Abstract
The composition and permeability of the cuticle has a large influence on its ability to protect the plant against various forms of biotic and abiotic stress. WAX INDUCER1 (WIN1) and related transcription factors have recently been shown to trigger wax production, enhance drought tolerance, and modulate cuticular permeability when overexpressed in Arabidopsis thaliana. We found that WIN1 influences the composition of cutin, a polyester that forms the backbone of the cuticle. WIN1 overexpression induces compositional changes and an overall increase in cutin production in vegetative and reproductive organs, while its downregulation has the opposite effect. Changes in cutin composition are preceded by the rapid and coordinated induction of several genes known or likely to be involved in cutin biosynthesis. This transcriptional response is followed after a delay by the induction of genes associated with wax biosynthesis, suggesting that the regulation of cutin and wax production by WIN1 is a two-step process. We demonstrate that at least one of the cutin pathway genes, which encodes long-chain acyl-CoA synthetase LACS2, is likely to be directly targeted by WIN1. Overall, our results suggest that WIN1 modulates cuticle permeability in Arabidopsis by regulating genes encoding cutin pathway enzymes.
INTRODUCTION
During their life cycle, plants are subjected to a variety of stresses, including pathogen and insect attack, exposure to UV light, or water deficit. An important part of their protection against these aggressions comes from chemicals they produce at their surface, in particular the cuticle. Cuticular compounds form a layer that is deposited on the outer epidermal cell wall of aerial organs and is composed of cutin and cutan polymers overlaid with a mixture of hydrophobic waxes. Wax components are deposited both within and on the surface of the cutin polymer. This layered, semirigid hydrophobic structure makes the cuticle an efficient barrier that waterproofs and protects internal tissues (Riederer and Schreiber, 2001; Jeffree, 2006; Riederer, 2006).
Most cuticular and epicuticular components are derived from fatty acid precursors. Cutin is a polyester typically formed by polymerization of C16 and C18 ω-hydroxy fatty acids and partial cross-linking of secondary hydroxyl functions (Kolattukudy, 1996; Stark and Tian, 2006). In Arabidopsis thaliana, as in other Brassica species, cutin is more similar in composition to the related polyester suberin. It is mostly composed of dioic acids that may be connected through glycerol moieties (Bonaventure et al., 2004; Franke et al., 2005; Pollard et al., 2006). Although high levels of dioic acid residues are unusual in plant cutins, the presence of acylated glycerol units may be a common occurrence, as it has been reported in several unrelated species (Graca et al., 2002). Waxes are also mostly composed of fatty acid derivatives, although they also often contain other classes of compounds, for example, terpenoids (Jetter et al., 2006).
The identification of enzymes associated with the cutin and wax pathways has been facilitated by the availability, in Arabidopsis and in other species, of mutants that are defective in wax and/or cutin accumulation. Enzymes catalyzing some of the steps in the wax pathway have been characterized or their function has been proposed based on the phenotype of the corresponding mutants. These include condensing enzymes, reductases, and putative decarbonylases (Aarts et al., 1995; Xu et al., 1997; Millar et al., 1999; Todd et al., 1999; Kurata et al., 2003; Zheng et al., 2005; Kunst et al., 2006).
A picture of the cutin biosynthesis pathway in Arabidopsis is emerging with the identification of a few enzymes involved in cutin production. One of these enzymes is LACERATA (LCR), a cytochrome P450 thought to be involved in the production of ω-hydroxy fatty acid components of the cutin polymer (Wellesen et al., 2001). This enzyme is similar to CYP86A2, which is, like LCR, required for the maintenance of cuticle integrity. Another recently identified enzyme is the long-chain acyl-CoA synthetase LACS2, which is thought to convert ω-hydroxy fatty acids to their CoA thioesters (Schnurr et al., 2004). ADHESION OF CALYX EDGES/HOTHEAD (ACE/HTH) is proposed to be an oxidase catalyzing the formation of dioic acids from ω-hydroxy acyl-CoAs (Krolikowski et al., 2003; Kurdyukov et al., 2006a). Enzymes related to the putative glycerol 3-phosphate acyl transferase GPAT5 may be required for the incorporation of glycerol moieties into cutin, as GPAT5 is required for the production of a cutin-like polymer in the seed coat (Beisson et al., 2007). The role of this enzyme in the production of glycerol-containing monomers is further supported by the presence of monoacylglycerol in the cuticular lipids of plants overexpressing GPAT5 (Li et al., 2006).
The intracellular trafficking and secretory mechanisms that mediate extracellular lipid production are less understood, although the involvement of specialized membrane transporters has recently been demonstrated (Pighin et al., 2004).
Epicuticular wax deposition is tightly regulated both developmentally and environmentally and involves the coordinated induction of several pathway genes (Suh et al., 2005; Shepherd and Griffiths, 2006). It is therefore likely that transcription factors play an important role in controlling this process, as these regulators have been shown to coordinate the expression of gene networks involved in complex metabolic pathways in plants (Broun, 2004). A transcription factor of the ethylene response factor (ERF) family, WAX INDUCER1/SHINE1 (WIN1/SHN1), has recently been shown to induce the production of epidermal waxes when overexpressed in Arabidopsis plants. Analysis of 35S:WIN1 overexpressors suggests that WIN1 influences wax accumulation through the direct or indirect regulation of metabolic pathway genes and that the phenotype of transgenic plants may result from a more permeable cuticular structure (Aharoni et al., 2004; Broun et al., 2004).
In this study, we have investigated the mode of action of WIN1 and the identity of its early and immediate gene targets using reverse genetic and molecular approaches. Our results suggest that WIN1 controls cuticle formation by directly or collaboratively activating the transcription of genes encoding enzymes of the cutin biosynthesis pathway.
RESULTS
WIN1 Overexpression Strongly Affects Cutin Production in Arabidopsis
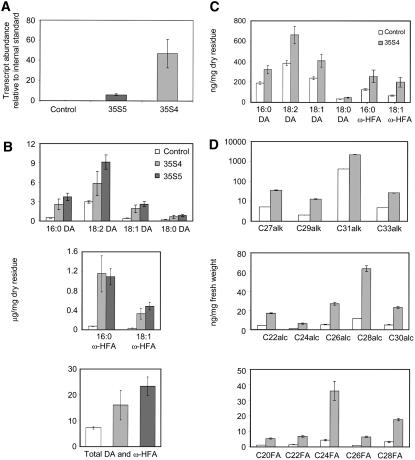
The rapid chlorophyll loss of 35S:WIN1/SHN1 leaves in ethanol indicated that the cuticle of WIN1/SHN1 overexpressors is more permeable to certain classes of chemicals (Aharoni et al., 2004). To determine whether increased cuticle permeability is associated with changes in cutin composition and/or amount, we subjected rosette leaves of 35S:WIN1 plants to extensive delipidation and cuticular material to hydrolysis and derivatization and analyzed residual insoluble lipids by gas chromatography–mass spectrometry (Kurdyukov et al., 2006a). Cutin composition was determined in the leaves of two independent lines (35S4 and 35S5) that overexpressed WIN1 to different degrees (Figure 1A). WIN1 overexpression strongly affected the accumulation of ω-hydroxy fatty acids and 16-hydroxy hexadecanoic and octadecenoic acids (16:0 and 18:1 ω-HFA). It also significantly impacted the most abundant cutin constituents, which, in Arabidopsis, are mono- and polyunsaturated dioic acids (Bonaventure et al., 2004; Franke et al., 2005) (Figure 1B). Specifically, there was a significant increase in the amounts of octadecadiene-1,18-, octadecene-1,18-, octadecane-1,18-, and hexadecane-1,16-dioic acids (18:2 DA, 18:1 DA, 18:0 DA, and 16:0 DA) (Figure 1B). By contrast, the amounts of minor constituents and other residual insoluble lipids were not significantly different in overexpressors compared with control plants (data not shown). The total amount of major constituents dioic and ω-hydroxy fatty acids (representing >50% of leaf cutin components) was up to twofold higher in 35S:WIN1 than in control plants, suggesting a significant increase in cutin accumulation in WIN1 overexpressors (Figure 1B). This result was consistent with the increase in cuticle thickness that we had previously observed in leaf epidermal sections (Broun et al., 2004).
Figure 1.
Cuticular Lipid Composition of WIN1 Overexpressors.
(A) WIN1 expression in 35S4 and 35S5 transgenic lines and in control wild-type Col-0 plants, measured in leaves by quantitative PCR (y axis scale: fold-change relative to the internal standard, UBQ10).
(B) Major cutin monomers in leaves of WIN1 overexpressors: dioic acid monomers (DA) (top panel), ω-hydroxy fatty acids (middle panel), sum of all dioic acids and ω-hydroxy fatty acids (ω-HFA; bottom panel).
(C) Cutin monomer composition in 35S:WIN1 flowers. The levels of each of the compounds in (C) and (D) are calculated on a dry weight basis of delipidated material (residue).
(D) Wax composition of 35S:WIN1 flowers. Scale of chart for alkanes is logarithmic. Values represent averages and standard errors from five biological replicates. alc, long-chain alcohols; alk, long-chain alkanes; FA, long-chain fatty acid.
To determine whether WIN1 overexpression has the same effect in different organs and if this effect is dependent on the level at which the gene is normally expressed, we also analyzed cutin composition in flowers, where the background level of WIN1 expression is higher than in leaves (Aharoni et al., 2004; Broun et al., 2004). We found that cutin was affected in similar ways in the flowers of 35S:WIN1 plants, with a strong increase in dioic acid content (Figure 1C). This reflected an even more dramatic change in flower than in leaf cutin, as dioic acids and ω-hydroxy acids represented ∼75% of all flower cutin components (as opposed to 50% in leaves). As in the case of leaves, the alterations in cutin content and composition were accompanied by a large increase in wax load (Figure 1D). These observations suggested that the mechanism of action of WIN1 is similar in leaves and in flowers.
