Abstract
Microspore-derived embryo (MDE) cultures are used as a model system to study plant cell totipotency and as an in vitro system to study embryo development. We characterized and compared the transcriptome and proteome of rapeseed (Brassica napus) MDEs from the few-celled stage to the globular/heart stage using two MDE culture systems: conventional cultures in which MDEs initially develop as unorganized clusters that usually lack a suspensor, and a novel suspensor-bearing embryo culture system in which the embryo proper originates from the distal cell of a suspensor-like structure and undergoes the same ordered cell divisions as the zygotic embryo. Improved histodifferentiation of suspensor-bearing MDEs suggests a new role for the suspensor in driving embryo cell identity and patterning. An MDE culture cDNA array and two-dimensional gel electrophoresis and protein sequencing were used to compile global and specific expression profiles for the two types of MDE cultures. Analysis of the identities of 220 candidate embryo markers, as well as the identities of 32 sequenced embryo up-regulated protein spots, indicate general roles for protein synthesis, glycolysis, and ascorbate metabolism in the establishment of MDE development. A collection of 135 robust markers for the transition to MDE development was identified, a number of which may be coregulated at the gene and protein expression level. Comparison of the expression profiles of preglobular-stage conventional MDEs and suspensor-bearing MDEs identified genes whose differential expression may reflect improved histodifferentiation of suspensor-bearing embryos. This collection of early embryo-expressed genes and proteins serves as a starting point for future marker development and gene function studies aimed at understanding the molecular regulation of cell totipotency and early embryo development in plants.
Microspore embryogenesis describes the process in which the immature male gametophyte is induced to form a haploid embryo in vitro. Microspore embryo culture is a valuable tool for plant breeders because the haploid embryos that are produced (microspore-derived embryos [MDEs]) can be converted to homozygous doubled haploids using chromosome-doubling agents. The ability to obtain a homozygous population of plants in a single generation not only significantly reduces the time needed to develop inbred lines, but also facilitates selection for recessive and polygenic traits and speeds up the development of mapping populations and molecular markers for marker-assisted breeding (Forster and Thomas, 2005). Microspore embryogenesis has been described for more than 250 plant species and efficient protocols are available for about 20 of these (Maluszinsky et al., 2003). As with other regeneration processes, many species still remain recalcitrant for microspore embryogenesis and, even within a successful species, there are usually recalcitrant genotypes. There is, therefore, considerable interest in identifying the molecular genetic factors that define and control this process.
MDE culture is also receiving increasing attention as a model system to study cell totipotency and for studies on early embryo development (Boutilier et al., 2005; Maraschin et al., 2005; Chan et al., 2006). The appeal of this system is 2-fold. First, large numbers of microspores are easily isolated and irreversibly induced by simple and defined stress treatment to undergo embryogenesis at high frequency. Second, in contrast to most in vitro organogenesis and somatic embryogenesis systems, hormones are usually not required to induce MDE development. As a result, MDEs develop directly from the microspores without an intervening callus phase. This direct transition from microspore to embryo development coupled with the efficiency of embryo induction makes it possible to produce large amounts of embryos from the few-celled stage onward for physiological, cell biological, and molecular analysis (Yeung, 2002; Boutilier et al., 2005; Chan and Pauls, 2007).
A large repository of data has been compiled concerning the early cellular and morphological changes that take place during the transition from male gametophyte to haploid embryo development in culture and commonalities among different species have been identified (for review, see Aionesei et al., 2005). The molecular mechanism underlying the induction of microspore embryogenesis is not known, although a number of studies have provided insight into this developmental process through the identification of genes and proteins that are differentially expressed during the transition from the gametophytic to embryonic cell state (Hosp et al., 2006). One of these differentially expressed genes, BABY BOOM, marks and stimulates early embryo formation (Boutilier et al., 2002).
Increasing use and availability of functional genomics tools have brought with it renewed interest in unraveling the molecular pathways that underlie cell totipotency. Identification of genes whose expression marks or controls cell totipotency is of major interest to the applied research community because such genes could be used to overcome recalcitrance for regeneration in crop species. Likewise, genes whose expression reflects the normal progression of embryo development can be used as markers to optimize critical points in the tissue culture process.
MDEs, although single cell in origin, do not exhibit the ordered cell divisions that occur during early zygotic embryogenesis. Multicellular embryos of 40 to 60 cells are formed through random (unorganized) cell divisions and only then develop into globular embryos with a distinct protoderm through a hypothetical self-organizing mechanism. Suspensor development, which takes on many forms in various plant species, is also absent in microspore embryos, although a short or malformed rudimentary suspensor is occasionally observed (Hause et al., 1994; Ilic-Grubor et al., 1998; Straatman et al., 2000; Yeung, 2002). Interestingly, we observed that when embryos with long uniseriate suspensors are formed, the embryo proper usually recapitulates the typical zygotic embryo pattern of cell division and histodifferentiation. In this study, we optimized our rapeseed (Brassica napus) MDE culture conditions to obtain a high frequency of zygotic-like embryos with long uniseriate suspensors. We then used microarray analysis and two-dimensional (2-D) gel electrophoresis/protein sequencing to examine the global and specific changes that take place at both the transcriptome and proteome level during the switch from male gametophyte to embryo development in conventional (suspensorless) MDE cultures and the new suspensor-bearing MDE cultures. Our analysis focused on the developmental events that take place between the two- to four-celled stage and the globular/heart stage of MDE development. Here, we identify metabolic pathways that are active during early MDE development, identify a collection of mRNA and protein markers that can be used at a practical level to define and optimize MDE cultures, and identify genes that are differentially expressed between suspensor-bearing and conventional MDEs.
RESULTS
Development of Rapeseed MDE Cultures
Conventional MDEs Differ from Zygotic Embryos
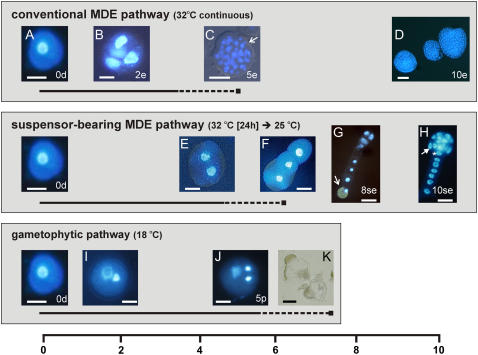
Our transcriptome and proteome analyses focused on the changes that occur as heat-stressed rapeseed microspores are reprogrammed to embryo development. We made use of two types of embryo cultures, conventional MDE cultures and a new system referred to as suspensor-bearing MDE cultures (Fig. 1). Conventional MDE development is induced by exposing microspores for 2 d or longer to heat stress at 32°C to 32.5°C. The embryos that develop after this heat-stress treatment largely resemble zygotic embryos (Yeung et al., 1996), with two major differences: (1) conventional MDEs do not initially display the typical ordered cell divisions and tissue differentiation observed during early zygotic embryo development (Tykarska 1976, 1979); and (2) conventional MDEs usually lack a well-formed (i.e. long and uniseriate) suspensor. In contrast to zygotic embryos, conventional MDEs first undergo a series of random divisions to form an unorganized cell mass. The protoderm layer of conventional MDEs is only established at the globular stage versus the eight-cell stage of zygotic embryos and is followed by histodifferentiation of the procambium and shoot meristem (Tykarska, 1976, 1979). A rudimentary root meristem develops, but is not well-defined, most likely due to the delay in polarity establishment and the absence of a well-defined suspensor (Yeung et al., 1996). Although suspensor development is routinely observed in conventional MDE cultures, it usually takes the form of short protuberances that develop near the future radicle pole of the embryo proper (Yeung et al., 1996; Ilic-Grubor et al., 1998). The occurrence of normal-looking suspensors in rapeseed MDE cultures has been reported sporadically in the literature (Pechan et al., 1991; Hause et al., 1994; Ilic-Grubor et al., 1998; Straatman et al., 2000). After critical evaluation of the culture conditions used in these studies, we hypothesized that the formation of suspensor-bearing embryos could be attributed to application of a shorter or milder heat-stress treatment than is usually applied. Furthermore, when long suspensors are formed, we noticed that the attached embryos proper undergo the same distinct cell division pattern and histodifferentiation as zygotic embryos (Fig. 1).
Figure 1.
Developmental fate of isolated rapeseed microspores. The culture temperature controls the developmental fate of isolated microspores (A). Conventional MDEs are obtained by continuous culture at 32°C (B–D). Suspensor-bearing MDEs are obtained by culturing for 24 h at 32°C followed by transfer to 25°C (E–H). Developing pollen are obtained by continuous culture at 18°C (I–K). The timeline at the bottom of the figure indicates the day of culture. Illustrated developmental stages are representative of the samples analyzed in this study; however, the timing of their appearance varies depending on the individual culture. The solid and dashed lines under each developmental pathway represent, respectively, the period of viable pollen formation and the period of pollen collapse. A, Freshly isolated late unicellular microspore. B, Four-celled embryo after 2 d of culture. C, Randomly divided multicellular embryo bursting out of the exine wall (white arrow) after 5 d of culture. Note the absence of a defined protoderm layer. D, Globular to early heart-stage embryos formed after 10 d of culture. E, Two-celled embryo at 4 to 5 d of culture. F, Uniseriate suspensor formed at day 6 to day 7 of culture. G, Longitudinal divisions in the distal tip cell of the filament leading to a four-cell embryo proper after 8 d of culture. Two additional nuclei are present behind the two nuclei in focus. The original microspore wall (white arrow) is visible at the opposite end of the cell file. H, Preglobular-stage embryo after 9 to 10 d of culture showing differentiation of the protoderm (white arrow) and hypophysal cell (asterisk). I, Bicellular pollen with a large vegetative nucleus and a smaller generative nucleus. J, Tricellular pollen with a large vegetative nucleus and two smaller sperm nuclei. K, Collapsed pollen-like structures. The developmental stages of the samples used for microarray analysis are as follows: 0d, starting microspore culture; 5p, 5-d pollen culture; 2e, 2-d conventional MDE culture; 5e, 5-d conventional MDE culture; 10e, 10-d conventional MDE culture; 8se, 8-d suspensor-bearing MDE culture; 10se, 10-d suspensor-bearing MDE culture. Photographs A to J are digital images from 4′,6-diamidino-2-phenylindole-stained material observed using a Zeiss Axioskop microscope or with an IM Zeiss microscope. Bars = 10 μm for A, B, E, F, I, and J; 30 μm for C, D, G, H, and K.
Optimized Protocol for Production of Zygotic-Like MDEs
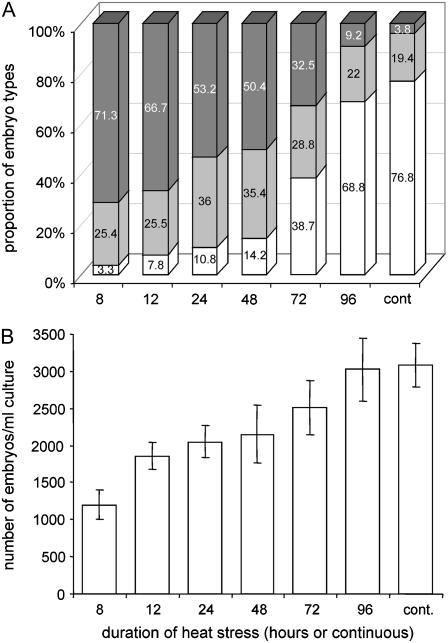
Based on the above observations, we optimized the culture conditions to obtain a high frequency of MDEs with long suspensors and zygotic embryo-like morphology. The critical aspects of the new culture system are a more accurate selection of the microspore starting population using narrower increments in floral bud size (Supplemental Table S1) and the use of a shorter and more accurate heat-stress treatment (Fig. 2). Cytological analyses showed that cultures that were most responsive in the development of normal-looking suspensors were always very uniform with respect to the stage of the starting microspore population, consisting of at least 50% to 60% late unicellular microspores and 30% to 40% early bicellular pollen. To better understand the relation between formation of MDEs with normal-looking suspensors and those with aberrant suspensors, we carried out a more thorough analysis of the duration of the 32°C heat-stress treatment (Fig. 2). Microspores from highly responsive buds were treated for various periods at 32°C and then transferred to 25°C. After 16 d of culture, the quality of suspensor formation was determined based on three morphological classes: (1) embryos with long, normal-looking suspensors (≥30 μm uniseriate); (2) embryos with short, irregular-shaped suspensors (<30 μm uniseriate); and (3) embryos without a suspensor. As shown in Figure 2, a 32°C heat-stress treatment applied for 8 to 12 h led to production of a long suspensor in more than two-thirds of the embryos (Fig. 2A). Increasing the duration of 32°C heat stress from 8 h to continuous treatment resulted in a gradual decrease in the frequency of embryos with a long suspensor to close to 0 and a parallel increase in embryos lacking suspensors. The second class of embryos, those with short appendant structures, either uniseriate or swollen and irregular, were found in all treatments, but the frequencies of such embryos were significantly higher when the microspores were treated at 32°C for 24 or 48 h as compared to shorter or longer heat-stress treatments (Fig. 2A). Although the modified heat-stress treatment positively affected the production of embryos with long, normal-looking suspensors, it had a negative effect on the total number of embryos formed (Fig. 2B).
