Abstract
Cyclin D (CYCD) plays an important role in cell cycle progression and reentry in response to external signals. Here, we demonstrate that Arabidopsis thaliana CYCD4 is associated with specific cell divisions in the hypocotyl. We observed that cycd4 T-DNA insertion mutants had a reduced number of nonprotruding cells and stomata in the hypocotyl epidermis. Conversely, CYCD4 overexpression enhanced cell division in nonprotruding cell files in the upper region of the hypocotyls, where stomata are usually formed in wild-type plants. The overproliferative cells were of stomatal lineage, which is marked by the expression of the TOO MANY MOUTHS gene, but unlike the meristemoids, most of them were not triangular. Although the phytohormone gibberellin promoted stomatal differentiation in the hypocotyl, inhibition of gibberellin biosynthesis did not prevent CYCD4 from inducing cell division. These results suggested that CYCD4 has a specialized function in the proliferation of stomatal lineage progenitors rather than in stomatal differentiation. We propose that CYCD4 controls cell division in the initial step of stomata formation in the hypocotyl.
INTRODUCTION
During postembryonic development in plants, organs are formed not only from undifferentiated cells in the meristem but also from differentiated cells; for example, lateral roots are derived from root pericycle cells (Charlton, 1996). The onset of lateral root formation coincides with a series of anticlinal asymmetric divisions in the xylem pole pericycle (Malamy and Benfey, 1997a). Another example is stomata that are generated from differentiated protodermal cells of aerial organs. In Arabidopsis thaliana, stomatal development requires three different precursor cells, namely, the meristemoid mother cell (MMC), the meristemoid, and the guard mother cell (GMC) (Nadeau and Sack, 2003). The MMC divides asymmetrically to produce a small triangular meristemoid and a neighbor cell. Some meristemoids directly differentiate into GMCs, while others further divide one to three times before they are converted into GMCs (Geisler et al., 2000). GMCs always divide symmetrically to produce two guard cells (GCs) that surround the pore.
For continuous functioning of the meristematic organization and the formation of new organs, cell division must be stringently controlled by machinery that regulates the cell cycle. Signaling pathways that regulate cell cycle progression ultimately converge to control the activity of cyclin-dependent protein kinases (CDKs). The activity and substrate specificity of CDKs depend on their binding to cyclins (Morgan, 1997). In plants, A-, B-, and D-type cyclins are assumed to play a major role in cell cycle control (de Jager et al., 2005). The A- and B-type cyclins are expressed from the S to the M phase, and they control DNA replication, the G2/M transition, and mitosis; the D-type cyclin is assumed to be a sensor of external signals and to play an essential role in cell cycle progression and in the reentry of quiescent cells into the cell cycle. In animals, cyclin D forms active kinase complexes with CDK4 and CDK6, which phosphorylate the retinoblastoma (Rb) protein and inactivate its suppressor function on the transcription factors E2F and DP; this leads to progression from the G1 to the S phase (Harbour and Dean, 2000). Recent studies have revealed that a similar Rb/E2F/DP pathway is also conserved in plants (Nakagami et al., 1999; Shen, 2002). Mitogen-induced signals stimulate cyclin D-CDK complexes at multiple levels, including those of gene transcription, translation, protein stability, and assembly and import of these complexes into the nucleus (Sherr and Roberts, 2004). Subsequently, active cyclin D-CDK complexes promote progression from the G1 to the S phase and thus enhance cell proliferation.
Plant cyclin D (CYCD) has been classified into the following six groups based on similarities in amino acid sequences: CYCD1, CYCD2/CYCD4, CYCD3, CYCD5, CYCD6, and CYCD7 (Wang et al., 2004). Recent studies have demonstrated the promotive effect of CYCD expression on cell division in plants; for example, overexpression of Antirrhinum majus CYCD1;1 in tobacco (Nicotiana tabacum) BY-2 cells accelerated cell entry into the S phase and into mitosis, and it was associated with CDK activity on histone H1 and Rb proteins (Koroleva et al., 2004). Overexpression of Arabidopsis CYCD2;1 in tobacco plants stimulated meristematic division by reducing the G1 phase (Cockcroft et al., 2000). Menges et al. (2006) reported that constitutive expression of CYCD3;1 in Arabidopsis cell suspension cultures reduced the proportion of G1-phase cells but extended the G2 phase, suggesting that CYCD3;1 dominantly drives the G1/S transition. In planta overexpression of CYCD3;1 caused hyperproliferation of leaf cells, inhibited cell differentiation, and reduced DNA ploidy, indicating that this gene may play an important role in the switch from cell proliferation to differentiation (Dewitte et al., 2003). However, the manner in which each CYCD is associated with the temporal and spatial control of cell division in the context of plant development is unclear. Recently, Masubelele et al. (2005) reported that during seed germination, transcription of Arabidopsis CYCD1;1 and CYCD4;1 was upregulated prior to the activation of cell division in the root meristem. In cycd1;1 and cycd4;1 mutants, the onset of cell proliferation was significantly delayed, while overexpression of CYCD1;1 resulted in a rapid increase in the number of cycling cells, which led to accelerated germination.
Arabidopsis has 10 CYCD genes; however, information regarding their molecular functions is quite limited. It was demonstrated that CYCD2;1 and CYCD3;1 were induced by the plant hormone cytokinin and/or sucrose at the mRNA level (Soni et al., 1995; Riou-Khamlichi et al., 1999, 2000). Furthermore, CYCD3;1 is a highly unstable protein that is degraded via the ubiquitin-proteasome pathway (Planchais et al., 2004). Healy et al. (2001) reported that CYCD2;1 and CYCD3;1 interact with CDKA;1, which is an ortholog of yeast Cdc2/Cdc28p. In Arabidopsis, CYCD4 includes two genes, namely, CYCD4;1 and CYCD4;2. We have recently demonstrated that both these CYCD4s form active kinase complexes with CDKA;1, whereas only CYCD4;1 can bind and activate CDKB2;1, which is a plant-specific CDK that is expressed from the G2 to the M phase (Kono et al., 2003, 2006). CYCD4;2 is unique in its amino acid sequence in that it lacks the Rb binding motif and the PEST sequence that are hallmarks of CYCDs. However, it was able to rescue G1 cyclin-deficient yeast, and its overexpression in hypocotyl explants caused faster callus induction than that in wild-type explants (Kono et al., 2006). This indicates that CYCD4;2 promotes cell division regardless of its unique amino acid sequence. Here, we observed knockout mutants and plants that overexpress CYCD4 genes and demonstrated that CYCD4 is involved in stomata formation in the hypocotyl. Our results suggest a specific requirement of CYCD control of cell divisions in populations outside of the meristems.
RESULTS
cycd4 Mutants Have Fewer Nonprotruding Cells in the Hypocotyl
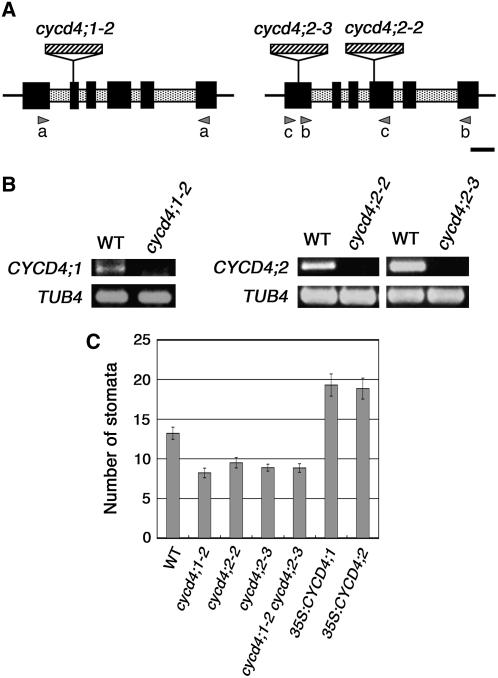
To elucidate the in planta function of CYCD4, we isolated loss-of-function mutants of the CYCD4;1 and CYCD4;2 genes from T-DNA insertion collections (Figure 1A). Since a cycd4;1 mutant has been independently isolated by Masubelele et al. (2005), our cycd4;1 mutant was designated as cycd4;1-2. In this mutant, the T-DNA was inserted into the 2nd exon of CYCD4;1 that was 288 bp downstream of the start codon. We identified three cycd4;2 mutant lines; of these, we eliminated cycd4;2-1 from our analyses due to the complexity of T-DNA integration. In cycd4;2-2 and cycd4;2-3, the T-DNA was inserted into the 4th and 1st exons of CYCD4;2 that were 616 and 141 bp downstream of the start codon, respectively. Total RNA was isolated from the wild-type and mutant seedlings that were homozygous for T-DNA insertions; the RNA was subjected to RT-PCR (Figure 1B). The cDNAs that encompassed each T-DNA insertion site could be amplified from the wild-type seedlings but not from the mutant seedlings; this indicated that the CYCD4 genes were knocked out in the mutants. Note that the expression level of CYCD4;2 in cycd4;1-2 and vice versa did not change compared with the wild-type seedlings (data not shown).
Figure 1.
T-DNA Insertion Mutants of CYCD4.
(A) Schematic diagrams of the CYCD4;1 (left) and CYCD4;2 (right) genes. Exons and introns are represented as solid and shaded boxes, respectively. The T-DNA insertion sites on each gene have been indicated. The arrowheads represent the primers that were used for RT-PCR: primer set (a), for cycd4;1-2; (b), for cycd4;2-2; and (c), for cycd4;2-3. Bar = 250 bp.