WIN1 Downregulation Affects Cutin Composition and Epicuticular Lipid Production
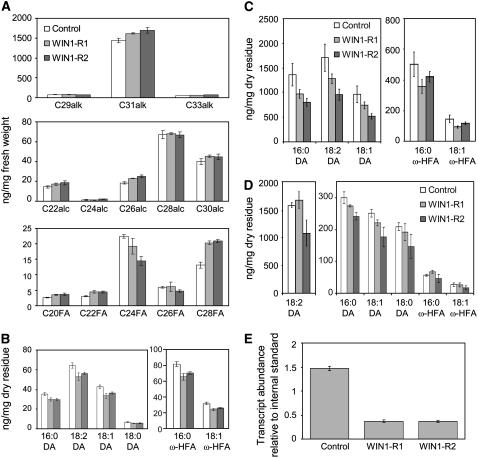
The modified cutin content of WIN1 overexpressors suggested that the transcription factor regulates cutin production in Arabidopsis. To determine whether the phenotype of 35S:WIN1 plants reflected the true biological role of WIN1, we examined the effect of WIN1 downregulation on cuticle formation. Searches of public mutant databases did not identify transgenic lines containing an insertion in WIN1. We therefore produced transgenic plants (WIN1-R), in which WIN1 was specifically downregulated by RNA interference (RNAi). As the gene is normally strongly expressed in floral organs (Aharoni et al., 2004; Broun et al., 2004), we first verified that WIN1 was silenced in the flowers of these plants and selected for our analysis two lines in which expression was most reduced (lines WIN1-R1 and WIN1-R2, which showed a reduction in WIN1 transcript levels to approximately one-third of those in the wild type) (Figure 2E).
Figure 2.
Effect of WIN1 Silencing on Flower Cuticular Lipids.
(A) Wax production on flowers of plants in which WIN1 is silenced.
(B) to (D) Effect of WIN1 silencing on cutin composition in whole flowers (B), petals (C), and leaves (D). The levels of each of the compounds are calculated on a dry weight basis of delipidated material (residue); values represent averages and standard errors from five biological replicates. alk, alkane; alc, alcohol; FA, fatty acid; DA, dioic acid; HFA, hydroxy fatty acid.
(E) WIN1 expression in flowers of silenced lines; the values represent ratios of gene expression relative to UBQ10, which was used as an internal standard.
To determine the effect of WIN1 silencing on extracellular lipid production, we first measured wax accumulation on floral organs of the RNAi lines. Downregulation of WIN1 expression caused a small increase in the production of major wax component hentriacontane (C31 alk, Figure 2A) and had a significant impact on the accumulation of some of the minor constituents. In particular, we observed that C24 fatty acid content decreased, while levels of C28 fatty acid increased (Figure 2A). As a second step, we asked whether changes in epicuticular lipid production were associated, as in the overexpressing lines, with changes in cutin composition. To answer this question, we first determined the composition of residual insoluble lipids in whole flowers and found that WIN1 silencing slightly but consistently reduced dioic acid and ω-hydroxy fatty acid content (Figure 2B). Since WIN1 is not expressed at the same level in all floral organs (Aharoni et al., 2004), the larger impact of silencing on cutin production could have been missed through a dilution effect. WIN1 downregulation could also have been compensated for by the activity of redundant transcription factor SHN3, as SHN3 is also expressed in flower organs (Aharoni et al., 2004). We therefore repeated this analysis using only petals, as WIN1 expression is highest in this part of the flower (Aharoni et al., 2004). We found that WIN1 silencing caused more significant changes in petal cutin composition, which were consistent with the whole-flower phenotype and opposite to modifications caused by overexpression. In particular, the dioic acid content in cutin was strongly reduced, and with it the total cutin content (Figure 2C).
In complement to this analysis, we also measured cutin production in leaves of the RNAi lines. We found that downregulation of WIN1 also caused a small decrease in cutin production in these organs, although the effect was not as pronounced and not as consistent as in petals (Figure 2D).
In summary, the effects of WIN1 silencing on extracellular lipid production indicated that the overexpression phenotype does reflect the true function of WIN1 and suggested that the transcription factor plays a role in the regulation of both wax and cutin production.
WIN1 Overexpression Has No Major Effect on Cell Wall Metabolism but Affects Leaf Membrane Fatty Acid Composition
The dramatic changes in the accumulation of extracellular lipids could have been caused in part by structural and/or compositional changes in the cell wall of WIN1 overexpressors. To consider this possibility, we analyzed the cell wall monosaccharide composition of 35S:WIN1 roots and shoots and used Fourier transform infrared (FTIR) microspectroscopy to detect any large structural changes in the cell wall in seedlings. Our analysis did not reveal significant compositional changes in matrix cell wall polysaccharides (see Supplemental Figure 1 online). Furthermore, the cell walls of 35S:WIN1 seedlings were most similar to those of wild-type controls when comparing them through an FTIR microspectroscopy-based cluster analysis with a range of known cell wall mutants (see Supplemental Figure 2 online). These results suggested that WIN1 overexpression has no effect on cell wall composition or structure or that these effects are limited spatially and/or in scope.
We also asked whether high levels of cutin and wax production could have, directly or indirectly, affected fatty acid precursor pools and, ultimately, membrane fatty acid production in vegetative organs. To this end, we first measured the accumulation of acyl-CoAs, which are intermediates in the biosynthesis of extracellular lipids and non-extra-plastidial membrane fatty acids and membrane fatty acid content in the leaves of 35S:WIN1 plants. Overexpression had no major effect on the accumulation of different acyl-CoAs in whole leaves, although we could detect a small but significant decrease in the levels of 16:0-CoA and 18:0-CoA (see Supplemental Figure 3A online). By contrast, we observed a strong effect of WIN1 overexpression on membrane fatty acid composition (see Supplemental Figure 3B online). In particular, the levels of 16:0 and 18-carbon fatty acids (especially 18:3) were reduced by up to 25%. This effect, taken together with changes in acyl CoA profiles, could suggest that competition for precursors shared between wax, cutin, and membrane fatty acid biosyntheses is dramatically increased in WIN1 overexpressors. To affect membrane fatty acid production, the increase in epidermal lipid metabolism would, however, have to affect precursor availability throughout the leaf. Alternatively, this effect may be indirect. For example, alterations in cuticular structure could result in the oxidation of 18:3, leading to a decrease in its accumulation.
Glucocorticoid-Inducible Expression of WIN1 in Transgenic Plants
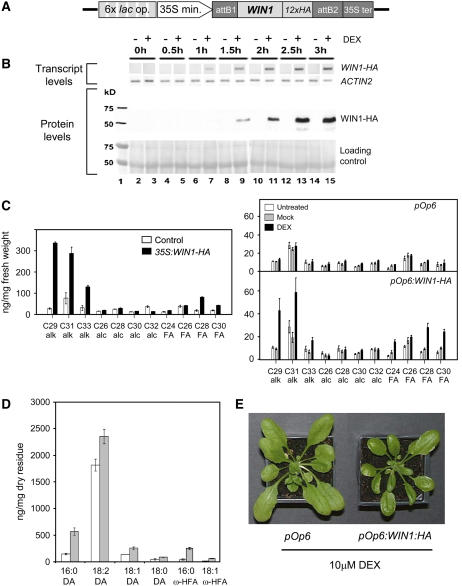
As described above, changes in WIN1 expression levels affected many aspects of extracellular lipid production in the epidermis. To separate direct from indirect effects and to determine the molecular and metabolic sequence of events immediately downstream of WIN1, we generated transgenic plants in which the gene is chemically inducible. Dexamethasone (DEX) treatments of lines overexpressing WIN1 fused at its C terminus with the rat glucocorticoid receptor domain (GR) did not induce glossiness or an increase in the production of wax, suggesting that the fusion protein was not active (data not shown). As an alternative approach, we used a two-component system that allows the induction of the gene of interest by DEX (Craft et al., 2005). In the corresponding transgenic lines (termed LhGR), the bacterial activator lacI is constitutively expressed as a fusion protein to the GAL4 activation domain and the GR domain. The transgene of interest is introduced on a separate T-DNA, under the control of tandem copies of the lac operator (pOp6) to which lacI can bind (Figure 3A). Treatment of these lines with DEX triggers binding of lacI-GAL4AD-GR to pOp6 and induction of the transgene. To measure WIN1 protein levels upon DEX induction, an LhGR line was transformed with a cassette encoding a C-terminal fusion of WIN1 to the HA epitope under the control of pOp6 (pOp6:WIN1-HA) (Figure 3A). Control lines were also generated that constitutively overexpressed WIN1-HA under the control of the 35S promoter.
Figure 3.
Inducible Control of WIN1 Expression.
(A) Schematic representation of the inducible WIN1-HA construct. 6x lac op., six tandem copies of the lac operator; 35S min., 35S minimum promoter; 12x HA, 12 tandem copies of a gene fragment encoding the HA epitope; 35S ter, 35S terminator.
(B) Induction of the transgene and protein in pOp6:WIN-HA-inducible overexpressors; transcripts and protein levels measured over time are presented in comparison with internal standards: ACT2 transcripts or ribulose-1,5-bisphosphate carboxylase/oxygenase small subunit (protein).
(C) Wax production on leaves of 35S:WIN1-HA and DEX-induced pOp6:WIN1-HA plants. Leaves were sampled from plants that were spayed continuously for 2 weeks.
(D) Composition of cutin components dioic acids (DA) and ω-hydroxy fatty acids (ω-HFA) upon DEX induction in leaves of pOp6:WIN1-HA plants. In this separate experiment, newly expanded leaves were harvested after spraying. “Residue” refers to tissue that was extensively delipidated before monomer analysis. In (C) and (D), values represent averages and standard errors from five biological replicates.
(E) pOp6:WIN1-HA (right) and empty vector control plants (left) after treatment with DEX. Glossiness was visible 60 h after treatment (wax production was not measured in these plants).
Applications of DEX to pOp6:WIN1-HA lines resulted in the induction of WIN1-HA expression, whereas we could not detect WIN1-HA transcripts in untreated or mock-treated leaves (Figure 3B). WIN1-HA expression was detectable by RT-PCR as early as 1 h after DEX treatment, and the protein was detectable by protein gel blot analysis 30 min later (Figure 3B). To test whether WIN1-HA induction by DEX was sufficient to trigger significant metabolic changes, we measured wax production on the leaves of inducible overexpressors that had been regularly treated with DEX from the seedling stage. DEX-treated WIN1-HA lines showed a marked increase in the production of several wax components, in particular C29, C31, and C33 alkanes, as in the case of 35S:WIN1-HA and 35S:WIN1 (Figure 3C). Increases in free fatty acid production, which we had only previously observed in 35S:WIN1 stems but had been reported for shn leaves, were also visible (Aharoni et al., 2004; Broun et al., 2004). As little as one DEX application could induce wax production, as glossiness was detectable as early as 60 h after that treatment (Figure 3E). By contrast, wax production was not affected in DEX-treated or in untreated controls.
