Abstract
Chlorophyll degradation is an aspect of leaf senescence, which is an active process to salvage nutrients from old tissues. non-yellow coloring1 (nyc1) is a rice (Oryza sativa) stay-green mutant in which chlorophyll degradation during senescence is impaired. Pigment analysis revealed that degradation of not only chlorophylls but also light-harvesting complex II (LHCII)–bound carotenoids was repressed in nyc1, in which most LHCII isoforms were selectively retained during senescence. Ultrastructural analysis of nyc1 chloroplasts revealed that large and thick grana were present even in the late stage of senescence, suggesting that degradation of LHCII is required for the proper degeneration of thylakoid membranes. Map-based cloning of NYC1 revealed that it encodes a chloroplast-localized short-chain dehydrogenase/reductase (SDR) with three transmembrane domains. The predicted structure of the NYC1 protein and the phenotype of the nyc1 mutant suggest the possibility that NYC1 is a chlorophyll b reductase. Although we were unable to detect the chlorophyll b reductase activity of NYC1, NOL (for NYC1-like), a protein closely related to NYC1 in rice, showed chlorophyll b reductase activity in vitro. We suggest that NYC1 and NOL encode chlorophyll b reductases with divergent functions. Our data collectively suggest that the identified SDR protein NYC1 plays essential roles in the regulation of LHCII and thylakoid membrane degradation during senescence.
INTRODUCTION
The final step of leaf development is senescence, which is an active process to salvage nutrients from old leaves. Leaf yellowing, which is caused by unmasking of preexisting carotenoids by chlorophyll degradation, is a good indicator of senescence (Matile, 2000). Most chlorophyll exists in protein complexes in leaves, because free chlorophyll photooxidatively damages cells. Chlorophyll a is a component of several protein complexes, including the photosystem I (PSI) and photosystem II (PSII) reaction center complexes and the cytochrome b6f complex. Chlorophyll b exists only in the light-harvesting chlorophyll a/b–protein complex (LHCP). LHCP binds chlorophyll a, chlorophyll b, and carotenoids (neoxanthin, violaxanthin, and lutein) (Liu et al., 2004). Chlorophyll b is thought to be important for the stability of LHCP (Bellemare et al., 1982). PSI-associated light-harvesting complex I (LHCI) and PSII-associated LHCII proteins are encoded by the Lhca and Lhcb gene families, respectively. LHCPs are localized in the thylakoid membrane. Lhcb1, -2, and -3 are major LHCII proteins and form trimers, but Lhcb4, -5, and -6 occur as monomers. LHCII is localized predominantly in grana, the stacking region of the thylakoid membrane. LHCII has been thought to play an important role in the formation of grana (Allen and Forsberg, 2001).
The chlorophyll synthesis pathway has been well characterized, and most, if not all, genes encoding enzymes involved in chlorophyll synthesis have been isolated (Nagata et al., 2005). On the other hand, the chlorophyll degradation pathway is less well understood (Hörtensteiner, 2006). To avoid the photooxidative toxicity of free chlorophylls and their colored catabolites, plant cells must degrade them rapidly. The first step of chlorophyll a degradation is dephytylation by chlorophyllase, resulting in the formation of chlorophyllide a (Chlide a). After Mg2+ removal, pheophorbide a, which is the last green compound in the chlorophyll-degrading pathway, is converted to red chlorophyll catabolite (RCC) by pheophorbide a oxygenase. RCC is then catabolized to primary fluorescent chlorophyll catabolites by RCC reductase (RCCR). In higher plants, chlorophyll b is degraded only after conversion to chlorophyll a. This reduction process is executed by two distinct enzymes, chlorophyll b reductase and hydroxymethyl chlorophyll a reductase (Ito et al., 1993; Scheumann et al., 1998; Rüdiger, 2002). Among the genes for enzymes involved in chlorophyll degradation, only chlorophyllase, pheophorbide a oxygenase, and RCCR have been isolated (Tsuchiya et al., 1999; Wüthrich et al., 2000; Pružinská et al., 2003; Tanaka et al., 2003). After hydrolysis of chlorophyll a, the resultant Chlide a can be used for the formation of chlorophyll b via the chlorophyll b synthesis pathway. This interconversion between chlorophyll a and chlorophyll b, called the chlorophyll cycle, is thought to be important in adjustment of the chlorophyll a/b ratio in various physiological conditions (Ito and Tanaka, 1996; Rüdiger, 2002).
Mutants that retain green leaves during senescence are called stay-green mutants. They are classified into several types (Thomas and Howarth, 2000). Several genes regulating the whole process of leaf senescence have been identified, including genes for hormonal and proteosomal regulation (Lim et al., 2003). Mutants primarily affecting chlorophyll degradation have also been reported. The most extensively studied mutant in this group is senescence-induced degradation (sid) in Festuca pratensis (Thomas and Howarth, 2000). In sid, the degradation of photosynthetic proteins, including LHCII and the ferredoxin binding protein of PSI, is inhibited, and chlorophyll a is more stable than chlorophyll b during leaf senescence (Thomas et al., 1999). cytG in soybean (Glycine max) is a cytoplasmic mutation in which degradation of LHCII is selectively inhibited and chlorophyll b is more stable than chlorophyll a (Guiamét et al., 2002). Very recently, Lethal Leaf Spot1 (LLS1)/Accelerated Cell Death1 was identified as pheophorbide a oxygenase. The lls1 mutant, originally isolated as a lesion-mimic mutant in maize (Zea mays), remains green during senescence under dark but not light conditions (Gray et al., 1997; Pružinská et al., 2003; Tanaka et al., 2003; Yang et al., 2004).
Here, we report the molecular cloning of Non-Yellow Coloring1 (NYC1). nyc1 is a recessive mutant in rice (Oryza sativa), and the nyc1 plant shows a stay-green phenotype during senescence. In nyc1, most LHCII members are selectively retained during senescence, in contrast with the wild type. nyc1 retains granal structure during senescence, suggesting that degradation of LHCII is required for proper degeneration of the thylakoid membrane during senescence. NYC1 shows similarity to a short-chain dehydrogenase/reductase and is localized in chloroplasts. The NYC1-like (NOL) protein, the most closely related protein to NYC1, showed chlorophyll b reductase activity in vitro. Although we were unable to demonstrate the chlorophyll b reductase activity of NYC1 in vitro, it would be reasonable to assume that NYC1 is also a chlorophyll b reductase. Thus, our data collectively indicate that chlorophyll b degradation is a key step in the degradation of LHCII and the thylakoid membrane during senescence.
RESULTS
nyc1 Has a Defect in Chlorophyll Degradation
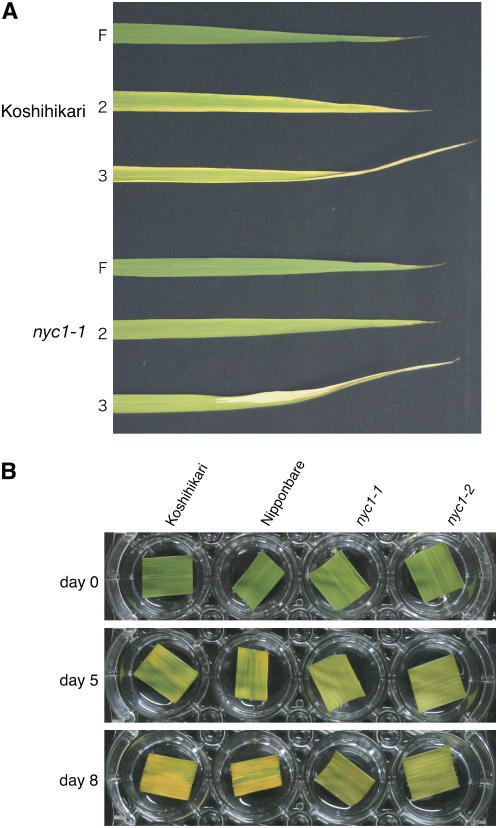
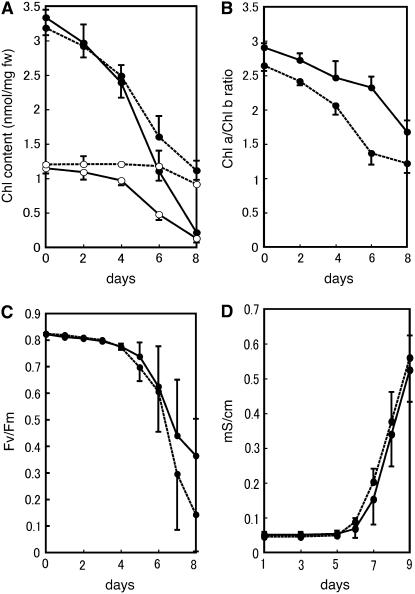
nyc1 was isolated as a stay-green mutant in the paddy field. nyc1 remains green until just before death in natural senescence and does not show yellowing (Figure 1A). To examine the effect of the nyc1 mutation in dark-induced senescence, we detached nyc1 and wild-type leaves and kept them in the dark at 27°C. After 7 d of incubation, the wild-type leaves turned yellow, but the nyc1 leaves remained green (Figure 1B). To examine the correlation between greenness and leaf functionality, we measured the change of chlorophyll content, the value of Fv/Fm (for the ratio of variable fluorescence to maximum fluorescence), and the membrane ion leakage over time (Figure 2). Fv/Fm reflects how much of the light energy absorbed by PSII is used for photosynthesis. In the wild type, chlorophyll a and chlorophyll b contents began to decrease by 2 d of dark incubation (2 DDI) and continued to decrease until 8 DDI (Figure 2A). In nyc1, reduction of the chlorophyll b content was suppressed (t test, P < 0.01): only a slight reduction at 6 to 8 DDI was observed. In nyc1, reduction of chlorophyll a content was less affected than reduction of chlorophyll b, but final chlorophyll a content (8 DDI) was higher than that of the wild type (t test, P < 0.01). In nyc1, the chlorophyll a content and the chlorophyll b content were 5.1 and 7.0 times the corresponding wild-type values at 8 DDI. The chlorophyll a/b ratio was reduced predominantly in the late stage of senescence in the wild type (Figure 2B), but the ratio was reduced earlier in nyc1, reaching ∼1.2 at 8 DDI. The value of Fv/Fm was 0.82 in both nyc1 and the wild type before dark incubation. During dark incubation, Fv/Fm values fell to 0.37 in the wild type and to 0.14 in nyc1 at 8 DDI, indicating that the decrease of photosynthetic efficiency of PSII in nyc1 is not slower than that in the wild type. The membrane ion leakage from detached leaves began to increase at 6 DDI and increased similarly in the wild type and nyc1 during dark incubation (Figure 2D). These results suggest that the higher chlorophyll content in nyc1 does not imply a higher photosynthesis ability and that NYC1 is involved in chlorophyll degradation, not in leaf longevity.