Figure 2.
Effect of different periods of heat-stress treatment on suspensor formation in rapeseed microspore culture. A, The frequencies of embryos with long, normal-looking suspensors ≥30 μm (dark gray bars), embryos with short suspensor-like structures (irregular-shaped appendant structures or uniseriate structures <30 μm; light gray bars), or embryos without a suspensor (white bars), B, Total embryo yield. All cultures were started with a density of 40,000 microspores per milliliter. After the heat-stress treatment, cultures were kept at 25°C. For the determination of total embryo yield (B), embryos larger than 0.4 mm were counted after 16 d of culture when the largest embryos had already reached 3 mm. Data are the means of five replicate experiments, each with three 6-cm petri dishes per treatment and 3 mL of original microspore suspension per dish. Bar = ±se.
Comparison of Conventional and Suspensor-Bearing MDE Cultures
The major developmental events that take place in the conventional and suspensor-bearing MDE cultures that were used in our transcriptome and proteome analyses are summarized in Figure 1. Conventional MDE cultures were induced from freshly isolated microspores (Fig. 1A) by continuous 32°C heat-stress treatment. MDE development begins with swelling of the microspore, followed by symmetric division of the vegetative nucleus. Approximately 15% to 25% of the microspores undergo embryonic divisions within the first 2 d of culture (Fig. 1B). By the fifth day of culture, randomly oriented cell divisions in a subset of these few-celled embryos generate preglobular-stage embryo clusters that can be seen breaking out of the microspore exine wall (Fig. 1C). Globular- to heart-stage embryos are usually present after 10 d of culture (Fig. 1D).
Suspensor-bearing MDEs were induced by culturing microspores for 1 d at 32°C, followed by transfer to 25°C. Suspensor-bearing MDEs do not originate by random division of the microspore, but rather by formation of a suspensor-like filament, followed by embryo-proper development from the distal cell. The first cell divisions take place at around the fourth day of culture (Fig. 1E) with a nose-like suspensor emerging from the swollen microspore around the seventh day of culture (Fig. 1F). The suspensor develops as a single file of cells, elongating by transverse divisions, with one end connected to the microspore. Around the eighth day of culture, the distal cell of the three- to eight-celled suspensor undergoes transverse divisions to form the embryo proper (Fig. 1G). Thereafter, cell division and histodifferentiation of the embryo proper follow the same regular pattern as in zygotic embryos, and preglobular-stage embryos with a defined protoderm and hypophysis can be seen after 10 d of culture (Fig. 1H). Cell division in the suspensor generally ceases once the embryo reaches the late-globular stage. Suspensor-bearing MDE cultures develop more slowly than conventional cultures, with the result that two- to four-celled embryos are found at around day 2 in conventional cultures and at day 8 in suspensor-bearing cultures (compare Fig. 1, B and G), whereas preglobular embryos are found at around day 5 in conventional cultures and day 10 in suspensor-bearing cultures (compare Fig. 1, C and H).
The heat-stress treatment used to induce conventional and suspensor-bearing MDE development is not sufficient to block gametophyte development in all of the donor microspores. As a result, both pollen and embryo developmental pathways initially occur side by side in the same cultures. The large amount of developing pollen in MDE cultures can confound attempts to identify early embryo-expressed genes and proteins; therefore, we used control pollen cultures to discriminate between embryo and pollen expression profiles in MDE cultures. Cultures with up to 90% viable tricellular pollen can be obtained by culturing isolated microspores at 18°C instead of 32°C (Fig. 1, I and J, gametophytic pathway; Custers et al., 1994). These pollen grains collapse a few days after reaching the tricellular stage (Fig. 1K). The developmental time course for pollen development and the subsequent collapse of mature pollen grains is temperature dependent and therefore differs between pollen cultures, conventional MDE cultures, and suspensor-bearing MDE cultures analyzed in this study (Fig. 1).
Transcriptome Analysis of MDE Cultures
Microarray Enriched for MDE Culture cDNAs
Microarray analysis was used to characterize the changes in gene expression that take place during the transition from gametophytic to conventional and suspensor-bearing MDE development and during subsequent outgrowth of the embryos. A custom microarray containing cDNA elements originating predominantly from MDE cultures was used for these analyses. The suspensor-bearing MDE culture protocol was not optimized at the time of microarray construction; therefore, only probes from conventional MDE cultures were spotted on the array. Complementary DNA libraries from three key time points in rapeseed MDE culture were used for gene isolation. These time points correspond to the swollen unicellular microspore stage (24 h), the 20- to 40-celled embryo stage (4 d), and the globular- to heart-shaped embryo stage (10 d). We also applied suppression subtraction hybridization (SSH; Diatchenko et al., 1996) to the 20- to 40-celled embryo stage and the globular- to heart-shaped embryo stage samples to normalize mRNA levels and to enrich for genes that are up- or down-regulated at each of these time points relative to other culture samples or plant tissues (see “Materials and Methods”). In total, 1,920 probes were spotted on the array, of which 88% correspond to cDNAs isolated from MDE cultures (Supplemental Table S2). Contig analysis showed that 73% of MDE culture probes are represented only once on the array (data not shown). The sequences and identities of the arrayed probes are available as Supplemental Table S2.
The custom cDNA microarray was hybridized to targets derived from two independent microspore cultures. The developmental time points that were analyzed are shown in Figure 1. For each of the two cultures, one sample was harvested immediately (0d; Fig. 1A); one sample was cultured to promote pollen development (5p; Fig. 1J), and five samples were cultured to induce embryo development. Of the five samples cultured for embryo development, three samples were induced for conventional MDE development (two- to four-celled stage [Fig. 1], preglobular stage [Fig. 1C], and globular to heart stage [Fig. 1D]), and two samples were induced for suspensor-bearing MDE development (two- to four-celled embryo-proper stage [Fig. 1G] and preglobular embryo-proper stage [Fig. 1H]). The percentage of embryo and viable pollen present in each culture at the successive sampling time points is shown in Table I. Each of the amplified targets was hybridized using a common reference strategy (“Materials and Methods”). Array hybridizations with amplified leaf and flower bud targets were performed to obtain an indication of the relative expression levels of the probes in non-embryo samples.
Table I.
Composition of rapeseed MDE cultures used for the transcriptome analysis
Values are expressed as percentages of structures showing pollen or embryo development at the time of harvest relative to the number of microspores present at the start of culture. The 10-d embryo cultures were sieved through nylon filters to obtain pure populations of embryos. Note that the total embryo yield generally decreases as the MDE cultures age. 0d, Freshly isolated microspores; 5p, 5-d pollen culture; 2e, 2-d conventional MDE culture; 5e, 5-d conventional MDE culture; 10e, 10-d conventional MDE culture; 8se, 8-d suspensor-bearing MDE culture; 10se, 10-d suspensor-bearing MDE culture.
| Culture | Culture 1
|
Culture 2
|
||
|---|---|---|---|---|
| % Embryo | % Pollen | % Embryo | % Pollen | |
| 0d | – | – | – | – |
| 5p | 0 | 75 | 0 | 75 |
| 2e | 14 | 27 | 12 | 40 |
| 5e | 17 | 5 | 11 | 13 |
| 10e | 7 | 0 | 5 | 0 |
| 8se | 17 | 0 | 12 | 0 |
| 10se | 14 | 0 | 14 | 0 |
Principal Component Analysis
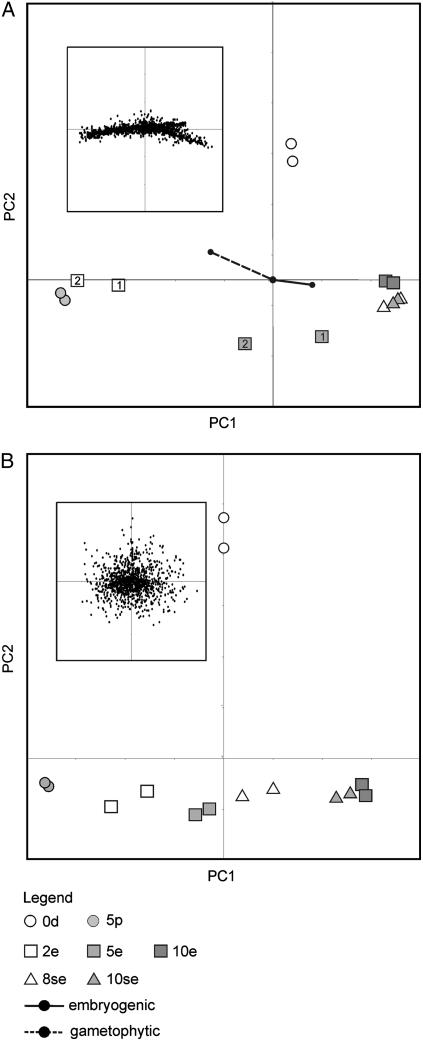
Principal component analysis (PCA) was used to analyze gene expression profiles obtained after microarray hybridization of amplified pollen and MDE culture samples. PCA helps to interpret overall patterns in gene expression by dimension reduction. A plot of the samples on the first two principal components, together explaining 82% of the observed variability, is shown in Figure 3A. Corresponding distribution of the individual genes is shown in the inset. Information on the composition of the culture samples in terms of the relative contribution of pollen and embryo-like structures, as shown in Table I, is added in the plot. The first principal component (PC1), which explains 70% of the variance, can be interpreted as describing the transition in the cultures from a pollen-dominated gene expression state to an embryo-dominated gene expression state. The 2-d conventional embryo culture samples (2e), which contain both embryogenic cells and a high proportion of viable pollen, are clustered toward one side of the axis together with the 5-d pollen culture samples, whereas the conventional (10e) and suspensor-bearing embryo or culture samples that do not contain viable pollen (8se and 10se) are clustered at the other end of the axis (Table I; Fig. 3). Microarray analysis of Arabidopsis (Arabidopsis thaliana) pollen gene expression profiles show that mature pollen grains contain proportionately more abundant mRNAs than do sporophytic tissues (Honys and Twell, 2003). The relative contribution of embryo-expressed genes to the entire mRNA pool in 2-d conventional embryo cultures might therefore be diluted by abundant pollen mRNAs, giving these cultures an overall pollen-like identity despite the presence of a large amount of developing embryos.
Figure 3.
PCA of gene and protein expression profiles detected in rapeseed pollen and MDE cultures. PCA plots of gene (A) and protein (B) expression profiles are shown. For both plots, the replicated culture samples/pools are indicated. For the PCA plot of the gene expression profiles, culture 1 and culture 2 are indicated as 1 and 2, respectively. For each plot, the corresponding distribution of genes or proteins is shown in the inset. The composition of the analyzed samples in terms of the percentage of gametophytic and embryogenic structures was incorporated into the plot of the gene expression profiles (vectors). 0d, Starting microspore culture; 5p, 5-d pollen culture; 2e, 2-d conventional MDE culture; 5e, 5-d conventional MDE culture; 10e, 10-d conventional MDE culture; 8se, 8-d suspensor-bearing MDE culture; 10se, 10-d suspensor-bearing MDE culture.