We also checked whether DEX induction of WIN1-HA expression resulted in changes in cutin composition. To this end, pOp6:WIN1-HA plants were sprayed once at the 9th leaf stage, and cutin composition was measured in the 10th leaf 2 weeks after treatment. DEX applications significantly affected the monomer composition and the amount of cutin produced in pOp6:WIN1-HA plants. Similarly to 35S:WIN1 plants, inducible overexpressors incorporated more dioic and ω-hydroxy fatty acid monomers into cutin than control plants, and the total monomer content suggested that more cutin was being produced (Figure 3D). The results of this experiment indicated that inducing WIN1 in a developing leaf causes changes in its cuticular makeup at maturity.
In summary, these experiments established that WIN1-HA had a similar activity to WIN1 and that pOp6:WIN1-HA plants can be used to study the near-term effects of WIN1 induction on metabolism and gene expression.
WIN1-HA Induction Causes a Rapid Increase in the Expression of Genes with a Known or Likely Involvement in Cutin Biosynthesis
To obtain a comprehensive view of the regulatory networks that are affected by WIN1 activity, we investigated short-term changes in gene expression triggered by the upregulation of WIN1-HA in pOp6:WIN1-HA plants upon DEX treatment using oligonucleotide arrays representing ∼22,000 Arabidopsis genes (Affymetrix ATH1). Our strategy was to profile gene expression soon after the WIN1-HA protein becomes detectable while allowing enough time for the expression of downstream genes to build up. We focused our analysis on leaves, where the gene is expressed at early stages in development (Aharoni et al., 2004; Broun et al., 2004). Since levels of endogenous WIN1 expression are lower in leaves than in flowers, performing the assays on leaves should facilitate the detection of gene expression changes that result from WIN1-HA activation. Furthermore, our experiments showed that WIN1 overexpression affects cuticle production in these organs and that WIN1 downregulation impacts on leaf cutin composition (Figures 1 and 2).
Since both WIN1-HA expression and protein accumulation could be detected by RT-PCR and protein gel blot analysis, respectively, as early as 1.5 h after induction, we performed our microarray experiment using tissue from pOp6:WIN-HA plants harvested 3 h after DEX treatment. Triplicate leaf samples were obtained from DEX-treated pOp6:WIN1-HA and pOp6 empty-vector control plants and used to generate materials for microarray hybridizations.
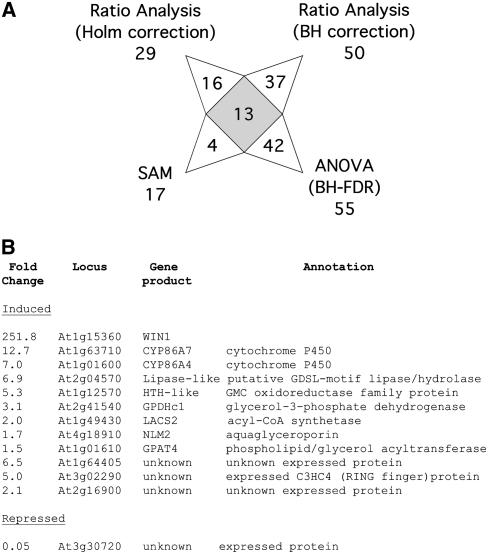
To analyze the microarray data and identify genes showing significant differences in expression, several algorithms and methods were used, and a short list of genes that were predicted by all contrasting methods was generated (Millenaar et al., 2006). Methods included the use of the MAS5 algorithm followed by significance analysis of microarrays (SAM) analysis and various data processing and statistical analysis pipelines implemented in the Resolver system, in particular, ratio analysis and analysis of variance (ANOVA) analysis (Figure 4A; see Methods for details). A comparison of the gene lists obtained with the different methods is presented in Supplemental Table 1 online. We identified, at the intersection of all lists, a set of 12 genes (not including WIN1 itself), with 11 of which significantly induced by WIN1 and one that was significantly repressed (Figure 4B).
Figure 4.
Changes in Gene Expression in pOp6:WIN-HA Plants upon DEX Induction.
(A) Diagram of the output of different microarray analyses. The number of genes (including WIN1) identified in each analysis and at the intersection of all the results are presented. Genes that were identified by several, but not all, of the analyses are not signaled out in the diagram, but the complete results on which this diagram is based are provided in Supplemental Table 1 online. BH, Benjamini-Hochberg correction for multiple testing; BH-FDR, Benjamini-Hochberg and false discovery rate correction; SAM, significance analysis of microarrays.
(B) Consensus list of genes that were identified as differentially expressed upon WIN1-HA induction by all the methods used for microarray data analysis.
Sequence analysis showed that eight of the 11 induced genes had similarity with or corresponded to genes of known function and that seven of those were predicted to encode enzymes with a known or possible involvement in lipid biosynthesis (Figure 4B). In particular, five of those genes were identified with a known or possible involvement in cutin biosynthesis. One of them was LACS2 (Schnurr et al., 2004). Two other genes encoded cytochrome P450 enzymes CYP86A4 and CYP86A7, which are known to have fatty acid ω-hydroxylase activity and are highly similar to Arabidopsis CYP86A2 and LCR (CYP86A8) (Wellesen et al., 2001; Xiao et al., 2004; Duan and Schuler, 2005). Another induced gene, At1g12570, encoded a protein containing a glucose-methanol-choline oxidoreductase domain with strong similarity to ACE/HTH (see Supplemental Figures 4 and 5 online) (Kurdyukov et al., 2006a). We also identified a gene encoding GPAT4, which is related to glycerol 3-phosphate acyl transferase GPAT5 (see Supplemental Figures 4 and 6 online) (Beisson et al., 2007). Interestingly, the induction of CYP86A4, CYP86A7, LACS2, and HTH-like was consistent with the phenotype of inducible and constitutive overexpressors: the plants produce higher amounts of dioic and ω-hydroxy fatty acid cutin monomers, and the production of these compounds from fatty acid substrates could likely occur in a series of reactions catalyzed by these enzymes (see Discussion).
Two more induced genes were also expected to play a role in lipid biosynthesis. GPDHc1 (At2g41540) encodes a cytosolic glycerol 3-phosphate dehydrogenase that plays an important role in modulating NADH/NAD+ levels in Arabidopsis under stress conditions (Shen et al., 2006). This enzyme converts dihydroxyacetone phosphate from the glycolytic pathway to glycerol-3-phosphate, which is a possible substrate for GPAT. The second gene, At2g04570, was predicted to encode a putative lipase containing a GDSL domain of unknown function. This protein bears similarity with a group of putative extracellular lipases (EXL1-3) that are present in the pollen coat (Mayfield et al., 2001).
The last known gene induced by WIN1, At4g18910, encoded NLM2, an enzyme that has glycerol permease activity when expressed in yeast cells (Weig and Jakob, 2000). The coinduction of this gene with GPAT4 and GPDHc1 was intriguing given its putative role in glycerol transport.
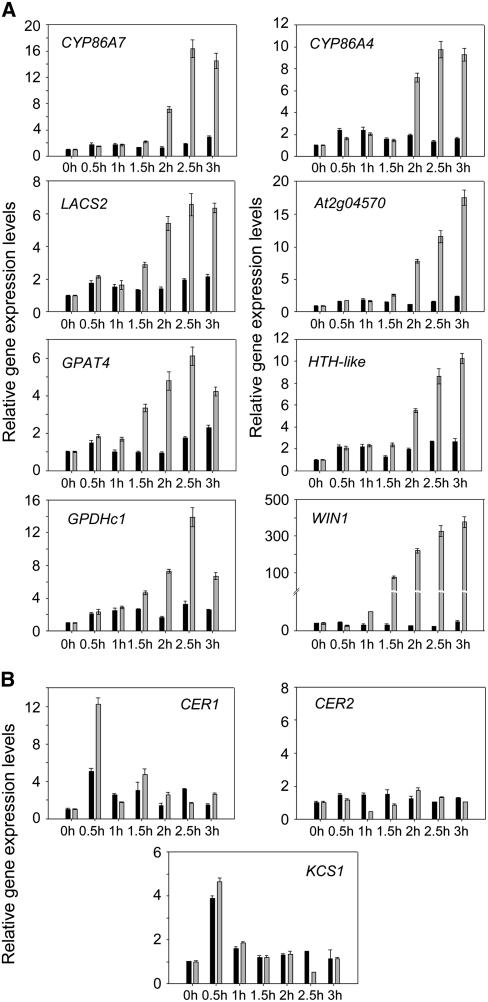
To verify the results of our microarray analysis, we measured the expression of the seven genes relevant to lipid biosynthesis using real-time RT-PCR, before and after DEX-induction of WIN1-HA. The short-term increase in the expression levels of these genes was monitored in leaves of pOp6:WIN1-HA lines at regular intervals following DEX treatment. We found that all the genes were induced within 2 h of DEX applications and that their expression initially increased in a coordinated fashion, in most cases within 30 min of the surge in WIN1-HA expression (Figure 5A).
Figure 5.
Effect of WIN1-HA Induction on Lipid Biosynthetic Pathway Gene Expression.
Short-term response to WIN1 induction of genes associated with cutin (A) and wax biosynthesis (B) after DEX application to pOp6:WIN1-HA plants, measured by real-time RT-PCR. Expression levels (fold change) are quantified relative to the expression level at 0 h. Gray bars, DEX treatment; black bars; mock treatment. Values represent averages and standard errors (n = 3).
These results indicated that WIN1 acts as a transcriptional activator that may directly control genes of the cutin biosynthesis pathway.
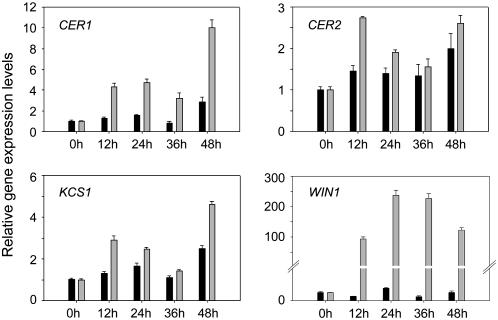
Late Induction of Wax Biosynthesis Genes CER1, CER2, and KCS1 by WIN1-HA
One of the most dramatic effects of overexpressing WIN1 is the large increase it causes in the production of cuticular wax in transgenic plants. Previous experiments had also shown that genes encoding enzymes of the wax biosynthesis pathway, in particular KCS1, CER1, and CER2, were significantly upregulated in WIN1-overexpressing plants (Broun et al., 2004). These genes, however, were not found to be upregulated within the time frame of the microarray experiment described above. To estimate the length of time required for the induction by WIN1 of wax biosynthetic genes, we measured the transcript levels of KCS1, CER1, and CER2 by real-rime RT-PCR in pOp6:WIN1-HA plants over an extended period of time after DEX treatment. We found no consistent increase in their expression within the first 3 h after WIN1 induction, thus confirming the microarray results (Figure 5B). However, when their expression was monitored over a 48-h time frame at 12-h intervals, KCS1, CER1, and CER2 induction could be detected at 12 h after induction (Figure 6). Thus, these genes are induced between 3 and 12 h after the DEX treatment of pOp6:WIN1-HA plants. Based on the late induction of CER1, KCS1, and CER2, in contrast with the early induction of cutin pathway genes, it is possible that the regulation of wax biosynthesis requires additional transcription factors acting downstream of WIN1. However, we cannot exclude, based on these data, the possibility that these genes and cutin pathway genes are similarly regulated by WIN1, with the wax biosynthesis genes being more slowly induced.
