Abstract
Viral infection leads to activation of the transcription factors interferon regulatory factor-3 and NF-κB, which collaborate to induce type I IFNs. The RNA helicase proteins RIG-I and MDA5 were recently identified as two cytoplasmic viral RNA sensors that recognize different species of viral RNAs produced during viral replication. In this study, we identified DAK, a functionally unknown dihydroacetone kinase, as a specific MDA5-interacting protein. DAK was associated with MDA5, but not RIG-I, under physiological conditions. Overexpression of DAK inhibited MDA5- but not RIG-I- or TLR3-mediated IFN-β induction. Overexpression of DAK also inhibited cytoplasmic dsRNA and SeV-induced activation of the IFN-β promoter, whereas knockdown of endogenous DAK by RNAi activated the IFN-β promoter, and increased cytoplasmic dsRNA- or SeV-triggered activation of the IFN-β promoter. In addition, overexpression of DAK inhibited MDA5- but not RIG-I-mediated antiviral activity, whereas DAK RNAi increased cytoplasmic dsRNA-triggered antiviral activity. These findings suggest that DAK is a physiological suppressor of MDA5 and specifically inhibits MDA5- but not RIG-I-mediated innate antiviral signaling.
Keywords: DAK, type I IFN, interferon regulatory factor-3, NF-κB, virus
Viral infection results in transcriptional induction of type I IFNs, including IFN-β and IFN-α family cytokines. Type I IFNs induce the expression of a set of IFN-stimulated genes that have inhibitory effects on viral replication in infected and neighboring uninfected cells (1–6). Transcriptional activation of the promoters of type I IFN genes requires the coordinated activation of multiple transcription factors and their cooperative assembly into transcriptional enhancer complexes in vivo. For example, the enhancer of the IFN-β gene contains a κB site recognized by NF-κB, a site for ATF-2/c-Jun, and two IFN-stimulated response elements (ISREs) recognized by phosphorylated interferon regulatory factor (IRF)-3 and/or IRF-7. It has been shown that transcriptional activation of the IFN-β gene requires coordinated and cooperative assembly of an enhanceosome that contains all of these transcription factors (7, 8).
The innate immune system has developed at least two distinct mechanisms for the recognition of viral RNAs (2, 3, 9). One is mediated by Toll-like receptor 3 (TLR3), which recognizes viral dsRNA released by infected cells (10). Engagement of TLR3 by dsRNA triggers TRIF-mediated signaling pathways, leading to IRF-3 and NF-κB activation (11–16). The second mechanism involves two RNA helicase proteins, RIG-I and MDA5, which function as cytoplasmic viral RNA sensors (17, 18). Both RIG-I and MDA5 contain two CARD modules at their N terminus and a DexD/H-box helicase domain at their C terminus. The helicase domains of RIG-I and MDA5 serve as intracellular viral RNA receptors, whereas the CARD modules are responsible for transmitting signals to downstream CARD-containing adaptor VISA/MAVS/IPS-1/Cardif, which in turn activates TAK1-IKKβ and TBK1/IKKε kinases, leading to activation of NF-κB and IRF-3 and induction of type I IFNs (19–24).
Although MDA5 and RIG-I share a similar structural architecture, and their signaling pathways converge at the adaptor level, gene-knockout studies indicate that the two proteins are required for responding to distinct species of RNA viruses. RIG-I responds to in vitro-transcribed dsRNA, vesicular stomatitis virus (VSV), Newcastle disease virus, and influenza virus in mice. In contrast, MDA5 recognizes poly(I:C) and is essential for the antiviral response to the picornavirus encephalomyocarditis virus (25, 26). Recently, it was demonstrated that RIG-I, but not MDA5, recognizes single-strand RNA bearing 5′ phosphate (27, 28). In addition, it has been shown that signaling mediated by RIG-I- and MDA5-mediated signaling is differentially regulated. For example, the V protein of paramyxovirus appears to selectively target MDA5- but not RIG-I-mediated IFN response (29–31).
In the present study, we identified DAK as an MDA5-interacting protein in yeast two-hybrid screens. DAK is associated with MDA5 but not RIG-I under physiological condition and dissociated with MDA5 upon viral infection in 293 cells. We also show that DAK selectively inhibits MDA5- but not RIG-I-mediated type I IFN signaling and innate antiviral response. These findings provide insight into the mechanisms of differential regulation of MDA5- and RIG-I-mediated signaling by cellular proteins.
Results
Identification of DAK as an MDA5-Associated Protein.
It is well established that MDA5 is essential for innate immune response to certain viruses. However, the regulatory mechanism of MDA5-mediated signaling has not been adequately characterized. To identify potential proteins that interact with MDA5 and regulate its signaling, we performed yeast two-hybrid screens of a 293 cell cDNA library using full-length MDA5 as bait. Among ≈1 × 106 independent clones screened, four β-galactosidase-positive clones encoded DAK. DAK is a member of the evolutionarily conserved family of dihydroxyacetone kinases. In bacteria, DAK phosphorylates dihydroxyacetone to produce Dha phosphate, an obligatory precursor for the biogenesis of glyceryl ether phospholipids. In mammals, DAK displays dual activities as FMN cyclase and ATP-dependent Pha kinase (32, 33). However, the physiological functions of DAK in higher eukaryotes are unknown.