Figure 1.
Naturally and Dark-Induced Senescent Leaves of nyc1.
(A) Naturally senescent leaves of Koshihikari and nyc1-1 at 21 d after flowering. F, the flag leaf (the last leaf); 2, the second leaf; 3, the third leaf.
(B) Wild-type (Koshihikari and Nipponbare) and nyc1 (nyc1-1 and nyc1-2) leaves were incubated in the dark. The same leaves are shown at different incubation times (0, 5, and 8 d).
Figure 2.
Physiological Changes in nyc1 Leaves during Dark-Induced Senescence.
(A) Change of chlorophyll content during dark incubation. Detached leaves of the same fresh weight were extracted with the same volume of 80% acetone. Chlorophyll contents were measured spectrophotometrically. Solid lines, Koshihikari; dotted lines, nyc1-1; closed circles, chlorophyll a; open circles, chlorophyll b. Error bars indicate sd. n = 3.
(B) Chlorophyll a/b ratio of the same samples as in (A). Solid lines, Koshihikari; dotted lines, nyc1-1. Error bars indicate sd.
(C) Fv/Fm values. Solid lines, Koshihikari; dotted lines, nyc1-1. Error bars indicate sd. n = 3.
(D) Membrane ion leakage. Solid lines, Koshihikari; dotted lines, nyc1-1. Error bars indicate sd. n = 6.
In the physiological change during natural senescence, results similar to those in dark-induced senescence were obtained (see Supplemental Figure 1 online). Lower leaves are more senescent than upper leaves during natural senescence in rice. The flag leaf (the last leaf before flowering), the second leaf, and the third leaf were examined at 21 d after flowering. Both chlorophyll a and chlorophyll b in the third leaf were more abundant in nyc1-1 than in the wild type (see Supplemental Figure 1A online; t test, P < 0.05), and chlorophyll b degradation was more strongly inhibited in nyc1-1 (see Supplemental Figures 1A and 1B online). The Fv/Fm value of nyc1-1 was lower than that of the wild type (see Supplemental Figure 1C online; t test, P < 0.05), and the membrane ion leakage values were similar between nyc1-1 and the wild type in the third leaf, suggesting that nyc1-1 did not retain the activity of the cell in spite of the higher content of chlorophyll. These observations suggest that the phenotype of nyc1 in natural senescence is essentially identical to that in dark-induced senescence.
The reason for the lower Fv/Fm value in nyc1 during senescence may be explained as follows. During senescence, the nyc1 mutant harvests more light than the wild type, owing to its higher LHCII content, but cannot use the absorbed light energy fully for photosynthesis because of the reduced amount of the PSII core complexes (Figure 3). The higher ratio of LHCII to the PSII core complexes in nyc1 lowers the Fv/Fm value.
Figure 3.
Stability of Photosynthetic Proteins in nyc1 during Dark Incubation.
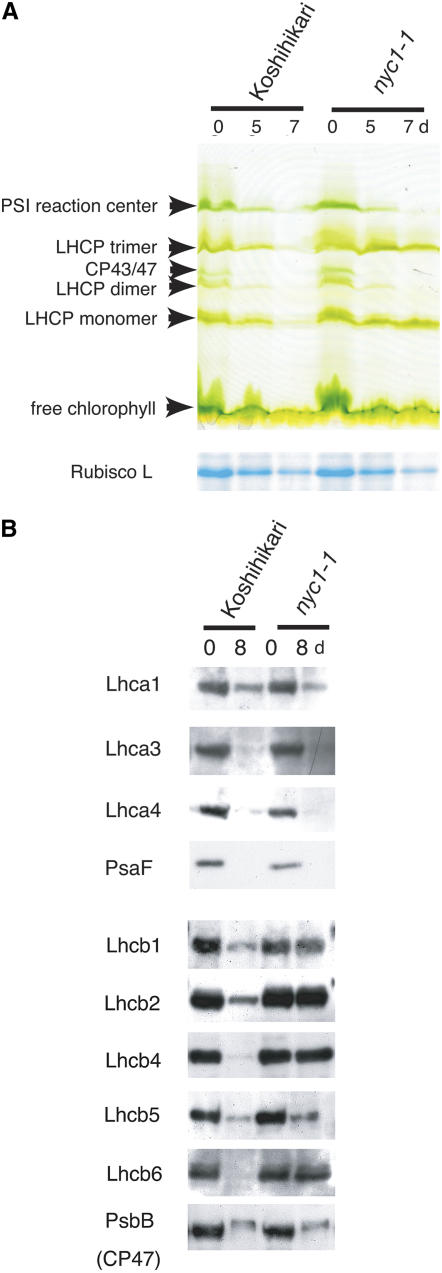
(A) SDS-PAGE analysis of leaf proteins. The top panel shows the change of chlorophyll binding proteins during dark incubation. The gel was visualized by chlorophyll bound to proteins (green gel). Total proteins were extracted from 100 mg (fresh weight) of leaves with 250 μL of the extraction buffer for green gel analysis. Each lane was loaded with 10 μL of non-heat-treated extract. In the bottom panel (Rubisco L [for ribulose-1,5-bis-phosphate carboxylase/oxygenase]), the gel was stained with Coomassie Brilliant Blue G 250. A total of 100 mg (fresh weight) of leaves was extracted with 4 mL of the extraction buffer for SDS-PAGE analysis. Each lane was loaded with 5 μL of heat-treated extract.
(B) Protein gel blot analysis of proteins from nonsenescent (0-d dark incubation) and fully senescent (8-d dark incubation) leaves. The extracts were prepared as in the bottom panel of (A). Each lane was loaded with 5 μL of extract.
Inhibition of LHCP Degradation in nyc1
To examine the behavior of chlorophyll–protein complexes in nyc1 during senescence, we performed green gel analysis (Figure 3A). Similar amounts of PSI reaction center complexes, LHCP trimer, dimer, and monomer, and CP43/47 were observed in mature leaves of both nyc1 and the wild type. In wild-type senescent leaves, the abundance of these protein complexes was reduced remarkably at 5 DDI, and only a trace was detected at 7 DDI. In nyc1, the abundances of chlorophyll a binding proteins, LHCP dimer (whose major component is LHCI), PSI reaction center complex, and CP43/47 were reduced to similar levels as in the wild type at 5 and 7 DDI, but the LHCP trimer and monomer were apparent even at 7 DDI. The abundance of the large subunit of ribulose-1,5-bis-phosphate carboxylase/oxygenase, a stromal protein that does not bind chlorophylls, was reduced similarly in the wild type and nyc1 during dark-induced senescence.
We performed protein gel blot analysis to examine the abundance of LHC apoproteins (Figure 3B). All LHCI proteins examined (Lhca1, Lhca3, and Lhca4) were degraded similarly in nyc1 and the wild type, consistent with the observation that the LHCP dimer disappeared in the green gel analysis in both lines. PsaF, a component of the PSI core complex, was degraded in the late stage of senescence in both the wild type and nyc1, suggesting that not only peripheral complexes of PSI but also the PSI core complex degraded during senescence in both the wild type and nyc1. The situation is more complex in LHCII than in LHCI. The major trimeric LHCII proteins, Lhcb1 and Lhcb2, were much more stable in nyc1 than in the wild type. Among the monomeric LHCII proteins, Lhcb4 and Lhcb6 were also stable in nyc1, but the amount of Lhcb5 was reduced in nyc1. The amount of CP47, a PSII core antenna, was also reduced during senescence in the wild type and nyc1. These observations suggest that the inhibition of protein degradation in nyc1 is limited to a particular type of LHCP, including the major trimeric LHCII. In several stay-green mutants, N-terminal truncation of the remaining LHCII was observed during senescence (Thomas and Howarth, 2000; Oh et al., 2003). However, no change in LHCII molecular weight was observed in nyc1, indicating that the N-terminal region of LHCII was retained in nyc1 during senescence.
