Abstract
Rice (Oryza sativa) seeds can germinate in the complete absence of oxygen. Under anoxia, the rice coleoptile elongates, reaching a length greater than that of the aerobic one. In this article, we compared and investigated the transcriptome of rice coleoptiles grown under aerobic and anaerobic conditions. The results allow drawing a detailed picture of the modulation of the transcripts involved in anaerobic carbohydrate metabolism, suggesting up-regulation of the steps required to produce and metabolize pyruvate and its derivatives. Sugars appear to play a signaling role under anoxia, with several genes indirectly up-regulated by anoxia-driven sugar starvation. Analysis of the effects of anoxia on the expansin gene families revealed that EXPA7 and EXPB12 are likely to be involved in rice coleoptile elongation under anoxia. Genes coding for ethylene response factors and heat shock proteins are among the genes modulated by anoxia in both rice and Arabidopsis (Arabidopsis thaliana). Identification of anoxia-induced ethylene response factors is suggestive because genes belonging to this gene family play a crucial role in rice tolerance to submergence, a process closely related to, but independent from, the ability to germinate under anoxia. Genes coding for some enzymes requiring oxygen for their activity are dramatically down-regulated under anoxia, suggesting the existence of an energy-saving strategy in the regulation of gene expression.
Higher plants are aerobic organisms that rapidly die when oxygen availability is limited due to soil flooding (Voesenek et al., 2006). Species originating from semiaquatic environments are, however, able to cope with flooding stress. They can survive complete submergence for weeks and some even have the capacity to grow vigorously and produce flowers and seeds in permanently water-saturated soils. In this context, a well-known crop is rice (Oryza sativa), which produces high yields even when it is grown in water-logged rice paddies. A broad range of metabolic and morphological adaptations characterizes these tolerant species. Flood-tolerant plants have developed the capacity to generate ATP without the presence of oxygen (fermentative metabolism) and/or to develop specific morphologies (e.g. air channels, enhanced shoot elongation) that improve the entrance of oxygen (Jackson, 1985; Crawford, 1992; Perata and Alpi, 1993; Armstrong et al., 1994; Drew et al., 2000; Sauter, 2000; Colmer, 2003; Gibbs and Greenway, 2003; Voesenek et al., 2006). Gene expression studies on plants exposed to low oxygen revealed the up-regulation of genes coding for transcription factors (Hoeren et al., 1998; Liu et al., 2005), signal transduction components (Baxter-Burrell et al., 2002), nonsymbiotic hemoglobin (Dordas et al., 2004), ethylene biosynthesis (Vriezen et al., 1999), nitrogen metabolism (Mattana et al., 1994), and cell wall loosening (Saab and Sachs, 1996). At the protein level, low oxygen selectively induces the synthesis of proteins known as anaerobic proteins, most of which are enzymes involved in sugar metabolism, glycolysis, and fermentation pathways (Sachs et al., 1980; Huang et al., 2005).
Despite knowledge on adaptive mechanisms and on regulation at the gene and protein level, our understanding of the mechanisms behind plant responses to anaerobiosis is very limited. Even flood-intolerant species, such as Arabidopsis (Arabidopsis thaliana), switch on many genes that are generally associated with responses to flooding (Klok et al., 2002; Branco-Price et al., 2005; Gonzali et al., 2005; Loreti et al., 2005). This strongly suggests that the regulation of flooding tolerance in plants is far more complex than anticipated for many years.
In rice, flooding tolerance—defined as the survival after several days of complete submergence of the entire plant—is strongly affected by a locus (Sub1) on chromosome 9 (Siangliw et al., 2003). This locus explains a large proportion of the variation in flooding tolerance between ‘indica’ (tolerant) and ‘japonica’ (intolerant; Toojinda et al., 2003). Two ethylene response factors (ERFs) on this locus inhibit ethylene production and underwater elongation and stimulate glycolysis and fermentation (Fukao et al., 2006). The importance of low elongation rates for the flooding tolerance of rice was already established by Setter and Laureles (1996). Introduction of the Sub1 locus into the background of ‘japonica’ significantly increased its flooding tolerance, thus demonstrating the importance of the Sub1 locus for flooding tolerance (Fukao et al., 2006). Identification of the Sub1 locus and the elucidation of its role in the adaptation of rice to submergence is a breakthrough in plant adaptation to anaerobiosis. It must be highlighted, however, that the ability of rice seeds to germinate under complete anoxia (Alpi and Beevers, 1983) is not likely to be explained in terms of Sub1 genes. Ethylene, the trigger for Sub1A expression, is not produced under anoxia and, indeed, ‘M202’ and ‘Nipponbare,’ both lacking the Sub1A gene (Xu et al., 2006), display better germination under anoxia when compared with the FR13A genotype (P. Perata, unpublished data) that possesses the Sub1A gene (Xu et al., 2006). Molecular mechanisms allowing rice to elongate the coleoptile in complete absence of oxygen—a behavior not observed in any other cereal—are thus largely unknown. In this article, we present transcript profiling of the anoxic rice coleoptile compared to its aerobic counterpart.
RESULTS AND DISCUSSION
Anaerobic Responses in Rice
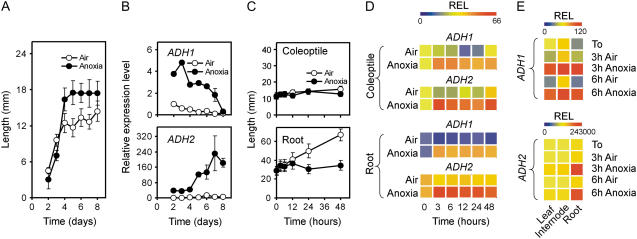
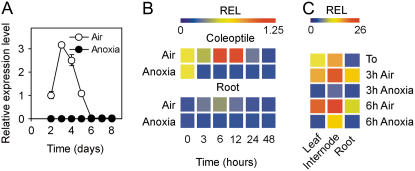
The ability of rice to germinate under anoxia is comparable to that in air, but only the coleoptile elongates (Fig. 1A), whereas both the root and the primary leaf fail to grow (Alpi and Beevers, 1983). Rice coleoptiles show enhanced expression of alcohol dehydrogenase (ADH) genes and distinct expression patterns are observed for ADH1 and ADH2, the latter being induced at a higher level (Fig. 1B). When air-germinated seedlings are transferred to anoxia, the coleoptile fails to elongate further and only the aerobic root continues to grow (Fig. 1C). Both ADH1 and ADH2 are rapidly induced in the coleoptile and root from aerobic seedlings transferred to anoxia (Fig. 1D). Twenty-day-old rice plants transferred to anoxia for 3 h show induction of ADH1 in leaves, internodes, and roots, whereas after 3 to 6 h of anoxia strong expression of ADH2 is observed, mostly in the roots (Fig. 1E). We chose to use 4-d-old coleoptiles for exploring the anoxic coleoptile transcriptome. Four days after germination, both the anoxic and aerobic coleoptiles are close to their maximal length (Fig. 1A) and the aerobic coleoptile is large enough to allow fast and clean removal of the primary leaf, avoiding extraction of leaf mRNA that would otherwise contaminate coleoptile mRNA preparations. Analysis of the microarray results (Supplemental Table S2) indicate that anoxia exerted a dramatic effect on the coleoptiles, with 1,364 probe sets showing increased expression and 1,770 probe sets indicating decreased expression after data filtering and selection of differentially expressed genes using a low (1%) false discovery rate (FDR) threshold (Supplemental Table S2).
Figure 1.
Effects of anoxia on rice coleoptile growth and ADH gene expression. A, Growth pattern of rice coleoptiles in air and anoxia; rice seeds were germinated in air and anoxia and the coleoptile length recorded daily. Data are means of 20 replicates ±sd. B, Pattern of expression of ADH1 (Os11g10480) and ADH2 (Os11g10510) genes in coleoptiles from rice seeds germinated in air and anoxia. Relative expression level (REL), measured by real-time reverse transcription (RT)-PCR, is shown as a heat map (REL, 1 = expression data from the aerobic coleoptile at day 2). Data are means of three replicates ±sd. C, Growth pattern of rice coleoptiles and roots from seedlings germinated under aerobic conditions for 4 d and then transferred to anoxia for an additional 48-h period; the coleoptile and root length were recorded during 48-h anoxic treatment as well as in air. Data are means of 20 replicates ±sd. D, Pattern of expression of ADH1 and ADH2 genes in the coleoptiles and roots from rice seeds germinated in air for 4 d and then transferred to anoxia for an additional 48-h period; REL, measured by real-time RT-PCR, is shown as a heat map (REL, 1 = expression data from the aerobic coleoptile at the beginning of the experiment, t = 0); data are means of three replicates (see Supplemental Table S3 for ±sd values). E, Pattern of expression of ADH1 and ADH2 genes in the leaves, internodes, and roots from rice plants grown in air for 20 d and then transferred to anoxia for an additional 3- to 6-h period; REL, measured by real-time RT-PCR, is shown as a heat map (REL, 1 = expression data from the aerobic leaf at the beginning of the experiment, t = 0); data are means of three replicates (see Supplemental Table S3 for ±sd values).