(B) Expression analysis of the mutants. RT-PCR was conducted for the total RNA obtained from the wild-type or mutant seedlings using the primers shown in (A). The amplified cDNAs were stained with ethidium bromide. TUBULIN4 (TUB4) was used as the control.
(C) Number of stomata visible on one surface of the entire hypocotyl. The stomata in the seedlings were counted at 11 d after germination. Data are presented as mean ± se of 20 samples.
All three mutants grew normally and did not exhibit any distinct macroscopic phenotype. However, in the hypocotyl, a significant reduction was observed in the number of stomata, that is, 62, 72, and 67% of the number in the wild-type plants in cycd4;1-2, cycd4;2-2, and cycd4;2-3, respectively (Figure 1C). The double mutants of cycd4;1-2 and cycd4;2-3 also exhibited fewer stomata. Introducing genomic fragments containing the CYCD4 genes restored the number of stomata visible on one surface of the entire hypocotyl (13.6 ± 2.6; mean ± se), thus demonstrating that the deficiency in stomata formation had been overcome. In leaves, both the single and double mutants did not exhibit a significant change in the size and number of epidermal cells and the number of stomata (Table 1). This suggests that CYCD4;1 and CYCD4;2 are not essential for mitotic division or stomata formation in leaves.
Table 1.
Cell Size, Cell Number, and Number of Stomata in the First Leaves of Wild-Type, cycd4 Mutant, and CYCD4-OE Seedlings
| Line | Leaf Blade Area (mm2) | Cell Area (μm2) | Cell Number | Number of Stomata | Stomatal Index (%) |
|---|---|---|---|---|---|
| Columbia wild type | 40.02 ± 2.58 | 1878 ± 98 | 21487 ± 1104 | 4125 ± 314 | 19.12 ± 0.84 |
| cycd4;1-2 | 35.41 ± 2.11 | 1865 ± 89 | 19224 ± 1240 | 3551 ± 241 | 18.38 ± 0.44 |
| cycd4;2-2 | 36.95 ± 1.34 | 1959 ± 47 | 18918 ± 745 | 3380 ± 214 | 17.82 ± 0.77 |
| cycd4;2-3 | 37.62 ± 1.61 | 2094 ± 116 | 18292 ± 795* | 3424 ± 229 | 18.59 ± 0.75 |
| cycd4;1-2 cycd4;2-2 | 37.44 ± 1.65 | 1789 ± 47 | 21244 ± 1381 | 3835 ± 264 | 18.09 ± 0.41 |
| cycd4;1-2 cycd4;2-3 | 36.96 ± 2.48 | 1732 ± 58 | 21639 ± 1646 | 3981 ± 367 | 18.19 ± 0.41 |
| 35S:CYCD4;1 (F12) | 40.55 ± 2.86 | 1295 ± 63*** | 31662 ± 2223*** | 5206 ± 317* | 16.64 ± 0.54* |
| 35S:CYCD4;2 (O3) | 40.14 ± 2.72 | 1554 ± 48** | 28785 ± 1838** | 4276 ± 350 | 16.27 ± 0.38** |
The cell size, cell number, and number of stomata were estimated for the abaxial epidermal cells. All measurements were performed using seedlings at 20 d after germination. Data are presented as mean ± se (n = 12). Significant differences between the wild-type and mutant or transgenic plants are as follows: *P < 0.05, **P < 0.01, and ***P < 0.001; the other values are not significant (P > 0.05).
The hypocotyl epidermis comprises two types of alternating cell files along the apical-basal axis, namely, protruding and nonprotruding cell files. The number of cells in the nonprotruding cell files was significantly reduced in the single and double mutants of cycd4;1 and cycd4;2 compared with the wild-type plants (Table 2). By contrast, there was no change in the number of protruding cells, that is, 15 to 16 cells in both the wild-type plants and the cycd4 mutants. We observed that the number of nonprotruding cells adjacent to the six most apical protruding cells (hereafter termed “upper nonprotruding cells”) in the cycd4 hypocotyls decreased to 60 to 70%. By contrast, the number of nonprotruding cells adjacent to the remaining 9 to 10 protruding cells near the hypocotyl-root junction was almost identical in the wild-type plants and cycd4 mutants (Table 2). Stomata are known to develop only in the upper region of nonprotruding cell files (Berger et al., 1998). In fact, we observed that almost all stomata were produced in the upper nonprotruding cells and that the number of these stomata in the cycd4 mutants was half that in the wild-type plants (Table 2). These results indicate that the destruction of CYCD4 genes reduced cell division in the nonprotruding cell files, thus reducing the number of stomata. In the hypocotyl epidermis, postembryonic cell proliferation is mainly restricted to the region associated with stomata formation (Gendreau et al., 1997). Consistent with this report, we observed that in the cycd4 mutants, cell division was compromised only in the upper nonprotruding cells where stomata are usually formed.
Table 2.
Number of Epidermal Cells and Stomata in the Hypocotyls of Wild-Type, cycd4 Mutant, and CYCD4-OE Seedlings
| Line | Protruding Cells | Nonprotruding Cells | Nonprotruding Cells (Lower) | Nonprotruding Cells (Upper) | Stomata |
|---|---|---|---|---|---|
| Columbia wild type | 15.4 ± 0.38 | 29.8 ± 0.85 | 11.9 ± 0.85 | 17.9 ± 0.63 | 2.44 ± 0.18 |
| cycd4;1-2 | 15.2 ± 0.22 | 23.6 ± 0.87*** | 11.8 ± 0.74 | 11.8 ± 0.78*** | 1.22 ± 0.15*** |
| cycd4;2-2 | 16.1 ± 0.31 | 24.1 ± 1.1*** | 11.4 ± 0.73 | 12.7 ± 0.60*** | 1.33 ± 0.17*** |
| cycd4;1-2 cycd4;2-2 | 15.7 ± 0.33 | 22.3 ± 0.69*** | 10.9 ± 0.48 | 11.4 ± 0.65*** | 1.11 ± 0.20*** |
| 35S:CYCD4;2 | 19.8 ± 0.43*** | 76.4 ± 2.3*** | 27.8 ± 1.5*** | 48.7 ± 2.4*** | 4.89 ± 0.59** |
The epidermal cells were counted in each protruding and nonprotruding cell file in the hypocotyl. The nonprotruding cells that lay adjacent to the six most apical protruding cells (upper) or those adjacent to the remaining 9 to 10 protruding cells near the hypocotyl-root junction (lower) were also counted. The stomata in the nonprotruding cell files adjacent to the six most apical protruding cells were counted. All the counting was performed using seedlings at 10 d after germination. Data are expressed as mean ± se (n = 12). Significant differences between the wild-type and mutant or transgenic plants are as follows: **P < 0.01 and ***P < 0.001; the other values are not significant (P > 0.05).
CYCD4 Expression in Seedlings
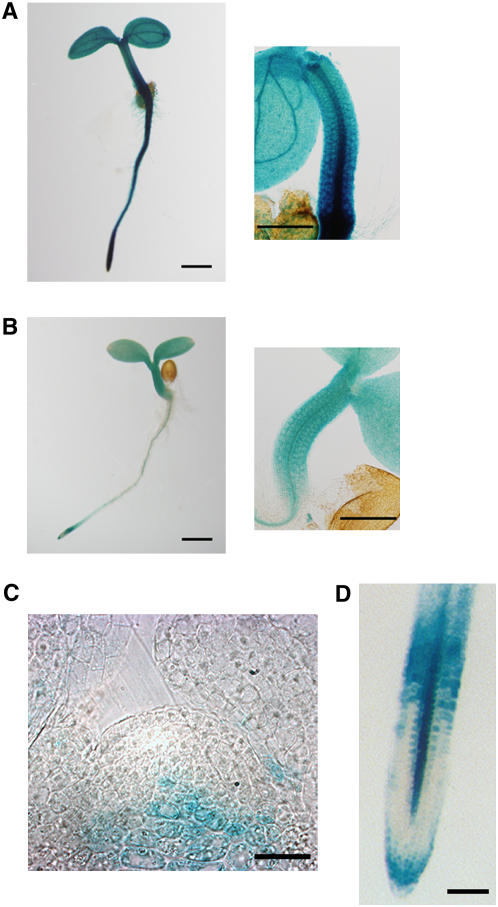
We examined the spatial expression patterns of CYCD4 during the early stage of seedling development using promoter-β-glucuronidase (GUS) fusion genes. The 1st exon and intron of each CYCD4 gene were included in the fusion constructs because these constructs produced stronger signals than those carrying only the promoter fragment that was upstream of the start codon. In fact, GUS expression driven by either one of the CYCD4 promoters was observed in the hypocotyls, although CYCD4;1 exhibited stronger expression than CYCD4;2 (Figures 2A and 2B). Furthermore, CYCD4;1 expression was observed in a broad range of tissues, including shoot and root apices, cotyledons, and vascular cylinders. Interestingly, CYCD4;2 promoter activity was not observed in the shoot and root meristems (Figures 2C and 2D).
Figure 2.
Expression of CYCD4;1 and CYCD4;2 in Arabidopsis Seedlings.
Transgenic plants harboring the ProCYCD4:CYCD4-GUS constructs were subjected to GUS staining. The 1st exon and intron of each CYCD4 gene were included into the fusion gene such that they were in frame with GUS.