Identification of Differentially Expressed Probes
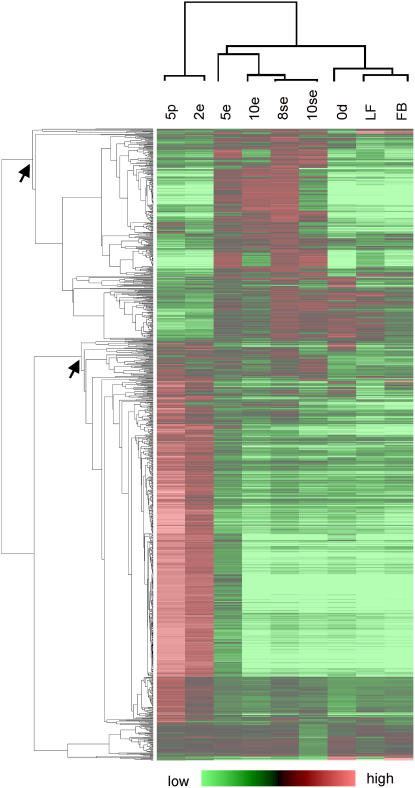
Cluster analysis of the microarray expression data was used to identify groups of similarly expressed genes in different culture samples. As shown in Figure 4, two main gene expression clusters were identified: a large cluster comprising genes primarily up-regulated in the 5-d pollen and 2-d conventional embryo culture samples, and a smaller cluster comprising genes up-regulated in the remaining conventional and suspensor-bearing embryo samples. A large number of known late pollen-expressed genes (Twell, 2002) are up-regulated in the 5-d pollen and conventional 2-d embryo culture samples, suggesting, as observed in the PCA plots, that the mRNA pool in the 2-d conventional MDE culture is dominated by abundant pollen transcripts. We refer to the larger gene expression cluster as pollen up-regulated and to the smaller gene expression cluster as embryo up-regulated. We made use of the ANOVA test statistic (F value) and lsd values (“Materials and Methods”) to identify the pollen and embryo up-regulated probes in these two clusters. The dominance of pollen-expressed genes in 2-d conventional embryo cultures confounds microarray-based attempts to identify embryogenesis-associated transcripts in this sample; therefore, the data for this time point, although presented, was omitted from this analysis. We identified 220 probes that are significantly up-regulated during one or more of the embryo cultures as compared to 5-d pollen cultures and 363 probes that are significantly up-regulated in 5-d pollen cultures as compared to all of the embryo cultures (Supplemental Table S3). Expression profiles of four selected embryo up-regulated probes and one pollen up-regulated probe were validated by quantitative reverse transcription (RT)-PCR (Supplemental Fig. S1; Supplemental Table S4).
Figure 4.
Hierarchical cluster analysis of the expression profiles of rapeseed probes expressed in freshly isolated microspores, pollen cultures, or MDE cultures. The columns represent the samples and the rows the individual probes. Probes that are up- or down-regulated compared to the common reference are indicated in red and green, respectively. The intensity of the colors increases with increasing expression differences, as shown in the bar at the bottom. The pollen and embryo up-regulated clusters are indicated with arrows. Only probes for which an expression ratio for both culture samples could be calculated were included in the analysis (1,059 probes). 0d, Starting microspore culture; 5p, 5-d pollen culture; 2e, 2-d conventional MDE culture; 5e, 5-d conventional MDE culture; 10e, 10-d conventional MDE culture; 8se, 8-d suspensor-bearing MDE culture; 10se, 10-d suspensor-bearing MDE culture. LF, Mature leaves; FB, flower buds (3–4 mm).
Analysis of the identities of the pollen and embryo up-regulated probes and their accompanying expression patterns in freshly isolated microspores, pollen, and embryo cultures provides insight into the developmental processes that take place as microspores develop into either pollen grains or haploid embryos (Supplemental Fig. S2; Supplemental Table S3). The progression from microspores to tricellular pollen grains is accompanied by a dramatic increase in the expression of cell wall-loosening enzymes needed for pollen germination. There is also a switch from expression of genes for glyocolysis-based carbohydrate metabolism in microspores to genes that play key roles in the glyoxylate cycle and gluconeogenesis in pollen (Supplemental Fig. S2; Supplemental Table S3). The corresponding enzymes are likely to be involved in the mobilization of lipid reserves by glyoxysomes to produce sugars to sustain pollen growth (Zhang et al., 1994). There is also a sharp decrease in the expression of probes for components of protein synthesis machinery (Supplemental Fig. S2; Supplemental Table S3). In contrast, expression of probes coding for glycolytic enzymes is maintained or increased during the switch from microspore to embryo development (Supplemental Table S3), whereas the majority of probes encoding components of the protein synthesis machinery are expressed at the same level during the transition from microspore to haploid embryo development (Supplemental Table S3).
Robust Markers for MDE Induction
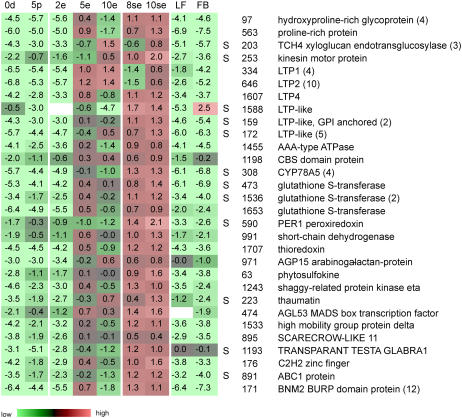
Conventional MDE cultures are used in practical settings to generate a large amount of haploid embryos for breeding purposes. Robust expression markers associated with the switch from microspore to embryo development in conventional MDE culture should not only discriminate between the development pathways associated with freshly isolated microspores and embryos, but also discriminate the pathways associated with pollen grains that develop simultaneously in the same culture. To identify these markers, we used the calculated F- and lsd values to search for probes showing significant up-regulated expression in two- to four-celled suspensor-bearing embryos (Fig. 1G, 8se) as compared to both freshly isolated microspores and pollen grains. The equivalent two- to four-celled conventional MDE samples (Fig. 1B, 2e) could not be analyzed due to the presence of abundant pollen-expressed mRNAs. Using this approach, we identified 135 robust expression markers for the commitment to haploid embryogenesis (Supplemental Table S3), a selection of which is shown in Figure 5 and described below.
Figure 5.
Robust markers for MDE development. The number listed beside each marker corresponds to the assigned probe number. The expression profile of a representative probe is presented when a marker is represented by multiple probes (number of multiple probes indicated in parentheses). The expression ratio (log2 scale) of each probe or probe set relative to the common reference is indicated. Color coding of the expression ratios is as in Supplemental Figure S4. S, Probes that are expressed at significantly higher levels in preglobular suspensor-bearing MDEs as compared to preglobular conventional MDEs. 0d, Starting microspore culture; 5p, 5-d pollen culture; 2e, 2-d conventional MDE culture; 5e, 5-d conventional MDE culture; 10e, 10-d conventional MDE culture; 8se, 8-d suspensor-bearing MDE culture; 10se, 10-d suspensor-bearing MDE culture; LF, mature leaves; FB, flower buds (3–4 mm).
The vast majority of these markers encodes proteins of unknown function or corresponds to functionally annotated proteins for which neither an expression profile nor a function during embryogenesis have been assigned. Twelve of these probes encode the BNM2 BURP domain protein, which was previously identified in two independent screens for embryo markers in MDE cultures (Boutilier et al., 2002; Tsuwamoto et al., 2006). BURP domain proteins are an abundant class of secreted proteins of unknown function that are defined by their modular structure and conserved C-terminal BURP domain (Hattori et al., 1998). The BNM2 BURP domain gene, although expressed in 0-d cultures and 5-d pollen cultures, is expressed at a 100 times higher level in two- to four-celled embryos (Fig. 5; Supplemental Table S4).
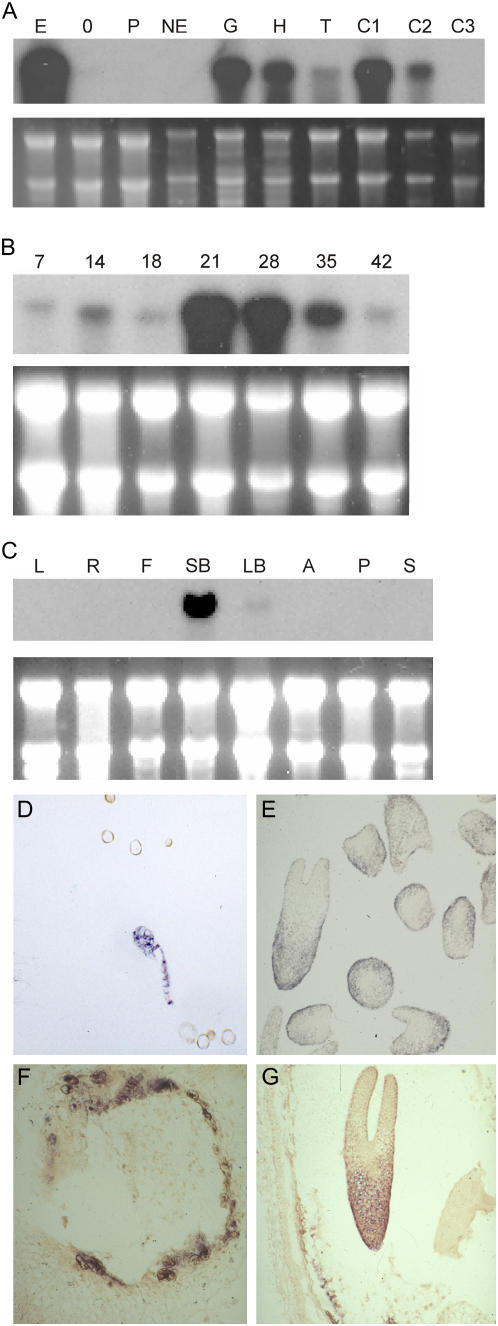
We analyzed the temporal and spatial expression pattern of BNM2 in independent MDE cultures, as well as a range of seed and nonseed samples. RNA gel-blot analysis of BNM2 expression confirmed our microarray and RT-PCR data (Fig. 6) and further showed that BNM2 is temporally regulated in a similar manner during MDE and seed development, with expression peaking around the midcotyledon stage of embryo development (Fig. 6, A and B). Messenger RNA in situ hybridizations on rapeseed MDEs and seeds showed that spatial expression of BNM2 is also conserved in MDEs and zygotic embryos; BNM2 is initially expressed throughout the preglobular embryo and becomes confined to the epidermal and ground tissues as the embryo differentiates (Fig. 6, D, E, and G). BNM2 is not an embryo-specific gene because it is expressed in flower buds (Fig. 6C) and in the integument and endosperm of developing seeds (Fig. 6, F and G).
Figure 6.
BNM2 BURP domain expression analysis. A, RNA gel-blot analysis of BNM2 gene expression in rapeseed pollen and MDE cultures. Total RNA was isolated from microspores at the start of culture (0); from embryo cultures after 4 d in culture at 32°C (E); from nonembryogenic heat-stressed microspore cultures (NE) obtained by culturing microspores for 1 d at 25°C, followed by 3 d at 32°C; from pollen cultures after 4 d at 25°C (P); and from purified MDEs at the globular (G), heart (H), torpedo (T), 21-d cotyledon (C1), 28-d cotyledon (C2), and 28-d cotyledon stage (C3). B, RNA gel-blot analysis of BNM2 transcripts in developing seeds. RNA samples were isolated from whole seeds collected at specific days after pollination (DAP). These developmental time points correspond approximately to the globular (7 DAP), heart/torpedo (14 DAP), early cotyledon (21 DAP), midcotyledon (28 DAP), and mid-to-late cotyledon (35 and 42 DAP) stages of development. C, RNA gel-blot analysis of BNM2 transcripts in nonseed tissues. RNA samples were isolated from mature leaves (L), roots (R), and stem (S), flowers at anthesis (F), small flower buds (SB, <3 mm), large flower buds (LB, 5–7 mm), and anthers (A) and pistils (P) and from flowers at anthesis. For A, B, and C, 10 μg of total RNA was loaded per sample. Ethidium bromide staining of RNA was used to compare sample loading. D to G, mRNA in situ hybridization analysis of BNM2 expression in MDEs (D and E) and seeds (F and G). The probe used for RNA gel-blot analysis and mRNA in situ hybridization detects at least two duplicated copies of the BNM2 gene in the amphidiploid rapeseed genome.