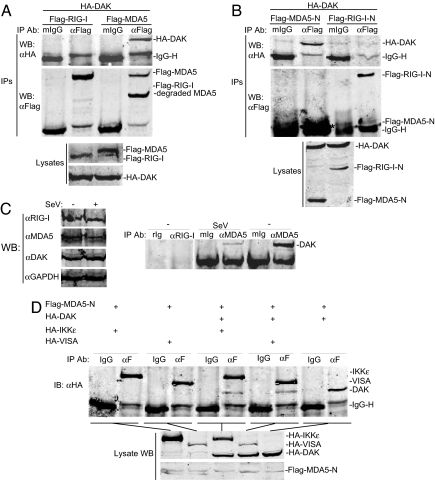
Because DAK interacts with MDA5 in yeast two-hybrid systems, we determined whether DAK interacts with MDA5 in mammalian cells. We transfected 293 cells with expression plasmids for hemagglutinin (HA)-tagged DAK and Flag-tagged MDA5 and performed coimmunoprecipitation experiments. The results indicated that DAK could interact with MDA5 (Fig. 1A). In the same experiment, DAK did not interact with RIG-I, which is structurally and functionally related to MDA5 (Fig. 1A). DAK also did not interact with VISA, TBK1, or IKKε, which are downstream signaling proteins of MDA5 and RIG-I (data not shown). Furthermore, coimmunoprecipitation experiments indicated that the N-terminal CARD modules of MDA5 (MDA5-N), but not RIG-I (RIG-I-N), were sufficient for its interaction with DAK (Fig. 1B). Immunofluorescent staining experiments suggest that DAK is a cytoplasmic protein and has a similar distribution pattern with MDA5 (data not shown).
Fig. 1.
Identification of DAK as an MDA5-associated protein. (A) DAK interacts with MDA5 but not RIG-I in mammalian overexpression system. The 293 cells (2 × 106) were cotransfected with the indicated plasmids (5 μg each), and cell lysates were immunoprecipitated with anti-Flag antibody (αFlag) or control mouse IgG (mIgG). The immunoprecipitates were analyzed by Western blots with anti-HA (Top) or anti-Flag (Middle) antibody. Expression of the transfected proteins was analyzed by Western blots with anti-HA and anti-Flag antibodies (Bottom). (B) DAK interacts with N-terminal domain of MDA5. The experiments were similarly performed as in A. The asterisk (Middle) indicates the band of Flag-MDA5-N, which is close to the IgG light-chain band. (C) Endogenous association of DAK with MDA5. The 293 cells (1 × 108) were infected with SeV or left uninfected for 4 h. Cell lysate was immunoprecipitated with mouse anti-MDA5 or rabbit anti-RIG-I antiserum or control IgG. The immunoprecipitates were analyzed by Western blots with anti-DAK antibody (Right). The expression levels of the endogenous proteins were analyzed by Western blots with anti-MDA5, anti-DAK, and anti-RIG-I antibodies (Left). Each lane was loaded with whole-cell lysate from 5 × 106 cells. (D) Interactions between MDA5-N and DAK, IKKε and VISA. The 293 cells (2 × 106) were cotransfected with the indicated plasmids (5 μg of each). Coimmunoprecipitation and Western blot analysis were performed as in A. WB, Western blot; IP, immunoprecipitation; IB, immunoblot.
To determine whether DAK interacts with MDA5 under physiological condition, we performed endogenous coimmunoprecipitation experiments. The results indicated that endogenous DAK was associated with endogenous MDA5 but not RIG-I in untransfected 293 cells, and the association between DAK and MDA5 was diminished after infection of 293 cells with Sendai virus (SeV) (Fig. 1C). The diminishing of MDA5-associated DAK upon viral infection was not due to down-regulation of protein levels of MDA5 and DAK as suggested by Western blot analysis (Fig. 1C). Taken together, these data suggest that DAK is associated with MDA5 under physiological condition, and this association is disrupted upon viral infection.
Because MDA5 signals through VISA, we investigated the relationship of interactions of MDA5 with VISA and DAK1. In transient transfection and coimmunoprecipitation experiments, MDA5-N could interact with VISA and DAK (Fig. 1D). However, when MDA5-N, VISA, and DAK were cotransfected, MDA5-N interacted more strongly with VISA than DAK, despite the fact that DAK was expressed at a higher level than VISA (Fig. 1D). Similarly, MDA5-N also interacted more strongly with IKKε than DAK (Fig. 1D). The simplest explanation for these observations is that MDA5-N has a higher affinity for VISA and IKKε than DAK. Because overexpression of MDA5-N potently activates IRF-3 and NF-κB and mimics the activation of MDA5 (26, 30), it is possible that the higher affinity of active MDA5 with VISA and IKKε allows MDA5's disassociation with DAK and recruitment of VISA and IKKε upon viral infection.
DAK Inhibits MDA5- but Not RIG-I-Mediated Activation of ISRE and the IFN-β Promoter.