(A) and (B) Seedlings harboring ProCYCD4;1:CYCD4;1-GUS (A) or ProCYCD4;2:CYCD4;2-GUS (B) at 3 d after germination. A magnified view around the hypocotyl is shown in the right panel. Bars = 0.5 mm.
(C) A seedling harboring ProCYCD4;2:CYCD4;2-GUS at 9 d after germination was stained, and sections (10 μm), including the shoot apical meristem, were observed. Bar = 5 μm.
(D) A root apex harboring ProCYCD4;2:CYCD4;2-GUS at 9 d after germination. Bar = 0.5 mm.
CYCD4 Overexpression Stimulates Cell Division in the Nonprotruding Cell File
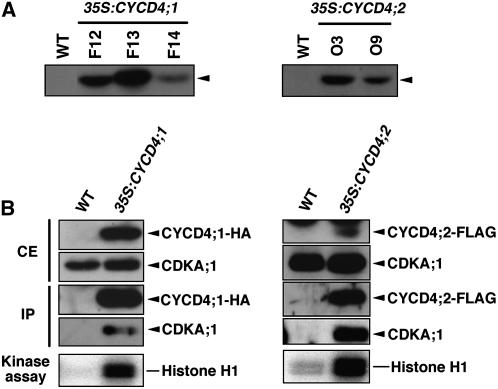
We observed that CYCD4 overexpression in tobacco BY-2 cells did not alter cell cycle progression (A. Kono and M. Umeda, unpublished data). This is in contrast with other CYCD genes, whose overexpression accelerated cell cycle progression in cell suspension cultures (Koroleva et al., 2004; Menges et al., 2006). To determine whether CYCD4 is involved in in planta cell division, we generated transgenic Arabidopsis plants that overexpress hemagglutinin (HA)-tagged CYCD4;1 and FLAG-tagged CYCD4;2 under the control of the cauliflower mosaic virus (CaMV) 35S promoter. Immunoblot analysis revealed several independent cell lines that overexpressed CYCD4;1-HA or CYCD4;2-FLAG (Figure 3A). Immunoprecipitation of protein extracts with the anti-HA or anti-FLAG antibody revealed that CDKA;1, the ortholog of yeast Cdc2/Cdc28p, was coprecipitated with either CYCD4;1-HA or CYCD4;2-FLAG (Figure 3B). The immunoprecipitates exhibited histone H1 kinase activity, indicating that both CYCD4;1-HA and CYCD4;2-FLAG were functional in activating CDKA;1 in planta. The CYCD4-overexpressing (CYCD4-OE) plants exhibited no distinct macroscopic phenotype (data not shown), and their DNA ploidy was almost identical to that of the wild-type plants (see Supplemental Figure 1 online). This is in contrast with CYCD3;1 overexpression that caused hyperproliferation of leaf epidermal cells, altered leaf architecture, and reduced DNA ploidy (Dewitte et al., 2003).
Figure 3.
CYCD4 Binds and Activates CDKA;1 in Arabidopsis Plants.
CYCD4;1 and CYCD4;2, which were tagged with HA and FLAG, respectively, were overexpressed under the control of the CaMV 35S promoter.
(A) Immunoblotting of total protein obtained from the wild-type and transgenic plants (CYCD4;1-OE lines F12, F13, and F14 and CYCD4;2-OE lines O3 and O9). The protein extract (20 μg) was immunoblotted with the anti-HA or anti-FLAG antibody to detect CYCD4;1-HA or CYCD4;2-FLAG (arrowheads), respectively.
(B) Kinase assay of CYCD4-CDKA;1 complexes. Total protein was extracted from the wild-type plants and CYCD4-OE lines F12 and O3, and 20 μg of crude extract (CE) was immunoblotted with the anti-HA or anti-FLAG antibody and the anti-CDKA;1 antibody. The protein extracts (1 mg) were immunoprecipitated with the anti-HA or anti-FLAG antibody, and the immunoprecipitates (IP) were examined by immunoblotting with the anti-HA or anti-FLAG antibody and the anti-CDKA;1 antibody. The immunoprecipitates were subjected to kinase assays using histone H1 as the substrate.
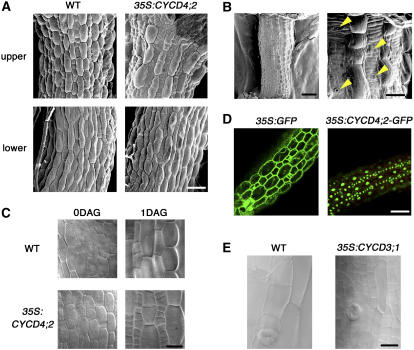
We observed a drastic change in the hypocotyls of the transgenic seedlings. In the CYCD4-OE seedlings, many small cells were generated in the nonprotruding cell files a few days after germination (Figure 4A). Cell division was also enhanced in the protruding cell files (∼130% of that in the wild-type plants); however, a considerably higher enhancement was observed in the nonprotruding cell files (∼260%) (Table 2). Accordingly, the numbers of nonprotruding cells and stomata in the upper hypocotyl region were significantly elevated (Table 2, Figure 1C). The stomatal index (i.e., the fraction of stomata in the upper nonprotruding cells, including the GCs) was 0.136 ± 0.007 (mean ± se) in the wild-type plants; however, it decreased to 0.099 ± 0.009 in the CYCD4;2-OE hypocotyls due to the prominent increase in the number of nonprotruding cells. This suggested that CYCD4 overexpression affected cell division but not stomatal differentiation. This enhanced cell division finally generated a line of short cells that were flanked by elongated protruding cells (Figure 4B). The data obtained for the transgenic plants carrying CYCD4;1 and CYCD4;2 were almost identical. Next, we report the representative results obtained for the CYCD4;2-OE plants.
Figure 4.
CYCD4 Overexpression Enhances Cell Division in the Hypocotyl.
(A) Hypocotyls of the wild-type and CYCD4;2-OE seedlings at 4 d after germination. The upper and lower hypocotyl regions were observed under a scanning electron microscope. Bar = 50 μm.
(B) The CYCD4;2-OE hypocotyl at 20 d after germination. The right panel shows a magnified view. The yellow arrowheads indicate stomata in the nonprotruding cell file. Bars = 200 μm (left) and 50 μm (right).
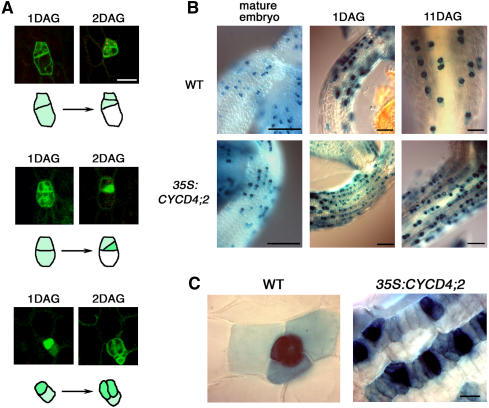
(C) The hypocotyl epidermis of the wild-type and CYCD4;2-OE plants. A mature embryo prior to germination (0DAG) and a seedling at 1 d after germination (1DAG) were observed under a light microscope. Bar = 20 μm.
(D) Localization of GFP and GFP-fused CYCD4;2 in the hypocotyl epidermis. The seedlings were harvested at 1 d after germination and observed under a confocal laser scanning microscope. Bar = 50 μm.
(E) Hypocotyls of the wild-type and CYCD3;1-OE seedlings at 12 d after germination. Bar = 25 μm.
No difference was observed between the hypocotyls of the wild-type and CYCD4;2-OE plants immediately after germination. However, 1 d after germination, small cells were aligned in tandem in the CYCD4;2-OE hypocotyls (Figure 4C), indicating that CYCD4;2 overexpression stimulated cell division after germination. We then overexpressed green fluorescence protein (GFP)–tagged CYCD4;2 under the control of the CaMV 35S promoter. Once again, cell division was enhanced in the hypocotyl of the transgenic plants (data not shown), and GFP fluorescence was detected only in the nucleus (Figure 4D). The above results indicate that CYCD4 is associated with the division of nonprotruding cells that is initiated after germination to produce stomata.
Specific Function of CYCD4 in the Hypocotyl
Furthermore, we also observed the hypocotyl of Pro35S:CYCD3;1 plants that had been reported previously (Dewitte et al., 2003). Small cells appeared in both the nonprotruding and protruding cell files; thus, they disrupted the uniform alignment in both cell files (Figure 4E). A similar but slightly stronger phenotype was observed in the hypocotyl that overexpressed E2Fa-DPa heterodimeric transcription factors (De Veylder et al., 2002). E2Fa-DPa is assumed to function downstream of CYCD3;1 and promote the G1/S transition. Therefore, it is probable that CYCD3;1 might induce a wide spectrum of divisions in the hypocotyl via E2Fa-DPa activation. This is in contrast with the observations for the CYCD4-OE plants, suggesting that CYCD4 plays a specific role in promoting cell division in the nonprotruding cell files.
