlquist, P. (2004). Alternate, virus-induced membrane rearrangements support positive-strand RNA virus genome replication. Proc. Natl. Acad. Sci. USA 101 11263–11268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel, R.W., Adkins, S., and Kao, C.C. (1997). Sequence-specific recognition of a subgenomic RNA promoter by a viral RNA polymerase. Proc. Natl. Acad. Sci. USA 94 11238–11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivakumaran, K., Choi, S.K., Hema, M., and Kao, C. (2004). Requirements for brome mosaic virus subgenomic RNA synthesis in vivo and replicase-core promoter interactions in vitro. J. Virol. 78 6091–6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjölund, R.D. (1997). The phloem sieve element: A river runs through it. Plant Cell 9 1137–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strebel, K. (2003). Virus-host interactions: Role of HIV proteins Vif, Tat, and Rev. AIDS 17(Suppl 4): S25–S34. [DOI] [PubMed] [Google Scholar]
- Takamatsu, N., Ishiakwa, M., Meshi, T., and Okada, Y. (1987). Expression of bacterial chloramphenicol acetyltransferase gene in tobacco plants infected by TMV-RNA. EMBO J. 6 307–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda, A., Nakamura, W., Sasaki, N., Goto, K., Kaido, M., Okuno, T., and Mise, K. (2005). Natural isolates of Brome mosaic virus with the ability to move from cell to cell independently of coat protein. J. Gen. Virol. 86 1201–1211. [DOI] [PubMed] [Google Scholar]
- Taliansky, M., Roberts, I.M., Kalinina, N., Ryabov, E.V., Raj, S.K., Robinson, D.J., and Oparka, K.J. (2003). An umbraviral protein, involved in long-distance RNA movement, binds viral RNA and forms unique, protective ribonucleoprotein complexes. J. Virol. 77 3031–3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taliansky, M.E., and Robinson, D.J. (2003). Molecular biology of umbraviruses: Phantom warriors. J. Gen. Virol. 84 1951–1960. [DOI] [PubMed] [Google Scholar]
- Tang, H., Xu, Y., and Wong-Staal, F. (1997). Identification and purification of cellular proteins that specifically interact with the RNA constitutive transport elements from retrovirus D. Virology 228 333–339. [DOI] [PubMed] [Google Scholar]
- Voinnet, O., Rivas, S., Mestre, P., and Baulcombe, D.C. (2003). An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33 949–956. [DOI] [PubMed] [Google Scholar]
- Voinnet, O., Vain, P., Angell, S., and Baulcombe, D.C. (1998). Systemic spread of sequence-specific transgene RNA degradation in plants is initiated by localized introduction of ectopic promoterless DNA. Cell 95 177–187. [DOI] [PubMed] [Google Scholar]
- Waigmann, E., Ueki, S., Trutnyeva, K., and Citovsky, V. (2004). The ins and outs of non-destructive cell-to-cell and systemic movement of plant viruses. Crit. Rev. Plant Sci. 23 195–250. [Google Scholar]
- Wassarman, K.M. (2002). Small RNAs in bacteria: Diverse regulators of gene expression in response to environmental changes. Cell 109 141–144. [DOI] [PubMed] [Google Scholar]
- Yang, S.J., Carter, S.A., Cole, A.B., Cheng, N.H., and Nelson, R.S. (2004). A natural variant of a host RNA-dependent RNA polymerase is associated with increased susceptibility to viruses by Nicotiana benthamiana. Proc. Natl. Acad. Sci. USA 101 6297–6302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo, B.C., Kragler, F., Varkonyi-Gasic, E., Haywood, V., Archer-Evans, S., Lee, Y.M., Lough, T.J., and Lucas, W.J. (2004). A systemic small RNA signaling system in plants. Plant Cell 16 1979–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, J., Gopinath, K., Murali, A., Yi, G., Hayward, S.D., Zhu, H., and Kao, C. (2007). RNA-binding proteins that inhibit RNA virus infection. Proc. Natl. Acad. Sci. USA 104 3129–3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Y., Green, L., Woo, Y.-M., Owens, R., and Ding, B. (2001). Cellular basis of potato spindle tuber viroid systemic movement. Virology 279 69–77. [DOI] [PubMed] [Google Scholar]
- Zhu, Y., Qi, Y., Xun, Y., Owens, R., and Ding, B. (2002). Movement of potato spindle tuber viroid reveals regulatory points of phloem mediated RNA traffic. Plant Physiol. 130 138–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.