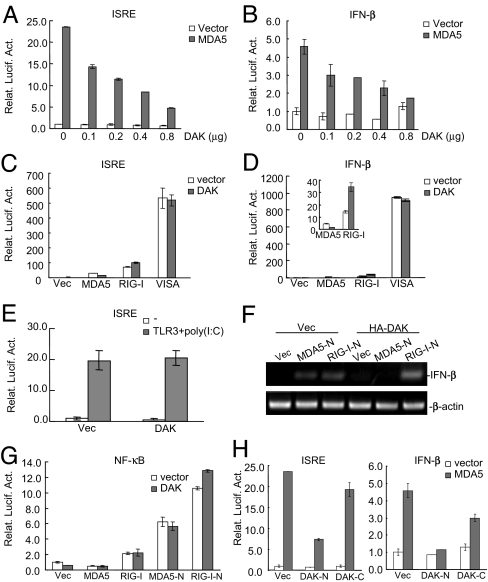
Because DAK is specifically associated with MDA5, we examined whether DAK is involved in regulation of MDA5-mediated signaling. In reporter assays, DAK inhibited MDA5-mediated activation of ISRE and the IFNβ promoter in a dose-dependent manner (Fig. 2 A and B). In these assays, DAK did not inhibit RIG-I- and TLR3-mediated activation of ISRE and the IFNβ promoter (Fig. 2 C–E). Biochemically, DAK selectively inhibited MDA5- but not RIG-I-mediated dimerization of IRF-3 [supporting information (SI) Fig. 6], which is a hallmark of IRF-3 activation. To determine whether DAK inhibits expression of bona fide type I IFN genes, we performed RT-PCR analysis. The results indicated that DAK inhibited MDA5-N- but not RIG-I-N-mediated expression of IFN-β (Fig. 2F). Taken together, these data suggest that DAK specifically inhibits MDA5- but not RIG-I-mediated IRF-3 signaling.
Fig. 2.
DAK inhibits MDA5- but not RIG-I-mediated activation of ISRE and the IFN-β promoter. (A and B) DAK inhibits MDA5-mediated activation of ISRE (A) and the IFN-β promoter (B) in a dose-dependent manner. The 293 cells (1 × 105) were transfected with ISRE (A) or the IFN-β promoter (B) luciferase reporter plasmid (0.05 μg), an expression plasmid for Flag-MDA5 (0.4 μg, gray bars) or an empty control plasmid (0.4 μg, white bars) and increased amounts of an expression plasmid for HA-DAK (0–0.8 μg as indicated). Luciferase assays were performed 16 h after transfection. (C–E) DAK has no significant effects on activation of ISRE and the IFN-β promoter mediated by RIG-I, TLR3, and VISA. The 293 cells (1 × 105) were transfected with the indicated reporter plasmid (0.05 μg) and expression plasmids (0.4 μg of each). Reporter assays were performed 16 h after transfection. In E, transfected cells were treated with poly I:C (40 μg/ml) for 6 h before reporter assays were performed. (F) DAK inhibits MDA5-N- but not RIG-I-N-induced expression of IFN-β. The 293 cells (5 × 105) were transfected with Flag-MDA5-N or Flag-RIG-I-N plasmid (1 μg) and HA-DAK or empty control plasmid (2 μg). RT-PCR was performed with human IFN-β or β-actin primers 18 h after transfection. (G) DAK does not inhibit MDA5-N and RIG-I-N-mediated activation of NF-κB. The 293 cells (1 × 105) were transfected with NF-κB luciferase reporter (0.1 μg) and the indicated plasmids (0.4 μg of each). Reporter assays were performed 16 h after transfection. (H) DAK-N was sufficient to inhibit MDA5-mediated activation of ISRE and the IFN-β promoter. The experiments were performed as in C and D. Relat. Lucif. Act., relative luciferase activity; Vec, vector.
In reporter assays, DAK did not inhibit MDA5- and RIG-I-mediated NF-κB activation (Fig. 2G), suggesting that DAK selectively inhibits MDA5-mediated IRF-3 but not NF-κB activation. DAK had no significant effects on activation of ISRE and the IFNβ promoter triggered by overexpression of VISA, TBK1, IKKε, and IRF3 (Fig. 2 C and D and data not shown), which are downstream signaling components of MDA5.
Structural studies indicate that the bacterium DAK contains two domains. The N-terminal domain forms a substrate-binding pocket and the C-terminal domain binds ATP (32, 33). Based on sequence homology between human and bacterium DAK, we constructed two human DAK mutants that contain its N- (DAK-N, aa 1–380) and C terminus (DAK-C, aa 340–575), respectively. Reporter assays indicated that DAK-N was sufficient to inhibit MDA5-mediated activation of ISRE and the IFNβ promoter, whereas DAK-C had minimal effect (Fig. 2H). Because DAK-N lacks the putative ATP-binding site, these data suggest that the kinase activity of DAK is not required for its inhibitory ability in MDA5-mediated signaling.
DAK Inhibits IRF-3 Activation Triggered by Cytoplasmic poly(I:C) and SeV in 293 Cells.