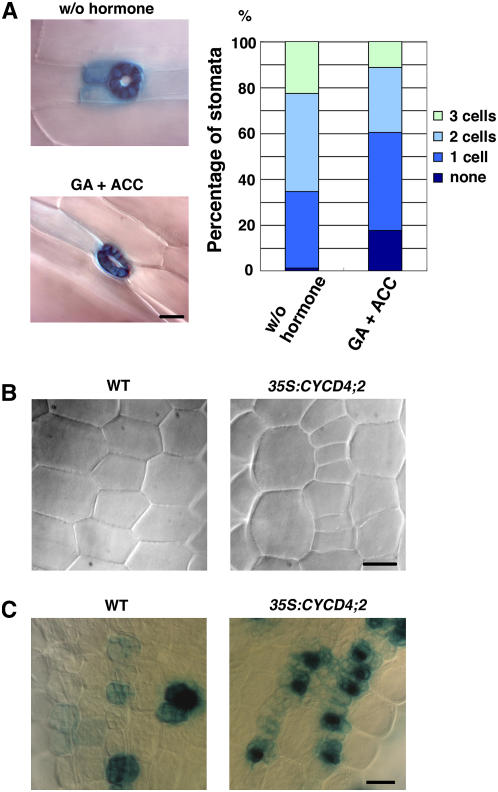
To determine whether CYCD4 overexpression affects stomata formation in leaves, we observed the fully expanded first leaves of the CYCD4-OE plants (Table 1). The number of abaxial epidermal cells increased (CYCD4;1, 147%; CYCD4;2, 134%), while the cell area decreased (CYCD4;1, 69%; CYCD4;2, 73%) in the leaves of these plants (see Supplemental Figure 2 online). Consequently, the leaf area was almost identical to that of the wild-type leaves. The number of stomata did not differ significantly; therefore, the stomatal index decreased only slightly in both the CYCD4-OE plants (CYCD4;1, 87%; CYCD4;2, 85%). These results indicate that CYCD4 overexpression stimulated cell division in leaves but did not promote stomata formation.
CYCD4 Overexpression Increases the Number of Cells of Stomatal Lineage
TOO MANY MOUTHS (TMM) encodes a Leu-rich repeat receptor-like protein that is assumed to function in stomatal spacing patterning (Geisler et al., 2000; Nadeau and Sack, 2002). In leaves, TMM is expressed in cells of stomatal lineage, and its expression begins in MMCs that undergo formative asymmetric division (Nadeau and Sack, 2002). Here, we first investigated TMM expression in wild-type hypocotyls using the GFP gene under the control of the TMM promoter. To identify each cell type during the development of stomatal cells, GFP fluorescence was monitored from 1 to 2 d after germination. We observed that the promoter was already active in the cells that were to divide perpendicular to the apical-basal axis to produce an MMC or its precursor (a cell that continues to divide symmetrically before generating an MMC) (Figure 5A, top panel). GFP fluorescence was retained in the MMCs and daughter meristemoids that had a characteristic triangular shape and were completely filled with cytoplasm (Figure 5A, middle panel). It is noteworthy that TMM expression disappeared rapidly in cells that did not divide further and that deviated from the stomatal lineage to differentiate into epidermal cells (Figure 5A, top and middle panels). GFP expression persisted in GCs that were generated by symmetric GMC divisions (Figure 5A, bottom panel). These results indicate that TMM is expressed in the early stages when protodermal cells enter the stomatal lineage to develop into MMC precursors, and its expression continues until GC differentiation. In leaves, satellite meristemoids are created by divisions of the larger daughter cell of the MMC division (Geisler et al., 2000). However, we studied the division patterns of ∼30 sister cells of the meristemoid and observed that they did not generate satellite meristemoids in the hypocotyl; instead, they developed into GMCs that divided symmetrically to produce GCs. These results indicate that in the hypocotyl, the division of MMC precursors and not the sister of the meristemoid influences the cell population of stomatal lineage and thus controls the number of stomata in the epidermis.
Figure 5.
TMM Expression in Hypocotyls.
(A) GFP expression driven by the TMM promoter in the wild-type hypocotyls. The GFP fluorescence derived from particular epidermal cells was monitored from 1 to 2 d after germination (DAG). Three independent expression patterns are shown. The cell outlines are schematically presented below the photographs. Bar = 25 μm.
(B) TMM expression in the upper hypocotyls of the wild-type and CYCD4;2-OE seedlings. GUS expression driven by the TMM promoter was observed in mature embryos prior to germination and in seedlings at 1 or 11 d after germination. Bars = 100 μm.
(C) Magnified views of the seedlings at 11 d after germination. Bar = 20 μm.
We then examined TMM expression in CYCD4-OE plants using the ProTMM:GUS construct. A patchy pattern of TMM expression could be observed in the hypocotyl of mature embryos from both the wild-type and CYCD4;2-OE lines (Figure 5B). However, 1 d after germination, the CYCD4;2-OE seedlings exhibited small TMM-expressing cells that were aligned in tandem, and this expression pattern persisted for >10 d after germination (Figures 5B and 5C). The TMM expression observed here might include a carryover of the GUS protein due to the rapid cell division induced by CYCD4;2 overexpression; however, it is evident that the overproliferative cells in the nonprotruding cell files were derived from the TMM-expressing cells of stomatal lineage. The triangular meristemoids and GCs exhibited a high level of TMM expression, while most of the other TMM-expressing cells were rectangular in shape. This suggested that they were MMCs or their precursors. By contrast, 11 d after germination, the wild-type seedlings expressed TMM only in the stomata and surrounding cells that were dispersed in the epidermis (Figures 5B and 5C). Hence, the number of TMM-expressing cells in the CYCD4-OE hypocotyls increased to twofold to threefold of that in the wild-type plants (see Supplemental Figure 3A online). Conversely, the number of these cells was significantly reduced in the single and double mutants of CYCD4, as shown in Supplemental Figure 3B online. The TMM expression in leaves was almost the same between the wild-type and CYCD4;2-OE leaves (see Supplemental Figure 4 online); this fact supports the above-mentioned assumption that CYCD4 is involved in proliferation of stomatal precursors in hypocotyls but not in leaves.
CYCD4 Overexpression Promotes Cell Division before Meristemoid Differentiation
Gibberellin (GA) is known to promote stomata formation in the hypocotyl, and this effect is pronounced when it is combined with ethylene (Saibo et al., 2003). Conversely, stomata are eliminated from the hypocotyls of the GA-deficient mutant ga1-3 and wild-type plants treated with the GA biosynthesis inhibitor paclobutrazol (PAC) (Saibo et al., 2003). When Arabidopsis seedlings were grown in the presence of GA and the ethylene precursor 1-aminocyclopropane-1-carboxylic acid (ACC), the number of stomata in the hypocotyl increased more than twofold (Table 3). However, we noticed that the number of upper nonprotruding cells also increased to 150% (Table 3). When TMM expression was observed in the absence of an exogenous phytohormone, 65% of the hypocotyl stomata were surrounded by two or three TMM-expressing cells (Figure 6A). However, GA and ACC treatment reduced the number of stomatal neighbor cells, such that 60% of the stomata were accompanied by one or no TMM-expressing cells (Figure 6A). These results indicate that GA not only enhanced cell division in the nonprotruding cells but also promoted stomatal differentiation, resulting in the rapid disappearance of TMM expression in the neighbor cells. As a consequence, GA treatment increased the stomatal index slightly to 150% (Table 3).
Table 3.
Number of Nonprotruding Cells and Stomata in Wild-Type Hypocotyls Treated with GA or PAC
| Treatment | Number of Nonprotruding Cells | Number of Stomata | Stomatal Index (%) |
|---|---|---|---|
| −GA, ACC | 14.1 ± 0.59 | 1.67 ± 0.17 | 11.7 ± 0.91 |
| +GA, ACC | 20.7 ± 0.69*** | 3.56 ± 0.32*** | 17.2 ± 1.4** |
| −PAC | 12.3 ± 0.63 | 1.67 ± 0.19 | 13.7 ± 1.5 |
| +PAC | 8.92 ± 0.38*** | 0.250 ± 0.13*** | 2.57 ± 1.4*** |
The nonprotruding cells adjacent to the six most apical protruding cells and the stomata in the same region were counted; the stomatal index was calculated. Wild-type plants were grown either in the presence of 10 μM GA and 50 μM ACC for 9 d or in the presence of 0.5 μM PAC for 7 d. Data are presented as mean ± se (n = 12). Significant differences between the nontreated and treated plants are as follows: **P < 0.01 and ***P < 0.001; the other values are not significant (P > 0.05).
Figure 6.
Cell Division and TMM Expression in Response to GA.
(A) TMM expression in the GA-treated hypocotyls. Transgenic plants expressing GUS under the control of the TMM promoter were grown in the absence of phytohormones (w/o hormone) or in the presence of 10 μM GA and 50 μM ACC (GA + ACC) for 9 d. Bar = 20 μm. In the graph, the stomata that lay adjacent to the indicated number of TMM-expressing cells are expressed in terms of percentage: w/o hormone, n = 154; GA + ACC, n = 84.
(B) and (C) Effect of the GA biosynthesis inhibitor PAC. Wild-type or CYCD4;2-OE seedlings were grown on medium supplemented with 0.5 μM PAC for 4 d, and the upper hypocotyl region was observed (B). GUS expression driven by the TMM promoter was also observed (C). Bars = 20 μm.