Probes encoding proteins involved in transcriptional regulation and signaling were also identified as robust markers for the transition to haploid embryo development (Supplemental Table S3; Fig. 5). Among the transcriptional regulators is the MADS-box transcription factor AGL53, which is also expressed in somatic embryo cultures (Lehti-Shiu et al., 2005). Two probes encoding a PHYTOSULFOKINE protein and an arabinogalactan protein, AGP15, are noteworthy due to the role of such proteins in promoting cell proliferation and in vitro embryogenesis (Yang et al., 2000; Matthys-Rochon, 2005; Letarte et al., 2006).
We identified a number of glutathione S-transferases (GSTs) that are up-regulated in embryos as compared to pollen and freshly isolated microspores (Supplemental Table S3; Fig. 5). GSTs have been identified as embryo markers in barley (Hordeum vulgare) MDE cultures (Vrinten et al., 1999; Maraschin et al., 2006), as well as in soybean (Glycine max; Thibaud Nissen et al., 2003) and chicory (Cichorium intybus) somatic embryo cultures (Galland et al., 2001). The three unique GSTs identified here belong to the plant-specific tau class of GSTs (Wagner et al., 2002).
We also identified 23 lipid transfer proteins (LTPs)/LTP-like proteins corresponding to six different groups among the robust MDE markers (Supplemental Table S3; Fig. 5). LTP/LTP-like proteins are small, basic, abundant proteins with an eight-Cys residue region that is also found in a larger group of structurally related proteins (Jose-Estanyol et al., 2004). LTPs are versatile markers for embryogenic cell formation because they have been identified in both somatic embryo and MDE cultures (Sterk et al., 1991; Vrinten et al., 1999).
Comparison of Suspensor-Bearing and Conventional MDE Gene Expression Profiles
The structural pattern of conventional and suspensor-bearing embryos differs significantly. To determine whether these differences are reflected at the gene expression level, we identified genes among the set of embryo up-regulated probes that are differentially expressed between preglobular-stage suspensor-bearing and conventional embryos (5 d and 8se samples). Using the calculated F values and lsd values as criteria, 43 of the 220 embryo up-regulated probes were found to be differentially expressed between the two types of embryos (Supplemental Table S3; Fig. 5). The majority of the differentially expressed probes were also identified as robust markers for MDE development. Only two of these probes, encoding a Leu-rich protein kinase and an AAA-like ATPase, are expressed at a higher level in conventional embryos (Supplemental Table S3). Many of the genes identified have tissue-specific expression patterns in other organisms. For example, LTP proteins are generally expressed in the L1 layer of the plant, whereas the Arabidopsis CYTOCHROME P450 CYP78A5 is expressed on the flanks of the embryonic meristem (Zondlo and Irish, 1999). The differential expression of these genes may therefore mark the underlying differences in histodifferentiation between suspensor- and non-suspensor-bearing embryos of the same developmental age.
Proteome Analysis
We used the same developmental time points studied in the microarray analysis to compile proteome maps of rapeseed MDE cultures. A larger amount of starting material was needed for proteome analysis using 2-D gel electrophoresis as compared to microarray analysis; therefore, biological replicates were made for each of the developmental time points by pooling samples derived from multiple independent cultures. The composition of these cultures, in terms of the percentage embryo development as measured at the end of the culture period, is shown in Table II. Protein extracts from each of these cultures were run on 2-D gels and analyzed with PDQuest software. Table II shows the number of spots detected per gel, which varies between, on average, 528 spots in the 0-d starting culture gels to 1,160 spots in the 10-d suspensor embryo culture gels. The same amount of protein was loaded for each sample; thus, the large increase in the number of detected protein spots in the embryogenic cultures after day 5 might reflect the increase in complexity of the developing embryos.
Table II.
Composition of rapeseed MDE cultures used for proteome analysis
The percentage of embryo formation relative to the number of microspores at the start of culture was determined in subsamples of the starting cultures grown continuously for 10 d at 32°C. The 10-d embryo cultures (10e) were sieved as described in Table I to obtain pure populations of embryos. The number of spots detected in each of the 2-D gels was determined by PDQuest software. 0d, Freshly isolated microspores; 5p, 5-d pollen culture; 2e, 2-d conventional MDE culture; 5e, 5-d conventional MDE culture; 10e, 10-d conventional MDE culture; 8se, 8-d suspensor-bearing MDE culture; 10se, 10-d suspensor-bearing MDE culture.
| Stage | % Embryo (Pool 1/Pool 2) | % Embryo (Average) | No. Spots (Gel 1/Gel 2) | Average |
|---|---|---|---|---|
| 0d | – | – | 576/479 | 528 |
| 5p | – | – | 1,239/883 | 1,061 |
| 2e | 7/4 | 5.5 | 722/673 | 698 |
| 5e | 5/5 | 5 | 639/538 | 589 |
| 10e | 2/4 | 3 | 1,007/908 | 958 |
| 8se | 2/5 | 3.5 | 884/1,053 | 969 |
| 10se | 2/6 | 4 | 1,046/1,273 | 1,160 |
PCA
PCA was used to identify and give biological meaning to the protein expression profiles that are characteristic for the pollen and MDE cultures. A plot of the pollen and MDE culture samples and the corresponding distribution of protein spots (inset) are shown in Figure 2B. The first two principal components explain 38% of the observed variability in protein expression between the different culture samples, with PC1 explaining 22% and PC2 16% of the variability. As observed for the microarray data, PC1 appears to describe a transition in the culture material from a pollen-dominated to an embryo-dominated state, whereas the 0-d culture is assigned a unique position PC2. Distribution of the pollen and embryo culture samples along PC1 is more gradual than was observed for the mRNA expression profiles (Fig. 3B). Together, these observations point to the greatly reduced contribution of pollen-expressed proteins as compared to pollen-expressed mRNAs to the sample identities.
Distribution of the expression profiles for the individual protein spots is centered on the intersection of the PC1 and PC2 axes, a position that reflects proteins with low variance or low information among the different samples. Thus, although PCA uncovered the most important structure in the 2-D gel data, the amount of variance not explained by the first two principal components is large (62%). A combination of technical and biological factors may contribute to the low variance of the protein expression data. For example, the levels of lower abundance proteins may change more than those of higher abundance proteins, but will not be detected by 2-D gel electrophoresis. Other factors contributing to the low variance of the protein dataset might include the use of pooled samples.
Protein Sequence Identification
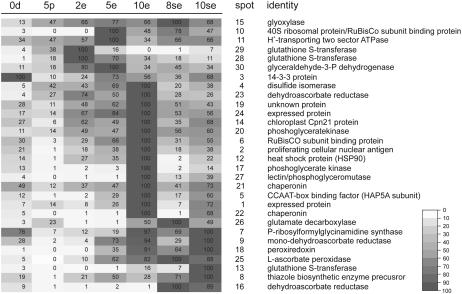
Our interest was to select a set of protein spots for subsequent sequencing that were up-regulated in the embryogenic cultures compared to the pollen cultures, rather than to exhaustively sequence all differentially expressed spots. Groups of spots were created with statistically significant expression profiles using PDQuest analysis tools (see “Materials and Methods”). In total, 45 spots were selected whose intensity was significantly increased in one or more of the embryogenic cultures compared to pollen. These spots were processed by in-gel tryptic digestion and the peptides were separated by nano-liquid chromatography (LC) and analyzed online by quadrupole time-of-flight (Q-TOF)-tandem mass spectrometry (MS/MS). For 30 of the selected 45 spots, sequence hits (35) were obtained corresponding to unique proteins (32) that show the highest similarity to Brassica or Arabidopsis proteins (Table III). For the remaining spots, either no hits were found after querying the nonredundant (Nr) database or their identification was not possible due to low-quality MS/MS spectra. A representative 2-D gel image showing the position of the 30 protein spots whose sequence was identified with confidence is shown as Supplemental Figure S3. For most of the identified spots, the molecular mass and pI values as determined from the 2-D gel correspond with the theoretical values as obtained from the corresponding protein hits in the database (Table III). The relative expression profiles of the identified protein spots in the pollen and MDE culture samples are shown in Figure 7.
Table III.
Differentially expressed proteins identified in rapeseed MDE cultures
Each entry contains a number that corresponds to the protein spots indicated in Figure 4. Matched Peptides gives the number of peptide sequences identified by LC-MS/MS that showed homology to the best-matching gene product. The molecular mass and isoelectric point (IEP) as estimated from the 2-D gel are given, and in parentheses the deduced molecular mass and IEP of the best-matching gene product. The last column represents the percentage of sequence coverage by the matched peptides to the most similar protein in the database. At, Arabidopsis; Bj, Brassica juncea; Bn, rapeseed; Bo, Brassica oleracea; Br, Brassica rapa.