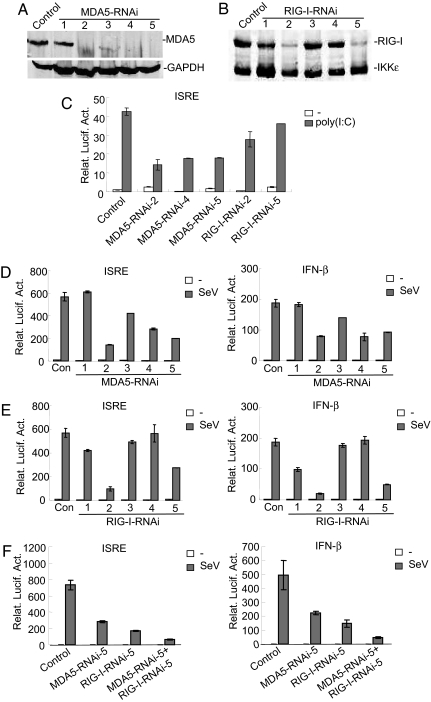
Because DAK specifically inhibits MDA5-mediated IRF-3 activation, we investigated whether it inhibits type I IFN signaling triggered by cytoplasmic poly(I:C) or viral infection. Previously, it had been demonstrated that, in mouse embryonic fibroblasts, MDA5 is involved in IFN signaling triggered by poly(I:C) transfected into the cytoplasm, whereas RIG-I responds to infection of a mutant strain of SeV (26). To determine the roles of MDA5 and RIG-I in signaling triggered by cytoplasmic poly(I:C) and SeV infection, we constructed five RNAi plasmids for each of them (Fig. 3 A and B). In reporter assays, knockdown of MDA5 significantly inhibited cytoplasmic poly(I:C)-induced ISRE activation, whereas knockdown of RIG-I had minimal inhibitory effect (Fig. 3C), suggesting that cytoplasmic poly(I:C)-induced IRF-3 activation is mostly mediated by MDA5 in 293 cells. In similar experiments, the efficiencies of knockdown of either MDA5 or RIG-I by RNAi plasmids are correlated with their abilities to inhibit activation of ISRE and the IFN-β promoter triggered by SeV infection (Fig. 3 D–F). Similar results were also obtained with HeLa cells (data not shown). In addition, cotransfection of MDA5 and RIG-I RNAi plasmids had accumulative inhibitory effects on the activation of ISRE and the IFN-β promoter triggered by SeV infection (Fig. 3F). In coimmunoprecipitation experiments, MDA5 and RIG-I did not interact with each other in the presence or absence of SenV infection (data not shown). Previously, it had been shown that either MDA5 or RIG-I knockout mice remain responsive to certain viruses. Taken together, these data suggest that both MDA5 and RIG-I participate in SeV-induced IFN signaling in 293 cells. They may be redundant or detect different structures of the viral RNAs, rather than acting together as heterodimers.
Fig. 3.
Both MDA5 and RIG-I are involved in cytoplasmic poly(I:C)- and SeV-triggered signaling. (A) Effects of MDA5 RNAi plasmids on the expression of endogenous MDA5. The 293 cells (1 × 106) were transfected with a control or MDA5 RNAi plasmids (10 μg of each). Forty-eight hours after transfection, cell lysates were analyzed by Western blots with anti-MDA5 (Upper) or anti-GAPDH (Lower) antibody. (B) Effects of RIG-I RNAi plasmids on the expression of RIG-I. The 293 cells (5 × 105) were transfected with HA-tagged RIG-I and IKKε plasmid (1 μg) and a control or RIG-I RNAi plasmids (2 μg of each). Forty-eight hours after transfection, cell lysates were analyzed by Western blot with anti-HA antibody. (C) Effects of MDA5 and RIG-I RNAi plasmids on cytoplasmic poly(I:C)-triggered ISRE activation. The 293 cells (1 × 105) were transfected with ISRE luciferase reporter plasmid (0.05 μg), control, or MDA5 or RIG-I RNAi plasmids (0.5 μg of each) as indicated. Thirty-six hours after transfection, cells were further transfected with poly(I:C) (4 μg, gray bars) or buffer (white bars) by Lipofectamine for 12 h before luciferase assays were performed. (D) Effects of MDA5 RNAi plasmids on SeV-triggered activation of ISRE and the IFN-β promoter. The 293 cells (1 × 105) were transfected with ISRE or the IFN-β promoter luciferase reporter plasmid (0.05 μg) as indicated and MDA5 RNAi plasmids (0.5 μg of each). Twenty-four hours after transfection, cells were infected with SeV (gray bars) or left untreated (white bars) for 12 h before luciferase assays were performed. (E) Effects of RIG-I RNAi plasmids on SeV-triggered activation of ISRE and the IFN-β promoter. The experiments were performed as in D. (F) Effects of a combination of MDA5 and RIG-I RNAi plasmids on SeV-triggered activation of ISRE and the IFN-β promoter. The experiments were performed as in D. The 293 cells were transfected with 0.5 μg of MDA5 or RIG-I RNAi plasmid alone, or a combination of 0.25 μg of each. Relat. Lucif. Act., relative luciferase activity.
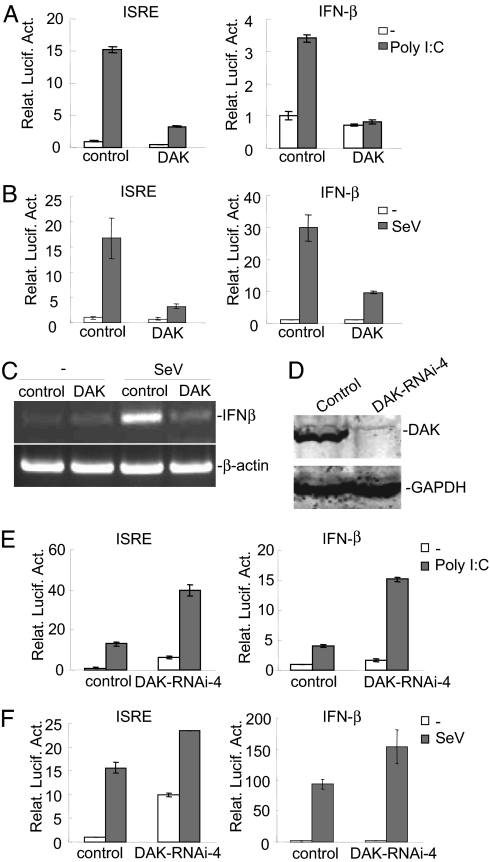
Because DAK specifically inhibits MDA5, which is involved in signaling triggered by cytoplasmic poly(I:C) and SeV infection, we determined the roles of DAK in signaling triggered by these stimuli. In reporter assays, overexpression of DAK potently inhibited activation of ISRE and the IFNβ promoter triggered by cytoplasmic poly(I:C) (Fig. 4A) and SeV (Fig. 4B) in 293 cells. Furthermore, DAK also inhibited endogenous expression of IFN-β triggered by SeV (Fig. 4C).