Next, we observed the hypocotyls of seedlings grown on medium containing PAC. As expected, the number of upper nonprotruding cells had decreased, and stomata formation was severely inhibited, as reported by Saibo et al. (2003); this caused a drastic decrease in the stomatal index to 19% (Table 3). As shown in Figure 6C, MMCs (or their precursors) and meristemoids could be identified as TMM-expressing cells in the PAC-treated hypocotyls, but only one or two stomata were found on one surface of the entire hypocotyl. These results indicate that GA is required for stomatal differentiation after cells have acquired meristemoid characteristics.
Even in the CYCD4;2-OE seedlings, stomata formation was severely inhibited by PAC treatment, and almost no stomata developed on the hypocotyl. However, we observed that small cells accumulated in the nonprotruding cell files (Figure 6B), and in fact, the number of upper nonprotruding cells reached 18.8 ± 1.2 (mean ± se), which is more than twice that in the wild-type seedlings (8.92 ± 0.38; see Table 3). This suggests that CYCD4 overexpression enhanced cell division in the nonprotruding cell files regardless of the inhibitory effect of PAC on cell division and stomatal differentiation. Moreover, the overproliferative cells expressed TMM, as was the case in the nontreated hypocotyls (Figure 6C). These results support our assumption that CYCD4 functions in the division of MMC precursors but not in the later process of stomata formation.
DISCUSSION
Previous reports have demonstrated that CYCDs promote cell cycle progression (Koroleva et al., 2004; Menges et al., 2006). However, overexpression of the CYCD4 genes in tobacco BY-2 cells did not affect cell cycle progression. Moreover, CYCD4 overexpression in Arabidopsis plants did not alter the morphology or DNA ploidy. In leaves, the number of epidermal cells increased, while the cell area decreased; this suggests that CYCD4 overexpression stimulated cell division. However, neither cell division nor cell growth was significantly affected in the leaves of single or double mutants of CYCD4, indicating that CYCD4 is not essential for the mitotic division of proliferating cells or that the other CYCDs may be redundant in function. In this regard, it is noteworthy that the CYCD4;2 promoter activity was eliminated from the shoot and root meristems. During seed germination in the cycd4;1-1 mutant, the onset of cell proliferation was significantly delayed in the root tips, and the overall number of dividing cells was reduced (Masubelele et al., 2005). This suggests that CYCD4;1 may play a distinct role in regulating the extent of cell division that occurs during germination. Schnittger et al. (2002) have reported that CYCD3;1 induced cell division in trichomes when it was expressed under the control of the GLABRA2 (GL2) promoter, while CYCD4;1 had no effect on the trichomes when it was expressed under the control of the same promoter. This further supports the assumption that CYCD4 function is distinct from that of other CYCDs.
In this study, we concluded that CYCD4 is associated with stomatal precursor formation in hypocotyls based on the following observations. (1) The cycd4 mutants had a reduced number of upper nonprotruding cells. As a result, the numbers of TMM-expressing cells and stomata were reduced. (2) CYCD4 overexpression enhanced cell division in the hypocotyl, particularly in the upper region of the nonprotruding cell file. This occurred after germination when stomata formation was initiated. (3) The overproliferative cells accumulated the GUS protein that was expressed under the control of the TMM promoter, suggesting that these cells were generated by reiterative symmetric divisions in the cells of stomatal lineage. Several reports have demonstrated that stomata formation in leaves was affected by the up- or downregulation of cell cycle–related genes. Overexpression of CDC6 or CDT1, both of which are required for DNA replication, elevated the density of stomata on Arabidopsis leaves (Castellano et al., 2004). Expression of a dominant-negative type of CDKB1;1 disturbed cell division and reduced the stomatal density (Boudolf et al., 2004). However, we demonstrated that in leaves, CYCD4 overexpression reduced the stomatal density and that the TMM expression pattern did not change at all. These results are not surprising because some organ-specific rules may operate in stomata formation. For instance, tmm mutants have stomatal clusters on leaves but no stomata in the hypocotyl and inflorescence stem, suggesting the indispensability of TMM in stomata formation in the hypocotyl (Yang and Sack, 1995; Geisler et al., 1998). Moreover, it is known that GA is essential for stomatal development in hypocotyls but not in leaves (Sun et al., 1992; Saibo et al., 2003). Our results revealed that up- or downregulation of CYCD4 had pronounced effects in the upper nonprotruding cells that produce stomata. GL2, which encodes a homeodomain transcription factor, is only expressed in the protruding cell files of the hypocotyl, and in gl2 mutants, stomata formation, but not ectopic cell division, is observed in the protruding cell files (Berger et al., 1998; Hung et al., 1998). This indicates that GL2 may play an inhibitory role in GC differentiation in the protruding cell files. CYCD4 overexpression in gl2 mutants enhanced cell division only in the nonprotruding cell files, as observed in the wild-type background (data not shown), suggesting that some factor(s) other than GL2 may modify the CYCD4 function that is specific to the nonprotruding cell files. Therefore, differential mechanisms may operate to drive extra cell divisions in stomatal precursors and to produce the final differentiated GCs.
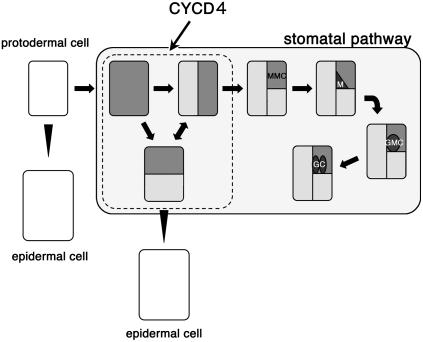
Using the GFP marker under the control of the TMM promoter, we noticed that typically, secondary meristemoids were not generated from existing meristemoids in hypocotyls; instead, the MMC precursors reiterated cell divisions before they acquired MMC characteristics. This indicates that the divisions of the MMC precursors increased the cell population and the number of stomata in the nonprotruding cell files. The number of nonprotruding cells in the cycd4 mutants was significantly reduced, while that in mature embryos was almost identical to that in the wild-type plants (data not shown). Therefore, we assume that the loss of CYCD4 function might cause a defect in the postembryonic divisions of the nonprotruding cells. This assumption was supported by the fact that CYCD4 overexpression caused the accumulation of many rectangular cells in tandem; this also occurred in the presence of PAC, which is a GA biosynthesis inhibitor that inhibits stomatal differentiation. Based on these results, we propose that CYCD4 plays a principal role in the divisions of MMC precursors but not in the asymmetric divisions of MMCs or in GC differentiation (Figure 7). In cycd4;1 and cycd4;2 double mutants, stomata were not completely eliminated from the hypocotyls (Figure 1C), which indicates that stomata formation is ensured even in the absence of CYCD4 function. CYCD4 is likely to increase the number of stomata by amplifying the MMC population.
Figure 7.
The Stomatal Pathway in Hypocotyls.
Some protodermal cells enter the stomatal pathway to develop into precursors of MMCs and begin to express TMM; this is indicated by gray color. The precursors reiterate cell divisions before they generate MMCs, which then divide asymmetrically to produce triangular meristemoids (M). Meristemoids are converted into GMCs, which then divide symmetrically to generate GCs. The other cells produced by the division of MMC precursors differentiate into epidermal cells.
In addition to TMM, a single loss-of-function mutation in the Arabidopsis STOMATAL DENSITY AND DISTRIBUTION1 (SDD1) or YODA (YDA) gene also induces increased stomatal density and clusters in leaves (Berger and Altmann, 2000; Von Groll et al., 2002; Bergmann et al., 2004). SDD1 and YDA encode a putative subtilisin-like extracytoplasmic protease and a mitogen-activated protein kinase kinase kinase, respectively (Berger and Altmann, 2000; Lukowitz et al., 2004). A model has been proposed in which SDD1 modifies a ligand for TMM, and the activated receptor signals downstream mitogen-activated protein kinase cascades via YDA to repress stomata formation (Bergmann et al., 2004). In contrast with TMM, loss of SDD1 or YDA increased the stomatal density not only in leaves but also in stems and hypocotyls (Berger and Altmann, 2000; Bergmann et al., 2004). Recently, Shpak et al. (2005) reported that ERECTA (ER) family Leu-rich repeat receptor-like kinases negatively control stomatal development in leaves and stems. In particular, ERECTA LIKE1 (ERL1) is important for maintaining stem cell activity and for preventing the terminal differentiation of meristemoids into GMCs. Genetic studies have suggested that TMM may inhibit ERL1 activity in the stem and promote stomatal differentiation; however, the mechanism by which TMM regulates the ER family is unknown. When CYCD4 was overexpressed in tmm mutants, overproliferation of the nonprotruding cells was observed as in the wild-type plants, but stomata were not formed (data not shown). This indicates that TMM is not a prerequisite for the divisions of the MMC precursors but is required for stomatal differentiation. Nevertheless, it is possible that the TMM/ER/YDA pathway controls CYCD4 function. Further genetic and molecular studies will clarify the mechanism by which the number of stomata in the hypocotyl epidermis is maintained.