| No. | Best-Matching Gene Product (Organism) | ID | Process | Matched Peptides (n) | Molecular Mass Hit (kD) | IEP Hit (pI) | Coverage (%) |
|---|---|---|---|---|---|---|---|
| 1 | Expressed protein (At) | At5g48480 | Unknown | 3 | 20.1 (17.6) | 4.60 (4.80) | 16.8 |
| 2 | Proliferating cellular nuclear antigen (At) | At2g29570 | DNA replication | 5 | 35.7 (29.2) | 4.49 (4.39) | 36.7 |
| Proliferating cellular nuclear antigen (Bn) | gi15222379 | DNA replication | 5 | 35.7 (29.2) | 4.49 (4.42) | 36.7 | |
| 3 | 14-3-3 protein (Bn) | gi13447112 | Signaling | 5 | 33.5 (18.2) | 4.64 (5.19) | 20.8 |
| 4 | Disulfide isomerase-like protein (At) | At1g21750 | Redox processes | 14 | 61.3 (55.6) | 4.66 (4.61) | 25.9 |
| 5 | CCAAT-box binding factor Hap5a subunit (At) | At1g08970 | Transcription | 4 | 31.2 (25.6) | 5.02 (4.91) | 16.4 |
| 6 | Rubisco subunit binding protein (Bn) | gi464727 | Protein modification/processing | 8 | 62.7 (61.6) | 4.81 (4.95) | 18.7 |
| 7 | P-ribosylformylglycinamidine synthase (At) | At1g74260 | Secondary metabolism | 2 | 126.1 (151.7) | 4.97 (4.99) | 2.1 |
| 8 | Thiazole biosynthetic enzyme precursor (At) | At5g54770 | Secondary metabolism | 3 | 34.6 (36.6) | 5.06 (5.78) | 9.7 |
| 9 | Monodehydroascorbate reductase (At) | At5g03630 | Redox processes | 2 | 49.3 (47.5) | 5.11 (5.06) | 6.7 |
| 10a | 40S ribosomal protein (Bn) | gi15214300 | Protein synthesis | 3 | 42.3 (32.0) | 5.12 (4.94) | 13.7 |
| 10b | Gln synthetase (Bn) | gi1084350 | Amino acid metabolism | 3 | 42.3 (39.1) | 5.12 (5.13) | 9.5 |
| 11 | H+-transporting two-sector ATPase (At) | At3g28710 | Electron transport | 3 | 42.4 (40.7) | 5.16 (4.81) | 12.8 |
| 12 | HSP90 heat shock protein (At) | At2g04030 | Protein modification/processing | 2 | 97.7 (88.6) | 5.17 (4.74) | 3.5 |
| 13 | GST (At) | gi15218518 | Redox processes | 2 | 26.5 (18.8) | 5.39 (5.64) | 13.5 |
| 14 | Chloroplast Cpn21 protein (At) | At5g20720 | Protein modification/processing | 5 | 28.7 (26.8) | 5.46 (9.35) | 21.3 |
| 15 | Lactoylglutathione lyase/glyoxylase I (Bo) | gi2494843 | Redox processes | 11 | 34.2 (31.6) | 5.31 (4.97) | 24.8 |
| 16 | Dehydroascorbate reductase (Bj) | gi22653413 | Redox processes | 3 | 30.7 (28.5) | 5.47 (8.40) | 11.7 |
| Dehydroascorbate reductase (Bj) | gi22653415 | Redox processes | 3 | 30.7 (24.3) | 5.47 (5.19) | 13.8 | |
| 17 | Phoshoglycerate kinase (At) | At1g56190 | Carbohydrate metabolism | 7 | 44.2 (49.9) | 5.32 (6.23) | 22.4 |
| 18 | Peroxiredoxin (Br) | gi4928472 | Redox processes | 6 | 21.5 (17.4) | 5.48 (5.23) | 41.3 |
| 19 | Expressed protein (At) | At5g02240 | Unknown | 10 | 33.9 (27.1) | 5.57 (6.20) | 19.3 |
| 20 | Phosphoglycerate kinase (At) | At1g79550 | Carbohydrate metabolism | 10 | 42.5 (42.1) | 5.48 (5.33) | 23.2 |
| 21 | Chaperonin (At) | At3g13860 | Protein modification/processing | 13 | 60.5 (60.4) | 5.54 (5.74) | 13.8 |
| 22 | Chaperonin; T-complex protein (At) | At3g03960 | Protein modification/processing | 2 | 60.8 (58.9) | 5.63 (5.07) | 2.7 |
| 23 | Dehydroascorbate reductase (Br) | gi33285914 | Redox processes | 4 | 29.6 (12.0) | 5.86 (6.16) | 33.3 |
| 24 | Expressed protein (At) | At5g02240 | Unknown | 6 | 32.9 (27.1) | 5.87 (6.20) | 6.3 |
| 25 | l-Ascorbate peroxidase (Bn) | gi1890354 | Redox processes | 13 | 30.9 (27.7) | 5.96 (5.83) | 42.4 |
| 26 | Glutamate decarboxylase (At) | At2g02010 | Signaling | 4 | 57.3 (55.9) | 6.09 (6.26) | 6.4 |
| 27a | Lectin (At) | At3g16400 | Carbohydrate binding | 2 | 65.5 (51.6) | 5.81 (5.18) | 5.6 |
| 27b | 2,3-Biphosphoglycerate-independent phosphoglycerate mutase (At) | At3g08590 | Carbohydrate metabolism | 3 | 65.5 (60.7) | 5.81 (5.43) | 6.3 |
| 28 | GST (GST6; At) | At2g47730 | Redox processes | 3 | 28.3 (19.7) | 6.30 (5.22) | 26.0 |
| 29 | GST (Bj) | gi2204102 | Redox processes | 2 | 34.1 (15.4) | 6.40 (5.17) | 15.1 |
| GST (At) | At1g78340 | Redox processes | 2 | 34.1 (25.2) | 6.40 (4.99) | 5.5 | |
| 30 | Glyceraldehyde-3-P dehydrogenase (At) | At1g13440 | Carbohydrate metabolism | 10 | 41.5 (36.9) | 6.56 (6.79) | 27.2 |
Figure 7.
Heat map of the relative signal intensities of proteins expressed in rapeseed pollen and MDE cultures. Signal intensities for each protein spot were normalized to the highest signal intensity for that protein spot (100%) in the culture series. Values shown are relative (i.e. they do not reflect the absolute expression levels of the proteins in the samples). The heat map was created in GeneMaths, version 2.01 (Applied Maths). The color-code bar is shown on the right. Sample legend as in Figure 3.
Sequenced proteins were selected based on their preferential expression in MDE cultures as compared to pollen cultures. Although many of the corresponding protein spots are still detected in pollen cultures, their expression is up to 20 times higher in the earliest embryo culture samples analyzed (2-d sample) as compared to pollen cultures, and their expression generally increases during subsequent stages of development. Most of the proteins that we identified are expressed during multiple stages of embryo development, as was observed for the embryo up-regulated probes identified in the transcriptome analysis; however, two of the identified protein spots show stage-enhanced expression (Fig. 7). Protein spot 29 corresponds to a tau class GST that is preferentially expressed in 2-d conventional embryo cultures. A second protein (spot 16) encodes a dehydroascorbate reductase and is preferentially expressed in suspensor-bearing MDE cultures.
The identified proteins are involved in a range of processes; however, proteins active in carbohydrate metabolism and redox processes are particularly abundant among sequenced proteins. Three consecutive enzymes of the glycolytic pathway (glyceraldehyde-3-P dehydrogenase, 3-phosphoglycerate kinase, and phosphoglycerate mutase) were up-regulated at various stages of embryo development (Fig. 7).
Genes and proteins involved in protecting cells against oxidative stress caused by reactive oxygen species (ROS) are well represented in both the pollen and embryo transcriptome; however, proteome data suggest a specific role for ascorbate metabolism during early embryo development. Ascorbate peroxidase, as well as ascorbate-recycling enzymes, dehydroascorbate reductase, and monodehydroascorbate reductase, are expressed (Fig. 7). The presence of these three enzymes suggests that high levels of ascorbate are required during the early stages of embryo development.
Identification of Candidate Transcript-Protein Pairs
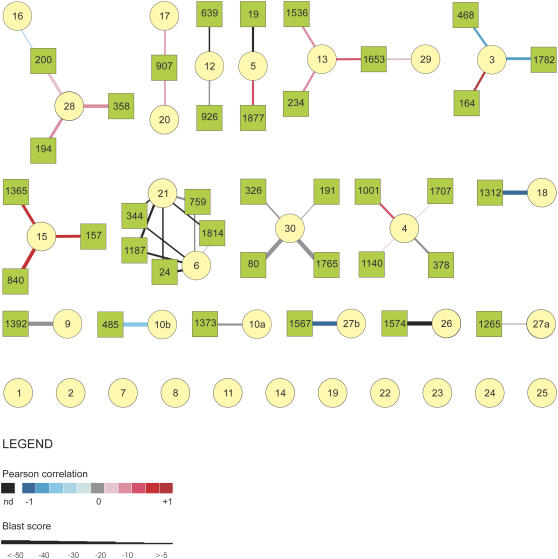
Performing expression analysis at both the mRNA and protein level can provide insight into the relationship between the timing of gene expression and protein function during development. Here, we examined the relationship between the temporal expression patterns of matched probes on the microarray and sequenced protein spots to identify coregulated transcript-protein pairs that can be used as robust markers and candidates for functional analysis. Matching the probe and protein sequences had to be performed indirectly because many of the probes on the array correspond to partial cDNA fragments, especially those obtained using SSH, making them difficult to align with the peptides derived from protein sequencing. Matched transcript-protein pairs were therefore identified by using BLASTX to compare all of the cDNA probe sequences on the microarray with the best hit obtained for each of the protein spots. The Pearson correlation of the transcript and protein expression profiles was then calculated for each of the resulting links (Supplemental Table S5). The resulting network is represented by the graph in Figure 8 in which cDNA probes (squares) and protein spots (circles) form the nodes, whereas the sequence similarity and the correlation between the mRNA and protein expression profiles across the different culture samples are encoded by, respectively, the thickness and the color of the connecting lines. Of the 32 proteins identified, 11 proteins did not show sequence similarity to any of the translated cDNA probes. The remaining 21 proteins showed sequence similarity to one or more of the translated cDNA probes. The sequence similarity between 14 of these 21 proteins and their corresponding translated cDNA probes was very high. Differences in the strength and direction of the correlation between the expression profiles of the matched protein-cDNA probes were observed. The expression profiles of the majority of the matched protein-transcript pairs showed either no correlation or only weak correlation. A strong negative expression correlation, which corresponds to an opposite expression pattern at the mRNA and protein level, was observed for cDNAs matched to proteins 3, 18, and 27b. The corresponding cDNA probes are all pollen expressed (Supplemental Tables S2 and S3). Of particular interest for this study are the expression patterns of four of the highly similar transcript-protein pairs that show a strong positive expression correlation, namely, those of a glyoxylase (spot 15, probes 157, 840, 1,365), a CCAAT-box binding factor (CBF) HAP5a subunit (spot 5, probes 19 and 1,877), a 14-3-3 protein (spot 3, probe 164), and a phosphoglycerate kinase (spot 20, probe 907). All of these matched DNA probes, with the exception of the two probes encoding the CBF HAP5a subunit, are among the list of embryo up-regulated probes identified in the microarray analysis (Supplemental Table S3). The CBF HAP5 subunit probes were not identified among the embryo up-regulated probes due to missing data points; however, independent validation by quantitative RT-PCR confirms that the corresponding gene is expressed in embryogenic cultures (Supplemental Fig. S1).
Figure 8.
Integration of sequence and expression data obtained from transcriptome and proteome analyses of rapeseed pollen and MDE development. In the network, the nodes represent the cDNA probes (circles) and protein spots (squares), whereas the line connecting the nodes represents the sequence similarity between the protein and cDNA sequences. The sequence similarity is indicated by the thickness of the lines (see inset). The similarity in expression profiles is represented by the color of the connecting lines (see inset). The assigned probe and protein numbers are shown inside the nodes. The cytoscape output is shown in Supplemental Table S5.
DISCUSSION
The small size and inaccessibility of the developing zygotic embryo make it technically challenging to perform large-scale, stage-specific analyses of the transcripts and proteins associated with few-celled embryos. Laser capture microdissection has been used for transcript profiling of specific cell types, but this technique has only been applied starting from the globular stage of embryo development (Casson et al., 2005). Currently, most of our knowledge about the genes and developmental programs that operate during the very early stages of embryo development comes from mutagenesis screens aimed at identifying gametophytic and embryo-defective mutants in model species. However, loss-of-function mutations in many genes have little or no phenotypic effect due to compensation of gene function by related family members, and genes for transcription factors and other signaling components appear to be underrepresented among embryo-defective mutants (Tzafrir et al., 2004). Here, we have taken an alternative approach, namely, transcriptome and proteome analysis of B. napus MDE cultures, to identify approximately 220 genes and 32 proteins that are expressed between the two- to four-cell and globular/heart stages of embryo development.
Pathways Associated with Microspore Embryogenesis
Analysis of the proteome and transcriptome datasets identified a number of metabolic processes associated with the establishment of embryo development in MDE cultures that can be used to differentiate between cells undergoing pollen or embryo development in the same culture. The shift from microspore to pollen development is accompanied by a sharp decrease in expression of probes coding for components of the protein synthesis machinery, whereas the majority of probes coding for the same class of proteins are not differentially expressed between microspores and the earliest stages of haploid embryo development. A number of studies have shown a decrease in mRNAs involved in protein synthesis in mature pollen grains relative to sporophytic tissues (Honys and Twell, 2003; Pina et al., 2005). In in vitro pollen cultures, protein synthesis activity shows a more or less steady increase from 0 to 5 d of culture at 18°C, but then drops dramatically thereafter (Cordewener et al., 2000). A decrease in protein synthesis activity was observed at 2 d in MDE cultures incubated at 32°C (Cordewener et al., 2000). This drop in protein synthesis capacity may reflect the accelerated pollen development that takes place at 32°C.
Both the MDE transcriptome and proteome datasets are enriched for mRNAs/proteins involved in glycolysis, a process that uses Glc or other monosaccharides to generate ATP or metabolites for the biosynthesis of storage products (Plaxton, 1996). The large burst of storage product accumulation in rapeseed zygotic embryos and MDEs occurs at the cotyledon stage of development (Taylor et al., 1990), which is much later than the embryo stages analyzed here. This suggests that the glycolysis genes/proteins expressed during very early MDEs are activated to supply the energy needed for the rapid growth that occurs during this phase of embryo development, rather than for storage product biosynthesis.