Fig. 4.
DAK negatively regulates IFN-β signaling triggered by cytoplasmic poly(I:C) and SeV in 293 cells. (A) DAK inhibits cytoplasmic poly(I:C)-induced activation of ISRE and the IFNβ promoter. The 293 cells (1 × 105) were transfected with the indicated reporter (0.05 μg) and expression plasmids (0.5 μg of each). Sixteen hours after transfection, cells were further transfected with poly(I:C) (4 μg) (gray bars) or buffer (white bars) for 12 h before luciferase assays were performed. (B) DAK inhibits SeV-triggered activation of ISRE and the IFN-β promoter. The 293 cells (1 × 105) were transfected with the indicated reporter (0.05 μg) and expression plasmids (0.5 μg of each). Sixteen hours after transfection, cells were infected with SeV or left uninfected for 12 h before luciferase assays were performed. (C) DAK inhibits SeV-induced expression of IFN-β. The 293 cells (1 × 105) were transfected with empty control plasmid or an expression plasmid for DAK (0.5 μg of each). Twelve hours after transfection, cells were infected with SeV or left uninfected for 12 h before RT-PCR was performed with human IFNβ and β-actin primers. (D) Effect of DAK RNAi #4 plasmid on the expression of DAK. The 293 cells (1 × 106) were transfected with a control or DAK RNAi plasmid (5 μg) for 60 h before Western blot analysis was performed with the indicated antibodies. (E) DAK RNAi potentiates cytoplasmic poly(I:C)-induced activation of ISRE and the IFN-β promoter in 293 cells. The 293 cells (1 × 105) were transfected with the indicated reporter (0.05 μg) and DAK RNAi (0.5 μg) plasmid. Thirty-six hours after transfection, cells were further transfected with poly(I:C) (4 μg) (gray bars) or buffer (white bars) for 12 h before luciferase assays were performed. (F) DAK RNAi potentiates SeV-triggered activation of ISRE and the IFN-β promoter in 293 cells. The 293 cells were transfected as in A. The transfected cells were infected with SeV or left uninfected for 12 h before luciferase assays were performed. Relat. Lucif. Act., relative luciferase activity.
To determine the physiological role of DAK in innate antiviral response, we made a DAK RNAi construct that could significantly inhibit endogenous DAK expression (Fig. 4D). In reporter assays, down-regulation of DAK by RNAi alone was sufficient to activate ISRE and the IFNβ promoter (Fig. 4 E and F). In the same experiments, DAK RNAi also enhanced activation of ISRE and the IFN-β promoter triggered by cytoplasmic poly(I:C) and SeV infection (Fig. 4 E and F). These results were further confirmed with additional DAK RNAi plasmids targeting different sequences of human DAK cDNA. In these experiments, the knockdown efficiencies of these RNAi plasmids are correlated with their abilities to activate basal or potentiate SeV-induced ISRE and IFN-β promoter (SI Fig. 7). These data suggest that DAK is a physiological suppressor of IFN-β signaling triggered by cytoplasmic poly(I:C) and SeV in 293 cells.
DAK Plays a Role in Regulation of MDA5-Mediated Cellular Antiviral Response.
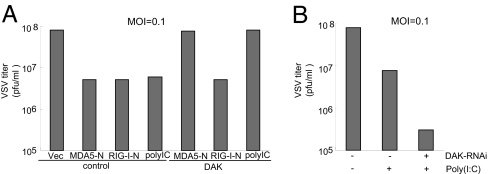
Because DAK is a physiological inhibitor of MDA-mediated IFN signaling, we determined whether DAK plays a role in regulation of MDA5-mediated cellular antiviral response. Previously, it had been shown that overexpression of MDA5-N and RIG-I-N induces type I IFNs and causes cellular antiviral response (17, 26, 28, 29). Using plaque assays, we found that overexpression of DAK completely abolished the inhibitory effect on viral replication mediated by overexpression of MDA5-N but not RIG-I-N (Fig. 5). Consistently, DAK RNAi enhanced the inhibitory effect on viral replication triggered by cytoplasmic poly(I:C) (Fig. 5). These data suggest that DAK is a physiological suppressor of MDA5-mediated cellular antiviral response.
Fig. 5.
Effects of DAK and DAK RNAi on MDA5-mediated antiviral response. (A) DAK inhibits MDA5-mediated antiviral response. The 293 cells (2 × 105) were transfected with the indicated expression plasmids (1 μg of each). Twenty-four hours after transfection, cells were infected with VSV [multiplicity of infection (MOI) = 0.1], and supernatants were harvested at 12 h after infection. Supernatants were analyzed for VSV production by using standard plaque assays. Plaques were counted, and titers were calculated as plaque-forming units per milliliter. (B) DAK RNAi potentiates cytoplasmic poly(I:C)-triggered antiviral response. The 293 cells (2 × 105) were transfected with a control or DAK RNAi plasmid (1 μg of each). Thirty-six hours after transfection, cells were further transfected with poly(I:C) (20 μg) or buffer. Twelve hours later, cells were infected with VSV (MOI = 0.1), and supernatants were harvested at 12 h after infection for plaque assays.
