Abstract
CHOP-10 (GADD153/DDIT-3) is a bZIP protein involved in differentiation and apoptosis. Its expression is induced in response to stresses such as nutrient deprivation, perturbation of the endoplasmic reticulum, redox imbalance, and UV exposure. Here we show that CHOP expression is induced in cultured pancreatic β-cells maintained in a basal glucose concentration of 5.5 mM and repressed by stimulatory glucose (≥11 mM). Both induction and repression of CHOP are dependent on the MAPKs ERK1 and ERK2. Two regulatory composite sites containing overlapping MafA response elements (MARE) and CAAT enhancer binding (CEB) elements regulate transcription in an ERK1/2-dependent manner. One site (MARE-CEB), from −320 to −300 bp in the promoter, represses transcription. The other site (CEB-MARE), from +2,628 to +2,641 bp in the first intron of the CHOP gene, activates it. MafA can influence transcription of both sites. The MARE-CEB is repressed by MafA, whereas the CEB-MARE site, which is homologous to the A2C1 component of the glucose-sensitive RIPE3b region of the insulin gene promoter, is activated by MafA. These results indicate that ERK1/2 have dual roles in regulating CHOP gene expression via both promoter and intronic regions, depending on environmental and metabolic stresses imposed on pancreatic β-cells.
Keywords: MafA, intron, extracellular signal-regulated kinase, cellular stress
CHOP-10/GADD153/DDIT-3 was originally cloned as one of several DNA damage-inducible transcripts (designated A153) that are rapidly expressed in mammalian cells in response to UV irradiation or alkylating agents (1). Further characterization showed that this particular gene product is also induced by serum reduction, for example, indicating a role for it in growth arrest or cell cycle progression. Thus, the gene was called GADD153 for growth arrest and DNA damage inducibility. Subsequently, the murine analog was cloned independently by Ron and Habener (2) using a radiolabeled protein probe containing C/EBP-β DNA-binding and dimerization domains in a screen for C/EBP-like interactors. The protein was then categorized as a bZIP C/EBP-like family transcription factor, C/EBP homologous protein 10 (CHOP-10), which has also been called C/EBP-ζ. CHOP is involved in a variety of stress response pathways, including endoplasmic reticulum (ER) stress (3–6), redox stress (7–9), and nutrient deprivation (10–13). Functional roles for CHOP have been described in apoptosis (14–17) and inhibition of adipocyte differentiation (4, 18,20).
CHOP has been implicated in the pathogenesis of diabetes mellitus by promoting β-cell destruction. CHOP expression can be induced by inflammatory cytokines via nitric oxide signaling (21). CHOP is induced via ER stress signals that promote β-cell death (22, 23). Disruption of the CHOP gene slows the onset of ER stress-induced diabetes, indicating that CHOP exacerbates the diabetic phenotype.
CHOP expression is itself regulated by other bZIP class transcription factors. The 5′ flanking sequence of the CHOP gene promoter contains overlapping cis-acting CAAT enhancer binding (CEB)-activating transcription factor (ATF)/cyclic AMP response element (CRE) DNA binding elements, which have been shown to bind to various complexes containing C/EBP-β, ATF-2, ATF-3, and ATF-4 in different cell types, depending on the mode and duration of induction (12, 20, 24–26). For example, C/EBP-β up-regulates the CHOP promoter in hepatocytes during the lipopolysaccharide-induced inflammatory acute phase response (20). Treatment of PC12 cells with arsenite results in binding of ATF-4 to the CEB-ATF/CRE site to enhance promoter activity; within 6 h ATF-4 is displaced by ATF-3 to repress CHOP transcription. Here we show that MafA can repress CHOP promoter activity via a Maf recognition element (MARE), which overlaps with the previously described CEB-ATF/CRE site (see Fig. 2A) (24, 27, 28). MafA also stimulates transcription through a CEB-MARE site 2.6 kb away from the transcription start site, within the first intron of the human CHOP gene. The elements are situated proximal to the second exon in the sequence 5′ of the coding sequence. This CEB-MARE site displays striking similarity to the glucose-responsive RIPE3b region of the insulin gene promoter. These findings provide insight into the cell-specific regulation of CHOP expression in pancreatic β-cells.
Fig. 2.