Inhibition of Degradation of Pigments Strongly Bound to LHCP in nyc1
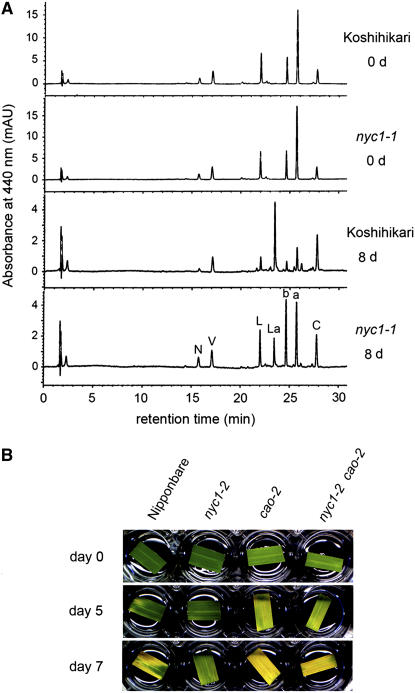
We examined the degradation of chlorophylls and other photosynthetic pigments during senescence by HPLC (Figure 4A). The chromatograms were very similar between the wild type and nyc1 before dark incubation. By 8 DDI, the amounts of most photosynthetic pigments except β-carotene were significantly reduced in the wild type. A major peak emerged between lutein and chlorophyll b. This novel substance is lutein 3-acetate, which we think is converted from lutein during senescence (M. Kusaba, T. Maoka, and S. Takaichi, unpublished data). In nyc1, the degradation of not only chlorophyll a and chlorophyll b but also neoxanthin and lutein was apparently inhibited. Interestingly, the latter two are LHCII binding pigments. The only exception was violaxanthin, which was reduced to a similar level as in the wild type. The binding of violaxanthin to LHCP is not strong (Ruban et al., 1999). These results and the observation that LHCII was stable during senescence in nyc1 suggest that the stability of LHCII in nyc1 inhibits the degradation of pigments strongly bound to it.
Figure 4.
Analysis of Photosynthetic Pigments.
(A) HPLC analysis of photosynthetic pigments. The same volume of extract was loaded in each injection. AU, absorption unit; N, neoxanthin; V, violaxanthin; L, lutein; La, lutein 3-acetate; b, chlorophyll b; a, chlorophyll a; C, β-carotene.
(B) Leaf color change in the nyc1 cao double mutant in dark-induced senescence. Leaves of Nipponbare (wild type), nyc1-2, cao-2, and nyc1-2 cao-2 at 0, 5, and 7 DDI are shown.
nyc1 cao Double Mutants Do Not Show the Stay-Green Phenotype
The key enzyme for chlorophyll b synthesis is chlorophyllide a oxygenase (CAO) (Tanaka et al., 1998; Espineda et al., 1999; Oster et al., 2000). chlorophyll b is important for LHCP stability, and no pigment-bound LHCP is detected in chlorophyll b–deficient mutants (Bellemare et al., 1982; Morita et al., 2005). To examine the relationship between the deficiency of chlorophyll b and the stay-green phenotypes in nyc1, we raised a double mutant from a cross between nyc1-2 and a chlorophyll b–deficient mutant, cao-2 (Morita et al., 2005) (Figure 4B). The double mutant showed a pale green phenotype like that of cao-2, suggesting that the double mutant and cao-2 had reduced chlorophyll contents. Detached leaves of the wild type, cao-2, and the nyc1-2 cao-2 double mutant turned yellow at 7 DDI, while the leaves of nyc1 retained greenness. Both cao-2 and nyc1-2 cao-2 began to turn yellow at 5 DDI and showed similar rates of reduction of chlorophyll contents (see Supplemental Figure 2 online), indicating that nyc1 does not have a significant effect on chlorophyll degradation during dark-induced senescence in the cao-2 background. This observation suggests that NYC1 is not involved in the degradation of chlorophyll in the chlorophyll–protein complexes except LHCP.
NYC1 Is Required for Proper Chloroplast Degeneration
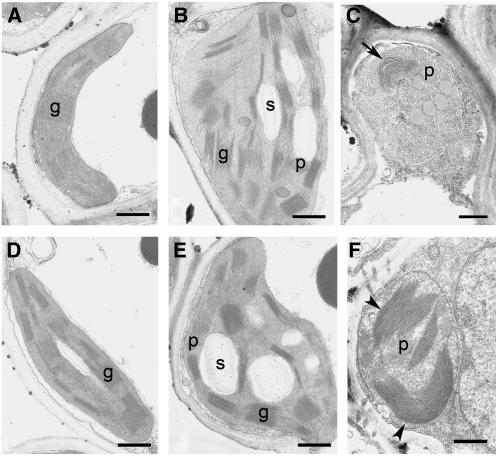
The ultrastructure of chloroplasts changed during senescence in the wild type and nyc1 (Figure 5). No obvious difference in the chloroplast structure was observed between the wild type and nyc1 before senescence (Figures 5A and 5D). At 5 DDI, chloroplasts swelled and became round in both the wild type and nyc1 (Figures 5B and 5E). At this stage, starch granules and plastoglobuli became obvious, and the array of grana stacks and intergrana became slightly disordered. At 8 DDI, chloroplasts shrank (Figures 5C and 5F). In wild-type chloroplasts, the volume of the thylakoid membrane was reduced significantly, suggesting the degradation of thylakoid membranes (Figure 5C). Surprisingly, very thick and wide grana stacks of thylakoid membranes were observed in nyc1 at this stage (Figure 5F). The grana structure was much larger and their number was smaller than before senescence, suggesting that the grana stacks were fused into large stacks. These observations collectively suggest that grana stacks are stable during senescence in nyc1, unlike in the wild type.
Figure 5.
Ultrastructures of Wild-Type and nyc1 Chloroplasts during Senescence.
(A) to (C) show the wild type, and (D) to (F) show nyc1-1. (A) and (D), 0 DDI; (B) and (E), 5 DDI; (C) and (F), 8 DDI. An arrow indicates thylakoid membranes with reduced volume. Arrowheads indicate the very large and thick grana stacks. g, grana stack; p, plastoglobule; s, starch granule. Bars = 500 nm.
Isolation of the NYC1 Gene
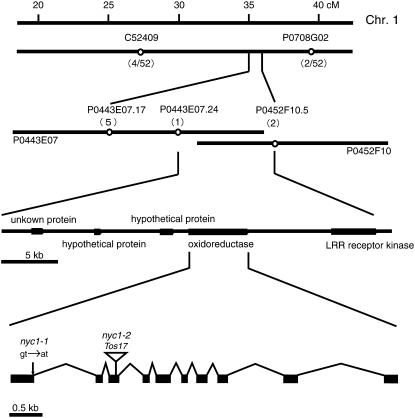
We performed map-based cloning to isolate NYC1 (Figure 6). We examined ∼1500 F2 seedlings of a cross between nyc1-1 and indica cv Kasalath for the stay-green phenotype. Initial mapping using 26 F2 plants homozygous for nyc1 mapped nyc1 between C52409 (27.3 to 28.4 centimorgans) and P0708G02 (39.0 centimorgans) on chromosome 1. Further mapping using the remaining F2 plants revealed that nyc1 was located between cleaved-amplified polymorphic sequence (CAPS) markers P0443E07.24 and P0452F10.5. Five genes were predicted in this ∼40-kb region. One of them seems to encode a member of the short-chain dehydrogenase/reductase (SDR) family (oxidoreductase in Figure 6). Sequence analysis of the nyc1-1 allele revealed a single base pair change, a G-to-A substitution, at the first exon/first intron junction, which may prevent splicing of the first intron and result in the early termination of translation. We found that another nyc1 allele, nyc1-2, which is derived from tissue culture mutagenesis, carries a Tos17 insertion in the third exon. To confirm that this putative SDR gene encodes NYC1, we performed a complementation experiment (see Supplemental Figure 3A online). An MluI-AflII genomic fragment (8.2 kb) containing the whole coding region of the putative SDR gene was introduced into line nyc1-2. Six independent transformants showed yellowing at 8 DDI (two of them are shown in Supplemental Figure 3A online), suggesting that the putative SDR gene is NYC1. One transformant showed a yellow stripe when incubated in the dark (see Supplemental Figure 3B online). We considered this to be due to a chimeric complementation by the NYC1 genomic fragment, suggesting that NYC1 acts cell-autonomously.
Figure 6.