Discussion
To prevent harmful effects resulting from spontaneous production in uninfected cells or overproduction of type I IFNs during an acute infection, the innate immune system has developed distinct strategies to regulate the production of type I IFNs. LGP2, a RNA helicase protein lacking CARD domains at its N terminus, acts as a negative regulator by sequestering viral RNA from RIG-I (30, 34). A20, a protein with dual deubiquitination and ubiquitin ligation activities, negatively regulates RIG-I-induced antiviral signaling through an unidentified mechanism (35, 36). Recently, a coiled-coil protein, SIKE, has been shown to be a physiological suppressor of virus-triggered type I IFN signaling through its constitutive association with TBK1 and IKKε (37). In addition to cellular inhibitors, the V protein of paramyxovirus has been shown to selectively target MDA5- but not RIG-I-mediated IFN signaling (29–31). In the present work, we identified the cellular protein DAK as a specific inhibitor of MDA5-mediated IFN signaling.
Although MDA5 and RIG-I share a similar structural architecture, DAK is associated with MDA5 but not RIG-I under physiological conditions in mammalian cells (Fig. 1). In transient transfection and coimmunoprecipitation experiments, DAK does not prevent MDA5-VISA interaction (Fig. 1). The simplest explanation for this observation is that activated MDA5 (such as viral RNA-bound or overexpressed MDA5) has a higher affinity for VISA than DAK. This is supported by our observation that MDA5 is disassociated with DAK upon viral infection.
Consistent with the observation that DAK is specifically associated with MDA5, overexpression of DAK inhibited MDA5- but not RIG-I-mediated activation of IRF-3 and the IFN-β promoter (Fig. 2). In similar experiments, DAK did not inhibit MDA5-meidated NF-κB activation. One explanation for this observation is that binding of DAK to MDA5 blocks the association of MDA5 with signaling components involved in the IRF-3 activation pathway but not those in the NF-κB activation pathway. It is possible that MDA5 is associated with another unidentified protein that blocks the association of MDA5 with components specifically involved in the NF-κB activation pathway.
Knockdown of DAK by RNAi activated IRF-3 and the IFN-β promoter and increased poly(I:C)- and SeV-triggered activation of IRF-3 and the IFN-β promoter (Fig. 4). Interestingly, the effects of DAK RNAi on activation of IRF-3 and the IFN-β promoter are more pronounced than on poly(I:C)- or SeV-triggered signaling (Fig. 4), suggesting that DAK functions to inhibit autoactivation of MDA5. This is confirmed by our observation that cotransfection of MDA5-RNAi inhibited DAK-RNAi-induced activation of ISRE and the IFN-β promoter, whereas cotransfection of RIG-I-RNAi had no significant inhibitory effects (SI Fig. 8). In plaque assays, overexpression of DAK abolished the inhibitory effect on viral replication mediated by overexpression of MDA5-N but not RIG-I-N, whereas DAK RNAi enhanced the inhibitory effect on viral replication triggered by cytoplasmic poly(I:C) (Fig. 5). Taken together, these results suggest that DAK is a physiological inhibitor of MDA5- but not RIG-I-mediated antiviral response.
Structural studies indicate that the bacterium DAK contains two domains. The N-terminal domain forms a substrate-binding pocket, and the C-terminal domain binds ATP (33). In mammals, DAK displays dual activities as FMN cyclase and ATP-dependent Pha kinase (32). Our results indicate that the N terminus of DAK, which lacks a C-terminal putative ATP-binding motif, is sufficient for inhibiting MDA5-mediated signaling (Fig. 2H). Therefore, the kinase activity of DAK is not required for its inhibition of MDA5-mediated IFN-β signaling.
Based on our findings, we propose the following model for the action of DAK. The N-terminal FMN cyclase domain of DAK, upon binding to the N-terminal CARDs of MDA5, is able to sequester MDA5 in an inactive form under physiological conditions. Upon viral infection, the RNA helicase domain of MDA5 binds to viral RNA, and this causes its conformational change and release of DAK, allowing MDA5 to recruit downstream adapter protein VISA and activating the type I IFN pathways. Although the detailed mechanisms on how MDA5 is inhibited by DAK need careful structural studies, the identification of a specific inhibitor of MDA5, but not RIG-I, provides important insight into how two related innate antiviral pathways are differentially regulated.
Materials and Methods
Reagents.
Mouse monoclonal antibodies against Flag and HA epitopes (Sigma, St. Louis, MO), poly(I:C) (InvivoGen, San Diego, CA), rabbit polyclonal antibodies against RIG-I (Jun Gu, Peking University, Peking, China), SeV (Congyi Zheng, Wuhan University, Wuhan, China), and VSV (Hong-Kui Deng, Peking University) were obtained from the indicated sources. Mouse anti-MDA5 and anti-DAK antibodies were obtained by injection of mice with the respective recombinant proteins.
Yeast Two-Hybrid Screens.
A human 293 cell cDNA library (Clontech, Palo Alto, CA) was screened with full-length MDA5 as bait, following protocols recommended by the manufacturer.
Constructs.
Mammalian expression plasmids for Flag- or HA-tagged MDA5, DAK, and their deletion mutants were constructed by standard molecular biology techniques. Mammalian expression plasmids for HA-TBK1, HA-IKKε, HA-TRIF, Flag-IRF3, Flag-RIG-1, and its deletion mutants were described (12, 22). ISRE-luciferase reporter plasmid was purchased from Stratagene (La Jolla, CA). NF-κB and the IFN-β promoter luciferase reporter plasmids were described (22).