Figure 6.
Induction of Wax Biosynthesis Pathway Genes by WIN1-HA Expression of Wax Biosynthetic Genes after DEX Induction of pOp6:WIN1-HA Plants Measured by Real-Time RT-PCR.
Time scale represents hours after DEX application. Expression levels (fold change) are quantified relative to the expression level at 0 h. Gray bars, DEX treatment; black bars, mock treatment. Values represent averages and standard errors (n = 3).
The Expression Patterns of WIN1 and WIN1-Induced Cutin Biosynthesis Genes Overlap
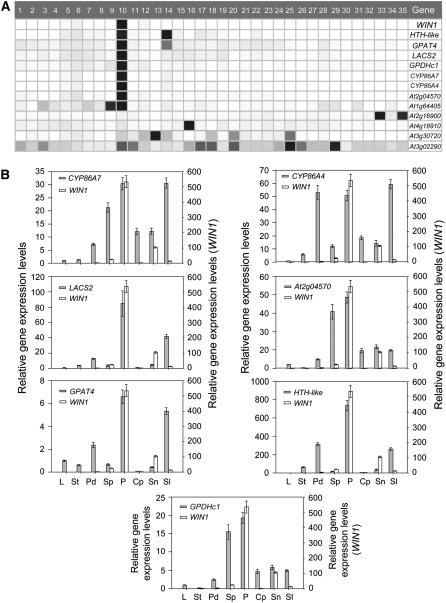
It is expected that genes that are targets of WIN1 should be expressed, at least partly, in the same tissues as the transcription factor. We therefore asked whether the expression of genes identified above overlaps with WIN1 expression. We first queried public repositories of microarray information, which contain data on expression of Arabidopsis genes in different tissues. We used Genevestigator to obtain a graphical representation of the expression of WIN1 and the genes in our consensus list of DEX-induced transcripts throughout the plant (Zimmermann et al., 2004). Strikingly, we found that the majority of the genes were, like WIN1, most highly expressed in petals but less or not at all in other plant tissues (Figure 7A). Some of the genes, however, had a distinct expression pattern, indicating that WIN1 is not the only transcription factor controlling their expression.
Figure 7.
Expression Pattern of Lipid Biosynthesis Genes Rapidly Induced by WIN1.
(A) Graphical representation of the expression of DEX-induced genes in public microarray experiments (Genevestigator): induced genes associated with cutin biosynthesis are most expressed in petals. 1, Seedlings; 2, cotyledons; 3, hypocotyl; 4, radicle; 5, inflorecsence; 6, flower; 7, carpel; 8, ovary; 9, stigma; 10, petal; 11, sepal; 12, stamen; 13, pollen; 14, pedicel; 15, silique; 16, seed; 17, stem; 18, node; 19, shoot apex; 20, cauline leave; 21, rosette; 22, juvenile leaf; 23, adult leaf; 24, petiole; 25, senescent leaf; 26, xylem; 27, cork; 28, roots; 29, lateral root; 30, root tip; 31, root elongation zone; 32, root hair zone; 33, root endodermis; 34, root endodermis + cortex; 35, lateral root cap. Darker shades of gray represent higher levels of gene expression.
(B) RT-PCR analysis of DEX-induced genes in different tissues relative to fully expanded leaves. Values represent the ratio of expression levels in a given organ to mature leaf levels. Values represent averages and standard errors (n = 3). L, rosette leaves; St, stems; Pd, flower pedicels; Sp, sepals; P, petals; Cp, carpels; Sn, stamens; Sl, siliques.
We then used real-time RT-PCR on seven genes found to be upregulated in the microarray experiment to verify the Genevestigator data and to determine their spatial pattern of expression. In this experiment, expression in the different tissues was normalized to message levels measured in fully expanded rosette leaves. The results indicated that these genes were all highly expressed in petals, but we also found that their expression was often more widespread and in some cases also quite high in other tissues. However, we could not find cases in which expression of any of these putative WIN1 targets is undetectable in organs where WIN1 is expressed (Figure 7B).
An inspection of recent microarray data obtained from stem epidermal tissue (Suh et al., 2005) indicated that all but one of the putative WIN1 target genes (At3g02290) are preferentially expressed in the epidermis, an observation that suggested that transcriptional changes caused by WIN1 induction preferentially occur in the epidermal layer where cutin biosynthesis also takes place.
In summary, the results of the expression analyses suggested that the putative cutin biosynthesis genes, which are upregulated rapidly after WIN1 induction, could be direct targets of the transcription factor.
The Promoters of WIN1-Induced Genes Do Not Contain GCC Boxes
To determine which among the genes induced upon DEX treatment of pOp6:WIN1-HA plants are most likely to be direct targets of WIN1, we screened sequences upstream of their coding regions for the presence of GCC boxes, which have been described in promoters bound by the ERF transcription factor (Nakano et al., 2006). For this analysis, promoter sequences up to 2 kb in length were examined using two online tools, PLACE and NSITE-PL, that are designed to recognize plant transcription factor–specific motifs (Higo et al., 1999; Shahmuradov et al., 2003). Surprisingly, none of the promoters contained identifiable GCC boxes, although some, including the LACS2 promoter, contained conserved motifs found in the promoter of some ethylene-induced genes (see Supplemental Table 2 online) (Rawat et al., 2005). By comparing the frequency of motifs detected by PLACE in these promoters and all upstream Arabidopsis gene sequences stored at The Arabidopsis Information Resource, we identified several AT-rich motifs that were more highly represented in the promoters of WIN1-induced genes but not known to be targeted by ERF transcription factors (see Supplemental Table 2 online). Based on these observations, we concluded that either WIN1 does not bind to most of the induced genes directly, or the sequences to which it binds are different from the cognate GCC box motif.
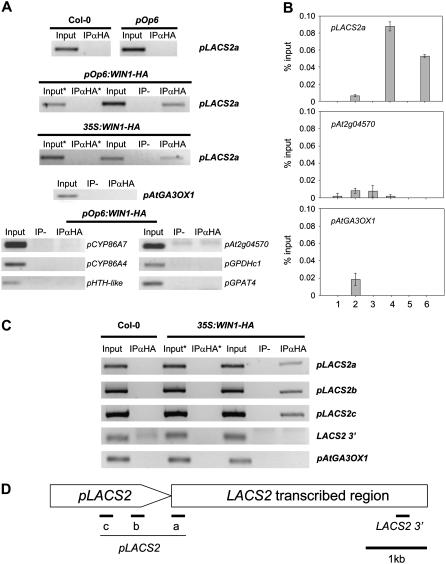
WIN1 Binds Genomic Fragments Containing the LACS2 Promoter in Planta
To determine if any of the putative WIN1 targets are bound by the transcription factor in planta, we performed chromatin immunoprecipitation (ChIP) experiments using specific anti-HA antibodies (Orlando and Paro, 1993).
In one experiment, we used leaves of control plants, a 35S:WIN1-HA line, and a pOp6:WIN1-HA line 6 h after DEX treatment. Negative controls were generated by omitting anti-HA antibodies or the cross-linking step. For the detection by PCR of DEX-induced gene promoter fragments, we used primers specific to seven genes on the ChIP DNA templates.
In most cases, we could not amplify promoter fragments from the cutin genes after ChIP. In some cases, the amplification of a promoter fragment occurred regardless of cross-linking or the presence of anti-HA antibodies (Figure 8A). In the case of LACS2, however, immunoprecipitated DNA was clearly enriched in promoter fragments, and this enrichment was conditional on the cross-linking step (Figures 8A and 8B).
Figure 8.
In Vivo Binding of WIN1-HA to LACS2.
(A) PCR amplification of promoter fragments (p) from LACS2 and other genes that were immunoprecipitated with anti-HA antibodies in leaf extracts of 35S:WIN1-HA and/or DEX-induced pOp6:WIN1-HA plants. The genetic background of the plants that were used for the experiments are indicated above each panel. The AtGA3OX1 gene, which is not induced upon DEX application to pOp6:WIN1-HA plants, was used as a negative control. Input, input DNA before immunoprecipitation; IPαHA, immunoprecipitated fragments with anti-HA antibodies (IP+Ab); IP-, IP performed without anti-HA antibodies (IP-Ab); asterisk, cross-linking step was omitted.
(B) Quantification of PCR amplification of the different templates by real-time PCR. 1, Col-0, IP+Ab; 2, pOp6, IP+Ab; 3, pOp6:WIN1-HA, IP-Ab; 4, pOp6:WIN1-HA, IP+Ab; 5, 35S:WIN1-HA, IP- Ab; 6, 35S:WIN1-HA, IP+Ab. Values represent averages and standard errors (n = 5).
(C) Immunoprecipitation with anti-HA antibodies of four LACS2 gene fragments from flower chromatin of Col-0 control plants (left) and 35S:WIN-HA plants (right). LACS2 promoter fragments denoted as pLACS2a, pLACS2b, and pLACS2c and the downstream gene fragment LACS2 3′ were detected by PCR as in (A), and the At GA3OX1 gene was used as a negative control.
(D) Positions in the LACS2 gene of the promoter and gene fragments that were amplified in (A) to (C). Sequence lengths are not drawn exactly to scale.
To test whether we could also detect WIN1-HA binding to LACS2 in flowers, where WIN1 is highly expressed, we conducted a second ChIP experiment using chromatin from flowers of 35S:WIN1-HA plants. As in the previous experiment, we could clearly detect LACS2 promoter fragments after immunoprecipitation, and this result was dependent on cross-linking and on the presence of anti-HA antibodies (Figure 8C).
To further validate this result and determine whether WIN1-HA binds, as our earlier results would suggest, to upstream or promoter regions in LACS2, we used additional primer pairs designed to amplify sequences in the promoter or the 3′ region of the gene (Figure 8D). Using ChIP-derived templates obtained from flowers, we were able to selectively amplify two additional promoter fragments but not the downstream gene fragment (Figure 8C), indicating that WIN1-HA is more likely to bind to the promoter or upstream region of LACS2 than to downstream sequences.