Anoxia Induces Genes Involved in Glycolysis, Pyruvate Metabolism, Fermentation, and a Futile Cycle of Starch Synthesis and Degradation
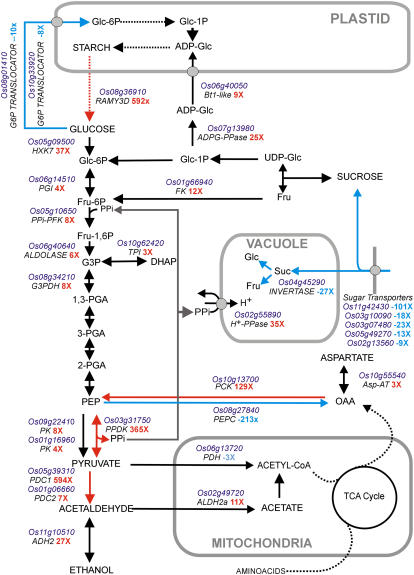
Several genes encoding glycolytic enzymes show enhanced mRNA accumulation, thus confirming the previous results based on enzymatic assays (Guglielminetti et al., 1995a). Hexokinase activity, which is crucial for channeling Glc into glycolysis, is enhanced under anoxia in rice (Guglielminetti et al., 1995a) and our microarray data reveal that OsHXK7 (Os05g09500) encodes for the previously measured anoxic hexokinase activity (Guglielminetti et al., 1995a). The transcripts coding for the other nine rice hexokinase genes (Cho et al., 2006) are expressed at low levels in the anoxic coleoptiles (see Supplemental Tables S1 and S2). Remarkably, OsHXK7 is a starvation-induced gene (Cho et al., 2006) and its induction under anoxia is likely a consequence of the lower sugar content of anoxic coleoptiles (Alpi and Beevers, 1983). Interestingly, genes involved in pyruvate metabolism are strongly up-regulated (Fig. 2). A transcript encoding phosphoenolpyruvate (PEP) carboxykinase (PCK) is among the genes showing high induction by anoxia, and pyruvate orthophosphate dikinase (PPDK) and pyruvate decarboxylase (PDC) are also strongly induced (Fig. 2). PPDK induction at the protein level in rice under hypoxia has been reported by Moons et al. (1998) and Huang et al. (2005). It is suggested that PPDK may provide pyrophosphate (PPi) for the PPi-dependent activity of phosphofructokinase (PPi-PFK), whose mRNA accumulation is increased under anoxia (Fig. 2). PPi is also needed for the activity of the PPi-dependent vacuolar H+ translocase (H+-PPase; Fig. 2; Carystinos et al., 1995). Activity of PEP carboxylase (PEPC) is repressed under anoxia (Fig. 2). PEPC is highly expressed in the aerobic coleoptile and would drain PEP from glycolysis in a reaction that, if not down-regulated under anoxia, would compete with PCK, which is absent under aerobic conditions, but strongly expressed under anoxia (Fig. 2; Supplemental Table S2). The role of PCK in the anaerobic metabolism is unexplored, but it is tempting to speculate that its activity may be useful to channel amino acids through oxaloacetate into glycolysis through a cataplerotic reaction (Owen et al., 2002). Expression of Asp aminotransferase (Asp-AT), an enzyme involved in cataplerosis (Owen et al., 2002), is high and enhanced under anoxia (Fig. 2; Supplemental Table S2).
Figure 2.
Effects of anoxia on carbohydrate metabolism in rice coleoptiles. Data mining of the transcriptome of aerobic and anoxic coleoptiles allowed the identification of the genes involved in starch and Suc metabolism, sugar transport, and glycolysis. Genes showing a statistically significant change in expression when the aerobic dataset was compared with the anoxic dataset (see “Materials and Methods”) are reported on the metabolic pathway shown in this figure. Dotted lines summarize metabolic steps catalyzed by several enzymes. Red arrows highlight the metabolic steps that, based on transcripts level changes, are strongly up-regulated under anoxia, whereas blue arrows indicate down-regulation. It should be remarked that increased mRNA level relative to an enzymatic step does not necessarily imply increased activity of the enzyme in vivo. The fold change (nX, air versus anoxia) is shown together with the gene abbreviation (see text). Fold changes are means of two biological replicates.
Genes coding for PDC and ADH2 are highly expressed in anoxic coleoptiles and a mitochondrial aldehyde dehydrogenase (ALDH2a), which is involved in the metabolism of the potentially toxic acetaldehyde (Perata and Alpi, 1991), is also induced under anoxia (Nakazono et al., 2000; Fig. 2). Expression of ALDH2a is usually associated with the ability to metabolize acetaldehyde during reaeration because, although mRNA accumulates during low-oxygen treatment and not upon reaeration, enzyme activity increases only when plants are transferred from submergence to aerobic conditions (Tsuji et al., 2003). ALDH activity correlates with anoxia tolerance in varieties of Echinocloa crus-galli and it has been proposed that ALDH may detoxify acetaldehyde formed through alcoholic fermentation during anaerobic germination (Fukao et al., 2003). Alternatively, PDC and ALDH2a may represent part of a pyruvate dehydrogenase (PDH) bypass (down-regulated under anoxia; Fig. 2) as proposed by Mellema et al. (2002).
Strong expression of α-amylase (RAMY3D) indicates that starch also plays a role in the coleoptiles, besides the known role of starch degradation in the seed endosperm under anoxia (Perata et al., 1992, 1993; Guglielminetti et al., 1995b). Anoxic rice coleoptiles contain amyloplasts with low starch content (Perata and Alpi, 1993), a likely consequence of active starch degradation. Interestingly, some genes involved in starch synthesis are also up-regulated by anoxia, which suggests that an apparently futile cycle of starch synthesis degradation is active in anoxic coleoptiles. The need to avoid starch-depleted amyloplasts, which would negatively affect the gravitropic response of the coleoptiles (Kutschera et al., 1991; Kutschera and Hoss, 1995), may explain the operation of a possible moderate flux of sugars to starch even under anoxia. Only a few genes involved in carbohydrate metabolism are repressed under anoxia. Suc transport appears to be severely impaired, with most translocators showing reduced expression under anoxia. The inability to transport sugars from the starchy endosperm to the growing coleoptile may explain why rice coleoptiles suffer from sugar starvation (Alpi and Beevers, 1983) despite the availability of starch degradation products in the endosperm (Perata et al., 1992). Suc degradation in the vacuole appears to be repressed, as well as translocation of Glc-6-P to the amyloplasts, a pathway that would drain Glc-6-P from glycolysis.
Effects of Anoxia on α-Amylases
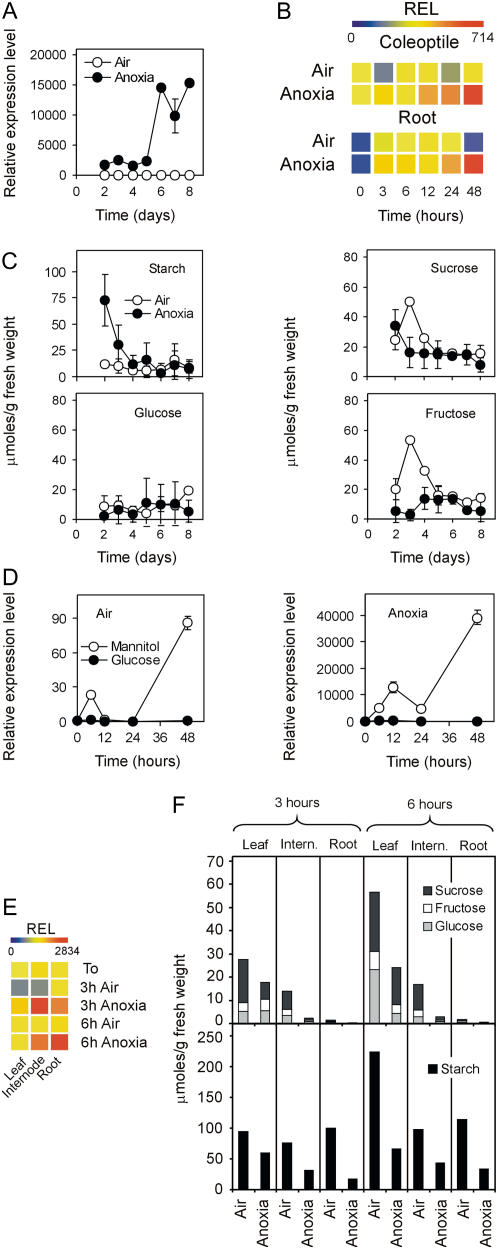
Seven α-amylase genes, out of a total of 10 genes in the rice genome (Huang et al., 1990), are represented on the rice GeneChip. Only RAMY3D is significantly modulated by anoxia (879-fold; see Supplemental Table S2). Detailed analysis of the expression pattern of RAMY3D in the anoxic rice coleoptile reveals that induction under anoxia is much stronger when anoxia is prolonged up to 6 to 8 d, with an anoxic mRNA level 15,000-fold higher than in air (Fig. 3A). Anoxia also induces RAMY3D in the coleoptiles and roots of seedlings transferred from air to anoxia (Fig. 3B). Because RAMY3D is known to be a starvation-induced gene (Yu et al., 1996; Loreti et al., 2003), we analyzed and compared the carbohydrate content of the aerobic and anoxic coleoptiles. Whereas starch is present at a relatively low and constant level in aerobic coleoptiles, this polysaccharide is present in the anoxic coleoptiles from 2-d-old seedlings, a likely consequence of slower use of starch in this tissue during the initial phases of germination under anoxia. Starch is, however, rapidly degraded during the subsequent days of anoxic germination (Fig. 3C), indicating that the expression of RAMY3D has relevant consequences on starch metabolism in anoxic coleoptiles. Whereas the aerobic coleoptile displays relatively high Suc and Fru content during the first 4 d of germination, the Suc, Glc, and Fru content of the anoxic coleoptile is low during all 8 d under analysis (Fig. 3C). Overall, anoxic coleoptiles are exposed, during the first 5 d under anoxia, to an average soluble sugar concentration 3.2-fold lower than the aerobic one, suggesting that anoxia may indirectly induce RAMY3D as a consequence of the lower sugar content of the anoxic coleoptiles and also in light of the results of Umemura et al. (1998), indicating that Glc, Fru, and Suc act equally as signaling molecules repressing RAMY3D expression. Strong induction of RAMY3D in 6- to 8-d-old coleoptiles may reflect the consequences of prolonged sugar starvation of the coleoptiles, rather than a response to the sugar level itself, because aerobic coleoptiles do not show RAMY3D induction at days 6 to 8 despite sugar content similar to that of anoxic coleoptiles. Furthermore, it must be highlighted that aerobic coleoptiles are flaccid and senescent at days 6 to 8, and this may prevent induction of RAMY3D despite the low sugar level observed in older aerobic coleoptiles.