Transfection and Reporter Assays.
The 293 cells were seeded on 24-well dishes and transfected the next day by standard calcium phosphate precipitation. To normalize for transfection efficiency, 0.05 μg of pRL-TK (Renilla luciferase) reporter plasmid was added to each transfection. Luciferase assays were performed by using a dual-specific luciferase assay kit (Promega, Madison, WI). Firefly luciferase activities were normalized based on Renilla luciferase activities. All reporter assays were repeated at least three times.
Coimmunoprecipitation and Western Blot Analysis.
Transient transfection, coimmunoprecipitation, and Western blotting experiments were performed as described (12, 22, 37). For endogenous coimmunoprecipitation experiments, cells (1 × 108) were lysed in 5 ml of lysis buffer, and the lysate was incubated with 1 μl of the indicated antiserum or control IgG. The subsequent procedures were carried out as described (12, 22, 37).
RNAi Experiments.
Double-strand oligonucleotides corresponding to the target sequences were cloned into the pSuper.retro RNAi plasmid (Oligoengine, Seattle, WA). In this study, the target sequences for human DAK cDNA are (i) AGCAGTCAAGAGTGCCGAA; (ii) CGCTCCTTATCGTGAAGAA; (iii) GGACTATGCTGGATTCTCT; (iv) CCGCCGATGAGATTGTGAA. The target sequences for human MDA5 cDNA are (i) TGACACAATTCGAATGATA; (ii) AGAAGTGTGCCGACTATCA; (iii) GAAGTGTGCCGACTATCAA; (iv) TGATAGATGCGTATACTCA; (v) GTGCATGAGGGAGGAACTG. The target sequences for human RIG-I cDNA are (i) CGATTCCATCACTATCCAT; (ii) AATTCATCAGAGATAGTCA; (iii) ATTCATCAGAGATAGTCAA; (iv) AGCCTTGGCATGTACACA; (v) GGAAGAGGTGCAGTATATT.
RT-PCR.
Total RNA was isolated from 293 cells by using TRIzol reagent (Tianwei Company, Beijing, China) and subjected to RT–PCR analysis to measure expression of IFN-β and β-actin. Gene-specific primer sequences were as follows. IFN-β, CACGACAGCTCTTTCCATGA (forward), AGCCAGTGCTCGATGAATCT (reverse); β-actin, GTCGTCGACAACGGCTCCGGCATG (forward), ATTGTAGAAGGTGTGGTGCCAGAT (reverse).
VSV Plaque Assay.
The experiments were performed as described (37).
Supplementary Material
Acknowledgments
We thank Jun Gu, Hongkui Deng, and Congyi Zheng for reagents. This work was supported by Chinese 973 Program (2006CB504301), the National Science Foundation of China (Project 30630019), and the Chinese 863 Program (2006AA02A306).
Abbreviations
- IRF
interferon regulatory factor
- ISRE
IFN-stimulated response element
- CARD
caspase recruitment domain
- TBK1
TANK-binding kinase 1
- IKKε
inhibitor of NF-κB kinase subunit ε.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. M.R. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/cgi/content/full/0700544104/DC1.
References
- 1.Durbin JE, Fernandez-Sesma A, Lee CK, Rao TD, Frey AB, Moran TM, Vukmanovic S, Garcia-Sastre A, Levy DE. J Immunol. 2000;164:4220–4228. doi: 10.4049/jimmunol.164.8.4220. [DOI] [PubMed] [Google Scholar]
- 2.Levy DE, Garcia-Sastre A. Cytokine Growth Factor Rev. 2001;12:143–156. doi: 10.1016/s1359-6101(00)00027-7. [DOI] [PubMed] [Google Scholar]
- 3.Levy DE, Marie IJ. Nat Immunol. 2004;5:699–701. doi: 10.1038/ni0704-699. [DOI] [PubMed] [Google Scholar]
- 4.Taniguchi T, Ogasawara K, Takaoka A, Tanaka N. Annu Rev Immunol. 2001;19:623–655. doi: 10.1146/annurev.immunol.19.1.623. [DOI] [PubMed] [Google Scholar]
- 5.Honda K, Takaoka A, Taniguchi T. Immunity. 2006;25:349–360. doi: 10.1016/j.immuni.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 6.Honda K, Taniguchi T. Nat Rev. 2006;6:644–658. doi: 10.1038/nri1900. [DOI] [PubMed] [Google Scholar]
- 7.Maniatis T, Falvo JV, Kim TH, Kim TK, Lin CH, Parekh BS, Wathelet MG. Cold Spring Harbor Symp Quant Biol. 1998;63:609–620. doi: 10.1101/sqb.1998.63.609. [DOI] [PubMed] [Google Scholar]
- 8.Wathelet MG, Lin CH, Parekh BS, Ronco LV, Howley PM, Maniatis T. Mol Cell. 1998;1:507–518. doi: 10.1016/s1097-2765(00)80051-9. [DOI] [PubMed] [Google Scholar]
- 9.tenOever BR, Sharma S, Zou W, Sun Q, Grandvaux N, Julkunen I, Hemmi H, Yamamoto M, Akira S, Yeh WC, et al. J Virol. 2004;78:10636–10649. doi: 10.1128/JVI.78.19.10636-10649.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Medzhitov R, Janeway CA., Jr Science. 2002;296:298–300. doi: 10.1126/science.1068883. [DOI] [PubMed] [Google Scholar]