Taken together, the immunoprecipitation of LACS2-WIN-HA complexes from both leaves and flowers, the early induction of LACS2 after WIN1 activation by DEX application, and its normal coexpression with WIN1 in the plant strongly suggested that LACS2 is a direct target of WIN1.
DISCUSSION
We reported the results of our investigation of the mechanism of action of transcription factor WIN1. We show that WIN1 overexpression and downregulation affect the amount and composition of cutin produced in transgenic plants. Using large-scale gene expression profiling, we also show that a group of genes known or likely to be involved in cutin biosynthesis are upregulated shortly after WIN1 is induced and that their domain of expression overlaps with that of WIN1. Finally, we demonstrate that WIN1 binds in planta to DNA fragments containing the promoter of one of these genes, LACS2. Our results indicate that WIN1 directly regulates the expression of one or more genes involved in cutin biosynthesis in Arabidopsis and suggest that it is through its influence on cutin composition that WIN1 modulates cuticle permeability. WIN1 is to our knowledge the first transcription factor identified to have such an immediate influence on a lipid pathway in plants.
WIN1 and the Control of Cuticular Lipid Production
The transcription factor WIN1 was recently found to induce the production of wax when overexpressed in transgenic Arabidopsis plants. We demonstrate here that it is also implicated in the regulation of cutin biosynthesis through its influence on the amount and composition of cutin monomers that are produced. Wax deposition in Arabidopsis is known to be influenced by cutin production not only because the two pathways share common precursors but also because the nature and amount of cutin produced impacts on wax deposition. For example, bodyguard mutants, which incorporate more cutin monomers into cell wall–associated lipids than wild-type plants, also produce more wax (Kurdyukov et al., 2006b). lacs2 mutants, which are thought to be defective in the activation of cutin monomers during the biosynthetic process, also accumulate more wax than wild-type plants (Schnurr et al., 2004). Similarly, transgenic plants that overexpress a fungal cutinase gene and in which the integrity of the cuticle layer is severely compromised accumulate significantly more alkanes in leaves than wild-type controls (Sieber et al., 2000). Wax accumulation may therefore be influenced by the amount of cutin that the plant produces and/or by compositional changes in the cutin polymer. Based on these observations and our findings, WIN1 may impact wax production not only through its effects on the expression of wax pathway genes but also by modulating cutin production and changing the physical properties of the cuticle.
WIN1-Regulated Gene Network Controlling Cutin Biosynthesis in Arabidopsis
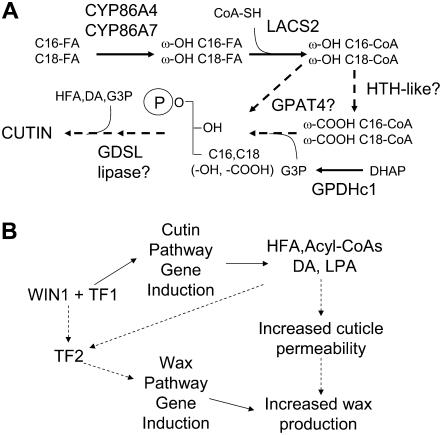
Gene expression profiling of plants in which WIN1 activity was induced by DEX led to the identification of putative target genes downstream of the transcription factor. Twelve of these genes were identified by all the various statistical methods that were used for microarray data analysis. Furthermore, a strong increase in expression, within 30 min to 1 h after WIN1 induction, could be detected by real-time RT-PCR for all of the genes within the subset that we tested. The products of five of these genes (LACS2, CYP86A7, CYP86A4, HTH-like, and GPAT4) are known or likely to be associated with cutin biosynthesis, and our findings suggest that they are part of a small WIN1-responsive regulatory network that operates primarily in flowers. Interestingly, these five genes can be ordered, based on the literature, in a possible pathway sequence leading to the production of cutin monomers (Figure 9A). In this pathway, CYP86A7 and A4 use free fatty acids as a preferred substrate (Wellesen et al., 2001) and add a hydroxyl group at the ω-carbon (Duan and Schuler, 2005). LACS2 activates the resulting hydroxy fatty acid, which becomes a substrate for HTH-like (Schnurr et al., 2004; Kurdyukov et al., 2006a). The dioic acyl-CoA may then be transferred to a glycerol backbone by GPAT4, or this enzyme could use ω-hydroxy acyl-CoAs as substrates to produce ω-hydroxy acyl-lyso phosphatidic acid. These alternative possibilities are supported by the phenotype of Arabidopsis gpat5 mutants, which have a strong reduction in both dioic acid and ω-hydroxy fatty acid constituents of the seed coat (Beisson et al., 2007). Our data further suggest the involvement of GPDHc1, which may be involved in providing glycerol 3-phosphate substrate for GPAT4. The downstream part of the proposed pathway is more hypothetical. Possibly, the putative GDSL lipase is involved in remodeling monoacyl-glycerol or transferring additional fatty acid moieties to the glycerol backbone, scenarios that we are currently investigating using a reverse genetic approach. The NLM2 permease may be involved in the transport of glycerol that is recycled from unused G3P or in the transport of G3P itself, although its affinity for this substrate has not been tested (Weig and Jakob, 2000). Based on the expression pattern of cutin biosynthesis genes, it would be expected that production of ω-hydroxy and dioic acids is most affected by WIN1 overexpression or silencing. We found that it is indeed the case, and these phenotypes come in support of our model.
Figure 9.
Regulation of Cutin and Wax Biosynthesis by WIN1.
(A) Model of cutin biosynthesis pathway regulated by WIN1. Hypothetical steps or steps for which in vivo substrates are unknown are represented by hatched arrows. ω-OH, ω-hydroxy; ω-COOH, ω-carboxy; FA, fatty acid; C16,C18 (-OH, -COOH), C16 or C18 hydroxyacyl or diacyl; HFA, hydroxy fatty acid; DA, dioic acid; G3P, glycerol 3-phosphate; DHAP, dihydroxyacetone phosphate.
(B) Regulation of cutin and wax biosynthesis by WIN1 in Arabidopsis. The hatched arrow between “increased cuticle permeability” and “increased wax production” indicates that additional factors may contribute to stimulate wax production. TF1, transcription factor 1 that interacts with WIN1 to activate immediate target genes; TF2, transcription factor 2 acting downstream of WIN1 and that activates genes of the wax biosynthesis pathway; HFA, hydroxy fatty acid; DA,dioic acid; LPA, lyso-phosphatidic acid.
The finding that WIN1 binds to the promoter region of one of the cutin pathway genes that it induces (LACS2), together with the coordinated and rapid induction of all the genes, strongly suggest that WIN1 is involved in the direct or collaborative regulation of this cutin pathway gene network (see below).
WIN1 Mode of Action: Direct or Indirect Binding?
Our results suggest that WIN1 does not generally bind directly to its gene targets in vivo: in only one of seven cases could we detect clear, cross-linking and antibody-dependent coprecipitation of WIN1-HA with target gene fragments. That WIN1 may not bind directly to most of its presumed targets was surprising. However, another ERF transcription factor, tomato (Solanum lycopersicum) Pti4, was found to only bind directly to some of the tested targets in ChIP experiments (Chakravarthy et al., 2003). It was suggested that Pti4 requires the action of a second transcription factor to induce some of its targets. This second regulator was proposed to act either downstream of Pti4 (a stepwise activation model) or to require interactions with the ERF factor for binding (a cooperative activation model). Similar models could apply to target regulation by WIN1. Our data (except for LACS2, which we show is bound by WIN1) do not allow us to conclusively distinguish between these possible modes of regulation. However, based on the short time frame within which most of the putative WIN1 targets are induced, we favor the interaction with a preexisting transcription factor as the model for WIN1-mediated target regulation (Figure 9B). The requirement for an interacting partner may also explain why we could not detect the binding of WIN1 to LACS2 promoter fragments in vitro in electrophoretic mobility shift assays (data not shown). It may also be an explanation for the absence of clear GCC box motifs in the promoters of WIN1-induced genes or in most Pti4 target promoters. Alternatively, WIN1 and Pti4 may bind to DNA sites that are distinct from those bound by GCC box binding ERFs. As the ERF motif of WIN1 has significantly diverged in sequence from that of the latter factors (Nakano et al., 2006), it is conceivable that its DNA binding properties are distinct.
WIN1 and the Coordination of Cutin and Wax Biosynthesis
The relative timing of cutin and wax pathway gene responses to WIN1 induction suggests that they are part of distinct, yet dependent, regulatory networks, the later upregulation of wax pathway genes implying that they are further downstream than the cutin pathway genes (Figure 9B). An alternative scenario, which seems less likely but that we cannot rule out, is that genes of the wax biosynthesis pathway are also directly or collaboratively regulated by WIN1 but are significantly more slowly induced. In either case, WIN1 may also cause, through its action on the cutin pathway, structural changes in the cuticle that promote wax transit to the surface of the plant (Figure 9B).
The strong expression of WIN1 and the relatively high wax and cutin content in flowers suggest that high levels of extracellular lipid production may be advantageous in these organs. In addition to offering increased protection against dehydration, a thick cuticle may prevent the adhesion of floral organs during flower development. In support of this hypothesis is the susceptibility of flowers to organ fusion in mutants that are defective in cuticle formation and/or wax production (Sieber et al., 2000; Wellesen et al., 2001). Connecting its mechanism of action to a precise biological role will require further characterization of the WIN/SHN clade using double, possibly triple, mutants to eliminate redundant activities.
METHODS
Plant Lines and Growth Conditions
Arabidopsis thaliana plants were grown in the greenhouse at 20 to 25°C, with 16-h daylength, and at light intensities of ∼120 μE·m−2·s−1. Seeds were sterilized and sown on 0.75% agar plates containing Murashige and Skoog (MS) medium with antibiotic selection where appropriate (30 to 50 mg/L hygromycin for plants containing pH2GW7-based constructs). Plants were transferred to 4-cm2 cells containing F2 compost (Levington) when they reached the four-leaf stage. For induction experiments, pOp6:WIN1-HA and pOp6 lines were grown in a climate-controlled cabinet (Sanyo) at 20°C, with 16-h daylength, 60% relative humidity, and at a light intensity of 60 μE·m−2·s−1.