Figure 3.
Effects of anoxia on α-amylase gene expression. A, Pattern of expression of the RAMY3D (Os08g36910) gene in coleoptiles from rice seeds germinated in air and anoxia; relative expression level (REL), measured by real-time reverse transcription (RT)-PCR, is shown (REL, 1 = expression data from the aerobic coleoptile at day 2); data are means of three replicates ±sd. B, Pattern of expression of the RAMY3D gene in the coleoptiles and roots from rice seeds germinated in air for 4 d and then transferred to anoxia for an additional 48-h period; REL, measured by real-time RT-PCR, is shown as a heat map (REL, 1 = expression data from the aerobic coleoptile at the beginning of the experiment, t = 0); data are means of three replicates (see Supplemental Table S3 for ±sd values). C, Starch, Suc, Fru, and Glc content in the coleoptiles from rice seeds germinated in air and anoxia. Data are mean ± sd of three replicates. D, Effects of Glc (100 mm) and mannitol (100 mm, used as an osmotic control) on the expression of the RAMY3D gene in the coleoptiles dissected from 4-d-old aerobically germinated seedlings; REL, measured by real-time RT-PCR, is shown in the graph (REL, 1 = expression data from the aerobic coleoptile at the beginning of the experiment, t = 0); data are means of three replicates ±sd. E, Pattern of expression of the RAMY3D gene in the leaves, internodes, and roots from rice plants grown in air for 20 d and then transferred to anoxia for an additional 3- to 6-h period; REL, measured by real-time RT-PCR, is shown as a heat map (REL, 1 = expression data from the aerobic leaf at the beginning of the experiment, t = 0); data are means of three replicates (see Supplemental Table S3 for ±sd values). F, Starch, Suc, Fru, and Glc content in leaves, internodes, and roots from 20-d-old rice plants treated under anoxia for 3 to 6 h. Data are means of three replicates (±sd did not exceed 20% of the reported data).
To obtain evidence of the ability of sugars to prevent anoxic induction of RAMY3D, we tested the effect of exogenous Glc and Suc on the expression of this α-amylase gene. The results, which are reported in Figure 3D, demonstrate that induction of RAMY3D under anoxia is completely abolished when coleoptiles are incubated in the presence of exogenous Glc, thus indicating that anoxia induces this gene indirectly as a consequence of the low sugar content of the coleoptile. Comparable results were obtained using Suc as exogenous sugar (data not shown). It must be highlighted that coleoptiles, which are dissected from the seedling and incubated without exogenous sugars, display a 40,000-fold induction of RAMY3D under anoxia in 48 h (Fig. 3D) compared to only 700-fold when the whole seedlings are subjected to anoxia (Fig. 3B), which indicates that flux of sugars from the endosperm to the coleoptile is also operating under anoxia and prevents an otherwise dramatic increase in the RAMY3D transcript level. This suggests that down-regulation of the sugar transporters shown in Figure 2 may reduce, but not abolish, the transfer of nutrients from the endosperm to the coleoptile. Anoxia induces RAMY3D also in the internodes and roots from 20-d-old rice plants treated under anoxia (Fig. 3E). Effects of anoxia on the carbohydrate content of leaves, internodes, and roots are in agreement with sugars playing a signaling role under anoxia. Starch is degraded under anoxia in all tissues investigated, including leaves that show only moderate induction of RAMY3D under anoxia, suggesting that, besides RAMY3D, other amylolytic activities may be involved in starch breakdown in anoxic leaves (Fig. 3F). Rice leaves display higher sugar content under anoxia when compared to internodes and roots (Fig. 3F). This likely hampers the anoxic induction of RAMY3D that is instead strongly induced in roots and internodes, where lower sugar content is observed under anoxia (Fig. 3F). The low level of sugars in the aerobic roots is, however, not sufficient for the induction of RAMY3D, although further reduction in sugar content following anoxic treatment correlates with the induction of RAMY3D. Overall, it appears that sugars act as a signaling molecule under anoxia, but the level of sugar itself appears to be required, but not sufficient, to activate the signaling pathway triggering induction of genes such as RAMY3D. We tested the effect of exogenous Glc on the expression of ADH1 and ADH2 genes and the results indicate that Glc is unable to counteract the anoxic induction of ADH genes, although the induction is reduced by about 40% when Glc is added to the incubation medium (data not shown). Therefore, Glc availability appears to play an important role in the modulation of some anoxia-induced genes, although the regulation of genes, such as ADH, is relatively independent of sugar availability.
Rice Expansins EXPA7 and EXPB12 Are Up-Regulated in the Anoxic Rice Coleoptile
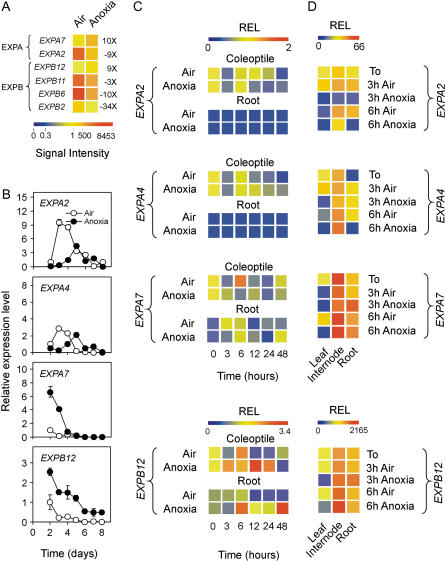
Coleoptile elongation under anoxia is due to cell expansion and expansins likely play a major role in this process (Cosgrove, 1999; Huang et al., 2000). Twenty-two α-expansins (EXPA) and 12 β-expansins (EXPB) are represented on the GeneChip, but only six are differentially expressed in the anoxic rice coleoptile (Fig. 4A). Only EXPA7 and EXPB12 appear to be more expressed under anoxia than in air (Fig. 4A). A previous report indicated EXPA2 and EXPA4 as possibly involved in anoxic coleoptile elongation (Huang et al., 2000). We therefore compared the patterns of expression of EXPA2, EXPA4, EXPA7, and EXPB12 in coleoptiles from aerobic and anaerobic seedlings. The results, which are shown in Figure 4B, indicate that, whereas expression of both EXPA2 and EXPA4 in air is compatible with the growth of the aerobic coleoptile, these mRNAs are comparatively less abundant under anoxia during the first 4 d of germination when most anoxic coleoptile growth is observed (Fig. 1A). The mRNA level of EXPA7 and EXPB12 are instead higher under anoxia at 2 to 3 d after germination, which coincides with the anoxic coleoptile elongation phase, making these expansins good candidates for explaining the anoxic growth of the rice coleoptile. EXPA2, EXPA4, and EXPA7 are not affected by the transfer of seedlings or 20-d-old rice plants from air to anoxia (Fig. 4, C and D), thus suggesting that EXPA7 is developmentally regulated in the anoxic rice coleoptile. EXPB12 is instead transiently up-regulated in coleoptiles and roots transferred from air to anoxia (Fig. 4C) and moderately induced in the internodes kept for 6 h under anoxia (Fig. 4D).
Figure 4.
Effects of anoxia on expansin gene expression. A, Pattern of expression of the expansin genes differentially regulated by anoxia; data are expressed as a heat map showing the signal intensity from the microarray experiment (data are means of two GeneChip experiments) performed on coleoptiles from rice seeds germinated in air and anoxia for 4 d. Fold change is shown on the right side of the heat map (FDR P value < 0.01). TIGR gene codes are as follows: EXPA2 = Os01g60770; EXPA7 = Os03g60720; EXPB2 = Os10g40710; EXPB6 = Os10g40700; EXPB11 = Os02g44108; and EXPB12 = Os03g44290. B, Pattern of expression of expansin genes EXPA2, EXPA4 (Os05g39990), EXPA7, and EXPB12 in the coleoptiles from rice seeds germinated in air and anoxia; relative expression level (REL), measured by real-time reverse transcription (RT)-PCR, is shown (REL, 1 = expression data from the aerobic coleoptile at day 2); data are means of three replicates ±sd. C, Pattern of expression of the expansin genes EXPA2, EXPA4, EXPA7, and EXPB12 in the coleoptiles and roots from rice seeds germinated in air for 4 d and then transferred to anoxia for an additional 48-h period; REL, measured by real-time RT-PCR, is shown as a heat map (REL, 1 = expression data from the aerobic coleoptile at the beginning of the experiment, t = 0); data are means of three replicates (see Supplemental Table S3 for ±sd values). D, Pattern of expression of the expansin genes EXPA2, EXPA4, EXPA7, and EXPB12 in the leaves, internodes, and roots from rice plants grown in air for 20 d and then transferred to anoxia for an additional 3- to 6-h period; REL, measured by real-time RT-PCR, is shown as a heat map (REL, 1 = expression data from the aerobic leaf at the beginning of the experiment, t = 0); data are means of three replicates (see Supplemental Table S3 for ±sd values).