The MDE proteome dataset also suggests an important role for ascorbate metabolism during early embryo development. Ascorbate peroxidase is a H2O2-scavenging enzyme that utilizes ascorbate to reduce H2O2 to water with the generation of monodehydroascorbate (Asada, 1992). Dehydroascorbate reductase and monodehydroascorbate reductase are ascorbate-recycling enzymes. Dehydroascorbate reductase converts dehydroascorbate to reduced ascorbate, whereas monodehydroascorbate reductase reduces monodehydroascorbate back to ascorbate. Recycling of dehydroascorbate and monodehydroascorbate to ascorbate is a rapid way to increase the ascorbate pool as de novo ascorbate biosynthesis requires activation and coordination of several enzymes (Noctor and Foyer, 1998). There are a number of possible roles for ascorbate during early MDE development. One function for ascorbate is as a scavenger to protect cellular compartments against ROS generated during aerobic metabolism and oxidative stress. Heat stress, such as is used to induce MDE development, has also been shown to generate oxidative burst and ROS production (Mittler et al., 2004). ROS are also known to play a role in many signaling processes, including response to pathogens, hormonal responses, cell division and growth, and metabolism (Gapper and Dolan, 2006). High ascorbate levels also promote cell division in different plant cell culture systems (Joy et al., 1988; De Pinto et al., 1999). Exogenous application of ascorbate to white spruce (Picea glauca) somatic embryos improves shoot formation and germination by promoting the division of meristematic cells (Stasolla et al., 2006). The ascorbate metabolism enzymes identified here are also expressed in suspensor-bearing embryos that only undergo a 1-d heat-stress treatment followed by 7- to 8-d culture at a lower temperature, suggesting that they have a developmental rather than a heat-stress-associated role during MDE development.
Robust Markers for Microspore Embryogenesis
One of the goals of this study, in addition to identifying a collection of early embryo-expressed genes, was to identify gene expression profiles associated with the initial transition from microspore to embryo development. Genes that are transiently expressed in embryogenic cells shortly after the heat-stress treatment could play an active role in the induction of embryogenesis, for example, in promoting stress tolerance or in the initial reprogramming of microspores to a new developmental state. Genes that are transcribed shortly after the initial heat-stress application but that are also transcribed at other stages of embryo development may play a role in the establishment of embryo identity, as was shown for the rapeseed BABY BOOM gene (Boutilier et al., 2002), or may simply reflect the embryo gene expression programs that are activated downstream of these key signaling pathways.
Identification of probes expressed as early as 2 d after the induction of conventional MDE cultures proved to be difficult due to the large contribution of pollen transcripts to the total mRNA pool at this time point. These pollen gene expression profiles could be correlated with the proportion of viable pollen grains in the cultures; however, we cannot rule out the possibility that pollen and embryo genes are coexpressed in the same embryogenic structures. The influence of the pollen genome at the proteome level was greatly reduced compared to the transcriptome, allowing us to identify a number of embryo up-regulated proteins in conventional 2-d embryo cultures. The reduced effect of the pollen proteome on the embryo culture identities as compared to the transcriptome may be due, in part, to an inverse correlation between the abundance of late pollen transcripts and their corresponding proteins (i.e. for some proteins, a large number of mRNAs may be present, but their corresponding proteins are less abundant in mature pollen [Holmes-Davis et al., 2005]).
One protein encoding a GST (spot 29) was identified as being transiently expressed in 2-d embryo cultures and is a good candidate for further analysis. GSTs are a family of enzymes that catalyze the conjugation of glutathione to lipophilic compounds. They are expressed at all stages of development and are involved in a range of processes, including detoxification of xenobiotics and protection against oxidative damage, and may also function as reversible ligands during hormone signaling (for review, see Dixon et al., 2002). GSTs have been identified as markers for the transition to embryo development in other in vitro embryo culture systems (Vrinten et al., 1999; Galland et al., 2001; Thibaud-Nissen et al., 2003; Maraschin et al., 2006). The GST gene isolated from induced barley microspore cultures was also shown to be up-regulated by stress treatments that do not lead to embryo formation, suggesting that it plays a general role in stress protection (Maraschin et al., 2006). It is not known whether the GSTs identified in this and other embryo marker screens protect against stress conditions during cell culture or whether they play a role in normal embryo development.
In contrast to the two- to four-celled conventional embryo cultures, we were able to identify a large number of probes that are up-regulated at the same stage of embryo development using the equivalent suspensor-bearing embryo cultures, most likely due to the absence of viable pollen grains in these cultures. The collection of probes expressed in two- to four-celled suspensor embryo cultures represents some of the earliest embryo-expressed genes identified in plants to date. Only a few of these probes encode functionally annotated proteins, making it difficult to identify underlying processes associated with the commitment to haploid embryo development. However, the robustness and utility of a number of the markers identified in this study are illustrated by their earlier identification as markers for embryogenic cells in tissue culture. The BURP domain protein BNM2 was independently identified here and in two subtractive screens of 4-d heat-stressed microspore embryo cultures (Boutilier et al., 2002; Tsuwamoto et al., 2006). BURP domain proteins are named for their founding members, BNM2, USP, RD22, and the POLYGALACTURONASE1 β-subunit (Hattori et al., 1998). They are secreted proteins that are characterized by their abundant expression, their modular structure, and their conserved C-terminal BURP domain (Hattori et al., 1998). BURP domain genes are expressed under a wide range of developmental and environmental conditions, which is reflected in the diverse processes in which they appear to participate, such as development (Batchelor et al., 2002; Wang et al., 2003) and stress (Yamaguchi-Shinozaki and Shinozaki, 1993; Yu et al., 2004). One BURP domain protein, the fava bean (Vicia faba) UNKNOWN SEED STORAGE PROTEIN (USP), is a marker for embryogenic cell types in wild tobacco (Nicotiana plumbaginifolia) cell cultures (Chesnokov et al., 2002). The BNM2 BURP domain gene identified in this study, in addition to being a marker for the earliest stages of MDE development, is also expressed in developing seeds and flower buds. As such, BNM2 is likely to be involved in developmental, rather than stress-related, processes associated with MDE culture.
We identified 23 LTPs belonging to six different classes as robust markers for the commitment to MDE development. LTPs have been previously identified as markers for embryogenic cells both in carrot (Daucus carota) somatic embryo cultures (Sterk et al., 1991; Toonen et al., 1997), as well as barley and rapeseed MDE cultures (Vrinten et al., 1999; Tsuwamoto et al., 2006). LTPs have been shown in vitro to exchange polar lipids between membranes; however, their specific functions are still a matter of speculation. Different roles have been attributed to LTPs and LTP-like proteins, including defense (Maldonado et al., 2002), cell wall extension (Nieuwland et al., 2005), programmed cell death (Eklund and Edqvist, 2003), and cutin synthesis (Sterk et al., 1991). During embryo development, LTPs have been shown to be expressed throughout the few-celled embryo, becoming localized to the protoderm upon formation (Sterk et al., 1991; Sabala et al., 2000). The Norway spruce (Picea abies) Pa18 LTP gene is expressed in both embryogenic and nonembryogenic cell clusters; however, in nonembryogenic clusters it fails to localize to the protoderm (Sabala et al., 2000). Likewise, failure to localize LTP expression to the embryo protoderm due to over and underexpression of the Pa18 gene leads to morphological alterations in the protoderm layer and to reduced embryo yields (Sabala et al., 2000; Hjortswang et al., 2002).
A New Zygotic Embryo-Like Culture System
The new suspensor-bearing microspore embryogenesis culture system is strikingly similar to the rapeseed zygotic embryogenesis pathway in that a short suspensor is formed prior to the establishment of the embryo-proper cell, followed by development of the embryo proper from the apical cell (Tykarska, 1976). Such embryos subsequently follow the same ordered pattern of cell division as rapeseed zygotic embryos. The rapeseed suspensor-bearing MDE system is the first in vitro embryo culture system that can efficiently produce high frequencies of morphologically normal embryos from single differentiated cells in culture. In vitro cultures, either starting from excised zygotic embryos (Liu et al., 1993) or fertilized ovules (Sauer and Friml, 2004) have been used as an alternative source of material for studies on early embryogenesis, but have their limitations due to the high frequency of embryo mortality when very young embryo stages are cultured and to developmental abnormalities induced by the isolation and/or culture procedures. This unique system now provides a tractable model for studying early embryogenesis under noninvasive conditions.
Key functions have been attributed to suspensors during seed development, such as the positioning of the embryo in close proximity to sources of nutrition in the embryo sac, absorption and transport of nutrients to the embryo, and the synthesis of hormones to support embryo growth (Raghavan, 2001). Our data suggest a new role for the suspensor in guiding the initial structural development of the embryo. Friml et al. (2003) reported that in Arabidopsis zygotic embryo polarity was established from the two-celled stage of embryogenesis onward. This polarity was built up via an auxin gradient in which auxin is transported by the PIN7 efflux carrier from the suspensor to the embryo proper. The authors propose that this basal-to-apical auxin gradient specifies the identity of the apical embryo structures. Following this hypothesis, a similar auxin gradient may be built up in the suspensor cell file of MDEs and this auxin gradient could contribute to the embryonic identity specification of the most distal cells in the file.
MATERIALS AND METHODS
Microspore Isolation and Culture
The procedures for microspore isolation and culture are provided as Supplemental Materials and Methods S1. Two independent microspore isolations, yielding 70- and 150-mL suspension, respectively, were performed for the microarray analysis. The microspores from each isolation were divided over three temperature treatments: (1) continuously at 18°C ± 1°C to allow pollen maturation; (2) 24 h at 32°C ± 0.2°C followed by transfer to 25°C ± 1°C for suspensor-bearing MDE development; and (3) continuously at 32°C ± 0.2°C for production of conventional MDE development. For microarray analysis, each of the two starting cultures was divided into seven samples. For each of the starting cultures, one sample was harvested immediately (0d), one sample was cultured for 5 d to obtain mature pollen (5e), two samples were cultured for 8 d (8se) and 10 d (10se) to obtain suspensor-bearing MDEs, and three samples were cultured for 2 d (2e), 5 d (5e), and 10 d (10e) to obtain conventional MDEs. The 10-d conventional embryo cultures were sieved (>50 and <200 μm) so that the sample contained only embryos, whereas all other samples were prepared directly from the crude culture suspensions. The percentages of embryo-forming microspores and pollen were determined at the points of harvest.
For the 2-D gel electrophoresis analyses, similar microspore cultures were initiated and samples were harvested at the indicated time points from pooled isolates. A small aliquot of each isolate was cultured continuously at 32°C ± 0.2°C to induce standard MDE development and used to determine the embryogenicity of the culture by counting the number of embryos that had developed after 10 d.
Microarray Preparation
The majority of the probe DNAs spotted on the array correspond to MDE cDNAs isolated from six different cDNA libraries: 24U, a 24-h 32°C heat-stressed MDE culture library in HybriZAP (Stratagene); 4U, a 4-d 32°C heat-stressed MDE culture library; 4F, a forward-subtracted SSH (CLONTECH) library made using 4-d 32°C heat-stressed MDE culture RNA as the tester and 0-d microspore and 4-d, 18°C pollen culture RNA as the driver; 4R, a reverse-subtracted SSH library made using 0-d microspore and 4-d, 18°C pollen culture RNA as the tester and 4-d, 32°C heat-stressed MDE RNA as the driver; 9U, a 9-d globular-stage MDE culture library from the SSH unsubtracted tester control; and 9F, a forward-subtracted SSH library from the 9-d-old globular-stage MDE RNA as the tester and leaf RNA as the driver. All clones were single-pass sequenced and queried against The Arabidopsis Information Resource (TAIR) AtH1seq and Brassica databases using BLASTN algorithms (Altschul et al., 1990). The best BLAST hit derived from the Brassica database was also used to requery the TAIR AtH1seq database. The last searches were performed in April 2006. Each clone was assigned an identity based on the best BLAST hits and used to group the proteins into functional categories using the Arabidopsis (Arabidopsis thaliana) functional catalog FunCat (Munich Information Center for Protein Sequences [MIPS]; Ruepp et al., 2004), Arabidopsis GO annotations (TAIR database), and literature searches.