All binary constructs were transformed into Arabidopsis accession Columbia (Col-0) using the floral dip method (Clough and Bent, 1998). The pH2WG7-pOp6 vector containing the WIN-12xHA construct was transformed into the activator line 4c-S5 described by Craft et al. (2005). Transgenic plants resistant to Basta were selected on soil by spraying with KASPAR (Certis). Transgenic plants resistant to kanamycin and hygromycin were selected on half-strength MS medium with 0.8% (w/v) agar containing 25 to 50 μg/mL of the appropriate antibiotics. Plants were grown at 20°C with a 16-h-light/8-h-dark photoperiod under white light (50 to 70 μE·m−2·s−1) from fluorescent bulbs.
Cutin and Wax Analysis
Cuticular wax analysis was performed as described previously (Broun et al., 2004), and the analysis of residual insoluble lipids, including cutin monomers also followed a published protocol (Kurdyukov et al., 2006a). For this analysis, we used leaves 9 to 12 of 6-week-old plants (that just started to bolt) or flowers of 7- to 8-week-old plants. As described above, the plants were either grown in the greenhouse or in a growth cabinet, and measurements were performed at different times of the year. Briefly, 150 to 300 mg of leaf tissue, 40 mg of flower tissue, or 3 mg of petals (∼200 petals) were extensively delipidated, the resulting residues dried, transmethylated, and then silylated in 50 to 100 μL BSTFA:TMCS (99:1). The standards added in the final extraction steps were either triacontane or dotriacontane, to a final concentration of 200 ng/μL injected. Gas chromatography–mass spectrometry analysis was performed using a Trace GC 2000 (ThermoQuest) connected to a GCQ Plus (ThermoQuest) and an AS2000 Autosampler (ThermoQuest), equipped with a Phenomenex Zebron ZB-1ms column (30 m × 0.25 mm; 0.5-μM film thickness) with helium as the carrier gas. Trimethylsilyl derivatives of wax components were separated after splitless injection of a 2-μL sample using the following temperature profile: (1) 150°C for 2 min, (2) increase at rate of 12°C/min up to 250°C, (3) increase at rate of 4°C/min up to 330°C, and (4) hold at 330°C for 2 min, (5) decrease at a rate of 70°C/min to 150°C. Trimethylsilyl derivatives of residual insoluble lipids were separated after splitless injection of 2 μL using the following conditions: (1) initial temperature 90°C, (2) increase at rate of 10°C/min to 300°C, (3) hold at 300°C for 10 min, and (4) decrease temperature at rate of 70°C/min to 90°C.
The amounts of cutin monomers we measured were generally higher than those previously reported in the literature, although published values can vary substantially and are often different from experiment to experiment, as different growth conditions, tissue types, and experiment setups may have been used (Bonaventure et al., 2004; Franke et al., 2005; Kurdyukov et al., 2006a, 2006b). Our results were generally consistent within and across experiments, and the relatively high monomer content we measured compared with published values could have a variety of causes, including differences in growth conditions, plant age, or GC detector response.
Analysis of Membrane Fatty Acids and Acyl-CoAs
Acyl-CoA and fatty acid analyses were performed as previously described (Browse et al., 1986; Larson and Graham, 2000). All CoA species were identified by comparison to true standards, and the identity of leaf fatty acid methyl esters was confirmed by mass spectral analysis and comparison to true fatty acid methylester standards.
Cell Wall Analysis
For monosaccharide analysis, shoot and root tissue was harvested from plants grown on MS medium without sucrose for 28 d under short-day conditions (8 h light, 16 h dark, 20°C). For the analysis of cell wall material matrix polysaccharides, cell wall material was hydrolyzed with trifluoracetic acid, and alditol acetates were obtained and analyzed by GC-MS as described (Albersheim et al., 1967). Uronic acids were quantified using the m-hydroxy-biphenyl assay (Blumenkrantz and Asboe-Hansen, 1973).
FTIR spectra were acquired on hypocotyls of 4-d-old dark-grown seedlings of WIN1 overexpressors, known cell wall mutants, and control plants. For each mutant, 20 spectra were collected from individual hypocotyls of seedlings from four independent cultures (five seedlings from each culture). Normalization of the data and the discriminant variable selection method were performed as described (Mouille et al., 2003). Based on the Mahalanobis distances calculated using the 32 selected wave numbers, a dendrogram was constructed using the Ward clustering algorithm (Mouille et al., 2003).
Preparation of Gene Expression Constructs
For the 35S:WIN1 construct, a WIN1 gene fragment was PCR amplified from leaf cDNA using primers 5′-GAGTTCGTCGACCATCAAGTTCCTACTTTCTCTC-3′ and 5′-TTAGTTTGTATTGAGAAGCTCCTCTATC-3′. The PCR product was cut with SalI and cloned into Gateway entry vector pENTR-1a (Invitrogen), which had been cut with NotI, blunt-ended, and then cut with SalI to produce recombinant vector pENTR-WIN1. The WIN1 fragment was subsequently transferred to the destination vector pH2GW7 (Karimi et al., 2002) using Gateway LR clonase enzyme mix (Invitrogen) to produce recombinant vector 35S:WIN1.
For the WIN1 RNAi construct (WIN1-R), a 141-bp specific fragment from WIN1 was PCR amplified using primers 5′-GAGTTCGTCGACCTTATTTGCTTATATATATGTACC-3′ and 5′-CTTAGTTACAAACACCAATACTTTAT-3′. The PCR product was cloned into pENTR-1a and subsequently transferred into the RNAi destination vector pH7GWIWG2(II) (Karimi et al., 2002), producing the WIN1-R vector using the same strategy as above.
For the 35S:WIN1-HA construct, a WIN1 cDNA fragment was PCR amplified using primers 5′-GAGTTCCTCGAGCATCAAGTTCCTACTTTCTCTC-3′ and 5′-CACCTTGGGCCCGTTTGTATTGAGAAGCTCCTC-3′. The PCR product was cut with XhoI and ApaI, purified, and ligated into a pGREEN plasmid derivative, pGTI0242, which contains a double 35S enhancer, a ligand binding domain of a rat GR, and a 12xHA epitope tag (T. Ito, unpublished data). Subsequently, the GR domain was excised using SmaI to produce the recombinant plasmid, pGTi0242ΔGR-WIN1.
To generate the DEX-inducible HA-tagged WIN1 (WIN1-HA), we amplified a WIN1-HA fragment using primers 5′-GCGCTCGAGAATTATCTAGATTAACTAGT-3′ and 5′-GCGGTCGACAATTACCACCATGGTACAGAC-3′. The PCR product was cut with XhoI and SalI, ligated into the Gateway entry vector, pENTR 1A (Invitrogen), and then recombined into the pH2WG7-pOp6 vector using the Gateway LR Clonase enzyme mix (Invitrogen). pH2WG7-pOp6 is a Gateway-compatible binary vector that is derived from pH2WG7 (Karimi et al., 2002) and in which the 35S promoter was replaced with six copies of a lac operator (pOp6) derived from the pH-TOP plasmid (Craft et al., 2005).
Induction of WIN1-HA Activity by DEX
Long-term induction was achieved by spraying plants every 3 d from cotyledon stage with a solution containing 10 μM DEX (Sigma-Aldrich) and 0.015% (v/v) Silwet. As a control, plants were sprayed with a mock solution containing 0.015% (v/v) Silwet and 0.033% (v/v) ethanol. Temporary induction was performed by spraying 3-week-old plants once with DEX.
Analysis of Gene Expression
For the analysis of the expression pattern of WIN1 and its putative targets, total RNA was extracted from various tissues from soil-grown plants using the RNeasy plant kit (Qiagen). Two micrograms of total RNA were treated with DNase I (Ambion) prior to reverse transcription by SuperScript II reverse transcriptase (Invitrogen) in a 20-μL reaction volume.
Quantitative real-time PCR assays were performed in triplicate on an ABI Prism 7000 sequence detection system (Applied Biosystems) using ACTIN2 expression as an endogenous control, except when WIN1 expression was measured in silenced and overexpressing lines. In this case, the UBQ10 gene was used as an internal standard. Fifteen to one hundred nanograms of cDNA were used as template in a 25-μL reaction containing 1× SYBR-green PCR master mix (Applied Biosystems) and 1 μM of each primer. The PCR thermal cycling parameters were 50°C for 2 min, followed by 95°C for 10 min, 40 cycles of 95°C for 15 s, and 60°C for 1 min. All calculations and statistical analyses were performed using the 2−ΔΔCt method from SDS RQ Manager 1.1 software (ABI Prism 7000 SDS software package; Applied Biosystems). The relative quantification RQmin/RQmax confidence level was set at 95%. Error bars display the calculated maximum (RQmax) and minimum expression levels (RQmin) that represent the se of the mean expression level (RQ). Amplification efficiencies were determined to be 99 to 100% using a dilution series of control cDNA ranging from 1 to 1000 ng per reaction. To determine the specificities of the primers, RT-PCR products were analyzed by gel electrophoresis to ensure that a single product of the expected size was detected in each reaction.
Primer sequences for quantitative PCR experiments are provided as part of Supplemental Table 3 online.
Protein Gel Blot Analysis of WIN1-HA Accumulation
Total protein was extracted from 300 mg of 3-week-old plant tissue. Protein extraction, separation, and blotting procedures were as described (Penfield et al., 2005). A rat anti-HA monoclonal antibody 3F10 (Roche) was applied in a dilution of 1:5000. The immunoreactive polypeptides were visualized with a 1:3000 dilution of a horseradish peroxidase–conjugated goat anti-rat polyclonal antibody (Santa Cruz Biotechnology) using enhanced chemiluminescence detection (Amersham Biosciences).
Microarray Experiments and Data Analysis
Transgenic 3-week-old pOp6:WIN1-HA and pOp6 plants were treated with DEX (sprayed once with 10 μM DEX, 0.015% [v/v] Silwet L-77, and 0.033% [v/v] ethanol). The success of the DEX treatment was determined by detection of WIN1-HA protein in protein gel blot analysis. Leaf total RNA was isolated using the RNeasy plant kit. RNA was treated with DNase I (Ambion), and the quality of the samples was checked using the RNA 6000 Nano LabChip kit (Agilent). RNA samples were then processed according to the Affymetrix Eukaryotic Sample and Array Processing protocol (https://www.affymetrix.com/support/downloads/manuals/expression_s2_manual.pdf). Hybridization of the in vitro–amplified RNA to Affymetrix Arabidopsis GeneChips (ATH1 arrays) and washing and scanning of the arrays were performed following standard Affymetrix protocols using a Hybridization Oven 640, a Fluidics Station 450, and a GeneChip Scanner 3000 7G. The experiment was performed in triplicate, preparing three independent biological replicates from six plants each.