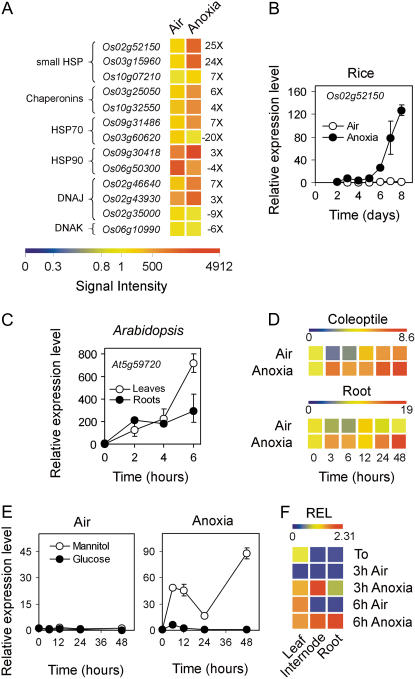
Genes Coding for Heat Shock Proteins Are Induced by Anoxia-Driven Sugar Starvation
Heat shock proteins (HSPs) are not only expressed in plants experiencing high-temperature stress, but also are expressed in response to a wide range of other environmental stresses, such as water, salinity, osmotic, cold, and oxidative stress (for review, see Wang et al., 2004). Indeed, HSPs play a crucial role in protecting plant cells against stressful conditions and in reestablishing cellular homeostasis (Wang et al., 2004). It has recently been demonstrated that heat pretreatment strongly enhances anoxia tolerance in Arabidopsis (Loreti et al., 2005), suggesting that HSPs may play a role in anoxia tolerance. A large number of genes coding for HSPs are represented on the rice GeneChip: 13 are differentially expressed under anoxia, nine are up-regulated, and four are down-regulated (Fig. 5A). Rice HSP20 (Os02g52150) shows higher expression under anoxia, with a peak in 6- to 8-d-old anoxic rice coleoptiles (Fig. 5B). A nearly identical pattern of expression was observed analyzing the pattern of expression of rice HSP17.4 (Os03g16030; data not shown). We compared expression of rice HSP20 with expression of an Arabidopsis small HSP known to be anoxia enhanced (Loreti et al., 2005). In Arabidopsis, induction of HSP25.3-P (At5g59720) by anoxia is rapid, with 700-fold induction in the leaves of seedlings after only 6 h under anoxia (Fig. 5C). All other anoxia-induced Arabidopsis small HSP genes (Loreti et al., 2005) show very similar fast induction under anoxia (data not shown). In rice, instead, induction of HSP20 (Os02g52150) is a slow process, with anoxic induction comparable to that observed in Arabidopsis after only 1 week of anoxic growth of the rice coleoptile (Fig. 5B). We checked whether the faster induction of HSP25.3-P observed in Arabidopsis (Fig. 5C) is a consequence of the air-to-anoxia transfer of the seedling. Aerobically germinated rice seedlings transferred to anoxia show an induction of HSP20 in both the coleoptile and roots (Fig. 5D) at a level well below that of Arabidopsis leaves and roots (Fig. 5C).
Figure 5.
Effects of anoxia on HSP gene expression. A, Pattern of expression of HSP genes differentially regulated by anoxia; data are expressed as a heat map showing the signal intensity from the microarray experiment (data are means of two GeneChip experiments) performed on coleoptiles from rice seeds germinated in air and anoxia for 4 d. HSP categories (left side of the heat map) are as described by Wang et al. (2004). Fold change is shown on the right side of the heat map (FDR P value < 0.01). B, Pattern of expression of Os02g52150 in the coleoptiles from rice seeds germinated in air and anoxia; relative expression level (REL), measured by real-time reverse transcription (RT)-PCR, is shown (REL, 1 = expression data from the aerobic coleoptile at day 2); data are means of three replicates ±sd. C, Pattern of expression of At5g59720 in Arabidopsis shoots and roots (10-d-old seedlings germinated in air and then transferred to anoxia for up to 6 h). REL, measured by real-time RT-PCR, is shown in the graph (REL, 1 = expression data from the Arabidopsis aerobic leaves at t = 0); data are means of three replicates ±sd. D, Pattern of expression of Os02g52150 in the coleoptiles and roots from rice seeds germinated in air for 4 d and then transferred to anoxia for an additional 48-h period; REL, measured by real-time RT-PCR, is shown in the heat map (REL, 1 = expression data from the aerobic coleoptile at the beginning of the experiment, t = 0); data are means of three replicates (see Supplemental Table S3 for ±sd values). E, Effects of Glc (100 mm) and mannitol (100 mm, used as an osmotic control) on the expression of the HSP gene Os02g52150 in the coleoptiles dissected from 4-d-old aerobically germinated seedlings; REL, measured by real-time RT-PCR, is shown in the graph (REL, 1 = expression data from the aerobic coleoptile at the beginning of the experiment, t = 0); data are means of three replicates ±sd. F, Pattern of expression of the HSP gene Os02g52150 in the leaves, internodes, and roots from rice plants grown in air for 20 d and then transferred to anoxia for an additional 3- to 6-h period; REL, measured by real-time RT-PCR, is shown as a heat map (REL, 1 = expression data from the aerobic leaf at the beginning of the experiment, t = 0); data are means of three replicates (see Supplemental Table S3 for ±sd values).
In Arabidopsis, anoxic induction of HSPs is enhanced by exogenous Suc (Loreti et al., 2005). When we tested the effect of exogenous sugars on the expression of HSP20 in rice, we realized that induction by anoxia is totally abolished in the presence both of Glc (Fig. 5E) and Suc (data not shown). It appears, therefore, that HSP20 is induced by anoxia as a consequence of sugar starvation occurring in the coleoptiles, which had been grown under anoxia for several days, rather than as a direct consequence of anoxia. This unveils a different effect of sugars on the anoxic induction of HSPs in Arabidopsis and rice. In Arabidopsis, a lipid-storing seed, exogenous Suc is needed to fuel the fermentative metabolism (P. Perata, unpublished data), but, although Suc feeding prolongs Arabidopsis survival to anoxia, it cannot prevent the rapid death of the seedlings. We speculate that, in Arabidopsis, Suc prevents an otherwise dramatically rapid metabolic collapse, allowing Arabidopsis to detect the protein dysfunction requiring the expression of HSPs (Wang et al., 2004). In rice, starch degradation delays sugar starvation (Perata et al., 1993), and the well-coordinated up-regulation of the fermentative metabolism (Fig. 2) allows rice to avoid the stressful condition triggering HSP induction. After only 1 week of anoxic germination, the coleoptile starts suffering from prolonged sugar starvation, the fermentative metabolism slows down, and the coleoptile experiences impaired metabolism triggering the induction of HSPs, a condition occurring only after a few hours of anoxia in Arabidopsis. Interestingly, HSP20 is also induced in tissues of 20-d-old plants treated under anoxia (Fig. 5F).
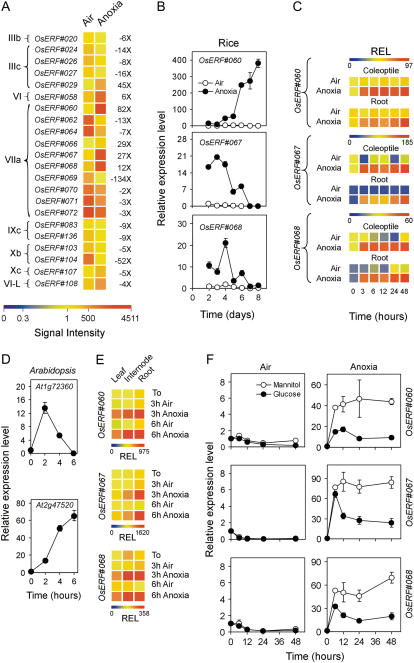
ERFs Are Induced by Anoxia in Both Rice and Arabidopsis
The mRNA level of several ERF-like transcription factors is enhanced by anoxia both in rice (Supplemental Table S2) and Arabidopsis (Loreti et al., 2005). Remarkably, the submergence tolerance of the rice variety FR13A is linked to a major quantitative trait locus, known as Submergence1 (Sub1), on chromosome 9 (Xu and Mackill, 1996). Xu et al. (2006) revealed that the Sub1 region encodes three transcription factors (Sub1A, Sub1B, and Sub1C) belonging to the B-2 subgroup of the ERFs/ethylene-responsive element-binding proteins (EREBPs)/APETALA2 (AP2)-like proteins. Transcription of both Sub1A and Sub1C is strongly up-regulated by submergence and down-regulated by desubmergence. The third ERF, Sub1B, is only slightly regulated upon submergence. It has to be pointed out that the ability of rice to germinate under complete anoxia cannot be explained in terms of Sub1A expression. Ethylene, the trigger for Sub1A expression, is not produced under anoxia. Furthermore, ‘Nipponbare’ lacks the Sub1A gene (Fukao et al., 2006; Xu et al., 2006), but shows vigorous germination under anoxia (Fig. 1A). This does not rule out the possibility that other ERFs may play a role in anoxia tolerance at the germination stage. A large number (111) of ERF family genes (Nakano et al., 2006) is represented on the rice GeneChip, six and 16 of them showing induction and repression, respectively, by anoxia (Fig. 6A). Sub1B (OsERF#063) is not expressed in rice coleoptiles (see Supplemental Table S1), whereas Sub1C (OsERF#073) is down-regulated under anoxia (FDR P value = 0.032; Supplemental Table S2), an expected result because this gene is up-regulated by ethylene (Fukao et al., 2006), which is a hormone requiring oxygen for its synthesis. Anoxia-induced ERFs are thus unlikely to be ethylene modulated and may play a role in the adaptation of plants to a complete lack of oxygen. To further confirm the induction of ERFs under anoxia, we selected three ERF genes (OsERF#060, OsERF#067, OsERF#068), all belonging to subgroup B-2 in the ERF gene family (Sakuma et al., 2002; corresponding to the group VIIa, according to the classification of Nakano et al. [2006]), similar to Sub1B and Sub1C genes (Xu et al., 2006). These three ERF genes were chosen because of their strong induction by anoxia and because of their homology with two Arabidopsis ERFs displaying induction by anoxia (Loreti et al., 2005; At1g72360 showing homology to OsERF#060; At2g47520 showing homology to OsERF#067/OsERF#068; Loreti et al., 2005). The three rice ERFs display distinct patterns of expression in the anoxic coleoptile (Fig. 6B), with OsERF#060 showing a pattern closely similar to that of ADH2 (Fig. 1B) and OsERF#067 mirroring the expression of ADH1 (Fig. 1B). The patterns of expression are instead very similar in the coleoptiles and roots from aerobically germinated seedlings transferred to anoxia (Fig. 6C). In Arabidopsis seedlings treated under anoxia for up to 6 h, At1g72360 is transiently induced by anoxia, whereas At2g47520 induction is prolonged during the 6-h anoxic treatment (Fig. 6D). In 20-d-old rice plants, the three ERFs are anoxia induced in all tissues, although the response in leaves is smaller (Fig. 6E). OsERF#060, OsERF#067, and OsERF#068 induction by anoxia is reduced in the presence of exogenous Glc (Fig. 6F), thus confirming that Glc availability improves the overall status of the seedlings, with consequent lower response in terms of anaerobic gene induction.