CYCD4;2 has a unique amino acid sequence in that it lacks the Rb binding motif (LXCXE) and the PEST sequence that is the hallmark of unstable proteins (Kono et al., 2006). However, in this study, CYCD4;2 formed active kinase complexes with CDKA;1 in plant cells, indicating its functionality as a cyclin. The absence of the Rb binding motif suggests that the divisions of MMC precursors in the hypocotyl can be stimulated independent of the Rb/E2F/DP pathway; for example, CYCD4-CDKA;1 may phosphorylate a substrate(s) other than Rb, which positively regulates cell division. Another possibility is that CYCD4 may interact with a gene-specific transcription factor(s) as reported in mammals; D-type cyclins control the transcription factors DMP1, C/EBPß/Nf-I16, and AML1 via CDK-independent mechanisms (Inoue and Sherr, 1998; Lamb et al., 2003; Peterson et al., 2005). It is also known that CYCD1 is directly associated with the estrogen or androgen receptor and up- and downregulates their transcriptional transactivation abilities, respectively (Zwijsen et al., 1997; Knudsen et al., 1999). In future studies, these possibilities will be examined for Arabidopsis CYCD4s. It has been reported that in Drosophila, the division of germline stem cells and their precursors (primordial germ cells) requires a specific function of cyclin B (Wang and Lin, 2005). Our study demonstrated a more specific requirement of CYCD in cell divisions associated with stomata formation. Recently, a gene named SPEECHLESS has been shown to direct the divisions of MMCs (MacAlister et al., 2007; Pillitteri et al., 2007), suggesting that it may control a specific set of cell cycle genes to induce the asymmetric divisions. The Arabidopsis genome includes >50 cyclin genes. Therefore, it is interesting to investigate the distinct functions of other cyclins that may be involved in the temporal and/or spatial control of cell division during postembryonic development in plants.
METHODS
Plant Materials and Treatments
Arabidopsis thaliana ecotype Col-0 was used for transformation. For in vitro cultivation, plants were grown on a medium containing 2.15 g/L of Murashige and Skoog basal salt mixture (Sigma-Aldrich) supplemented with 1% sucrose, 3 mg/L thiamine-HCl, 5 mg/L nicotinic acid, and 0.5 mg/L pyridoxine-HCl. GUS staining was conducted as described previously (Umeda et al., 2000). The DNA ploidy was measured using a flow cytometer (Ploidy Analyzer; Partec) according to the manufacturer's protocol.
Identification of T-DNA Insertion Mutants
We isolated cycd4;1 and cycd4;2 mutants from the collections obtained from the Max-Planck Institute for Plant Breeding Research (Ríos et al., 2002) and the Salk Institute, respectively. The cycd4;1-2 mutant was MPI1247, and the cycd4;2-2 and cycd4;2-3 alleles corresponded to SALK_85720 and SALK_127016, respectively. The insertions were examined by genomic PCR using Ex Taq DNA polymerase (Takara) and a set of primers that hybridize to the T-DNA and both CYCD4s; 5′-AGCCTCTCTCCTTCTCACACAATCTC-3′ and 5′-GGCTGCCAATGATAAACAAGCCACAG-3′ were used for cycd4;1-2, and 5′-CGACACAAGATCGATTTTTCAGATGGGT-3′ and 5′-GGAGGAGGAAACAGAAGCAGTAGAAAGA-3′ were used for cycd4;2-2 and cycd4;2-3, respectively. The nucleotide sequences of the amplified fragments were determined to identify the T-DNA insertion site. Each mutant line was backcrossed three times with the wild-type plants. RT-PCR was performed for the total RNA obtained from the seedlings using the Titanium One-Step RT-PCR kit (BD Biosciences Clontech). In a total reaction volume of 20 μL, 1 μg of RNA and the following primers were used: 5′-TTGGAAGAAGAGATGCCCTC-3′ and 5′-AGCTGAAACAACAAGCCGAT-3′ for cycd4;1-2, 5′-TCAGAATCCAAGCCCTTGGT-3′ and 5′-GAATGAGAATAACGACGACTC-3′ for cycd4;2-2, and 5′-CACCATGGCTGAATTTATGGAACCA-3′ and 5′-ACGGTGTCACTGCCCGTAAC-3′ for cycd4;2-3. As the control, β-TUBULIN4 (TUB4) cDNA was amplified using the primers 5′-CTCTGTGCATCAGCTTGTCGAAAACG-3′ and 5′-CCGAGGGAGCCATTGACAACATCTT-3′. The PCR conditions were one cycle at 50°C for 60 min and 20 cycles (for TUB4) or 30 cycles (for CYCD4;1 and CYCD4;2) at 94°C for 30 s, 65°C for 30 s, and 68°C for 1 min.
Plasmid Construction for CYCD4 Overexpression
The open reading frame of CYCD4;1 that lacked the stop codon was PCR amplified with a SalI site at the N terminus and an NcoI site at the C terminus, and it was subcloned into the SmaI site of pBluescript II SK– (Stratagene). Next, the plasmid was digested with HindIII and NcoI, and the resultant fragment was subcloned into pPily (Ferrando et al., 2000) that was digested with HindIII and NcoI to add a HA tag at the C terminus of CYCD4;1 under the control of the CaMV 35S promoter. The resultant plasmid was digested with KpnI, and the CYCD4;1-HA fragment was subcloned into the KpnI site of pSPTV20 (Becker et al., 1992). The open reading frame of CYCD4;2 that lacked the stop codon was PCR amplified and cloned into the pENTR-D/TOPO vector (Invitrogen). The resultant plasmid pENTR-CYCD4;2 was subjected to the LR reaction using the destination vector pGWB11 (T. Nakagawa, unpublished data) to produce a binary vector containing the C-terminal FLAG-tagged CYCD4;2 under the control of the CaMV 35S promoter. Furthermore, pENTR-CYCD4;2 was also subjected to the LR reaction using the destination vector pGWB5 (T. Nakagawa, unpublished data) to produce a binary vector containing the C-terminal GFP-tagged CYCD4;2 under the control of the CaMV 35S promoter.
Plasmid Construction for Expression Analysis
The CYCD4;1 promoter region, which extends from 2491 to 1 bp upstream of the start codon, was PCR amplified with SalI sites at the 5′ and 3′ ends and subcloned into pCR-XL-TOPO (Invitrogen) to produce pCR-CYCD4;1pro. The genomic region containing the 1st exon and intron of CYCD4;1, which extends from 270 bp upstream to 585 bp downstream of the start codon, was PCR amplified with a BamHI site at the C terminus, and it was subcloned into the SmaI site of pBluescript II SK– to produce SK–-CYCD4;1int. Furthermore, pCR-CYCD4;1pro was digested with SalI and HindIII that cuts it at 189 bp upstream of the start codon, and the SalI-HindIII fragment was subcloned into SK–-CYCD4;1int that was digested with SalI and HindIII. Next, the plasmid was digested with SalI and BamHI, and the resultant fragment was subcloned into the SalI-BamHI site of the binary vector pPCV812 (Koncz et al., 1994) to produce the GUS fusion gene.
The CYCD4;2 promoter region, which extends from 2567 to 1 bp upstream of the start codon, was PCR amplified with SalI sites at the 5′ and 3′ ends, and it was subcloned into pBluescript II SK– to produce SK–-CYCD4;2pro. The genomic region containing the 1st exon and intron of CYCD4;2, which extends from 1580 bp upstream to 402 bp downstream of the start codon, was PCR amplified with a BamHI site at the C terminus and subcloned into the SmaI site of pBluescript II SK– to produce SK–-CYCD4;2int. Furthermore, SK–-CYCD4;2int was digested with EcoRI that cuts it at 1562 bp upstream of the start codon and PstI that cuts within the multicloning sites of pBluescript II SK–; the resultant fragment was subcloned into pSK–-CYCD4;2pro that was digested with EcoRI and PstI. Next, the plasmid was digested with XhoI in the multicloning sites and with BamHI, and the XhoI-BamHI fragment was subcloned into the SalI-BamHI site of pPCV812 to produce the GUS fusion gene.
The promoter region of TMM, which encompasses 2026 bp upstream to 3 bp downstream from the start codon, was PCR amplified and subcloned into pENTR-D/TOPO to produce pENTR-TMMpro. Next, the plasmid was subjected to the LR reaction using the destination vector pDD333 (T. Nakagawa, unpublished data). The resultant plasmid pDD-TMMpro along with pENTR-TMMpro was further subjected to the LR reaction using the destination vector pGWB3450 (T. Nakagawa, unpublished data) to produce a binary vector carrying two copies of the TMM promoter, each of which was translationally fused to either GFP or GUS at the start codon.
Plasmid Construction for Complementation of cycd4 Mutants
The genomic fragment containing CYCD4;1, which extends from 2491 bp upstream of the start codon to 1000 bp downstream of the stop codon, was PCR amplified with a SacI site at the 3′ end and subcloned into pBluescript II SK–. Next, the plasmid was digested with SacI and XhoI that cuts it at 2390 bp upstream of the start codon, and the XhoI-SacI fragment was subcloned into the SalI/SacI site of the binary vector pSPTV20. The genomic fragment containing CYCD4;2, which extends from 2577 bp upstream of the start codon to 1105 bp downstream of the stop codon, was subcloned into pENTR/D-TOPO. Following this, the resultant plasmid was subjected to the LR reaction using the destination vector pGWB1 (T. Nakagawa, unpublished data). The binary plasmids were used for transformation of the T-DNA insertion mutants, and pGWB2 (T. Nakagawa, unpublished data) that lacked the ccdB gene was used as a vector control.