Map-Based Cloning of NYC1.
The numbers of recombination events between the molecular markers and the nyc1 locus are shown in parentheses. P0443E07 and P0452F10 are overlapped BACs. NYC1 consists of 10 exons and 9 introns. The positions of base pair substitution in nyc1-1 and Tos17 insertion in nyc1-2 are shown.
NYC1 Is a Putative Plastid SDR with Three Transmembrane Domains
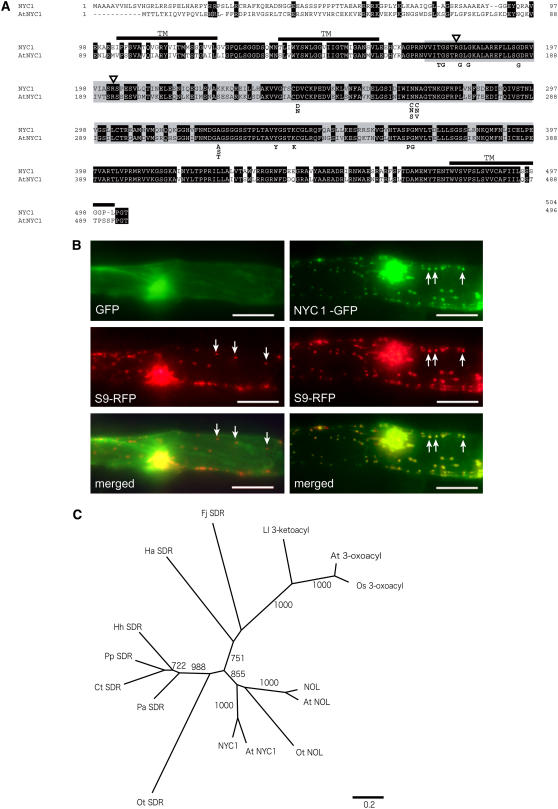
NYC1 encodes a protein with 504 amino acids belonging to the SDR family (Figure 7A). Kallberg et al. (2002) classified SDR into five families. NYC1 belongs to the classical SDR family, which encompasses oxidoreductase. The amino acid sequence of NYC1 contains all important amino acid residues conserved in the classical SDR family, including a dinucleotide binding motif (TGXXXGXG) and a catalytic site (YXXXK). NYC1 has an Arg at key position 15 (180th amino acid) and another at key position 37 (202nd amino acid), suggesting that the protein uses NADP(H) as a cofactor (Kallberg et al., 2002). The TMHMM server (http://www.cbs.dtu.dk/services/TMHMM-2.0/), which predicts the presence of membrane-spanning helices in a given protein sequence, predicted that NYC1 has three transmembrane domains. The putative SDR catalytic domain is located between the second and third transmembrane domains. NYC1 has an N-terminal region, which does not show high similarity to its putative ortholog in Arabidopsis thaliana (At4g13250). PSORT (http://psort.nibb.ac.jp/) analysis predicted that this region could be a chloroplast transit peptide but with low affirmativity.
Figure 7.
Characteristics of NYC1 Proteins.
(A) Amino acid alignment of NYC1 and its putative ortholog in Arabidopsis (At NYC1). Putative transmembrane domains are indicated as TM. Identical amino acid residues are shown in white letters. Regions exhibiting similarity to short-chain dehydrogenase/reductase are shown in gray. Triangles indicate the amino acid residues important for selection of cofactor (see text). Important amino acid residues conserved among the classical SDRs are shown below the alignment.
(B) Plastid localization of the GFP-NYC1 protein. The GFP and RFP proteins were transiently expressed in onion epidermal cells. The left panels show the same cell into which GFP-expressing and plastid-localizing RFP-expressing plasmids were cointroduced. The right panels show the same cells into which NYC1-GFP–expressing and plastid-localizing RFP-expressing plasmids were cointroduced. GFP, control GFP protein; S9-RFP, plastid-localizing S9-RFP protein; NYC1-GFP, GFP fused with a putative transit peptide of NYC1 at the N-terminal end; merged, merged images of the GFP and RFP images. Arrows indicate plastids. Bars = 100 μm.
(C) A neighbor-joining tree constructed using SDR domains of NYC1 and NYC1-related proteins. The tree is based on the alignment shown in Supplemental Figure 4 online. Bootstrap values of >700 with 1000 repeats are shown. 3-Oxoacyl-[acyl-carrier-protein] reductases are the second most similar SDR proteins to NYC1 in plants. Os 3-oxoacyl and At 3-oxoacyl are 3-oxoacyl-[acyl-carrier-protein] reductases in rice and Arabidopsis, respectively. At NYC1 and At NOL are NYC1 and NOL in Arabidopsis. Ot NOL is the most similar SDR to NYC1, and Ot SDR is the second most similar, in Ostreococcus tauri. Other SDRs are as follows: Fj SDR, Flavobacterium johnsoniae; Hh SDR, Halorhodospira halophila; Ct SDR, Chlorobium tepidum; Pa SDR, Prosthecochloris aestuarii; Pp SDR, Pelodictyon phaeoclathratiforme; Ha SDR, Herpetosiphon aurantiacus; Ll 3-ketoacyl, a 3-ketoacyl-(acyl-carrier-protein) reductase of Lactococcus lactis.
To examine the intracellular localization of NYC1, we fused the region encoding the putative transit peptide of NYC1 to the green fluorescent protein (GFP) gene and introduced the fusion construct into epidermal cells of onion (Allium cepa) by particle bombardment (Figure 7B). The signal was colocalized with that of red fluorescent protein (RFP) fused to the transit peptide from plastid-localizing rice S9 ribosomal protein, suggesting that NYC1 is localized in plastids. Specifically, NYC1 might be localized in the chloroplast membrane, because the protein has three putative transmembrane domains. This observation suggests that the substrate of NYC1 is located in chloroplasts. A database search (National Center for Biotechnology Information BLASTP; http://www.ncbi.nlm.nih.gov/blast/) revealed highly similar genes in Arabidopsis, maize, and Xerophyta humilis. In the catalytic domain, NYC1 shows 77.7% identity to NYC1 in Arabidopsis (At4g13250), 95.4% identity to NYC1 in maize, and 84.3% identity to NYC1 in Xerophyta. In rice and Arabidopsis, distinct putative SDRs highly similar to NYC1, named NOL, were found. NOL and NOL in Arabidopsis (At5g04900) showed 44.7 and 46.8% identity, respectively, to NYC1. Furthermore, an SDR similar to NYC1 was found in Ostreococcus tauri, which belongs to the Prasinophyceae, one of the most ancient groups in the green plant lineage (Derelle et al., 2006). This protein, named O. tauri NYC1-like, shows 36.9% amino acid identity to NYC1. These three NOL proteins also belong to the classical SDR family and contain all of the important amino acid residues conserved in this family (see Supplemental Figure 4 online). These NOL proteins have a Lys or an Arg at key position 15 and an Arg at key position 37, suggesting that the proteins use NADP(H) as a cofactor (Kallberg et al., 2002). They do not have a transmembrane domain and were predicted by PSORT to be localized in the stroma. A phylogenetic tree constructed using SDR domains of NYC1-related SDR sequences suggests that NYC1 and NOL proteins belong to the same clade (Figure 7C; see Supplemental Figure 4 online). Therefore, it is likely that NYC1 and NOL have the same or similar function.