Figure 6.
Effects of anoxia on ERF gene expression. A, Pattern of expression of ERF genes differentially regulated by anoxia; data are expressed as a heat map showing the signal intensity and fold change from the microarray experiment (data are means of two GeneChip experiments) performed on the coleoptiles from rice seeds germinated in air and anoxia for 4 d. ERF categories (left side of the heat map) are as described by Nakano et al. (2006). Fold change is shown on the right side of the heat map (FDR P value < 0.01). See Nakano et al. (2006) or Supplemental Table S2 for corresponding TIGR gene codes. B, Pattern of expression of the ERF genes in coleoptiles from rice seeds germinated in air and anoxia; relative expression level (REL), measured by real-time reverse transcription (RT)-PCR, is shown (REL, 1 = expression data from the aerobic coleoptile at day 2); data are means of three replicates ±sd. C, Pattern of expression of rice ERF genes in the coleoptiles and roots from rice seeds germinated in air for 4 d and then transferred to anoxia for an additional 48-h period; REL, measured by real-time RT-PCR, is shown as a heat map (REL, 1 = expression data from the aerobic coleoptile at the beginning of the experiment, t = 0); data are means of three replicates (see Supplemental Table S3 for ±sd values). D, Pattern of expression of the ERF genes in Arabidopsis seedlings (4-d-old seedlings germinated in air and then transferred to anoxia for up to 6 h). REL, measured by real-time RT-PCR, is shown in the graphs (REL, 1 = expression data from the Arabidopsis aerobic seedlings at t = 0); data are means of three replicates ±sd. E, Pattern of expression of rice ERF genes in the leaves, internodes, and roots from rice plants grown in air for 20 d and then transferred to anoxia for an additional 3- to 6-h period; REL, measured by real-time RT-PCR, is shown as a heat map (REL, 1 = expression data from the aerobic leaf at the beginning of the experiment, t = 0); data are means of three replicates (see Supplemental Table S3 for ±sd values). F, Effects of Glc (100 mm) and mannitol (100 mm, used as an osmotic control) on the expression of rice ERF genes in coleoptiles dissected from 4-d-old aerobically germinated seedlings; REL, measured by real-time RT-PCR, is shown in the graph (REL, 1 = expression data from the aerobic coleoptile at the beginning of the experiment, t = 0); data are means of three replicates ±sd.
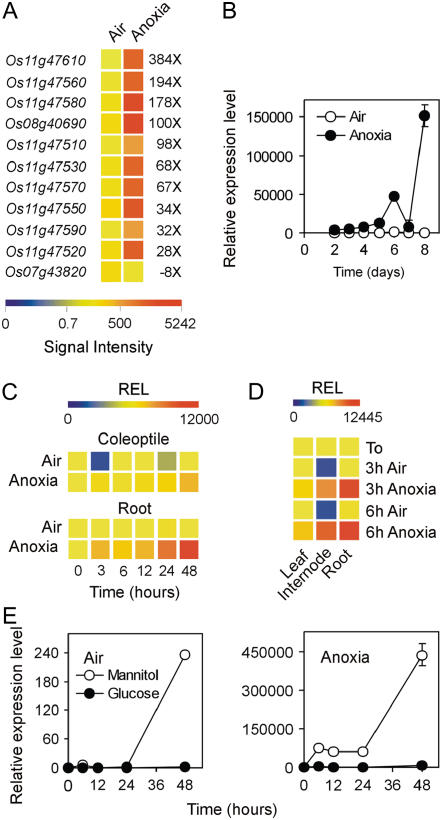
Xylanase Inhibitor Protein Genes Are Strongly Induced by Anoxia-Driven Sugar Starvation
Data mining of microarray data revealed that, surprisingly, xylanase inhibitor protein (XIP) genes were strongly up-regulated under anoxia. XIPs are known to be induced by pathogen-related pathways, as well as by wounding and methyl jasmonate (Igawa et al., 2005). Ten of the 31 genes represented on the GeneChip are induced under anoxia and only one is repressed (Fig. 7A). Some of these XIPs, such as Os11g47560, are among the genes showing the strongest induction under anoxia (Supplemental Table S2). Os11g47560 is indeed induced 150,000-fold in the coleoptile that elongated under anoxia for 8 d (Fig. 7B) and induction is also observed in the roots transferred from air to anoxia (Fig. 7C), as well as in 20-d-old rice plants (Fig. 7D). Treatment of coleoptiles with exogenous Glc revealed that Os11g47560, similar to RAMY3D, is a starvation-induced gene (Fig. 7E). Even in aerobic conditions, surgically dissected coleoptiles (and thus unable to take Glc from the seed endosperm) display induction of Os11g47560 when incubated in mannitol, but such induction is dramatic under anoxia, with a 450,000-fold induction value. The fold induction value for isolated anoxic coleoptiles (Fig. 7E) is much higher than that of coleoptiles sampled from intact seedlings (Fig. 7C), thus confirming the existence of Glc flow from the endosperm to the anoxic coleoptile. Anoxia may induce XIPs to protect seedlings from pathogen infection in the moist environment of flooded soil. Other pathogen or disease-related genes are not up-regulated under anoxia (Supplemental Table S2), and this hypothesis deserves further investigation.
Figure 7.
Effects of anoxia on XIP gene expression. A, Pattern of expression of XIP genes differentially regulated by anoxia; data are expressed as a heat map showing the signal intensity from the microarray experiment (data are means of two GeneChip experiments) performed on coleoptiles from rice seeds germinated in air and anoxia for 4 d. Fold change is shown on the right side of the heat map (FDR P value < 0.01). B, Pattern of expression of the Os11g47560 gene in coleoptiles from rice seeds germinated in air and anoxia; relative expression level (REL), measured by real-time reverse transcription (RT)-PCR, is shown in the graph (REL, 1 = expression data from the aerobic coleoptile at day 2); data are means of three replicates ±sd. C, Pattern of expression of the Os11g47560 gene in coleoptiles and roots from rice seeds germinated in air for 4 d and then transferred to anoxia for an additional 48-h period; REL, measured by real-time RT-PCR, is shown as a heat map (REL, 1 = expression data from the aerobic coleoptile at the beginning of the experiment, t = 0); data are means of three replicates (see Supplemental Table S3 for ±sd values). D, Pattern of expression of the Os11g47560 gene in the leaves, internodes, and roots from rice plants grown in air for 20 d and then transferred to anoxia for an additional 3- to 6-h period; REL, measured by real-time RT-PCR, is shown as a heat map (REL, 1 = expression data from the aerobic leaf at the beginning of the experiment, t = 0); data are means of three replicates (see Supplemental Table S3 for ±sd values). E, Effects of Glc (100 mm) and mannitol (100 mm, used as an osmotic control) on the expression of the Os11g47560 gene in the coleoptiles dissected from 4-d-old aerobically germinated seedlings; REL, measured by real-time RT-PCR, is shown in the graph (REL, 1 = expression data from the aerobic coleoptile at the beginning of the experiment, t = 0); data are means of three replicates ±sd.
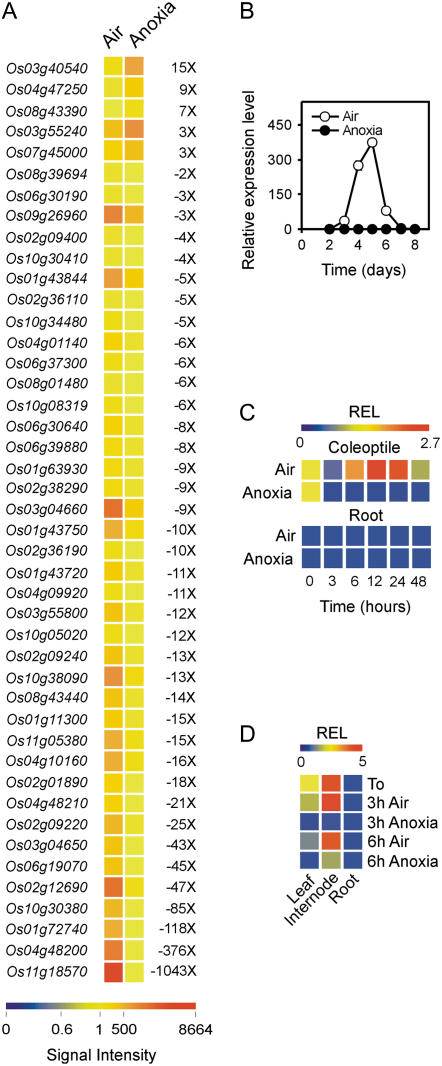
Genes Coding for P450 Are Down-Regulated under Anoxia
A survey of genes that are down-regulated by anoxia revealed that 39 genes coding for P450 enzymes are significantly down-regulated under anoxia, whereas only five genes are up-regulated (Fig. 8A). Cytochromes P450 are heme-containing oxygenases, which transfer a single oxygen atom from O2 to substrates and, therefore, have oxygen-binding capability (Schuler, 1996). Down-regulation of P450s by anoxia prompted us to analyze the detailed pattern of expression of the P450 gene (Os11g18570), which shows highest expression in the aerobic coleoptile and strongest down-regulation under anoxia. Os11g18570 is transiently expressed in the aerobic coleoptile, with a peak of expression coinciding with the end of coleoptile elongation (Fig. 8B). Under anoxia, Os11g18570 is not expressed. We observed rapid down-regulation of Os11g18570 when the aerobically germinated seedlings were transferred to anoxia (Fig. 8C). This P450 gene appears to be expressed in the aerobic coleoptile, but not in the root (Fig. 8C), of rice seedlings. In 20-d-old rice plants, Os11g18570 is predominantly expressed in the internodes, where down-regulation by anoxia can also be observed (Fig. 8D). Glc cannot prevent the rapid decline in the expression of Os11g18570 (data not shown). Due to the diversity of pathways requiring monooxygenase activity, a wide variety of environmental and developmental signals modulate the expression of CYP450 (Dixon and Paiva, 1995; Mizutani et al., 1998). It is tempting to speculate that the anoxia-driven down-regulation of CYP450 genes is linked to the need for avoiding energy waste related to transcription of genes whose products require oxygen.