- 11.Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 12.Han KJ, Su X, Xu LG, Bin LH, Zhang J, Shu HB. J Biol Chem. 2004;279:15652–15661. doi: 10.1074/jbc.M311629200. [DOI] [PubMed] [Google Scholar]
- 13.Hoebe K, Du X, Georgel P, Janssen E, Tabeta K, Kim SO, Goode J, Lin P, Mann N, Mudd S, et al. Nature. 2003;424:743–748. doi: 10.1038/nature01889. [DOI] [PubMed] [Google Scholar]
- 14.Oshiumi H, Matsumoto M, Funami K, Akazawa T, Seya T. Nat Immunol. 2003;4:161–167. doi: 10.1038/ni886. [DOI] [PubMed] [Google Scholar]
- 15.Yamamoto M, Sato S, Hemmi H, Hoshino K, Kaisho T, Sanjo H, Takeuchi O, Sugiyama M, Okabe M, Takeda K, et al. Science. 2003;301:640–643. doi: 10.1126/science.1087262. [DOI] [PubMed] [Google Scholar]
- 16.Yamamoto M, Sato S, Mori K, Hoshino K, Takeuchi O, Takeda K, Akira S. J Immunol. 2002;169:6668–6672. doi: 10.4049/jimmunol.169.12.6668. [DOI] [PubMed] [Google Scholar]
- 17.Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 18.Kang DC, Gopalkrishnan RV, Lin L, Randolph A, Valerie K, Pestka S, Fisher PB. Oncogene. 2004;23:1789–1800. doi: 10.1038/sj.onc.1207300. [DOI] [PubMed] [Google Scholar]
- 19.Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S. Nat Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 20.Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 21.Seth RB, Sun L, Ea CK, Chen ZJ. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 22.Xu LG, Wang YY, Han KJ, Li LY, Zhai Z, Shu HB. Mol Cell. 2005;19:727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 23.Sharma S, tenOever BR, Grandvaux N, Zhou GP, Lin R, Hiscott J. Science. 2003;300:1148–1151. doi: 10.1126/science.1081315. [DOI] [PubMed] [Google Scholar]
- 24.Fitzgerald KA, McWhirter SM, Faia KL, Rowe DC, Latz E, Golenbock DT, Coyle AJ, Liao SM, Maniatis T. Nat Immunol. 2003;4:491–496. doi: 10.1038/ni921. [DOI] [PubMed] [Google Scholar]
- 25.Kato H, Sato S, Yoneyama M, Yamamoto M, Uematsu S, Matsui K, Tsujimura T, Takeda K, Fujita T, Takeuchi O, et al. Immunity. 2005;23:19–28. doi: 10.1016/j.immuni.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 26.Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, et al. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 27.Hornung V, Ellegast J, Kim S, Brzozka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann KK, Schlee M, et al. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 28.Pichlmair A, Schulz O, Tan CP, Naslund TI, Liljestrom P, Weber F, Reis e Sousa C. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 29.Andrejeva J, Childs KS, Young DF, Carlos TS, Stock N, Goodbourn S, Randall RE. Proc Natl Acad Sci USA. 2004;101:17264–17269. doi: 10.1073/pnas.0407639101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoneyama M, Kikuchi M, Matsumoto K, Imaizumi T, Miyagishi M, Taira K, Foy E, Loo YM, Gale M, Jr, Akira S, et al. J Immunol. 2005;175:2851–2858. doi: 10.4049/jimmunol.175.5.2851. [DOI] [PubMed] [Google Scholar]
- 31.Childs K, Stock N, Ross C, Andrejeva J, Hilton L, Skinner M, Randall R, Goodbourn S. Virology. 2007;359:190–200. doi: 10.1016/j.virol.2006.09.023. [DOI] [PubMed] [Google Scholar]
- 32.Cabezas A, Costas MJ, Pinto RM, Couto A, Cameselle JC. Biochem Biophys Res Commun. 2005;338:1682–1689. doi: 10.1016/j.bbrc.2005.10.142. [DOI] [PubMed] [Google Scholar]
- 33.Bachler C, Flukiger-Bruhwiler K, Schneider P, Bahler P, Erni B. J Biol Chem. 2005;280:18321–18325. doi: 10.1074/jbc.M500279200. [DOI] [PubMed] [Google Scholar]
- 34.Rothenfusser S, Goutagny N, DiPerna G, Gong M, Monks BG, Schoenemeyer A, Yamamoto M, Akira S, Fitzgerald KA. J Immunol. 2005;175:5260–5268. doi: 10.4049/jimmunol.175.8.5260. [DOI] [PubMed] [Google Scholar]
- 35.Wang YY, Li L, Han KJ, Zhai Z, Shu HB. FEBS Lett. 2004;576:86–90. doi: 10.1016/j.febslet.2004.08.071. [DOI] [PubMed] [Google Scholar]
- 36.Lin R, Yang L, Nakhaei P, Sun Q, Sharif-Askari E, Julkunen I, Hiscott J. J Biol Chem. 2006;281:2095–2103. doi: 10.1074/jbc.M510326200. [DOI] [PubMed] [Google Scholar]
- 37.Huang J, Liu T, Xu LG, Chen D, Zhai Z, Shu HB. EMBO J. 2005;24:4018–4028. doi: 10.1038/sj.emboj.7600863. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.