Microscopy Observations
Plant tissues were fixed overnight at 4°C in a solution of 90% ethanol and 10% acetic acid, hydrated through a graded series of ethanol, and mounted in a chloral hydrate solution (71% chloral hydrate and 11% glycerol) overnight at room temperature. Embryos obtained from dry seeds were soaked in water for at least 1 h and subsequently fixed and cleared as described previously (Malamy and Benfey, 1997b). For scanning electron microscopy, specimens were prefixed with 2% glutaraldehyde in 0.1 M phosphate buffer at 4°C for 4 h and postfixed with 1% osmium tetroxide at 4°C overnight and 0.05% ruthenium tetroxide for 5 min at room temperature under dark conditions. The fixed specimens were dehydrated in an ethanol series and dried with liquid carbon dioxide in a critical-point dryer (HCP-2; Hitachi), coated with platinum-palladium for 2 min in an ion sputter (E-1030; Hitachi), and observed using a scanning electron microscope (S-800; Hitachi) at 7 kV. To observe GFP fluorescence, the seedlings were placed on a slide glass, immersed in 100 μL of water, and covered with a cover glass using adhesive tape as a spacer. At least one cotyledon was kept in contact with air to enable respiration. Seedlings were grown vertically in a Petri dish under continuous light conditions and observed using a confocal laser scanning microscope system (MicroRadiance MR/AG-2; Bio-Rad) with an argon-ion laser (488 nm).
Leaf Growth Analysis
The kinematic analysis of leaf growth was performed as described previously (De Veylder et al., 2001). Plants were grown in soil under continuous light conditions for 20 d. Healthy first/second leaves were harvested and fixed in a solution of 90% ethanol and 10% acetic acid at 4°C overnight, hydrated through a graded series of ethanol, and stored in water at 4°C. The samples were mounted on a slide glass and cleared with chloral hydrate solution during the overnight incubation. Data were collected by scanning images of the abaxial epidermis located at 50% the distance between the tip and the base of the leaf blade, halfway between the midrib and the leaf margin. The images, including at least 40 cells in focus, were excised using the photo-editing program Photoshop Elements 2.0 (Adobe). The epidermal cells, including GCs, in the excised image were counted, and the area of the excised image was measured using the image analysis program NIH image 1.63. The average cell area and stomatal index were determined based on these measurements. Next, the leaf blade area was measured, and the total number of epidermal cells on the abaxial side was estimated based on the average cell area. Finally, the total number of stomata was calculated based on the total cell number and stomatal index.
Immunoblotting and Kinase Assays
Total protein was extracted from the whole seedlings and subjected to immunoblotting using an ECL protein gel blotting detection system (Amersham Biosciences) with the anti-HA (Roche), anti-FLAG M2 (Sigma-Aldrich), and anti-CDKA;1 antibodies (Umeda et al., 2000). For the kinase assays, the protein extracts were immunoprecipitated using the anti-HA or anti-FLAG antibody, and the immunoprecipitates were subjected to a kinase reaction using 1 μg of histone H1 (Roche) as the substrate. A detailed protocol for the kinase assays has been described previously (Umeda et al., 1998).
Accession Numbers
Arabidopsis Genome Initiative locus identifiers for the genes mentioned in this article are as follows: At5g65420 (CYCD4;1), At5g10440 (CYCD4;2), At3g48750 (CDKA;1), At4g34160 (CYCD3;1), and At1g80080 (TMM).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. DNA Ploidy Distribution in Wild-Type and CYCD4-OE Seedlings.
Supplemental Figure 2. Epidermal Cells of Wild-Type and CYCD4-OE Leaves.
Supplemental Figure 3. Number of TMM-Expressing Cells in the Hypocotyls of Wild-Type, CYCD4-OE, and cycd4 Mutant Seedlings.
Supplemental Figure 4. TMM Expression in Wild-Type and CYCD4;2-OE Leaves.
Supplementary Material
Acknowledgments
We thank James A.H. Murray for providing the CYCD3;1-OE seeds. We also thank the ABRC at Ohio State University and the Max-Planck Institute for Plant Breeding Research for providing the seeds of the T-DNA insertion mutants. Furthermore, we appreciate the assistance provided by Nobuhiro Tsutsumi and Shin-ichi Arimura with regard to the confocal laser scanning microscope system. This work was supported by a Grant-in-Aid for Scientific Research on Priority Areas (Grants 17027007 and 18056006) and a Grant-in-Aid for Scientific Research (B) (Grant 16370019) from the Ministry of Education, Sports, Culture, Science, and Technology of Japan and by the Program for Promotion of Basic Research Activities for Innovative Biosciences.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Masaaki Umeda (mumeda@bs.naist.jp).
Online version contains Web-only data.
References
- Becker, D., Kemper, E., Schell, J., and Masterson, R. (1992). New plant binary vectors with selectable markers located proximal to the left T-DNA border. Plant Mol. Biol. 20 1195–1197. [DOI] [PubMed] [Google Scholar]
- Berger, D., and Altmann, T. (2000). A subtilisin-like serine protease involved in the regulation of stomatal density and distribution in Arabidopsis thaliana. Genes Dev. 14 1119–1131. [PMC free article] [PubMed] [Google Scholar]
- Berger, F., Linstead, P., Dolan, L., and Haseloff, J. (1998). Stomata patterning on the hypocotyl of Arabidopsis thaliana is controlled by genes involved in the control of root epidermis patterning. Dev. Biol. 194 226–234. [DOI] [PubMed] [Google Scholar]
- Bergmann, D.C., Lukowitz, W., and Somerville, C.R. (2004). Stomatal development and pattern controlled by a MAPKK kinase. Science 304 1494–1497. [DOI] [PubMed] [Google Scholar]
- Boudolf, V., Barrôco, R., de Almeida Engler, J., Verkest, A., Beeckman, T., Naudts, M., Inzé, D., and De Veylder, L. (2004). B1-type cyclin-dependent kinases are essential for the formation of stomatal complexes in Arabidopsis thaliana. Plant Cell 16 945–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano, M.M., Boniotti, M.B., Caro, E., Schnittger, A., and Gutierrez, C. (2004). DNA replication licensing affects cell proliferation or endoreduplication in a cell type-specific manner. Plant Cell 16 2380–2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlton, W.A. (1996). Lateral root initiation. In Plant Roots: The Hidden Half, 2nd ed, Y. Waisel, A. Eshel, and U. Kfkafi, eds (New York: Marcel Dekker), pp. 149–173.
- Cockcroft, C.E., den Boer, B.G.W., Healy, J.M.S., and Murray, J.A.H. (2000). Cyclin D control of growth rate in plants. Nature 405 575–579. [DOI] [PubMed] [Google Scholar]
- de Jager, S.M., Maughan, S., Dewitte, W., Scofield, S., and Murray, J.A.H. (2005). The developmental context of cell-cycle control in plants. Semin. Cell Dev. Biol. 16 385–396. [DOI] [PubMed] [Google Scholar]
- De Veylder, L., Beeckman, T., Beemster, G.T.S., de Almeida Engler, J., Ormenese, S., Maes, S., Naudts, M., Van Der Schueren, E., Jacqmard, A., Engler, G., and Inzé, D. (2002). Control of proliferation, endoreduplication and differentiation by the Arabidopsis E2Fa-DPa transcription factor. EMBO J. 21 1360–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder, L., Beeckman, T., Beemster, G.T.S., Krols, L., Terras, F., Landrieu, I., Van Der Schueren, E., Maes, S., Naudts, M., and Inzé, D. (2001). Functional analysis of cyclin-dependent kinase inhibidors of Arabidopsis. Plant Cell 13 1653–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewitte, W., Riou-Khamlichi, C., Scofield, S., Healy, J.M.S., Jacqmard, A., Kilby, N.J., and Murray, J.A.H. (2003). Altered cell cycle distribution, hyperplasia, and inhibited differentiation in Arabidopsis caused by the D-type cyclin CYCD3. Plant Cell 15 79–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrando, A., Farràs, R., Jásik, J., Schell, J., and Koncz, C. (2000). Intron-tagged epitope: A tool for facile detection and purification of proteins expressed in Agrobacterium-transformed plant cells. Plant J. 22 553–560. [DOI] [PubMed] [Google Scholar]
- Geisler, M., Nadeau, J., and Sack, F.D. (2000). Oriented asymmetric divisions that generate the stomatal spacing pattern in Arabidopsis are disrupted by the too many mouths mutation. Plant Cell 12 2075–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler, M., Yang, M., and Sack, F.D. (1998). Divergent regulation of stomatal initiation and patterning in organ and suborgan regions of the Arabidopsis mutants too many mouths and four lips. Planta 205 522–530. [DOI] [PubMed] [Google Scholar]
- Gendreau, E., Traas, J., Desnos, T., Grandjean, O., Caboche, M., and Höfte, H. (1997). Cellular basis of hypocotyl growth in Arabidopsis thaliana. Plant Physiol. 114 295–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbour, J.W., and Dean, D.C. (2000). The Rb/E2F pathway: Expanding roles and emerging paradigms. Genes Dev. 14 2393–2409. [DOI] [PubMed] [Google Scholar]
- Healy, J.M.S., Menges, M., Doonan, J.H., and Murray, J.A.H. (2001). The Arabidopsis D-type cyclins CycD2 and CycD3 both interact in vivo with the PSTAIRE cyclin-dependent kinase Cdc2a but are differentially controlled. J. Biol. Chem. 276 7041–7047. [DOI] [PubMed] [Google Scholar]
- Hung, C.-Y., Lin, Y., Zhang, M., Pollock, S., Marks, M.D., and Schiefelbein, J. (1998). A common position-dependent mechanism controls cell-type patterning and GLABRA2 regulation in the root and hypocotyl epidermis of Arabidopsis. Plant Physiol. 117 73–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue, K., and Sherr, C.J. (1998). Gene expression and cell cycle arrest mediated by transcription factor DMP1 is antagonized by D-type cyclins through a cyclin-dependent-kinase-independent mechanism. Mol. Cell. Biol. 18 1590–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen, K.E., Cavenee, W.K., and Arden, K.C. (1999). D-type cyclins complex with the androgen receptor and inhibit its transcriptional transactivation ability. Cancer Res. 59 2297–2301. [PubMed] [Google Scholar]
- Koncz, C., Martini, N., Szabados, L., Hrouda, M., Bachmair, A., and Schell, J. (1994). Specialized vectors for gene tagging and expression studies. In Plant Molecular Biology Manual, Vol. B2, S.B. Gelvin, R.A. Schilperoort, and D.P.S. Verma, eds (Dordrecht, The Netherlands: Kluwer), pp. 1–22.