Figure 8.
Effects of anoxia on P450 gene expression. A, Pattern of expression of P450 genes differentially regulated by anoxia; data are expressed as a heat map showing the signal intensity from the microarray experiment (data are means of two GeneChip experiments) performed on the coleoptiles from rice seeds germinated in air and anoxia for 4 d. Fold change is shown on the right side of the heat map (FDR P value < 0.01). B, Pattern of expression of the Os11g18570 gene in the coleoptiles from rice seeds germinated in air and anoxia; relative expression level (REL), measured by real-time reverse transcription (RT)-PCR, is shown in the graph (REL, 1 = expression data from the aerobic coleoptile at day 2); data are means of three replicates ±sd. C, Pattern of expression of the Os11g18570 gene in coleoptiles and roots from rice seeds germinated in air for 4 d and then transferred to anoxia for an additional 48-h period; REL, measured by real-time RT-PCR, is shown as a heat map (REL, 1 = expression data from the aerobic coleoptile at the beginning of the experiment, t = 0); data are means of three replicates (see Supplemental Table S3 for ±sd values). D, Pattern of expression of the Os11g18570 gene in the leaves, internodes, and roots from the rice plants grown in air for 20 d and then transferred to anoxia for an additional 6-h period; REL, measured by real-time RT-PCR, is shown as a heat map (REL, 1 = expression data from the aerobic leaf at the beginning of the experiment, t = 0); data are means of three replicates (see Supplemental Table S3 for ±sd values).
Anoxia Down-Regulates Catalase
A catalase-coding transcript is strongly down-regulated under anoxia (see Supplemental Table S2). A dramatic reduction in the activity of catalase was previously observed under anoxia (Alpi and Beevers, 1983), whereas hypoxia increased catalase activity (Ushimaru et al., 1999). The authors (Ushimaru et al., 1999) speculate that catalase may convert H2O2 (generated by glyoxysome metabolism) to oxygen under hypoxia, thus alleviating oxygen shortage conditions. We studied in more detail the effects of anoxia on catalase. Three genes coding for catalase are represented on the GeneChip. Os03g03910 is not expressed in rice coleoptiles, Os06g51150 is moderately expressed and unaffected by anoxia, whereas Os02g02400 is strongly expressed in the aerobic coleoptile but absent under anoxia (Fig. 9A). In light of the data of Alpi and Beevers (1983) indicating that over 90% of catalase activity is lost under anoxia, Os02g02400 is the gene that contributes to most of the catalase activity in the rice coleoptile. A time-course analysis shows that Os02g02400 is transiently expressed during aerobic germination, but never under anoxia (Fig. 9A). When aerobically germinated seedlings were transferred to anoxia, the Os02g02400 transcript disappeared within 3 h in the coleoptiles, the only tissues showing expression of this gene (Fig. 9B). Exogenous Glc is unable to counteract the decline in Os02g02400 transcript under anoxia (data not shown). Expression of Os02g02400, which is strong in the internodes of 20-d-old rice plants, is rapidly down-regulated under anoxia (Fig. 9C). It is unclear why anoxia drives rapid down-regulation of catalase. When submerged seedlings were exposed to air, the activities of catalase exceeded the level of catalase activity detected in aerobically grown controls (Ushimaru et al., 1994), which indicates that the H2O2 degradation system is rapidly reconstituted under aerobic conditions. The absence of catalase under anoxia does not appear to affect the development of postanoxic injuries because anoxia-germinated seedlings rapidly adapt to air with root and leaf development (data not shown). It is thus likely that, in the absence of oxygen, production of H2O2 is negligible, making catalase superfluous in the anoxic coleoptile. Down-regulation of catalase can therefore be part of a strategy of metabolism reorganization aimed at producing only the enzymes that are requested for housekeeping, as well as for anaerobic metabolism, with consequent energy saving, which is likely of great importance under anoxia (Perata and Alpi, 1993; Fukao and Bailey-Serres, 2004). An alternative hypothesis can be proposed, assuming that oxygen peroxide can act as a second messenger in low-oxygen signaling (Fukao and Bailey-Serres, 2004; Bailey-Serres and Chang, 2005). Although it is predicted that, under anoxia, H2O2 should be negligible, H2O2 was detected under anoxia (Blokhina et al., 2001), possibly as a consequence of traces of oxygen in the experimental system, thus suggesting that the decline in catalase activity may contribute to the buildup of an H2O2 level acting as a signal.
Figure 9.
Effects of anoxia on catalase gene expression. A, Pattern of expression of the Os02g02400 gene in the coleoptiles from rice seeds germinated in air and anoxia; relative expression level (REL), measured by real-time reverse transcription (RT)-PCR, is shown in the graph (REL, 1 = expression data from the aerobic coleoptile at day 2); data are means of three replicates ±sd. B, Pattern of expression of the Os02g02400 gene in the coleoptiles and roots from rice seeds germinated in air for 4 d and then transferred to anoxia for an additional 48-h period; REL, measured by real-time RT-PCR, is shown as a heat map (REL, 1 = expression data from the aerobic coleoptile at the beginning of the experiment, t = 0); data are means of three replicates (see Supplemental Table S3 for ±sd values). C, Pattern of expression of the Os02g02400 gene in the leaves, internodes, and roots from rice plants grown in air for 20 d and then transferred to anoxia for an additional 6-h period; REL, measured by real-time RT-PCR, is shown as a heat map (REL, 1 = expression data from the aerobic leaf at the beginning of the experiment, t = 0); data are means of three replicates (see Supplemental Table S3 for ±sd values).
CONCLUSION
The ability of the rice coleoptile to elongate under anoxia represents an unveiled enigma, whereas the mechanisms and genes involved in the adaptation of rice plants to submergence have recently been discovered (Fukao et al., 2006; Xu et al., 2006). When rice plants are flooded, the ethylene produced as a consequence of submergence acts as a trigger of Sub1 genes, down-regulating their elongation and thus preserving energy for their survival. This mechanism cannot explain the germination of rice under anoxia, a process that is observed also in rice varieties, such as ‘Nipponbare,’ which was used in our experiments and which lacks the Sub1A gene (Xu et al., 2006), although other anoxia ERFs (Fig. 6) may play a role in anoxia tolerance. The experiments described in this article contribute to a better understanding of the anoxic responses of rice to anoxia at the molecular level. Although the transcript steady-state level detected by microarray analysis does not take into account the possible regulation of transcript translation efficiency (Fennoy and Bailey-Serres, 1995), our results indicate that glycolysis is up-regulated under anoxia. This is supported by a previous work, based on enzyme activity measurements, indicating that anaerobic transcripts are actually translated (Guglielminetti et al., 1995a). The metabolism of pyruvate appears to play an important role beyond the expected induction of PDC, with PCK playing a cataplerotic role, and the production of PPi by PPDK supporting the ATP-independent activity of PPi-PFK and of vacuolar H+-PPase (Fig. 2). Sugar transporters are down-regulated under anoxia and anoxic rice coleoptiles experience sugar starvation (Alpi and Beevers, 1983). It is not unlikely that moderate sugar starvation may represent a signal for the activation of the key genes required for anaerobic metabolism, including hexokinase, regulating carbohydrate entry into glycolysis (Fox et al., 1998). We indeed observed an important role for sugar signaling in the modulation of several genes induced by anoxia, which indicates that sugars may represent important second messengers in low-oxygen signaling. The genes coding for some enzymes that require oxygen for their activity are dramatically down-regulated under anoxia, indicating a possible strategy aimed at preventing the production of enzymes that would not be operative under anoxia, thus saving energy for the expression of genes required for the adaptive response of the rice seedling. Elongation of the coleoptile is the most remarkable phenotype of the anoxic coleoptile, and identification of EXPA7 and EXPB12, rather than EXPA2 and EXPA4 (Huang et al., 2000), as candidates for the expansin involved in the anoxic coleoptile elongation provides the basis for future investigations in this field, including transgenic approaches, as well as the study of the germplasm showing a distinct ability to elongate the coleoptile under anoxia.