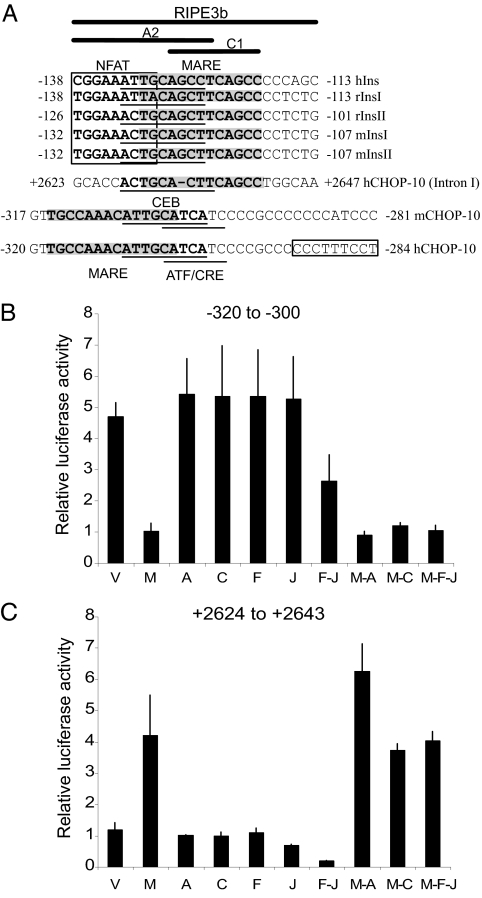
Regulation of elements from the CHOP gene by selected factors. (A) Schematic alignment of DNA sequences of conserved regions of the insulin gene promoter and CHOP gene promoter/intron I and relative DNA-binding sites for bZIP transcription factors. The boxed region is the NFAT consensus. The shaded regions indicate MARE consensus sequences. Underlined sequences include C/EBP-β and ATF-CRE sites. (B and C) Promoter–reporter assays of CHOP gene promoter regions 3× −320 to −300 bp (B) and +2,624 to +2,643 bp (C) in Min6 cells maintained in 25 mM glucose overexpressing indicated bZIP transcription factors. V, vector only; M, MafA; A, Atf-2; C, C/EBPβ; F, c-Fos; J, c-Jun. Promoter assays were repeated a minimum of three times.
Results
bZIP Transcription Factors Bind to the 5′ Flanking Promoter and Intronic Region of the CHOP Promoter.
We previously described differential regulation of the insulin gene promoter by changes in expression and interactions between the bZIP trans-acting factors MafA and C/EBP-β in pancreatic β-cells in low and high glucose (29). Thus, we hypothesized that other genes might also be similarly affected by the specific bZIP expression profile and signaling of the β cell in response to its environment. We found an effect of ERK1/2 on expression of the CHOP gene (data not shown). Thus, we scanned the 5′ flanking promoter as well as intronic and exonic regions and found that the CHOP gene contained elements in both the promoter and the first intron that might bind bZIP family proteins that differentially regulate the insulin gene in β-cells in low and high glucose. Schmitt-Ney and Habener (30) showed that the first exon of the CHOP gene could modulate CHOP gene expression in mouse fibroblasts.
By ChIP analysis of ≈200 bp segments, a ChIP print of proteins bound to DNA in vivo between base pairs −954 to +3,238 relative to the transcription start site was mapped for bZIP factors c-Fos, c-Jun, ATF-2, MafA, and C/EBP-β in isolated human pancreatic islets maintained in the presence of 5.5 or 16 mM glucose (Fig. 1A and Table 1). The relative abundance or enrichment of each specific 200-bp region of DNA in the CHOP promoter and intron pulled down by immunoprecipitation with the bZIP antibody was assessed by comparing their threshold of detection by PCR at 1-, 10-, and 100-fold dilutions. In the succeeding experiments, PCR was performed with 100-fold dilutions to identify the most strongly associated factors; thus, comparisons to subsequent experiments are made with PCR with the most dilute samples of Fig. 1A. Although numerous interactions were detected, we focused primarily on two regions that contain consensus Maf-binding elements, as discussed further below (Fig. 2A).
Fig. 1.
ChIP assays of the CHOP gene from human islets. Human islets were cultured in the indicated glucose concentrations, and binding of transcription factors to segments of the CHOP promoter was analyzed by ChIP assay. (A) PCR products from ChIP assays of regions −954 to +3,238 bp of the CHOP gene in islets in 5.5 mM or 16 mM glucose. Triangles reflect the 1-, 10-, and 100-fold dilution of material used as input in PCR reactions. (B) PCR products from ChIP assays of regions −472 to −226 or +2,514 to + 2,805 bp in islets cultured in indicated glucose in the presence or absence of U0126 with 100-fold dilution of material as input for the PCR reactions. n = 3. (C) PCR products from ChIP assays of regions −472 to −226 or +2,514 to + 2,805 bp in islets cultured in 5.5 mM glucose and exposed to 16 mM glucose for 30 min or 30 mM K+ for 5 min in the presence or absence of U0126 with 100-fold dilution of material as input for the PCR reactions. n = 3. (D) Immunoblot for MafA expression in Min6 cells, 293, and human islets in 5.5 mM or 16 mM glucose. All blots were performed at least three times.