- Kono, A., Ohno, R., Umeda-Hara, C., Uchimiya, H., and Umeda, M. (2006). A distinct type of cyclin D, CYCD4;2, involved in the activation of cell division in Arabidopsis. Plant Cell Rep. 25 540–545. [DOI] [PubMed] [Google Scholar]
- Kono, A., Umeda-Hara, C., Lee, J., Ito, M., Uchimiya, H., and Umeda, M. (2003). Arabidopsis D-type cyclin CYCD4;1 is a novel cyclin partner of B2-type cyclin-dependent kinase. Plant Physiol. 132 1315–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koroleva, O.A., Tomlinson, M., Parinyapong, P., Sakvarelidze, L., Leader, D., Shaw, P., and Doonan, J.H. (2004). CycD1, a putative G1 cyclin from Antirrhinum majus, accelerates the cell cycle in cultured tobacco BY-2 cells by enhancing both G1/S entry and progression through S and G2 phases. Plant Cell 16 2364–2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb, J., Ramaswamy, S., Ford, H.L., Contreras, B., Martinez, R.V., Kittrell, F.S., Zahnow, C.A., Patterson, N., Golub, T.R., and Ewen, M.E. (2003). A mechanism of cyclin D1 action encoded in the patterns of gene expression in human cancer. Cell 114 323–334. [DOI] [PubMed] [Google Scholar]
- Lukowitz, W., Roeder, A., Parmenter, D., and Somerville, C. (2004). A MAPKK kinase gene regulates extra-embryonic cell fate in Arabidopsis. Cell 116 109–119. [DOI] [PubMed] [Google Scholar]
- MacAlister, C.A., Ohashi-Ito, K., and Bergmann, D.C. (2007). Transcription factor control of asymmetric cell divisions that establish the stomatal lineage. Nature 445 537–540. [DOI] [PubMed] [Google Scholar]
- Malamy, J.E., and Benfey, P.N. (1997. a). Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124 33–44. [DOI] [PubMed] [Google Scholar]
- Malamy, J.E., and Benfey, P.N. (1997. b). Analysis of SCARECROW expression using a rapid system for assessing transgene expression in Arabidopsis roots. Plant J. 12 957–963. [DOI] [PubMed] [Google Scholar]
- Masubelele, N.H., Dewitte, W., Menges, M., Maughan, S., Collins, C., Huntley, R., Nieuwland, J., Scofield, S., and Murray, J.A.H. (2005). D-type cyclins activate division in the root apex to promote seed germination in Arabidopsis. Proc. Natl. Acad. Sci. USA 102 15694–15699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menges, M., Samland, A.K., Planchais, S., and Murray, J.A.H. (2006). The D-type cyclin CYCD3;1 is limiting for the G1-to-S-phase transition in Arabidopsis. Plant Cell 18 893–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan, D.O. (1997). Cyclin-dependent kinases: Engines, clocks, and microprocessors. Annu. Rev. Cell Dev. Biol. 13 261–291. [DOI] [PubMed] [Google Scholar]
- Nadeau, J.A., and Sack, F.D. (2002). Control of stomatal distribution on the Arabidopsis leaf surface. Science 296 1697–1700. [DOI] [PubMed] [Google Scholar]
- Nadeau, J.A., and Sack, F.D. (2003). Stomatal development: Cross talk puts mouths in place. Trends Plant Sci. 8 294–299. [DOI] [PubMed] [Google Scholar]
- Nakagami, H., Sekine, M., Murakami, H., and Shinmyo, A. (1999). Tobacco retinoblastoma-related protein phosphorylated by a distinct cyclin-dependent kinase complex with Cdc2/cyclin D in vitro. Plant J. 18 243–252. [DOI] [PubMed] [Google Scholar]
- Peterson, L.F., Boyapati, A., Ranganathan, V., Iwama, A., Tenen, D.G., Tsai, S., and Zhang, D.-E. (2005). The hematopoietic transcription factor AML1 (RUNX1) is negatively regulated by the cell cycle protein cyclin D3. Mol. Cell. Biol. 25 10205–10219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillitteri, L.J., Sloan, D.B., Bogenschutz, N.L., and Torii, K.U. (2007). Termination of asymmetric cell division and differentiation of stomata. Nature 445 501–505. [DOI] [PubMed] [Google Scholar]
- Planchais, S., Samland, A.K., and Murray, J.A.H. (2004). Differential stability of Arabidopsis D-type cyclins: CYCD3;1 is a highly unstable protein degraded by a proteasome-dependent mechanism. Plant J. 38 616–625. [DOI] [PubMed] [Google Scholar]
- Ríos, G., et al. (2002). Rapid identification of Arabidopsis insertion mutants by non-radioactive detection of T-DNA tagged genes. Plant J. 32 243–253. [DOI] [PubMed] [Google Scholar]
- Riou-Khamlichi, C., Huntley, R., Jacqmard, A., and Murray, J.A.H. (1999). Cyctokinin activation of Arabidopsis cell division through a D-type cyclin. Science 283 1541–1544. [DOI] [PubMed] [Google Scholar]
- Riou-Khamlichi, C., Menges, M., Healy, J.M., and Murray, J.A.H. (2000). Sugar control of the plant cell cycle: Differential regulation of Arabidopsis D-type cyclin gene expression. Mol. Cell. Biol. 20 4513–4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saibo, N.J.M., Vriezen, W.H., Beemster, G.T.S., and Van Der Straeten, D. (2003). Growth and stomata development of Arabidopsis hypocotyls are controlled by gibberellins and modulated by ethylene and auxins. Plant J. 33 989–1000. [DOI] [PubMed] [Google Scholar]
- Schnittger, A., Schöbinger, U., Bouyer, D., Weinl, C., Stierhof, Y.-D., and Hülskamp, M. (2002). Ectopic D-type cyclin expression induces not only DNA replication but also cell division in Arabidopsis trichomes. Proc. Natl. Acad. Sci. USA 99 6410–6415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, W.H. (2002). The plant E2F-Rb pathway and epigenetic control. Trends Plant Sci. 7 505–511. [DOI] [PubMed] [Google Scholar]
- Sherr, C.J., and Roberts, J.M. (2004). Living with or without cyclins and cyclin-dependent kinases. Genes Dev. 18 2699–2711. [DOI] [PubMed] [Google Scholar]
- Shpak, E.D., McAbee, J.M., Pillitteri, L.J., and Torii, K.U. (2005). Stomatal patterning and differentiation by synergistic interactions of receptor kinases. Science 309 290–293. [DOI] [PubMed] [Google Scholar]
- Soni, R., Carmichael, J.P., Shah, Z.H., and Murray, J.A.H. (1995). A family of cyclin D homologs from plants differentially controlled by growth regulators and containing the conserved retinoblastoma protein interaction motif. Plant Cell 7 85–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, T., Goodman, H.M., and Ausubel, F.M. (1992). Cloning the Arabidopsis GA1 locus by genomic subtraction. Plant Cell 4 119–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umeda, M., Bhalerao, R.P., Schell, J., Uchimiya, H., and Koncz, C. (1998). A distinct cyclin-dependent kinase-activating kinase of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 95 5021–5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umeda, M., Umeda-Hara, C., and Uchimiya, H. (2000). A cyclin-dependent kinase-activating kinase regulates differentiation of root initial cells in Arabidopsis. Proc. Natl. Acad. Sci. USA 97 13396–13400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Groll, U., Berger, D., and Altmann, T. (2002). The subtilisin-like serine protease SDD1 mediates cell-to-cell signaling during Arabidopsis stomatal development. Plant Cell 14 1527–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, G., Kong, H., Sun, Y., Zhang, X., Zhang, W., Altman, N., de Pamphilis, C.W., and Ma, H. (2004). Genome-wide analysis of the cyclin family in Arabidopsis and comparative phylogenetic analysis of plant cyclin-like proteins. Plant Physiol. 135 1084–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z., and Lin, H. (2005). The division of Drosophila germline stem cells and their precursors requires a specific cyclin. Curr. Biol. 15 328–333. [DOI] [PubMed] [Google Scholar]
- Yang, M., and Sack, F.D. (1995). The too many mouths and four lips mutations affect stomatal production in Arabidopsis. Plant Cell 7 2227–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwijsen, R.M.L., Wientjens, E., Klompmaker, R., Van der Sman, J., Bernards, R., and Michalides, R.J.A.M. (1997). CDK-independent activation of estrogen receptor by cyclin D1. Cell 88 405–415. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.