MATERIALS AND METHODS
Plant Material
Rice (Oryza sativa ‘Nipponbare’) and Arabidopsis (Arabidopsis thaliana; gl1) seeds were sterilized with diluted bleach (15-min incubation in 1.7% sodium hypochlorite, rinsing and washing in sterile water 10 times) and germinated on sterile filter paper at 28°C (23°C for Arabidopsis) in the dark. Three different treatments were performed: (1) seed germination under anoxia since imbibition; (2) transfer of the aerobically (4-d-old) germinated seedlings to anoxia; and (3) anoxic treatment of the surgically dissected coleoptiles from aerobically (4-d-old) germinated seedlings. RNA samples used for microarray experiments were produced from two independent biological replicates (anoxic treatment no. 1, see above), each resulting from pooling of at least five petri dishes containing 25 seedlings each. The primary leaf was removed from the coleoptiles of air-germinated seedlings (the leaf is absent in anoxia-germinated seedlings). All air/anoxia treatments were performed in the dark at 28°C for rice and 23°C for Arabidopsis. Twenty-day-old plants were used as a source of roots, internodes, and leaves as indicated in the figure legends. Rice plants were grown using a hydroponics system: Seeds were sown in rockwool hydroponic growing media in Hoagland solution. Twenty-day-old plants were treated under anoxia in the light, with the anoxic treatment starting at the beginning of the light treatment. An enclosed anaerobic work station (Anaerobic System model 1025; Forma Scientific) was used to provide an oxygen-free environment for seed germination or seedling incubation. This chamber uses palladium catalyst wafers and desiccant wafers to maintain strict anaerobiosis to <10 μg mL−1 O2 (according to the specifications provided by the manufacturer). High-purity N2 was used for purging the chamber initially and the working anaerobic gas mixture was N2:H2 at a ratio of 90:10.
RNA Isolation, cRNA Synthesis, and Hybridization to Affymetrix GeneChips
Total RNA was extracted from seedling samples using the Ambion RNAqueous kit. RNA quality was assessed by agarose gel electrophoresis and spectrophotometry. RNA was processed for use on Affymetrix Rice Genome GeneChip arrays, as previously described (Loreti et al., 2005). Hybridization, washing, staining, and scanning procedures were performed by Genopolis (University of Milano-Bicocca), as described in the Affymetrix technical manual. Microarray analysis was performed using R/Bioconductor (Gentlemen et al., 2004). Expression measures were obtained using GeneChip Robust Multi-Array (Wu and Irizarry, 2005), a multiarray analysis method estimating probe set signals taking into account the physical affinities between probes and targets. Normalization was done by a quantile method (Bolstad et al., 2003). To reduce the number of noninformative genes, two different filters were applied: The first removes the probe sets presenting an Affymetrix absent call (A) for both conditions; the second eliminates the probe sets showing, as maximum signal in the two conditions, a value ≤ the 95th percentile of the overall absent call signal distribution. This filtering yielded 21,484 probe sets. To identify a statistically reliable number of differentially expressed genes among the two conditions, a linear model was performed (Wettenhall and Smyth, 2004). For assessing differential expression, an empirical Bayesian method (Smyth, 2004) was used to moderate the se of the estimated log-fold changes. To control P values in a context of multiple testing problems, a Benjamini-Hochberg correction of the FDR (Reiner et al., 2003) was applied (adjusted P value ≤ 0.01), leading to 3,134 differentially expressed probe sets. Microarray datasets were deposited in a public repository with open access (accession no. GSE6908; http://www.ncbi.nlm.nih.gov/projects/geo).
Real-Time Reverse Transcription-PCR
RNA was extracted from the seedlings grown as indicated in the figure legends. Total RNA, extracted using the RNAqueous kit (Ambion), according to the manufacturer's instructions, was subjected to DNase treatment using the TURBO DNA-free kit (Ambion). Two micrograms of each sample were reverse transcribed into cDNA using the high-capacity cDNA archive kit (Applied Biosystems). Real-time PCR amplification was carried out with the ABI Prism 7000 sequence detection system (Applied Biosystems), using the primers described in Supplemental Table S4 and the default ABI Prism 7000 PCR program for PCR conditions. Glc-3-P dehydrogenase was used as an endogenous control for rice and Ubiquitin10 was used as an endogenous control for Arabidopsis. TaqMan probes specific for each gene were used. Probe sequences are reported in Supplemental Table S4. PCR reactions were carried out using 50 ng of cDNA and TaqMan Universal PCR master mix (Applied Biosystems), following the manufacturer's protocol. Relative quantitation of each single gene expression was performed using the comparative threshold cycle method, as described in the ABI PRISM 7700 Sequence Detection System User Bulletin Number 2 (Applied Biosystems).
Carbohydrate Analyses
Analysis of Suc, Fru, and Glc were carried out as previously described (Guglielminetti et al., 1995a). Samples (0.05–0.3 g fresh weight) were rapidly frozen in liquid nitrogen and ground to a powder. Samples were then extracted as described by Tobias et al. (1992). After centrifugation, the supernatant was used for the analysis of Suc, Fru, and Glc, whereas the starch-containing pellet was extracted using 10% KOH, centrifuged, and the neutralized supernatant treated with 2.5 units amyloglucosidase (from Rhizopus niger) for 3 h to release Glc. Samples were assayed by coupled enzymatic assay methods measuring the increase in A340. The efficiency of the methods was tested by using known amounts of carbohydrates. Incubations of the samples and standards were carried out at 37°C for 30 min. The reaction mixtures (1 mL) were as follows. Glc: 300 mm Tris-HCl, pH 7.6, 10 mm MgCl2, 2 mm ATP, 0.6 mm NADP, 1 unit hexokinase, 1 unit Glc-6-P dehydrogenase; Fru was assayed as described for Glc plus the addition of 2 units of phosphoglucoisomerase; the increase in A340 was recorded. Suc was first broken down using 85 units of invertase (in 150 mm sodium acetate, pH 4.6) and the resulting Glc was assayed as described above. Starch was quantified on the basis of the Glc units released after the amyloglucosidase treatment.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. Entire dataset of microarray experiments.
Supplemental Table S2. Differentially expressed genes list.
Supplemental Table S3. Real-time PCR data (heat maps in figures).
Supplemental Table S4. List of primers and TaqMan probes.
MIAME Checklist S1.
Supplementary Material
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Pierdomenico Perata (p.perata@sssup.it).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Alpi A, Beevers H (1983) Effects of O2 concentration on rice seedlings. Plant Physiol 71 30–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong W, Brändle R, Jackson MB (1994) Mechanisms of flood tolerance in plants. Acta Bot Neerl 43 307–358 [Google Scholar]
- Bailey-Serres J, Chang R (2005) Sensing and signalling in response to oxygen deprivation in plants and other organisms. Ann Bot (Lond) 96 507–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter-Burrell A, Yang ZB, Springer PS, Bailey-Serres J (2002) RopGAP4-dependent Rop GTPase rheostat control of Arabidopsis oxygen deprivation tolerance. Science 296 2026–2028 [DOI] [PubMed] [Google Scholar]
- Blokhina OB, Chirkova TV, Fagerstedt KV (2001) Anoxic stress leads to hydrogen peroxide formation in plant cells. J Exp Bot 52 1–12 [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP (2003) A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19 185–193 [DOI] [PubMed] [Google Scholar]
- Branco-Price C, Kawaguchi R, Ferreira RB, Bailey-Serres J (2005) Genome-wide analysis of transcript abundance and translation in Arabidopsis seedlings subjected to oxygen deprivation. Ann Bot (Lond) 96 647–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carystinos GD, Macdonald HR, Monroy AF, Dhindsa RS, Poole RJ (1995) Vacuolar H+-translocating pyrophosphatase is induced by anoxia or chilling in seedlings of rice. Plant Physiol 108 641–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho JI, Ryoo N, Ko S, Lee SK, Lee J, Jung KH, Lee YH, Bhoo SH, Winderickx J, An G, et al (2006) Structure, expression, and functional analysis of the hexokinase gene family in rice (Oryza sativa L.). Planta 224 598–611 [DOI] [PubMed] [Google Scholar]
- Colmer TD (2003) Long-distance transport of gases in plants: a perspective on internal aeration and radial oxygen loss from roots. Plant Cell Environ 26 17–36 [Google Scholar]
- Cosgrove DJ (1999) Enzymes and other agents that enhance cell wall extensibility. Annu Rev Plant Physiol Plant Mol Biol 50 391–417 [DOI] [PubMed] [Google Scholar]
- Crawford RMM (1992) Oxygen availability as an ecological limit to plant distribution. Adv Ecol Res 23 93–185 [Google Scholar]
- Dixon RA, Paiva NL (1995) Stress-induced phenylpropanoid metabolism. Plant Cell 7 1085–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dordas C, Hasinoff BB, Rivoal J, Hill RD (2004) Class-1 hemoglobins, nitrate and NO levels in anoxic maize cell-suspension cultures. Planta 219 66–72 [DOI] [PubMed] [Google Scholar]
- Drew MC, He CJ, Morgan PW (2000) Programmed cell death and aerenchyma formation in roots. Trends Plant Sci 5 123–127 [DOI] [PubMed] [Google Scholar]
- Fennoy SL, Bailey-Serres J (1995) Post-transcriptional regulation of gene expression in oxygen-deprived roots of maize. Plant J 7 287–295 [DOI] [PubMed] [Google Scholar]
- Fox TC, Green BJ, Kennedy RA, Rumpho ME (1998) Changes in hexokinase activity in Echinochloa phyllopogon and Echinochloa crus-pavonis in response to abiotic stress. Plant Physiol 118 1403–1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukao T, Bailey-Serres J (2004) Plant responses to hypoxia—Is survival a balancing act? Trends Plant Sci 9 449–456 [DOI] [PubMed] [Google Scholar]