Expression of NYC1 and NOL
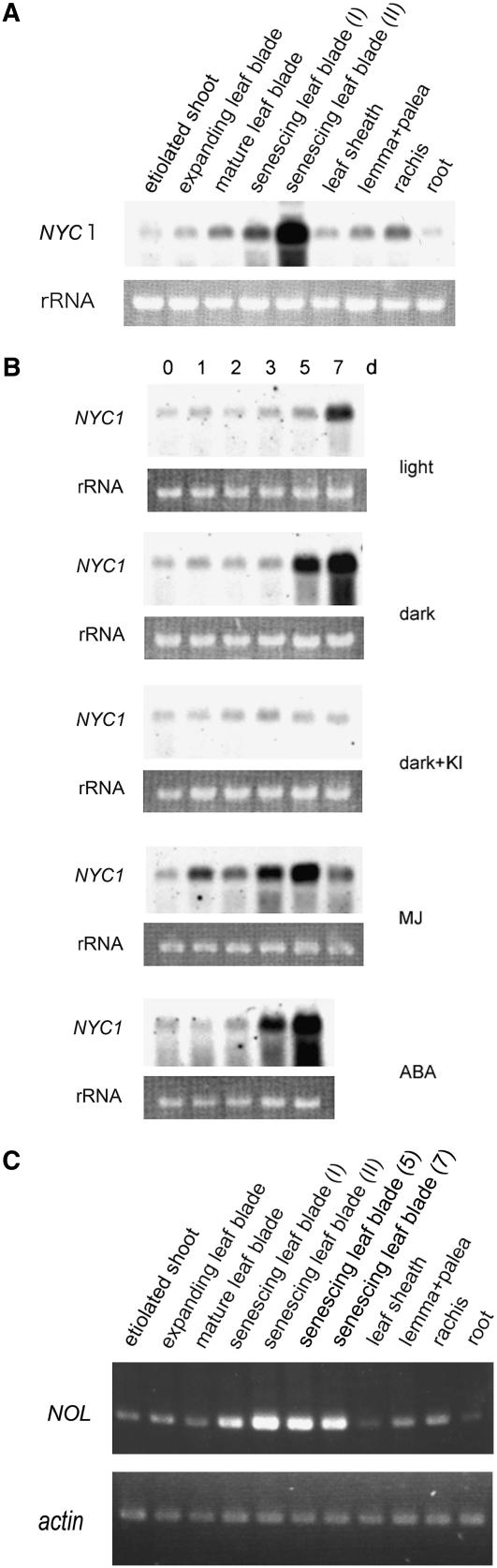
We examined the pattern of NYC1 expression in various tissues and under various conditions (Figure 8). NYC1 was expressed in most tissues to some extent but was very strongly expressed in late-senescing leaves (Figure 8A). Unexpectedly, some expression was detected in nonphotosynthetic tissues, such as etiolated shoots and roots. Plant hormones are known to affect leaf yellowing; for example, abscisic acid and methyl jasmonate enhance and cytokinin inhibits leaf yellowing in rice (see Supplemental Figure 5 online). Dark incubation also induces yellowing. On a parallel with leaf yellowing pattern, dark incubation and abscisic acid and methyl jasmonate treatment enhanced NYC1 expression in leaves, and kinetin inhibited dark enhancement of NYC1 expression (Figure 8B). Consistent with the abscisic acid inducibility of NYC1 expression, the NYC1 promoter has a dehydration-responsive element (GCCGAC) (Yamaguchi-Shinozaki and Shinozaki, 1994) at −746 bp and an abscisic acid–responsive element (CACGTGG) (Ingram and Bartels, 1996) at −720 bp from the initiation codon, although the response of NYC1 to abscisic acid appears relatively slow.
Figure 8.
Expression Patterns of NYC1 and NOL.
(A) RNA gel blot analysis of NYC1 expression in various tissues. The top panel shows the expression of NYC1. The bottom panel shows rRNA visualized with ethidium bromide staining. Senescing leaf blade (I), pale green naturally senescing leaf blade; senescing leaf blade (II), almost completely yellowing naturally senescing leaf blade.
(B) RNA gel blot analysis of NYC1 expression in dark- and hormone-treated leaves under the same condition as in (A). The top and bottom panels in the same treatment show the expression of NYC1 and rRNA visualized with ethidium bromide staining, respectively. ABA, abscisic acid; MJ, methyl jasmonate.
(C) Semiquantitative RT-PCR analysis of NOL expression in various tissues and in naturally and dark-induced senescent leaves. The bottom panel shows the expression of the actin gene as a reference. Senescing leaf blade (I), pale green naturally senescing leaf blade; senescing leaf blade (II), almost completely yellowing naturally senescing leaf blade. senescing leaf blade (5) and senescing leaf blade (7) show dark-induced senescent leaf blades at 5 and 7 d.
We examined the expression of NOL by semiquantitative RT-PCR (Figure 8C). NOL was strongly expressed in the naturally and dark-induced senescent leaves but was also expressed in all tissues examined, including nonphotosynthetic tissues. This expression pattern is very similar to that of NYC1 (Figure 8A).
NOL Has Chlorophyll b Reductase Activity
As discussed above, we think that the retention of high chlorophyll content in nyc1 during senescence is due to retention of LHCII. On the other hand, because chlorophyll b plays an important role in LHCII stability, NYC1 may be involved in chlorophyll b degradation (Bellemare et al., 1982; Horn and Paulsen, 2004). The first step of chlorophyll b degradation is reduction of chlorophyll b to 7-hydroxymethyl chlorophyll a, which is catalyzed by chlorophyll b reductase (Scheumann et al., 1998, 1999). In this scenario, NYC1 might be chlorophyll b reductase, and degradation of chlorophyll b by NYC1 would reduce LHCII stability, resulting in LHCII degradation.
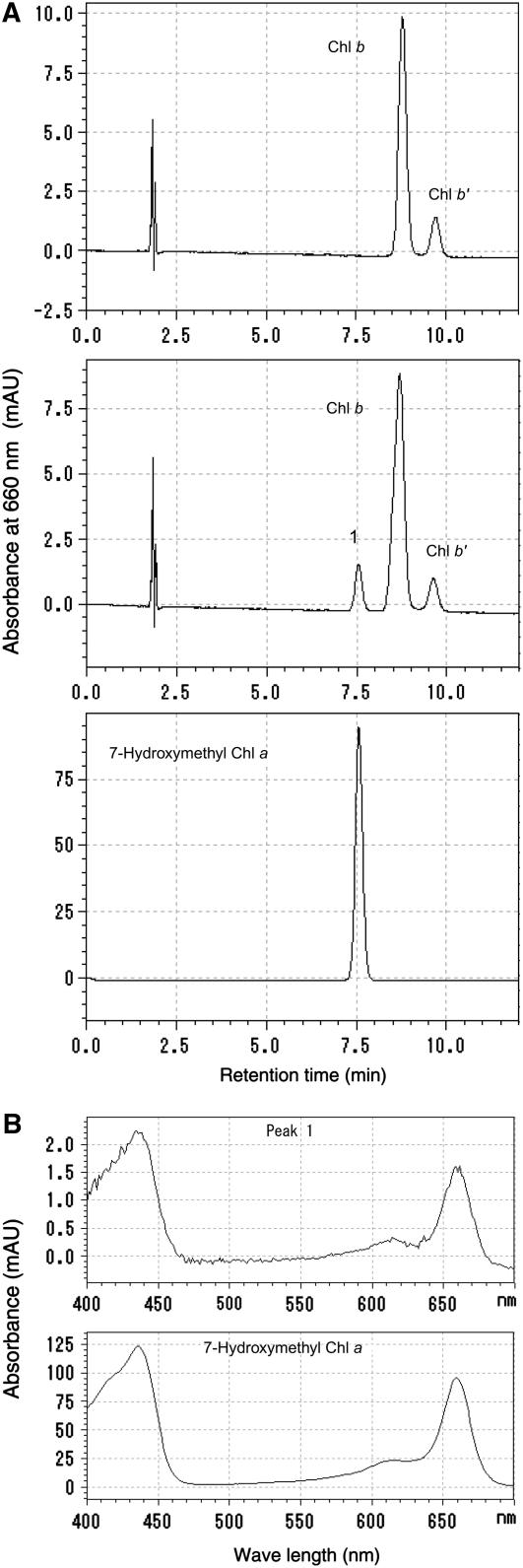
To examine this possibility, we attempted to detect chlorophyll b reductase activity of recombinant NYC1 protein produced in Escherichia coli. In spite of our efforts, we could not detect the activity of NYC1 in vitro even when we used the soluble chlorophyll b–related substance Chlide b, which could be a better substrate than chlorophyll b (Ito and Tanaka, 1996; Scheumann et al., 1998, 1999). So we tested the chlorophyll b reductase activity of NOL, the most similar SDR to NYC1 in rice. Pigment analysis by HPLC revealed that the peak corresponding to 7-hydroxymethyl chlorophyll a (Figures 9A, peak 1, and 9B) appeared when the recombinant NOL protein–containing extract was incubated with chlorophyll b, indicating that NOL has chlorophyll b reductase activity. We detected the chlorophyll b reductase activity of At NOL as well (H. Ito, unpublished data). These results suggest that NOL is a chlorophyll b reductase.
Figure 9.
Assay of Chlorophyll b Reductase Activity of NOL.
(A) Pigments extracted from the reaction mixture incubated with chlorophyll b and crude extract from E. coli harboring empty (top panel) or recombinant NOL-expressing (middle panel) plasmids were subjected to HPLC. Peak 1 newly appeared in the incubation of chlorophyll b and NOL. Bottom panel, 7-hydroxymethyl chlorophyll a prepared from chlorophyll b using NaBH4. The absorbance at 660 nm is shown.
(B) Top and bottom panels show the absorption spectrum of peak 1 in the middle panel in (A) and that of 7-hydroxymethyl chlorophyll a in the bottom panel in (A).
DISCUSSION
Pigments and LHCII Stability in nyc1
The green gel and protein gel blot analyses revealed that LHCII is selectively retained in nyc1 during senescence. This observation explains many characteristics of nyc1.
nyc1 does not have a significant effect on chlorophyll degradation in the cao background, in which little or no LHCII accumulated, suggesting that LHCII is required for the stay-green phenotype in nyc1 (Bellemare et al., 1982; Morita et al., 2005). In the late stage of senescence, retention of chlorophyll b was obvious in nyc1, but the chlorophyll a content in nyc1 was also higher than in the wild type, and the chlorophyll a/b ratio in the senescent nyc1 leaf was nearly 1. We think that this value reflects the fact that only LHCII, which contains a complex of eight chlorophyll a and six chlorophyll b (Liu et al., 2004), is retained in nyc1 in the late stage of senescence. nyc1 retained more neoxanthin and lutein, which are also components of LHCII, than did the wild type during senescence. The collective pigment contents in nyc1 were well correlated with the prolonged stability of LHCII in this mutant.