To analyze the microarray data and identify genes showing significant differences in expression, several algorithms and methods were used. First, MAS5 expression estimates were obtained using the GCOS software (Affymetrix) and further analyzed using SAM (Tusher et al., 2001). SAM analysis (which provides an estimate of the false discovery rate [FDR]) was performed using the software available at http://www-stat.stanford.edu/∼tibs/SAM/ as an Excel (Microsoft) add-in. A delta value of 1.39 was applied, and after 100 permutations resulted in an FDR of 24% and 17 significantly changing genes. The raw data (CEL files) from the Affymetrix hybridizations were also processed and analyzed using Resolver v5.1 (Rosetta Biosoftware). Resolver uses an error model–based approach to stabilize the variance estimation to improve the specificity and sensitivity in differential gene expression detection (Rajagopalan, 2003; Weng et al., 2006). Two approaches were used with the Resolver system, ratio analysis (creating ratios from the intensity profiles) and ANOVA, and in each case with (various methods of) multiple test corrections. For the ratio analysis, both the default and the pairwise methods for building the ratio that are implemented in Resolver were used (the default method averages the replicates before building the ratio, whereas the pairwise method first creates all possible pairwise ratios and then averages them). Since Resolver does not provide for multiple test correction of the built ratios, the P values generated by the system for each method were processed through Bioconductor's multtest package in R (http://www.bioconductor.org/packages/bioc/stable/src/contrib/html/multtest.html), using the Holm and Benjamini-Hochberg (BH) procedures. In these cases, genes were considered differentially expressed if they were assigned a corrected P value ≤0.05 in both the default and pairwise methods, but no fold-change cutoff was applied, which resulted in gene lists of 29 (Holm) and 50 (BH) members. Error-weighted ANOVA was performed in Resolver with BH-FDR adjustment. In this analysis, genes considered differentially expressed were those that had P values ≤0.01 (55 genes).
ChIP
Chromatin was isolated from 1.5 g of formaldehyde cross-linked or non-cross-linked tissue from 3-week-old plants or flowers from developing inflorescence as described (Bowler et al., 2004). The chromatin was resuspended in 500 μL of nuclei lysis buffer containing 50 mM Tris-HCl, pH 8, 10 mM EDTA, 1% (w/v) SDS, 1 mM PMSF, and Complete Protease Inhibitor Cocktail (Roche). Chromatin shearing was performed by sonication using a Bandolin Sonoplus HD 2070 with an MS73 probe. Sonication settings were four cycles at 10 s each, with a 40% duty cycle and power of 20%. Electrophoresis of sheared samples showed that an average fragment size of 200 to 2000 bp had been achieved. In each ChIP assay, 150 μL of sheared chromatin were diluted in 1350 μL of ChIP buffer containing 1.1% (v/v) Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl, pH 8.0, and 167 mM NaCl. The diluted chromatin was precleared using 40 μL of ChIP buffer–equilibrated sheared salmon sperm DNA/Protein G agarose beads (Update) for 2 h at 4°C. Precleared chromatin solution was collected by centrifugation. Sixty microliters were removed for an input sample and the rest split into 600-μL aliquots. One aliquot served as a negative control and was incubated without antibodies. Another aliquot was incubated with 1.5 μg of rat anti-HA monoclonal antibody 3F10 (Roche) overnight at 4°C. Collection of immune complexes on sheared salmon sperm DNA/Protein G agarose beads with subsequent washes, elution, reverse cross-linking, and proteinase K treatments were performed as in Bowler et al. (2004). The DNA fragments were cleaned up using a Qiaquick PCR DNA purification kit (Qiagen). Promoter and other gene fragments were amplified using 1 μL of purified DNA as template in each PCR reaction. The fragments were cloned into the pGEM-T vector (Promega) and sequenced to verify that the correct target had been amplified.
Primer sequences for ChIP PCR experiments are provided as part of Supplemental Table 3 online.
Phylogenetic Analysis
Phylogenetic trees were generated using alignments of complete predicted protein sequences using the ClustalX program (see Supplemental Figures 4 to 6 online) (Thompson et al., 1997). Alignment parameters were as follows: gap opening penalty = 10, and gap extension penalty = 0.2. Gonnet weight matrices were selected as a way to determine the similarity of nonidentical amino acids. The trees were generated using the neighbor-joining method (Saitou and Nei, 1987) with a number of bootstrap replicates set at 1000.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: WIN1, At1g15360; CYP86A7, At1g63710; CYP86A4, At1g01600; HTH-like, At1g12570; GPDHc1, At2g41540; LACS2, At1g49430; NLM2, At4g18910; GPAT4, At1g01610; At2g04570; At1g64405; At3g02290; At2g16900; and At3g30720.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Monosaccharide Composition of Shoot and Root Cell Walls in 35S:WIN1 Overexpressors.
Supplemental Figure 2. Dendogram Obtained by Hierarchical Cluster Analysis of 42 Arabidopsis Lines Based on FTIR Spectra Sampled from Dark-Grown Hypocotyls.
Supplemental Figure 3. Composition of Leaf Acyl-CoAs and Membrane Fatty Acids in 35S:WIN1 Plants.
Supplemental Figure 4. Phylogenetic Analysis of Genes Related to DEX-Induced GPAT4 and HTH-Like.
Supplemental Figure 5. Alignment of the Predicted Amino Acid Sequences of Proteins Related to HTH.
Supplemental Figure 6. Alignment of the Predicted Amino Acid Sequences of Proteins Related to GPAT4.
Supplemental Table 1. Short-Term Regulation of Gene Expression by WIN1-HA: Output of Different Statistical Analyses of Microarray Data.
Supplemental Table 2. Promoter Motifs in Genes That Were Significantly Upregulated after WIN1-HA Induction by DEX in pOp6:WIN-HA Plants.
Supplemental Table 3. Primer Sequences.
Supplementary Material
Acknowledgments
We thank Toshiro Ito for providing the pGTI0242 vector, Christiane Nawrath for her assistance with mass spectral analysis, Naveed Aziz for his technical support with microarray hybridizations and data analysis, and Uzma Urooj for her technical assistance with this project. This work was supported by Grant P20283 from the Biotechnology and Biological Sciences Research Council (UK).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Pierre Broun (pbroun@york.ac.uk).
Online version contains Web-only data.
References
- Aarts, M.G.M., Keijzer, C.J., Stiekema, W.J., and Pereira, A. (1995). Molecular characterization of the CER1 gene of Arabidopsis involved in epicuticular wax biosynthesis and pollen fertility. Plant Cell 7 2115–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aharoni, A., Dixit, S., Jetter, R., Thoenes, E., van Arkel, G., and Pereira, A. (2004). The SHINE clade of AP2 domain transcription factors activates wax biosynthesis, alters cuticle properties, and confers drought tolerance when overexpressed in Arabidopsis. Plant Cell 16 2463–2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albersheim, P., Nevins, D.J., English, P.D., and Karr, A. (1967). A method for the analysis of sugars in plant cell wall polysaccharides by gas-liquid chromatography. Carbohydr. Res. 5 340–345. [Google Scholar]
- Beisson, F., Li, Y., Bonaventure, G., Pollard, M., and Ohlrogge, J.B. (2007). The acyltransferase GPAT5 is required for the synthesis of suberin in seed coat and root of Arabidopsis. Plant Cell 19 351–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenkrantz, N., and Asboe-Hansen, G. (1973). New method for quantitative determination of uronic acids. Anal. Biochem. 54 484–489. [DOI] [PubMed] [Google Scholar]
- Bonaventure, G., Beisson, F., Ohlrogge, J., and Pollard, M. (2004). Analysis of the aliphatic monomer composition of polyesters associated with Arabidopsis epidermis: Occurrence of octadeca-cis-6, cis-9-diene-1,18-dioate as the major component. Plant J. 40 920–930. [DOI] [PubMed] [Google Scholar]
- Bowler, C., Benvenuto, G., Laflamme, P., Molino, D., Probst, A.V., Tariq, M., and Paszkowski, J. (2004). Chromatin techniques for plant cells. Plant J. 39 776–789. [DOI] [PubMed] [Google Scholar]
- Broun, P. (2004). Transcription factors as tools for metabolic engineering in plants. Curr. Opin. Plant Biol. 7 202–209. [DOI] [PubMed] [Google Scholar]
- Broun, P., Poindexter, P., Osborne, E., Jiang, C.Z., and Riechmann, J.L. (2004). WIN1, a transcriptional activator of epidermal wax accumulation in Arabidopsis. Proc. Natl. Acad. Sci. USA 101 4706–4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browse, J., McCourt, P.J., and Somerville, C.R. (1986). Fatty acid composition of leaf lipids determined after combined digestion and fatty acid methyl ester formation from fresh tissue. Anal. Biochem. 152 141–145. [DOI] [PubMed] [Google Scholar]
- Chakravarthy, S., Tuori, R.P., D'Ascenzo, M.D., Fobert, P.R., Despres, C., and Martin, G.B. (2003). The tomato transcription factor Pti4 regulates defense-related gene expression via GCC box and non-GCC box cis elements. Plant Cell 15 3033–3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Craft, J., Samalova, M., Baroux, C., Townley, H., Martinez, A., Jepson, I., Tsiantis, M., and Moore, I. (2005). New pOp/LhG4 vectors for stringent glucocorticoid-dependent transgene expression in Arabidopsis. Plant J. 41 899–918. [DOI] [PubMed] [Google Scholar]
- Duan, H., and Schuler, M.A. (2005). Differential expression and evolution of the Arabidopsis CYP86A subfamily. Plant Physiol. 137 1067–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke, R., Briesen, I., Wojciechowski, T., Faust, A., Yephremov, A., Nawrath, C., and Schreiber, L. (2005). Apoplastic polyesters in Arabidopsis surface tissues – A typical suberin and a particular cutin. Phytochemistry 66 2643–2658. [DOI] [PubMed] [Google Scholar]
- Graca, J., Schreiber, L., Rodrigues, J., and Pereira, H. (2002). Glycerol and glyceryl esters of omega-hydroxyacids in cutins. Phytochemistry 61 205–215. [DOI] [PubMed] [Google Scholar]
- Higo, K., Ugawa, Y., Iwamoto, M., and Korenaga, T. (1999). Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 27 297–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffree, C.E. (2006). The fine structure of the plant cuticle. In Biology of the Plant Cuticle, M. Riederer and C. Muller, eds (Oxford, UK: Blackwell Publishing), pp. 11–125.
- Jetter, R., Kunst, L., and Samuels, A.L. (2006). Composition of plant cuticular waxes. In Biology of the Plant Cuticle, M. Riederer and C. Muller, eds (Oxford, UK: Blackwell Publishing), pp. 145–181.