- Fukao T, Kennedy RA, Yamasue Y, Rumpho ME (2003) Genetic and biochemical analysis of anaerobically induced enzymes during seed germination of Echinochloa crus-galli varieties tolerant and intolerant of anoxia. J Exp Bot 54 1421–1429 [DOI] [PubMed] [Google Scholar]
- Fukao T, Xu KN, Ronald PC, Bailey-Serres J (2006) A variable cluster of ethylene response factor-like genes regulates metabolic and developmental acclimation responses to submergence in rice. Plant Cell 18 2021–2034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentlemen RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5 R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs J, Greenway H (2003) Mechanisms of anoxia tolerance in plants. I. Growth, survival and anaerobic catabolism. Funct Plant Biol 30 1–47 [DOI] [PubMed] [Google Scholar]
- Gonzali S, Loreti E, Novi G, Poggi A, Alpi A, Perata P (2005) The use of microarrays to study the anaerobic response in Arabidopsis. Ann Bot (Lond) 96 661–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guglielminetti L, Perata P, Alpi A (1995. a) Effect of anoxia on carbohydrate-metabolism in rice seedlings. Plant Physiol 108 735–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guglielminetti L, Yamaguchi J, Perata P, Alpi A (1995. b) Amylolytic activities in cereal seeds under aerobic and anaerobic conditions. Plant Physiol 109 1069–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeren FU, Dolferus R, Wu YR, Peacock WJ, Dennis ES (1998) Evidence for a role for AtMYB2 in the induction of the Arabidopsis alcohol dehydrogenase gene (ADH1) by low oxygen. Genetics 149 479–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Takano T, Akita S (2000) Expression of alpha-expansin genes in young seedlings of rice (Oryza sativa L.). Planta 211 467–473 [DOI] [PubMed] [Google Scholar]
- Huang N, Sutliff TD, Litts JC, Rodriguez RL (1990) Classification and characterization of the rice alpha-amylase multigene family. Plant Mol Biol 14 655–668 [DOI] [PubMed] [Google Scholar]
- Huang SB, Greenway H, Colmer TD, Millar AH (2005) Protein synthesis by rice coleoptiles during prolonged anoxia: implications for glycolysis, growth and energy utilization. Ann Bot (Lond) 96 703–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igawa T, Tokai T, Kudo T, Yamaguchi I, Kimura M (2005) Taxi-I is significantly induced by biotic and abiotic signals that trigger plant defense. Biosci Biotechnol Biochem 69 1058–1063 [DOI] [PubMed] [Google Scholar]
- Jackson MB (1985) Ethylene and the responses of plants to soil waterlogging and submergence. Annu Rev Plant Physiol 36 145–174 [Google Scholar]
- Klok EJ, Wilson IW, Wilson D, Chapman SC, Ewing RM, Somerville SC, Peacock WJ, Dolferus R, Dennis ES (2002) Expression profile analysis of low-oxygen response in Arabidopsis root cultures. Plant Cell 14 2481–2494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutschera U, Hoss R (1995) Mobilization of starch after submergence of air-grown rice coleoptiles—implications for growth and gravitropism. Bot Acta 108 266–269 [Google Scholar]
- Kutschera U, Siebert C, Masuda Y, Sievers A (1991) Effects of submergence on development and gravitropism in the coleoptile of Oryza-sativa L. Planta 183 112–119 [DOI] [PubMed] [Google Scholar]
- Liu FL, VanToai T, Moy LP, Bock G, Linford LD, Quackenbush J (2005) Global transcription profiling reveals comprehensive insights into hypoxic response in Arabidopsis. Plant Physiol 137 1115–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loreti E, Poggi A, Novi G, Alpi A, Perata P (2005) A genome-wide analysis of the effects of sucrose on gene expression in Arabidopsis seedlings under anoxia. Plant Physiol 137 1130–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loreti E, Yamaguchi J, Alpi A, Perata P (2003) Sugar modulation of alpha-amylase genes under anoxia. Ann Bot (Lond) 91 143–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattana M, Coraggio I, Bertani A, Reggiani R (1994) Expression of the enzymes of nitrate reduction during the anaerobic germination of rice. Plant Physiol 106 1605–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellema S, Eichenberger W, Rawyler A, Suter M, Tadege M, Kuhlemeier C (2002) The ethanolic fermentation pathway supports respiration and lipid biosynthesis in tobacco pollen. Plant J 30 329–336 [DOI] [PubMed] [Google Scholar]
- Mizutani M, Ward E, Ohta D (1998) Cytochrome p450 superfamily in Arabidopsis thaliana: isolation of cDNAs, differential expression, and RFLP mapping of multiple cytochromes p450. Plant Mol Biol 37 39–52 [DOI] [PubMed] [Google Scholar]
- Moons A, Valcke R, Van Montagu M (1998) Low-oxygen stress and water deficit induce cytosolic pyruvate orthophosphate dikinase (PPDK) expression in roots of rice, a C-3 plant. Plant J 15 89–98 [DOI] [PubMed] [Google Scholar]
- Nakano T, Suzuki K, Fujimura T, Shinshi H (2006) Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol 140 411–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazono M, Tsuji H, Li YH, Saisho D, Arimura S, Tsutsumi N, Hirai A (2000) Expression of a gene encoding mitochondrial aldehyde dehydrogenase in rice increases under submerged conditions. Plant Physiol 124 587–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen OE, Kalhan SC, Hanson RW (2002) The key role of anaplerosis and cataplerosis for citric acid cycle function. J Biol Chem 277 30409–30412 [DOI] [PubMed] [Google Scholar]
- Perata P, Alpi A (1991) Ethanol-induced injuries to carrot cells—the role of acetaldehyde. Plant Physiol 95 748–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perata P, Alpi A (1993) Plant responses to anaerobiosis. Plant Sci 93 1–17 [Google Scholar]
- Perata P, Geshi N, Yamaguchi J, Akazawa T (1993) Effect of anoxia on the induction of alpha-amylase in cereal seeds. Planta 191 402–408 [Google Scholar]
- Perata P, Pozueta-Romero J, Akazawa T, Yamaguchi J (1992) Effect of anoxia on starch breakdown in rice and wheat seeds. Planta 188 611–618 [DOI] [PubMed] [Google Scholar]
- Reiner A, Yekutieli D, Benjamini Y (2003) Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics 19 368–375 [DOI] [PubMed] [Google Scholar]
- Saab IN, Sachs MM (1996) A flooding-induced xyloglucan endo-transglycosylase homolog in maize is responsive to ethylene and associated with aerenchyma. Plant Physiol 112 385–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs MM, Freeling M, Okimoto R (1980) The anaerobic proteins of maize. Cell 20 761–767 [DOI] [PubMed] [Google Scholar]
- Sakuma Y, Liu Q, Dubouzet JG, Abe H, Shinozaki K, Yamaguchi-Shinozaki K (2002) DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem Biophys Res Commun 290 998–1009 [DOI] [PubMed] [Google Scholar]
- Sauter M (2000) Rice in deep water: How to take heed against a sea of troubles. Naturwissenschaften 87 289–303 [DOI] [PubMed] [Google Scholar]
- Schuler MA (1996) Plant cytochrome P450 monooxygenases. CRC Crit Rev Plant Sci 15 235–284 [Google Scholar]
- Setter TL, Laureles EV (1996) The beneficial effect of reduced elongation growth on submergence tolerance of rice. J Exp Bot 47 1551–1559 [Google Scholar]
- Siangliw M, Toojinda T, Tragoonrung S, Vanavichit A (2003) Thai jasmine rice carrying QTLch9 (SubQTL) is submergence tolerant. Ann Bot (Lond) 91 255–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK (2004) Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: Article 3 [DOI] [PubMed]
- Tobias RB, Boyer CD, Shannon JC (1992) Alterations in carbohydrate intermediates in the endosperm of starch-deficient maize (Zea mays L.) genotypes. Plant Physiol 99 146–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toojinda T, Siangliw M, Tragoonrung S, Vanavichit A (2003) Molecular genetics of submergence tolerance in rice: QTL analysis of key traits. Ann Bot (Lond) 91 243–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji H, Meguro N, Suzuki Y, Tsutsumi N, Hirai A, Nakazono M (2003) Induction of mitochondrial aldehyde dehydrogenase by submergence facilitates oxidation of acetaldehyde during re-aeration in rice. FEBS Lett 546 369–373 [DOI] [PubMed] [Google Scholar]
- Umemura T, Perata P, Futsuhara Y, Yamaguchi J (1998) Sugar sensing and α-amylase gene repression in rice embryos. Planta 204 420–428 [DOI] [PubMed] [Google Scholar]
- Ushimaru T, Kanematsu S, Shibasaka M, Tsuji H (1999) Effect of hypoxia on the antioxidative enzymes in aerobically grown rice (Oryza sativa) seedlings. Physiol Plant 107 181–187 [Google Scholar]
- Ushimaru T, Shibasaka M, Tsuji H (1994) Resistance to oxidative injury in submerged rice seedlings after exposure to air. Plant Cell Physiol 35 211–218 [Google Scholar]
- Voesenek LACJ, Colmer TD, Pierik R, Millenaar FF, Peeters AJM (2006) How plants cope with complete submergence. New Phytol 170 213–226 [DOI] [PubMed] [Google Scholar]
- Vriezen WH, Hulzink R, Mariani C, Voesenek LACJ (1999) 1-Aminocyclopropane-1-carboxylate oxidase activity limits ethylene biosynthesis in Rumex palustris during submergence. Plant Physiol 121 189–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Vinocur B, Shoseyov O, Altman A (2004) Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci 9 244–252 [DOI] [PubMed] [Google Scholar]
- Wettenhall JM, Smyth GK (2004) LimmaGUI: a graphical user interface for linear modeling of microarray data. Bioinformatics 20 3705–3706 [DOI] [PubMed] [Google Scholar]
- Wu Z, Irizarry RA (2005) Stochastic models inspired by hybridization theory for short oligonucleotide arrays. J Comput Biol 12 882–893 [DOI] [PubMed] [Google Scholar]
- Xu K, Xu X, Fukao T, Canlas P, Maghirang-Rodriguez R, Heuer S, Ismail AM, Bailey-Serres J, Ronald PC, Mackill DJ (2006) Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature 442 705–708 [DOI] [PubMed] [Google Scholar]
- Xu KN, Mackill DJ (1996) A major locus for submergence tolerance mapped on rice chromosome 9. Mol Breed 2 219–224 [Google Scholar]
- Yu SM, Lee YC, Fang SC, Chan MT, Hwa SF, Liu LF (1996) Sugars act as signal molecules and osmotica to regulate the expression of alpha-amylase genes and metabolic activities in germinating cereal grains. Plant Mol Biol 30 1277–1289 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.