Interestingly, the stabilization of LHCPs in nyc1 was limited to particular members of the LHC family. At least three Lhca proteins and Lhcb5 were not stable in nyc1 during senescence. This is consistent with the observation that chlorophyll b content was reduced slightly, and in the green gel analysis, the LHCP dimer band, whose major component is thought to be Lhca, disappeared during senescence in nyc1. The degradation of LHCPs is differently regulated among members of the LHC family.
Grana Stack and LHCII Stability in nyc1
The retention of LHCII seemed to affect grana stack stability during senescence in nyc1. LHCII is thought to play an important role in the formation or maintenance of grana stacks (Allen and Forsberg, 2001). A model predicts that the interaction between positively charged N-terminal regions and the negatively charged stromal surface of the LHCII trimer contributes to the cohesion of thylakoid membranes in grana stacks (Standfuss et al., 2005). In nyc1, dissociation of grana stacks was repressed in the late stage of senescence, suggesting that the degradation of LHCII is necessary for the dissociation of grana stacks during senescence. The retention of grana stacks also implies that a substantial part of thylakoid lipids were not degraded.
On the other hand, the complex structure of grana stacks was revealed very recently (Shimoni et al., 2005). It is possible that high connectivity between thylakoid membranes in grana might contribute physically to the maintenance of stacking. Our observation suggests an important role of LHCII in the proper degradation of stacking of the thylakoid membrane. Probably both LHCII-mediated cohesion and physical connection contribute to the maintenance of thylakoid membrane stacking. Furthermore, the fused very large grana were observed at the very late stage of leaf senescence in nyc1. This observation might provide some clues to understand how stacking of grana is regulated, including the influence of entropy (Kim et al., 2005).
SDR-Mediated Regulation of LHCII Degradation
Pigment-bound LHCII is resistant to trypsin digestion, suggesting that a particular protease that degrades pigment-bound LHCII acts in the process of LHCII degradation or that pigments are removed before LHCII degradation (Paulsen et al., 1993). Recently, a metalloprotease, At FtsH6, was reported as a candidate LHCII protease (Żelisko et al., 2005). Other LHCII protease candidates have also been reported (Yang et al., 2000; Georgakopoulos et al., 2002). The observation that NYC1 encodes an SDR, not a protease, raises the hypothesis that the oxidation/reduction of a particular substance decreases LHCII stability, resulting in LHCII degradation by proteases. Because chlorophyll b is directly involved in LHCII stability (Bellemare et al., 1982; Horn and Paulsen, 2004) and the first step of chlorophyll b degradation is the reduction of chlorophyll b into 7-hydroxymethyl chlorophyll a, NYC1 might encode chlorophyll b reductase and might regulate LHCII degradation. Chlorophyll b reductase activity requires NADPH as a cofactor and is located in the thylakoid membrane (Scheumann et al., 1998, 1999). These characteristics of chlorophyll b reductase are consistent with those of NYC1. NYC1 is predicted to be an SDR requiring NADPH as a cofactor and is located in chloroplasts, possibly in the thylakoid membrane.
This hypothesis is supported by the observation that NOL, an SDR closely related to NYC1, exhibited chlorophyll b reductase activity in vitro. NYC1 and NOL have similar SDR domains with the conserved amino acid residues important for the oxidoreductase activity (see Supplemental Figure 4 online). A phylogenetic analysis suggests that NYC1 and NOL belong to the same clade (Figure 7C). Taking into account the phenotype of the nyc1 mutant, we suggest that NYC1 encodes another chlorophyll b reductase, although we were not able to detect the activity with the recombinant protein produced in E. coli. Empirically, overexpression of a membrane protein in an active form in E. coli is difficult. Attempts to produce various membrane proteins in an active form in E. coli often fail, partly due to the formation of insoluble aggregates and to low-level expression (Wang et al., 2003). In this study, we were able to obtain a functional form of NOL, which does not contain a predicted membrane-spanning region, with an E. coli expression system. Production of a functional NYC1 protein is a future challenge using a different expression system such as recently developed cell-free systems (Klammt et al., 2004).
If NYC1 encodes chlorophyll b reductase, this implies that degradation of chlorophyll b might be an important regulatory point in the degradation of LHCII during senescence. As discussed above, LHCII may be digested by proteases only after chlorophyll b in LHCII is degraded. Furthermore, stabilization of LHCII may inhibit the dissociation of grana stacks and the degradation of thylakoid membranes during senescence. Importantly, the nyc1 phenotype was observed in the presence of another isoform of chlorophyll b reductase, NOL, in senescent leaves. It is possible that this observation is due to the functional divergence of NYC1 and NOL. For example, NYC1 may interact with LHCII directly in the thylakoid membrane and reduce chlorophyll b to 7-hydroxymethyl chlorophyll a. NOL may reduce the chlorophyll b of LHCI and Lhcb5 and/or free chlorophyll b in the stroma. It may be interesting to investigate the function of O. tauri NYC1-like, because no SDR with transmembrane domains belonging to the NYC1–NOL clade is found in O. tauri, even though its complete genome sequence has been revealed (Derelle et al., 2006).
We suggest that the reduction of chlorophyll b in LHCII by NYC1 is a key step in the regulation of several aspects of senescence, including the LHCII and thylakoid membrane degradation. It will be intriguing to investigate further how the predicted chlorophyll b reductases NOL and NYC1 function in senescence and other physiological conditions in which the chlorophyll cycle operates.
METHODS
Plant Materials
nyc1-1 was isolated from a rice (Oryza sativa cv Koshihikari) M2 population derived from seeds treated with ethylmethane sulfonate according to Iida et al. (1997). nyc1-2 was a tissue culture–derived mutant of cv Nipponbare obtained from the Rice Genome Resource Center, National Institute of Agrobiological Sciences, Japan. The chlorophyll b–deficient mutant, cao-2, was described by Morita et al. (2005).
The youngest fully expanded leaves were used for all analyses except that of tissue-specific expression. Detached leaves were incubated in 3 mM MES buffer, pH 5.8, at 27°C. A concentration of 50 μM abscisic acid, methyl jasmonate, or kinetin was added to the MES solution in the phytohormone treatments.
Analysis of Photosynthetic Pigments, Photochemical Efficiency, and Membrane Ion Leakage
For pigment extraction, plant tissues of the same fresh weight were ground in a mortar with liquid nitrogen and extracted with the same volume of 80% acetone. The HPLC apparatus used for the analysis was equipped with a Symmetry C8 column (Waters) and a photodiode-array detector (SPD-M10A; Shimadzu) according to Zapata et al. (2000) and Tanaka et al. (2003). The chlorophyll a and chlorophyll b contents were determined according to Porra et al. (1989). Fv/Fm was measured with an OS1-FL fluorometer (Opti-Sciences) according to the manufacturer's instructions. For the measurement of membrane ion leakage, three leaf discs of 6 mm diameter floated on 500 μL of water were incubated in the dark at 27°C. The conductivity was measured with a Twin Cond B-173 conductivity meter (Horiba). Leaf discs of naturally senescent leaves were incubated for 12 h at 27°C under light.
Protein and RNA Analyses
Green gel analysis, SDS-PAGE analysis, and protein gel blot analysis were performed according to Morita et al. (2005). For green gel analysis, 100 mg (fresh weight) of leaves was extracted with 250 μL of the extraction buffer (0.3 M Tris, pH 6.8, 1% SDS, 2% Triton X-100, and 10% glycerol). For SDS-PAGE and protein gel blot analyses, 100 mg (fresh weight) of leaves was extracted with 4 mL of 1× SDS buffer (62.5 mM Tris, pH 6.8, 2.5% SDS, 5% mercaptoethanol, and 10% glycerol). Antibodies against Lhca, Lhcb, and PsbB were purchased from Agrisera. Anti-PsaF antibody was provided by Y. Takahashi (Okayama University). Total RNA was prepared with an RNeasy kit (Qiagen). In the RNA gel blot analysis, 5 μg of total RNA was electrophoresed on a 1.2% agarose gel and transferred to a nylon membrane. mRNA was detected by a digoxigenin labeling system (Roche Diagnostics).
In the semiquantitative RT-PCR, 15 ng of cDNA was used for a 25-μL PCR. The PCR conditions were 1 min of denaturing at 94°C, followed by 27 cycles of 94°C for 30 s, 55°C for 1 min, and 72°C for 30 s. The linear amplification was confirmed with preliminary experiments including 25, 27, and 29 cycles of amplification. PCR products (5 μL) were run on a 1.5% gel and stained with ethidium bromide. The primers used for NOL and actin amplification were 5′-GAAAGGGTAGAATCTGCGGTG-3′/5′-CTGCAGAGATTTTGTAAGGTG-3′ and 5′-TCCATCTTGGCATCTCTCAG-3′/5′-GTACCCGCATCAGGCATCTG-3′, respectively.