- Karimi, M., De Meyer, B., and Hilson, P. (2002). Modular cloning and expression of tagged fluorescent protein in plant cells. Trends Plant Sci. 10 103–105. [DOI] [PubMed] [Google Scholar]
- Kolattukudy, P.E. (1996). Biosynthetic pathways of cutin and waxes, and their sensitivity to environmental stresses. In Plant Cuticles: An Integrated Functional Approach, G. Kerstiens, ed (Oxford, UK: BIOS Scientific Publishers), pp. 83–108.
- Krolikowski, K.A., Victor, J.L., Wagler, T.N., Lolle, S.J., and Pruitt, R.E. (2003). Isolation and characterization of the Arabidopsis organ fusion gene HOTHEAD. Plant J. 35 501–511. [DOI] [PubMed] [Google Scholar]
- Kunst, L., Jetter, R., and Samuels, A.L. (2006). Biosynthesis and transport of plant cuticular waxes. In Biology of the Plant Cuticle, M. Riederer and C. Muller, eds (Oxford, UK: Blackwell Publishing), pp. 182–215.
- Kurata, T., Kawabata-Awai, C., Sakuradani, E., Shimizu, S., Okada, K., and Wada, T. (2003). The YORE-YORE gene regulates multiple aspects of epidermal cell differentiation in Arabidopsis. Plant J. 36 55–66. [DOI] [PubMed] [Google Scholar]
- Kurdyukov, S., Faust, A., Nawrath, C., Bar, S., Voisin, D., Efremova, N., Franke, R., Schreiber, L., Saedler, H., Metraux, J.P., and Yephremov, A. (2006. b). The epidermis-specific extracellular BODYGUARD controls cuticle development and morphogenesis in Arabidopsis. Plant Cell 18 321–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurdyukov, S., Faust, A., Trenkamp, S., Bar, S., Franke, R., Efremova, N., Tietjen, K., Schreiber, L., Saedler, H., and Yephremov, A. (2006. a). Genetic and biochemical evidence for involvement of HOTHEAD in the biosynthesis of long-chain alpha-,omega-dicarboxylic fatty acids and formation of extracellular matrix. Planta 224 315–329. [DOI] [PubMed] [Google Scholar]
- Larson, T.R., and Graham, I.A. (2000). Application of a new method for the sensitive detection and quantification of acyl-CoA esters in Arabidopsis thaliana seedlings and mature leaves. Biochem. Soc. Trans. 28 575–577. [PubMed] [Google Scholar]
- Li, Y., Beisson, F., Ohlrogge, J., and Pollard, M. (2006). Ectopic expression of a suberin-associated acyltransferase gene produces monoacylglycerols as a major constituent of cuticular waxes in Arabidopsis. In Current Advances in the Biochemistry and Cell Biology of Plant Lipids, C. Benning and J. Ohlrogge, eds (Salt Lake City, UT: Aardvark Global Publishing Company), p. 149.
- Mayfield, J.A., Fiebig, A., Johnstone, S.E., and Preuss, D. (2001). Gene families from the Arabidopsis thaliana pollen coat proteome. Science 292 2482–2485. [DOI] [PubMed] [Google Scholar]
- Millar, A.A., Clemens, S., Zachgo, S., Giblin, E.M., Taylor, D.C., and Kunst, L. (1999). CUT1, an Arabidopsis gene required for cuticular wax biosynthesis and pollen fertility, encodes a very-long-chain fatty acid condensing enzyme. Plant Cell 11 825–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millenaar, F.F., Okyere, J., May, S.T., van Zanten, M., Voesenek, L.A., and Peeters, A.J. (2006). How to decide? Different methods of calculating gene expression from short oligonucleotide array data will give different results. BMC Bioinformatics 7 137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouille, G., Robin, S., Lecomte, M., Pagant, S., and Hofte, H. (2003). Classification and identification of Arabidopsis cell wall mutants using Fourier-Transform InfraRed (FT-IR) microspectroscopy. Plant J. 35 393–404. [DOI] [PubMed] [Google Scholar]
- Nakano, T., Suzuki, K., Fujimura, T., and Shinshi, H. (2006). Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 140 411–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlando, V., and Paro, R. (1993). Mapping Polycomb-repressed domains in the bithorax complex using in vivo formaldehyde cross-linked chromatin. Cell 75 1187–1198. [DOI] [PubMed] [Google Scholar]
- Penfield, S., Josse, E.M., Kannangara, R., Gilday, A.D., Halliday, K.J., and Graham, I.A. (2005). Cold and light control seed germination through the bHLH transcription factor SPATULA. Curr. Biol. 15 1998–2006. [DOI] [PubMed] [Google Scholar]
- Pighin, J.A., Zheng, H., Balakshin, L.J., Goodman, I.P., Western, T.L., Jetter, R., Kunst, L., and Samuels, A.L. (2004). Plant cuticular lipid export requires an ABC transporter. Science 306 702–704. [DOI] [PubMed] [Google Scholar]
- Pollard, M., Molina, I., Beisson, F., Li, Y., dos Santos, D., Suh, M.C., Bonaventure, G., and Ohlrogge, J. (2006). Deconstructing plant lipid polyester structures and biosynthesis. In Current Advances in the Biochemistry and Cell Biology of Plant Lipids, C. Benning and J. Ohlrogge, eds (Salt Lake City, UT: Aardvark Global Publishing Company), p. 147.
- Rajagopalan, D. (2003). A comparison of statistical methods for analysis of high density oligonucleotide array data. Bioinformatics 19 1469–1476. [DOI] [PubMed] [Google Scholar]
- Rawat, R., Xu, Z.F., Yao, K.M., and Chye, M.L. (2005). Identification of cis-elements for ethylene and circadian regulation of the Solanum melongena gene encoding cysteine proteinase. Plant Mol. Biol. 57 629–643. [DOI] [PubMed] [Google Scholar]
- Riederer, M. (2006). Introduction: Biology of the plant cuticle. In Biology of the Plant Cuticle, M. Riederer and C. Muller, eds (Oxford, UK: Blackwell Publishing), pp. 1–10.
- Riederer, M., and Schreiber, L. (2001). Protecting against water loss: Analysis of the barrier properties of plant cuticles. J. Exp. Bot. 52 2023–2032. [DOI] [PubMed] [Google Scholar]
- Saitou, N., and Nei, M. (1987). The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4 406–425. [DOI] [PubMed] [Google Scholar]
- Schnurr, J., Shockey, J., and Browse, J. (2004). The acyl-CoA synthetase encoded by LACS2 is essential for normal cuticle development in Arabidopsis. Plant Cell 16 629–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahmuradov, I.A., Gammerman, A.J., Hancock, J.M., Bramley, P.M., and Solovyev, V.V. (2003). PlantProm: A database of plant promoter sequences. Nucleic Acids Res. 31 114–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, W., Wei, Y., Dauk, M., Tan, Y., Taylor, D.C., Selvaraj, G., and Zou, J. (2006). Involvement of a glycerol-3-phosphate dehydrogenase in modulating the NADH/NAD+ ratio provides evidence of a mitochondrial glycerol-3-phosphate shuttle in Arabidopsis. Plant Cell 18 422–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd, T., and Griffiths, D.W. (2006). The effects of stress on plant cuticular waxes. New Phytol. 171 469–499. [DOI] [PubMed] [Google Scholar]
- Sieber, P., Schorderet, M., Ryser, U., Buchala, A., Kolattukudy, P., Metraux, J.P., and Nawrath, C. (2000). Transgenic Arabidopsis plants expressing a fungal cutinase show alterations in the structure and properties of the cuticle and postgenital organ fusions. Plant Cell 12 721–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark, R.E., and Tian, S. (2006). The cutin biopolymer matrix. In Biology of the Plant Cuticle, M. Riederer and C. Muller, eds (Oxford, UK: Blackwell Publishing), pp. 126–144.
- Suh, M.C., Samuels, A.L., Jetter, R., Kunst, L., Pollard, M., Ohlrogge, J., and Beisson, F. (2005). Cuticular lipid composition, surface structure, and gene expression in Arabidopsis stem epidermis. Plant Physiol. 139 1649–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., and Higgins, D.G. (1997). The ClustalX windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd, J., Post-Beittenmiller, D., and Jaworski, J.G. (1999). KCS1 encodes a fatty acid elongase 3-ketoacyl-CoA synthase affecting wax biosynthesis in Arabidopsis thaliana. Plant J. 17 119–130. [DOI] [PubMed] [Google Scholar]
- Tusher, V.G., Tibshirani, R., and Chu, G. (2001). Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98 5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weig, A.R., and Jakob, C. (2000). Functional identification of the glycerol permease activity of Arabidopsis thaliana NLM1 and NLM2 proteins by heterologous expression in Saccharomyces cerevisiae. FEBS Lett. 481 293–298. [DOI] [PubMed] [Google Scholar]
- Wellesen, K., Durst, F., Pinot, F., Benveniste, I., Nettesheim, K., Wisman, E., Steiner-Lange, S., Saedler, H., and Yephremov, A. (2001). Functional analysis of the LACERATA gene of Arabidopsis provides evidence for different roles of fatty acid omega-hydroxylation in development. Proc. Natl. Acad. Sci. USA 98 9694–9699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng, L., Dai, H., Zhan, Y., He, Y., Stepaniants, S.B., and Bassett, D.E. (2006). Rosetta error model for gene expression analysis. Bioinformatics 22 1111–1121. [DOI] [PubMed] [Google Scholar]
- Xiao, F., Goodwin, S.M., Xiao, Y., Sun, Z., Baker, D., Tang, X., Jenks, M.A., and Zhou, J.M. (2004). Arabidopsis CYP86A2 represses Pseudomonas syringae type III genes and is required for cuticle development. EMBO J. 23 2903–2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, X.J., Dietrich, C.R., Delledonne, M., Xia, Y.J., Wen, T.J., Robertson, D.S., Nikolau, B.J., and Schnable, P.S. (1997). Sequence analysis of the cloned glossy8 gene of maize suggests that it may code for a beta-ketoacyl reductase required for the biosynthesis of cuticular waxes. Plant Physiol. 115 501–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, H., Rowland, O., and Kunst, L. (2005). Disruptions of the Arabidopsis Enoyl-CoA reductase gene reveal an essential role for very-long-chain fatty acid synthesis in cell expansion during plant morphogenesis. Plant Cell 17 1467–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann, P., Hirsch-Hoffmann, M., Hennig, L., and Gruissem, W. (2004). GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136 2621–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.