A smaller number of probe DNAs comprised previously identified cDNA clones isolated from MDE cultures (Custers et al., 2001) and maturing seeds (Soeda et al., 2005), as well as gene-specific probes for Arabidopsis transcription factors (http://www2.mpiz-koeln.mpg.de/∼adis/tfarray/menu.php; de Folter et al., 2004). The complete list and putative identities of arrayed clones is available as Supplemental Table S2).
Information on microarray spotting, target amplification, labeling and hybridization, and data acquisition are provided in Supplemental Materials and Methods S1.
Microarray Data Analysis
Spot intensities were background corrected by subtracting the average over the 12 background control spots containing nonhybridizing yeast (Saccharomyces cerevisiae) DNA, as described above. Normalization for dye effects was performed using the intensities of the spiked Luciferase control spots described above. All the data are presented as log2-transformed ratios. For array 72-16, a pipetting error was made with the Luciferase spike control, resulting in a shift of the median normalized log2 ratio from 0 to −1.4. This shift was manually corrected.
ANOVA was used for analysis, per clone, of the background-corrected and normalized log ratio values. In the ANOVA model, a nested structure was used with the two cultures (biological replicates), arrays within cultures (the swapped dye pair or technical replicates), and duplicated spots within each array as the blocking factors (factors generating random variation), and the different samples (each sample is a combination of a time point and a biological tissue) as the single treatment factor. A weight factor was introduced in the ANOVA so that, in the averaging over the two cultures, each culture received equal weight despite the fact that two arrays were available for the samples from culture 1 (the swapped dye pair) and only one array was available for the samples from culture 2. For each clone, an F probability was separately calculated to test for significant differences between any of the samples. The F statistic is used to help identify whether the differences between treatment means are due to background noise or the actual treatment. If there are no differences between treatments, the F statistic will be less than or near 1. A maximum of 42 log-ratio values (seven samples × three arrays × two replicate spots within arrays) was available per clone if there were no missing values. lsd (α = 0.05) for between-sample means were calculated per probe using the se at the level of biological replicates.
lsd values were used to identify differential gene expression patterns among the probes that showed a significant expression pattern across the treatments based on the F-statistic analysis (P = 0.005, 577 probes). Two samples are considered statistically different if they differ by as much or more than the lsd. Pair-wise comparisons were made between the expression ratios of the signal intensities for each probe in the different cultures to identify probes that are differentially expressed. Probes that are differentially expressed will exhibit an absolute difference in expression ratios between the samples that is greater or equal to the lsd.
The normalized mean log2 signal ratios for all the microarray probes (excluding the controls) for each culture sample were used for PCA. Data were centered by subtracting the average of every row/column from every signal ratio. Composition of the analyzed samples in terms of the percentage of gametophytic and embryogenic structures was incorporated into the transcriptome plot using the position of the samples as weights.
Microarray Validation
Real-time RT-PCR experiments are described in Supplemental Materials and Methods S1. The RT-PCR primer sequences and threshold cycle values are available as Supplemental Table S4 and Supplemental Figure S2.
RNA gel-blot analysis and mRNA in situ hybridization was performed using a full-length BNM2 cDNA as described by Boutilier et al. (2002).
Protein Sample Preparation and 2-D Gel Electrophoresis
Protein isolation and subsequent 2-D gel electrophoresis were performed as described in Cordewener et al. (1994), with minor modifications as described in Supplemental Materials and Methods S1. For each of the culture extracts, 400 μg of protein were loaded on precast Immobiline DryStrip gels (pH 4–7, 18 cm; Amersham Biosciences). By using the Isodalt system (Amersham Biosciences), the second-dimension gels of all seven time points could be run in parallel in one experiment. Following 2-d gel electrophoresis, gels were stained with SYPRO Ruby (Bio-Rad) fluorescent stain.
2-D Gel Data Analysis
Digitized gel images were analyzed using PDQuest software, version 7.1.0 (Bio-Rad). In PDQuest, there are no default settings for spot detection so the spot detection parameters (sensitivity, minimum peak, size scale, large spot size, background removal, smoothing) were optimized for each group of replicates. Selection of protein spots of interest for MS analysis was performed by a combination of pairwise t-test analyses. From the combined set of the differential protein spots, a subset was chosen on the basis of spot intensity and quality of differential behavior.
PCA was performed after normalization as follows. Total spot volume per gel was used to normalize spot intensities to compensate for the considerable variation in the total volume of all matched spots per gel. For each gel, 0 values (representing absent spots) were replaced with 0.03 percentiles. The log-transformed data matrix was then used for PCA in GeneMaths.
Protein Identification by MS
Protein spots of interest were excised from the gel, dried, and subjected to in-gel trypsin (sequence grade modified porcine trypsin; Promega) digestion according to Shevchenko et al. (1996). Tryptic peptides were analyzed on a Q-TOF-2 mass spectrometer (Waters Corporation) as described by America et al. (2006). The MassLynx package, version 3.4, was used to process MS/MS data. First, the MaxEnt 3 module was used to deconvolute the data. MS/MS spectra containing good-quality collision-induced dissociation products were selected for further processing. The BioLynx de novo sequencing algorithm PepSeq was used to interpret MS/MS spectra and to generate peptide sequences in an automated fashion. The interpretation of MS/MS spectra was further scrutinized interactively using the PepSeq module. A list was compiled of possible peptide sequences per MS/MS spectrum. A BLASTP search against the National Center for Biotechnology Information (NCBI) Nr database was performed in batch mode with the compiled list of possible peptide sequences per protein sample. Output of the BLAST batch was sorted per hit in the database and based on PepSeq score and BLAST e value using an in-house-developed algorithm.
Integrated Transcript and Protein Analysis
All partial sequences from the cDNA fragments on the microarray were queried against a full-length protein database (n = 32) based on the protein identities derived from the 30 sequenced differentially expressed protein spots using the BLASTX program (at e value < 0.001 and a maximum of three hits). The Pearson correlation of the probe and protein expression profiles was determined using the mean log expression ratios (microarray) and the mean log intensities (2-D gel analysis). The resulting network was visualized using Cytoscape 1.1.1 (www.cytoscape.org).
Sequence data from this article can be found in Supplemental Table S2.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Quantitative real-time RT-PCR validation of pollen and embryo up-regulated genes.
Supplemental Figure S2. Comparison of the functional/process categories assigned to pollen and embryo up-regulated probes.
Supplemental Figure S3. Overview of protein spots chosen for sequencing.
Supplemental Table S1. Effect of temperature and bud size on suspensor formation.
Supplemental Table S2. Source, sequence, and expression ratios of the DNA probes spotted on the microarray.
Supplemental Table S3. Differentially expressed DNA probes.
Supplemental Table S4. Raw data for quantitative real-time RT-PCR validation of pollen and embryo up-regulated genes.
Supplemental Table S5. Cytoscape output: sequence and expression profile similarity between linked transcript-protein pairs.
Supplemental Materials and Methods S1. Protocols for microspore culture, microarray analyses, RT-PCR, and protein analysis.
Supplementary Material
Acknowledgments
We thank Ruud de Maagd for comments on the manuscript. We are grateful to Claudio Stasolla and Yves Gibon for providing insight into ascorbate metabolism and glycolysis, respectively.
This work was supported by the Dutch Ministry of Agriculture, Nature, and Food Quality (program no. DWK 281–392), a Natural Science and Engineering Research Council visiting fellowship in a Canadian government laboratory, and the Biotechnology Research Indonesia-Netherlands research program, with financial aid from the Royal Netherlands Academy of Arts and Sciences and the fellowship program Quality for Undergraduate Education, Bogor Agricultural University, Indonesia.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Kim Boutilier (kim.boutilier@wur.nl).
The online version of this article contains Web-only data.
References
- Aionesei T, Touraev A, Heberle Bors E (2005) Pathways to microspore embryogenesis. In CE Palmer, WA Keller, KJ Kasha, eds, Haploids in Crop Improvement II, Vol 56. Springer-Verlag, Heidelberg, pp 11–34
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215 403–410 [DOI] [PubMed] [Google Scholar]
- America AHP, Cordewener JHG, van Geffen MHA, Lommen A, Vissers JPC, Bino RJ, Hall RD (2006) Alignment and statistical difference analysis of complex peptide data sets generated by multidimensional LC-MS. Proteomics 6 641–653 [DOI] [PubMed] [Google Scholar]
- Asada K (1992) Ascorbate peroxidase—a hydrogen peroxide-scavenging enzyme in plants. Physiol Plant 85 235–241 [Google Scholar]
- Batchelor AK, Boutilier K, Miller SS, Hattori J, Bowman L, Hu M, Lantin S, Johnson DA, Miki BLA (2002) SCB1, a BURP domain protein gene from developing seed coats. Planta 215 523–532 [DOI] [PubMed] [Google Scholar]
- Boutilier K, Fiers M, Liu CM, van Der Geest AHM (2005) Biochemical and molecular aspects of haploid embryogenesis. In CE Palmer, WA Keller, KJ Kasha, eds, Haploids in Crop Improvement II, Vol 56. Springer-Verlag, Heidelberg, pp 73–95
- Boutilier K, Offringa R, Sharma VK, Kieft H, Ouellet T, Zhang L, Hattori J, Liu CM, van Lammeren AA, Miki BL, et al (2002) Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. Plant Cell 14 1737–1749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casson S, Spencer M, Walker K, Lindsey K (2005) Laser capture microdissection for the analysis of gene expression during embryogenesis of Arabidopsis. Plant J 42 111–123 [DOI] [PubMed] [Google Scholar]
- Chan J, Pauls KP (2007) Brassica napus Rop GTPases and their expression in microspore cultures. Planta 225 469–484 [DOI] [PubMed] [Google Scholar]
- Chan J, Woronuk G, Schulze D, Brazolot J, Pauls KP (2006) When microspores decide to become embryos—cellular and molecular changes. Can J Bot 84 668–678 [Google Scholar]
- Chesnokov YV, Meister A, Manteuffel R (2002) A chimeric green fluorescent protein gene as an embryogenic marker in transgenic cell culture of Nicotiana plumbaginifolia Viv. Plant Sci 162 59–77 [Google Scholar]
- Cordewener J, Bergervoet J, Liu CM (2000) Changes in protein synthesis and phosphorylation during microspore embryogenesis in Brassica napus. J Plant Physiol 156 156–163 [Google Scholar]
- Cordewener JHG, Busink R, Traas JA, Custers JBM, Dons HJM, Van Lookeren Campagne MM (1994) Induction of microspore embryogenesis in Brassica napus L. is accompanied by specific changes in protein synthesis. Planta 195 50–56 [Google Scholar]
- Custers JBM, Cordewener JHG, Fiers MA, van Maasen BTH, Van Lookeren Campagne MM, Liu CM (2001) Androgenesis in Brassica. In SS Bhojwani, WY Soh, eds, Current Trends in the Embryology of Angiosperms. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 451–470