Transmission Electron Microscopy
Detached leaves were soaked in primary fixation buffer (2.5% glutaraldehyde in 100 mM cacodylate buffer, pH 7.4) and postfixed for 2 h in secondary fixation buffer (1% OsO4 in 100 mM cacodylate buffer, pH 7.4). Specimens were subsequently stained and observed as described previously (Tanaka et al., 2003).
Map-Based Cloning
About 1500 F2 seedlings of a cross between nyc1-1 and indica cv Kasalath were examined for the stay-green phenotype. Detached leaves were incubated at 27°C in the dark for 8 d, and the greenness was judged by eye. The sequences of primers used are 5′-CAACCTAGAAGAAGATATTACTCCTCTTAG-3′ and 5′-TTCTTGTTCTTCTTGATATATAGATC-3′ (HinfI) for the dCAPS marker P0443E07.24 and 5′-ATGTCGTCGCCGTCGTCGCCGCCG-3′ and 5′-AGCCGATTCTTGGGATCATACAACAGC-3′ (BspT104 I) for the CAPS marker P0452F10.5. The 8.2-kb MluI-AflII genomic fragment, which contains the entire NYC1 gene, was cloned into pTH2 binary vector. nyc1-2 was transformed with this construct by Agrobacterium tumefaciens–mediated transformation according to Kusaba et al. (2003).
Construction and Visualization of GFP Fusion Protein
The plastid-localized RFP construct with the transit peptide from rice ribosomal protein S9 is the RFP derivative of Os PRS9TP-GFP (Arimura et al., 1999). NYC1-GFP was constructed by insertion of the NYC1 cDNA fragment encoding the first 67 amino acids into the XbaI-BamHI site of pJ4-GFP (Igarashi et al., 2001). Particle bombardment was performed with a PDS-1000/He particle gun (Bio-Rad). Three micrograms of plasmid precipitated onto 1.0-μm gold beads was introduced into onion (Allium cepa) epidermal cells. Twenty-four hours after bombardment, protein expression was observed and images were captured with a BZ-8000 fluorescence microscope (Keyence). The filters used were OP-66836 for GFP and OP-66838 for RFP.
Assay of Chlorophyll b Reductase Activity
To produce NOL recombinant protein, we amplified a NOL cDNA fragment (from the 55th amino acid to the C-terminal end) by RT-PCR from mature rice leaf RNA. Amplified fragments were inserted into the pGEX-3X plasmid (GE Healthcare) between the BamHI and EcoRI sites to produce a glutathione S-transferase fusion protein. A culture of Escherichia coli (BL21 DE3) containing either NOL or empty pGEX-3X plasmid was grown in Luria-Bertani broth containing 60 μg/mL ampicillin. Production of the recombinant protein was induced for 3 h at 37°C by the addition of isopropyl-β-d-thiogalactoside at a final concentration of 0.4 mM.
For the assay of enzymatic activity, 1.0 mL of E. coli culture (OD600 = 1.0) was harvested by centrifugation, and the pellet was suspended in 50 μL of BugBuster (Invitrogen). NADPH (1 μL) was added to the suspension at a final concentration of 1 mM. The suspension was incubated with 25 nmol of chlorophyll b at 37°C for 1 h. Pigments were analyzed by HPLC as described above. In this analysis, we used a VP-ODS column (Shimadzu) and methanol solvent (1.7 mL/min). 7-Hydroxymethyl chlorophyll a was prepared from chlorophyll b by reduction with NaBH4 (Holt, 1959).
Phylogenetic Analysis
Amino acid sequences of the conserved SDR domain were aligned using ClustalW (http://www.ddbj.nig.ac.jp/search/clustalw-j.html) and then manually adjusted. The phylogenetic tree was constructed using Phylip3.66 (http://evolution.genetics.washington.edu/phylip.html) and the alignment shown in Supplemental Figure 4 online. Bootstrap values were calculated from 1000 trials.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: NYC1, AB255025; NOL, AB255026; NOL in Arabidopsis, AB255027 (At5g49000); NYC1 in Arabidopsis, AB255028 (At4g13250); NYC1 in Xerophyta, AY566698; Zm NYC1, AY105189; Fj SDR, EAS60331; Hh SDR, EAR45242; Ct SDR, AAM72376; Pa SDR, EAN22525; Fj SDR, EAS60331; Pp SDR, EAN26287; Ot SDR, CAL55830; Ot NOL, CAL50202; Ha SDR, ZP01424628; At 3-oxoacyl, AAG40337 (At1g24360); Os 3-oxoacyl, CAE04594; Ll 3-ketoacyl, NP266930.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Physiological Changes in nyc1 Leaves during Natural Senescence.
Supplemental Figure 2. Chlorophyll Degradation in nyc1-2 cao-2.
Supplemental Figure 3. Complementation Tests of nyc1.
Supplemental Figure 4. Multiple Alignment of SDR Sequences.
Supplemental Figure 5. Dark and Hormonal Regulation of Leaf Senescence in Rice.
Supplementary Material
Acknowledgments
We thank Nobuhiro Tsutsumi, Yousuke Takahashi, and Yasuo Niwa for providing RFP and GFP constructs; Yuichiro Takahashi for providing anti-PsaF antibody; Mitsue Tokutomi-Miyao, Momoyo Ito, Koichiro Ushijima, Hidenori Sassa, Toshio Takyu, and Mikio Nakazono for helpful suggestions; Junko Kishimoto for helpful assistance; Yuko Ohashi and Ichiro Mitsuhara for providing pTH2 binary vector; and Yasuo Nagato for encouragement. This work was supported by grants from the Ministry of Agriculture, Forestry, and Fisheries of Japan (Rice Genome Project IP-1011) and a grant for nuclear research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Makoto Kusaba (akusaba@mail.ecc.u-tokyo.ac.jp).
Online version contains Web-only data.
References
- Allen, J.F., and Forsberg, J. (2001). Molecular recognition in thylakoid structure and function. Trends Plant Sci. 6 317–326. [DOI] [PubMed] [Google Scholar]
- Arimura, S., Takusagawa, S., Hatano, S., Nakazono, M., Hirai, A., and Tsutsumi, N. (1999). A novel plant nuclear gene encoding chloroplast ribosomal protein S9 has a transit peptide related to that of rice chloroplast ribosomal protein L12. FEBS Lett. 450 231–234. [DOI] [PubMed] [Google Scholar]
- Bellemare, G., Bartlett, S.G., and Chua, N.H. (1982). Biosynthesis of chlorophyll a/b-binding polypeptides in wild type and the chlorina-f2 mutant of barley. J. Biol. Chem. 257 7762–7767. [PubMed] [Google Scholar]
- Derelle, E., et al. (2006). Genome analysis of the smallest free-living eukaryote Ostreococcus tauri unveils many unique features. Proc. Natl. Acad. Sci. USA 103 11647–11652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espineda, C.E., Linford, A.S., Devine, D., and Brusslan, J.A. (1999). The AtCAO gene, encoding chlorophyll a oxygenase, is required for chlorophyll b synthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 96 10507–10511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgakopoulos, J.H., Sokolenko, A., Arkas, M., Sofou, G., Herrman, R.G., and Argyroudi-Akoyunoglou, J.H. (2002). Proteolytic activity against the light-harvesting complex and the D1/D2 core proteins of photosystem II in close association to the light-harvesting complex II trimer. Biochim. Biophys. Acta 1556 53–64. [DOI] [PubMed] [Google Scholar]
- Gray, J., Close, P.S., Briggs, S.P., and Johal, G.S. (1997). A novel suppressor of cell death in plants encoded by the Lls1 gene of maize. Cell 89 25–31. [DOI] [PubMed] [Google Scholar]
- Guiamét, J.J., Tyystjärvim, E., John, I., Kairavuo, M., Pichersky, E., and Noodén, L.D. (2002). Photoinhibition and loss of photosystem II reaction centre proteins during senescence of soybean leaves. Enhancement of photoinhibition by the ‘stay-green’ mutation cytG. Physiol. Plant. 115 468–478. [DOI] [PubMed] [Google Scholar]
- Holt, A.S. (1959). Reduction of chlorophyllides, chlorophylls and chlorophyll derivatives by sodium borohydride. Plant Physiol. 34 310–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn, R., and Paulsen, H. (2004). Early steps in the assembly of light-harvesting chlorophyll a/b complex. J. Biol. Chem. 279 44400–44406. [DOI] [PubMed] [Google Scholar]
- Hörtensteiner, S. (2006). Chlorophyll degradation during senescence. Annu. Rev. Plant Biol. 57 55–77. [DOI] [PubMed] [Google Scholar]
- Igarashi, D., Ishida, S., Fukazawa, J., and Takahashi, Y. (2001). 14-3-3 proteins regulate intracellular localization of the bZIP transcriptional activator RSG. Plant Cell 13 2483–2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida, S., Kusaba, M., and Nishio, T. (1997). Mutants lacking glutelin subunits in rice: Mapping and combination of mutated glutelin genes. Theor. Appl. Genet. 94 177–183. [Google Scholar]
- Ingram, J., and Bartels, D. (1996). The molecular basis of dehydration tolerance in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47 377–403. [DOI] [PubMed] [Google Scholar]
- Ito, H., and Tanaka, A. (1996). Determination of the activity of chlorophyll b to chlorophyll a conversion during greening of etiolated cucumber cotyledons by using pyrochlorophyllide b. Plant Physiol. Biochem. 34 35–40. [Google Scholar]
- Ito, H., Tanaka, Y., Tsuji, H., and Tanaka, A. (1993). Conversion of chlorophyll b to chlorophyll a by isolated cucumber etioplasts. Arch. Biochem. Biophys. 306 148–151. [DOI] [PubMed] [Google Scholar]
- Kallberg, Y., Oppermann, U., Jornvall, H., and Persson, B. (2002). Short-chain dehydrogenase/reductases (SDRs). Eur. J. Biochem. 269 4409–4417. [DOI] [PubMed] [Google Scholar]
- Kim, E.H., Chow, W.S., Horton, P., and Anderson, J.M. (2005). Entropy-assisted stacking of thylakoid membranes. Biochim. Biophys. Acta 1708 187–195. [DOI] [PubMed] [Google Scholar]
- Klammt, C., Löhr, F., Schäfer, B., Haase, W., Dötsch, V., Rüterjans, H., Glaubitz, C., and Bernhard, F. (2004). High level cell-free expression and specific labeling of integral membrane proteins. Eur. J. Biochem. 271 568–580. [DOI] [PubMed] [Google Scholar]
- Kusaba, M., Miyahara, K., Iida, S., Fukuoka, H., Takano, T., Sassa, H., Nishimura, M., and Nishio, T. (2003). Low glutelin content1: A dominant mutation that suppresses the glutelin multigene family via RNA silencing in rice. Plant Cell 15 1455–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, P.O., Woo, H.R., and Nam, H.G. (2003). Molecular genetics of leaf senescence in Arabidopsis. Trends Plant Sci. 8 272–278. [DOI] [PubMed] [Google Scholar]
- Liu, Z., Yan, H., Wang, K., Kuang, T., Zhang, J., Gui, L., An, X., and Chang, W. (2004). Crystal structure of spinach major light-harvesting complex at 2.72 Å resolution. Nature 428 287–292. [DOI] [PubMed] [Google Scholar]
- Matile, P. (2000). Biochemistry of Indian summer: Physiology of autumnal leaf coloration. Exp. Gerontol. 35 145–158. [DOI] [PubMed] [Google Scholar]
- Morita, R., Kusaba, M., Yamaguchi, H., Amano, E., Miyao, A., Hirochika, H., and Nishimura, M. (2005). Characterization of chlorophyllide a oxygenase (CAO) in rice. Breed. Sci. 55 361–364. [Google Scholar]
- Nagata, N., Tanaka, R., Satoh, S., and Tanaka, A. (2005). Identification of a vinyl reductase gene for chlorophyll synthesis in Arabidopsis thaliana and implications for the evolution of Prochlorococcus species. Plant Cell 17 233–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh, M.H., Moon, Y.H., and Lee, C.H. (2003). Increased stability of LHCII by aggregate formation during dark-induced leaf senescence in the Arabidopsis mutant, ore10. Plant Cell Physiol. 44 1368–1377. [DOI] [PubMed] [Google Scholar]
- Oster, U., Tanaka, R., Tanaka, A., and Rüdiger, W. (2000). Cloning and functional expression of the gene encoding the key enzyme for chlorophyll b biosynthesis (CAO) from Arabidopsis thaliana. Plant J. 21 305–310. [DOI] [PubMed] [Google Scholar]
- Paulsen, H., Finkenzeller, B., and Kühlein, N. (1993). Pigments induce folding of light-harvesting chlorophyll a/b-binding protein. Eur. J. Biochem. 215 809–816. [DOI] [PubMed] [Google Scholar]
- Porra, R.J., Thompson, W.A., and Kriedemann, P.E. (1989). Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: Verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta 975 384–394. [Google Scholar]
- Pružinská, A., Tanner, G., Anders, I., Roca, M., and Hörtensteiner, S. (2003). Chlorophyll breakdown: Pheophorbide a oxygenase is a Rieske-type iron-sulfur protein, encoded by the accelerated cell death 1 gene. Proc. Natl. Acad. Sci. USA 100 15259–15264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruban, A.V., Lee, P.J., Wentworth, M., Young, A.J., and Horton, P. (1999). Determination of the stoichiometry and strength of binding of xanthophylls to the photosystem II light harvesting complexes. J. Biol. Chem. 274 10458–10465. [DOI] [PubMed] [Google Scholar]
- Rüdiger, W. (2002). Biosynthesis of chlorophyll b and the chlorophyll cycle. Photosynth. Res. 74 187–193. [DOI] [PubMed] [Google Scholar]
- Scheumann, V., Schoch, S., and Rüdiger, W. (1998). Chlorophyll a formation in the chlorophyll reductase reaction requires reduced ferredoxin. J. Biol. Chem. 273 35102–35108. [DOI] [PubMed] [Google Scholar]
- Scheumann, V., Schoch, S., and Rüdiger, W. (1999). Chlorophyll b reduction senescence of barley seedlings. Planta 209 364–370. [DOI] [PubMed] [Google Scholar]
- Shimoni, E., Rav-Hon, O., Ohad, I., Brumfeld, V., and Reich, Z. (2005). Three-dimensional organization of higher-plant chloroplast thylakoid membranes revealed by electron tomography. Plant Cell 17 2580–2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Standfuss, J., van Scheltinga, A.C.T., Lamborghini, M., and Kühlbrandt, W. (2005). Mechanisms of photoprotection and nonphotochemical quenching in pea light-harvesting complex at 2.5 Å resolution. EMBO J. 24 919–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, A., Ito, H., Tanaka, R., Tanaka, N.K., Yoshida, K., and Okada, K. (1998). Chlorophyll a oxygenase (CAO) is involved in chlorophyll b formation from chlorophyll a. Proc. Natl. Acad. Sci. USA 95 12719–12723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, R., Hirashima, M., Satoh, S., and Tanaka, A. (2003). The Arabidopsis-accelerated cell death gene ACD1 is involved in oxygenation of pheophorbide a: Inhibition of the pheophorbide a oxygenase activity does not lead to the “stay-green” phenotype in Arabidopsis. Plant Cell Physiol. 44 1266–1274. [DOI] [PubMed] [Google Scholar]
- Thomas, H., and Howarth, J. (2000). Five ways to stay green. J. Exp. Bot. 51 329–337. [DOI] [PubMed] [Google Scholar]
- Thomas, H., Morgan, W.G., Thomas, A.M., and Ougham, H.J. (1999). Expression of the stay-green character introgressed into Lolium temulentum Ceres from a senescence mutant of Festuca pratensis. Theor. Appl. Genet. 99 92–99. [Google Scholar]
- Tsuchiya, T., Ohta, H., Okawa, K., Iwamatsu, A., Shimada, H., Masuda, T., and Takamiya, K. (1999). Cloning of chlorophyllase, the key enzyme in chlorophyll degradation: Finding of a lipase motif and the induction by methyl jasmonate. Proc. Natl. Acad. Sci. USA 21 15326–15367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, D.-N., Safferling, M., Lemieux, M.J., Griffith, H., Chen, Y., and Li, X.-D. (2003). Practical aspects of overexpressing bacterial secondary membrane transporters for structural studies. Biochim. Biophys. Acta 1610 23–36. [DOI] [PubMed] [Google Scholar]
- Wüthrich, K.L., Bovet, L., Hunziker, P.E., Donnison, I.S., and Hörtensteiner, S. (2000). Molecular cloning, functional expression and characterization of RCC reductase involved in chlorophyll catabolism. Plant J. 21 189–198. [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki, K., and Shinozaki, K. (1994). A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 6 251–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, D.-H., Paulsen, H., and Andersson, B. (2000). The N-terminal domain of the light-harvesting chlorophyll a/b-binding protein complex (LHCII) is essential for its acclimative proteolysis. FEBS Lett. 466 385–388. [DOI] [PubMed] [Google Scholar]
- Yang, M., Wardzala, E., Johal, G.S., and Gray, J. (2004). The wound-inducible Lls1 gene from maize is an orthologue of the Arabidopsis Acd1 gene, and the LLS1 protein is present in non-photosynthetic tissues. Plant Mol. Biol. 54 175–191. [DOI] [PubMed] [Google Scholar]
- Zapata, M., Rodrígues, F., and Garrido, J.L. (2000). Separation of chlorophylls and carotenoids from marine phytoplankton: A new HPLC method using a reversed phase C8 column and pyridine-containing mobile phases. Mar. Ecol. Prog. Ser. 195 29–45. [Google Scholar]
- Żelisko, A., García-Lorenzo, M., Jackowski, G., Jansson, S., and Funk, C. (2005). AtFtsH6 is involved in the degradation of the light-harvesting complex II during high-light acclimation and senescence. Proc. Natl. Acad. Sci. USA 102 13699–13704. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.