- Custers JBM, Cordewener JHG, Nöllen Y, Dons HJM, Van Lookeren Campagne MM (1994) Temperature controls both gametophytic and sporophytic development in microspore cultures of Brassica napus. Plant Cell Rep 13 267–271 [DOI] [PubMed] [Google Scholar]
- de Folter S, Busscher J, Colombo L, Losa A, Angenent GC (2004) Transcript profiling of transcription factor genes during silique development in Arabidopsis. Plant Mol Biol 56 351–366 [DOI] [PubMed] [Google Scholar]
- De Pinto MC, Francis D, De Gara L (1999) The redox state of the ascorbate-dehydroascorbate pair as a specific sensor of cell division in tobacco BY-2 cells. Protoplasma 209 90–97 [DOI] [PubMed] [Google Scholar]
- Diatchenko L, Lau YF, Campbell AP, Chenchik A, Moqadam F, Huang B, Lukyanov S, Lukyanov K, Gurskaya N, Sverdlov ED, et al (1996) Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc Natl Acad Sci USA 93 6025–6030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon DP, Lapthorn A, Edwards R (2002) Plant glutathione transferases. Genome Biol 3 reviews 3004.3001–3004.3010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eklund DM, Edqvist J (2003) Localization of nonspecific lipid transfer proteins correlate with programmed cell death responses during endosperm degradation in Euphorbia lagascae seedlings. Plant Physiol 132 1249–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml J, Vieten A, Sauer M, Weijers D, Schwarz H, Hamann T, Offringa R, Jürgens G (2003) Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426 147–153 [DOI] [PubMed] [Google Scholar]
- Forster BP, Thomas WTB (2005) Doubled haploids in genetics and plant breeding. Plant Breed Rev 25 57–88 [Google Scholar]
- Galland R, Randoux B, Vasseur J, Hilbert JL (2001) A glutathione S-transferase cDNA identified by mRNA differential display is upregulated during somatic embryogenesis in Cichorium. Biochim Biophys Acta 1522 212–216 [DOI] [PubMed] [Google Scholar]
- Gapper C, Dolan L (2006) Control of plant development by reactive oxygen species. Plant Physiol 141 341–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori J, Boutilier K, van Lookeren Campagne MM, Miki BL (1998) A conserved BURP domain defines a novel group of plant proteins with unusual primary structures. Mol Gen Genet 259 424–428 [DOI] [PubMed] [Google Scholar]
- Hause B, Van Veenendaal WLH, Hause G, Van Lammeren AAM (1994) Expression of polarity during early development of microspore-derived and zygotic embryos of Brassica napus L. cv. Topas. Bot Acta 107 407–415 [Google Scholar]
- Hjortswang HI, Filonova LH, Vahala T, von Arnold S (2002) Modified expression of the Pa18 gene interferes with somatic embryo development in Norway spruce. Plant Growth Regul 38 75–82 [Google Scholar]
- Holmes-Davis R, Tanaka CK, Vensel WH, Hurkman WJ, McCormick S (2005) Proteome mapping of mature pollen of Arabidopsis thaliana. Proteomics 5 4864–4884 [DOI] [PubMed] [Google Scholar]
- Honys D, Twell D (2003) Comparative analysis of the Arabidopsis pollen transcriptome. Plant Physiol 132 640–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosp J, Maraschin SF, Touraev A, Boutilier K (2006) Functional genomics of microspore embryogenesis. Euphytica doi/10.1007/s10681-006-9238-9
- Ilic-Grubor K, Attree SM, Fowke LC (1998) Comparative morphological study of zygotic and microspore-derived embryos of Brassica napus L. as revealed by scanning electron microscopy. Ann Bot (Lond) 82 157–165 [Google Scholar]
- Jose-Estanyol M, Gomis-Ruth FX, Puigdomenech P (2004) The eight-cysteine motif, a versatile structure in plant proteins. Plant Physiol Biochem 42 355–365 [DOI] [PubMed] [Google Scholar]
- Joy RW, Patel KR, Thorpe TA (1988) Ascorbic acid enhancement of organogenesis in tobacco callus. Plant Cell Tissue Organ Cult 13 219–228 [Google Scholar]
- Lehti-Shiu MD, Adamczyk BJ, Fernandez DE (2005) Expression of MADS-box genes during the embryonic phase. Plant Mol Biol 58 89–107 [DOI] [PubMed] [Google Scholar]
- Letarte J, Simion E, Miner M, Kasha KJ (2006) Arabinogalactans and arabinogalactan-proteins induce embryogenesis in wheat (Triticum aestivum L.) microspore cultures. Plant Cell Rep 24 691–698 [DOI] [PubMed] [Google Scholar]
- Liu CM, Xu ZH, Chua NH (1993) Proembryo culture: in vitro development of early globular-stage zygotic embryos from Brassica juncea. Plant J 3 291–300 [Google Scholar]
- Maldonado AM, Doerner P, Dixon RA, Lamb CJ, Cameron RK (2002) A putative lipid transfer protein involved in systemic resistance signalling in Arabidopsis. Nature 419 399–403 [DOI] [PubMed] [Google Scholar]
- Maluszinsky M, Kasha KJ, Szarejko I (2003) Published doubled haploid protocols in plant species. In M Maluszynski, KJ Kasha, BP Forster, I Szarejko, eds, Doubled Haploid Production in Crop Plants. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 309–335
- Maraschin SF, Caspers M, Potokinad E, Wülfert F, Granerd A, Spaink HP, Wang M (2006) cDNA array analysis of stress-induced gene expression in barley androgenesis. Physiol Plant 127 535–550 [Google Scholar]
- Maraschin SF, de Priester W, Spaink HP, Wang M (2005) Androgenic switch: an example of plant embryogenesis from the male gametophyte perspective. J Exp Bot 56 1711–1726 [DOI] [PubMed] [Google Scholar]
- Matthys-Rochon E (2005) Secreted molecules and their role in embryo formation in plants: a mini-review. Acta Biol Crac Ser Bot 47 23–29 [Google Scholar]
- Mittler R, Vanderauwer S, Gollery M, Van Breusegem F (2004) Reactive oxygen gene network of plants. Trends Plant Sci 9 490–498 [DOI] [PubMed] [Google Scholar]
- Nieuwland J, Feron R, Huisman BA, Fasolino A, Hilbers CW, Derksen J, Mariani C (2005) Lipid transfer proteins enhance cell wall extension in tobacco. Plant Cell 17 2009–2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor G, Foyer CH (1998) Ascorbate and glutathione: keeping active oxygen under control. Annu Rev Plant Physiol Plant Mol Biol 49 249–279 [DOI] [PubMed] [Google Scholar]
- Pechan PM, Bartels D, Brown DCW, Schell J (1991) Messenger-RNA and protein changes associated with induction of Brassica microspore embryogenesis. Planta 184 161–165 [DOI] [PubMed] [Google Scholar]
- Pina C, Pinto F, Feijó JA, Becker JD (2005) Gene family analysis of the Arabidopsis pollen transcriptome reveals biological implications for cell growth, division control, and gene expression regulation. Plant Physiol 138 744–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaxton WC (1996) The organization and regulation of plant glycolysis. Annu Rev Plant Physiol Plant Mol Biol 47 185–214 [DOI] [PubMed] [Google Scholar]
- Raghavan V (2001) Life and times of the suspensor of angiosperm embryos. In NS Rangaswamy, ed, Phytomorphology Golden Jubilee Issue 2001. International Society of Plant Morphologists, Delhi, India, pp 251–276
- Ruepp A, Zollner A, Maier D, Albermann K, Hani J, Mokrejs M, Tetko I, Guldener U, Mannhaupt G, Munsterkotter M, et al (2004) The FunCat, a functional annotation scheme for systematic classification of proteins from whole genomes. Nucleic Acids Res 32 5539–5545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabala I, Elfstrand M, Farbos I, Clapham D, von Arnold S (2000) Tissue-specific expression of Pa18, a putative lipid transfer protein gene, during embryo development in Norway spruce (Picea abies). Plant Mol Biol 42 461–478 [DOI] [PubMed] [Google Scholar]
- Sauer M, Friml J (2004) In vitro culture of Arabidopsis embryos within their ovules. Plant J 40 835–843 [DOI] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M (1996) Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal Chem 68 850–858 [DOI] [PubMed] [Google Scholar]
- Soeda Y, Konings MC, Vorst O, van Houwelingen AM, Stoopen GM, Maliepaard CA, Kodde J, Bino RJ, Groot SPC, van der Geest AHM (2005) Gene expression programs during Brassica oleracea seed maturation, osmopriming, and germination are indicators of progression of the germination process and the stress tolerance level. Plant Physiol 137 354–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stasolla C, Lam MSW, Yeung EC (2006) Exogenous applications of ascorbic acid enhance shoot apical meristem growth and induce shoot organogenesis in germinating white spruce (Picea glauca) somatic embryos. Int J Plant Sci 167 429–436 [Google Scholar]
- Sterk P, Booij H, Schellekens GA, Van Kammen A, De Vries SC (1991) Cell-specific expression of the carrot EP2 lipid transfer protein gene. Plant Cell 3 907–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straatman KR, Nijsse J, Kieft H, Van Aelst AC, Schel JHN (2000) Nuclear pore dynamics during pollen development and androgenesis in Brassica napus. Sex Plant Reprod 13 43–51 [Google Scholar]
- Taylor DC, Weber N, Underhill EW, Pomeroy MK, Keller WA, Scowcroft WR, Wilen RW, Moloney MM, Holbrook LA (1990) Storage-protein regulation and lipid accumulation in microspore embryos of Brassica napus L. Planta 181 18–26 [DOI] [PubMed] [Google Scholar]
- Thibaud-Nissen F, Shealy RT, Khanna A, Vodkin LO (2003) Clustering of microarray data reveals transcript patterns associated with somatic embryogenesis in soybean. Plant Physiol 132 118–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toonen MAJ, Verhees JA, Schmidt EDL, van Kammen A, de Vries SC (1997) AtLTP1 luciferase expression during carrot somatic embryogenesis. Plant J 12 1213–1221 [DOI] [PubMed] [Google Scholar]
- Tsuwamoto R, Fukuoka H, Takahata Y (2007) Identification and characterization of genes expressed in early embryogenesis from microspores of Brassica napus. Planta 225 641–652 [DOI] [PubMed] [Google Scholar]
- Twell D (2002) Pollen developmental biology. In SD O'Neill, JA Roberts, eds, Plant Reproduction: Annual Plant Reviews, Vol 6. Sheffield Academic Press, Sheffield, UK, pp 86–153
- Tykarska T (1976) Rape embryogenesis. I. The proembryo development. Acta Soc Bot Pol 45 3–15 [Google Scholar]
- Tykarska T (1979) Rape embryogenesis. II. Development of embryo proper. Acta Soc Bot Pol 48 391–421 [Google Scholar]
- Tzafrir I, Pena-Muralla R, Dickerman A, Berg MR, Hutchens R, Sweeney S, Mc TC, Elver J, Aux G, Patton D, et al (2004) Identification of genes required for embryo development in Arabidopsis. Plant Physiol 135 1206–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrinten PL, Nakamura T, Kasha KJ (1999) Characterization of cDNAs expressed in the early stages of microspore embryogenesis in barley (Hordeum vulgare) L. Plant Mol Biol 41 455–463 [DOI] [PubMed] [Google Scholar]
- Wagner U, Edwards R, Dixon DP, Mauch F (2002) Probing the diversity of the Arabidopsis glutathione S-transferase gene family. Plant Mol Biol 49 515–532 [DOI] [PubMed] [Google Scholar]
- Wang A, Xia Q, Xie W, Datla R, Selvaraj G (2003) The classical Ubisch bodies carry a sporophytically produced structural protein (RAFTIN) that is essential for pollen development. Proc Natl Acad Sci USA 100 14487–14492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K (1993) The plant hormone abscisic acid mediates the drought-induced expression but not the seed-specific expression of rd22, a gene responsive to dehydration stress in Arabidopsis thaliana. Mol Gen Genet 238 17–25 [DOI] [PubMed] [Google Scholar]
- Yang HP, Matsubayashi Y, Hanai H, Sakagami Y (2000) Phytosulfokine-α, a peptide growth factor found in higher plants: its structure, functions, precursor and receptors. Plant Cell Physiol 41 825–830 [DOI] [PubMed] [Google Scholar]
- Yeung EC (2002) The canola microspore-derived embryo as a model system to study developmental processes in plants. J Plant Biol 45 119–133 [Google Scholar]
- Yeung EC, Rahman MH, Thorpe TA (1996) Comparative development of zygotic and microspore-derived embryos in Brassica napus L. cv Topas. 1. Histodifferentiation. Int J Plant Sci 157 27–39 [Google Scholar]
- Yu S, Zhang L, Zuo K, Li Z, Tang K (2004) Isolation and characterization of a BURP domain-containing gene BnBDC1 from Brassica napus involved in abiotic and biotic stress. Physiol Plant 122 210–218 [Google Scholar]
- Zhang JZ, Laudencia-Chingcuanco DL, Comai L, Li M, Harada JJ (1994) Isocitrate lyase and malate synthase genes from Brassica napus L. are active in pollen. Plant Physiol 104 857–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zondlo SC, Irish VF (1999) CYP78A5 encodes a cytochrome P450 that marks the shoot apical meristem boundary in Arabidopsis. Plant J 19 259–268 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.