Table 1.
Effects of glucose and MAPK inhibition on CHOP
| Glucose, mM | Islets |
Islets/cells |
Cell lines |
||
|---|---|---|---|---|---|
| Promoter binding | Intron binding | Protein | Promoter assay | Intron assay | |
| 5.5 | Less Maf | More Maf | High | High | High |
| 25/16 | More Maf | Less Maf | Low | Low | High |
| 5.5 + U0126 | Inhibition | Inhibition | Reduction | No change | Low |
| 25 + U0126 | Inhibition | Inhibition | Increase | High | Low |
MafA expression is restricted to tissues involved in eye development and to pancreatic β-cells (9, 27, 31–34). The region from −472 to −226 bp of the 5′ flanking promoter was most readily detected in MafA-chromatin immunoprecipitates from human islets cultured in 16 mM glucose compared with the other promoter regions, suggesting its greater association with MafA. Less DNA was precipitated from this region if islets had been cultured in 5.5 mM glucose. This result indicates a higher relative affinity or abundance of MafA in the region from −472 to −226 bp in human islets exposed to high glucose.
The greatest relative association of C/EBP-β was observed much farther downstream of the transcriptional start site at +1,588 to +1,830 bp and +2,514 to +2,805 bp of the CHOP gene from islets in 16 mM glucose. This finding contrasted with what was observed in 5.5 mM glucose; under that condition, strong MafA and ATF-2 binding to the region from +2,514 to + 2,805 bp was noted, but no detectable C/EBP-β was found. These results suggest that MafA binds differentially to regions of the CHOP gene depending on the duration of exposure to fasting or high glucose. The data also suggest the idea that MafA/ATF-2 and C/EBP-β have opposing roles in regulating the CHOP gene via intronic regions.
ERK1/2 Activity Is Required for Binding of MafA and C/EBP-β to the CHOP Gene Promoter.
A comparison of binding conditions indicated that MafA binds to the CHOP promoter in the region from −472 to −226 bp in islets maintained in or acutely stimulated by high glucose. However, MafA bound to the intronic promoter element if islets were incubated in 5.5 mM glucose but not as well in islets maintained in high glucose. Preventing activation of ERK1/2 with U0126 blocked the association of MafA with both regions of the CHOP gene under all glucose conditions (Fig. 1B). C/EBP-β only bound to the intronic region if islets were maintained in 16 mM glucose, and its binding also depended on ERK1/2 activity. In contrast, U0126 had no effect on the association of ATF-2 with the CHOP gene. These results indicate that the binding of MafA and C/EBP-β to the CHOP gene requires ERK1/2 activity.
Because MafA is one of several factors that bind to the insulin gene promoter, we used ChIP to determine if some of these other factors also associated with the homologous region of the CHOP gene promoter (Figs. 1C and 2A). In this case, islets were stimulated briefly with 16 mM glucose or 30 mM K+, which depolarizes cells, stimulating insulin release and activating ERK1/2. Under these conditions, both glucose and depolarization promoted the binding of MafA to the intronic region of the CHOP gene, but Beta2, PDX-1, and NFAT were not detected on the CHOP intron.
Maintenance of β-cells in high glucose causes induction of the small inhibitory form of C/EBP-β (29, 35). To determine whether differences in MafA binding might also be related to changes in its expression, extracts of Min6 cells and human islets maintained in either high or low glucose were immunoblotted with an anti-Maf antibody. In contrast to what has been observed in HIT-T15 cells (36, 37), MafA expression was significantly higher in Min6 cells and in human islets maintained in high glucose (Fig. 1D). U0126 did not alter the amount of immunoreactive MafA. The protein was not detected in 293 cells treated in the same manner.
Overlapping MARE-CEB Consensus Sequences Lie at −318 to −305 bp Within the 5′ Flanking Promoter and at +2,628 to +2,641 bp Within the Intronic Region of the Human CHOP Gene Promoter.
The DNA sequences in the regions from −472 to −226 bp, from +1,588 to +1,830 bp, and from +2,514 to +2,805 bp in intronic regions of the human CHOP gene were analyzed for bZIP consensus sites. In addition to previously reported CEB, CRE/ATF, and AP-1 cis-acting elements in the region from −472 to −226 bp, we identified a MARE consensus site within −318 to −305 bp that overlaps with the CEBP-β binding site (Fig. 2A). This site is 100% conserved in human and mouse genes. A composite CEB-MARE site was also identified within the +2,514- to + 2,805-bp intronic region of the human CHOP gene at +2,628 to +2,641 bp and is homologous to the A2C1 element of the RIPE3b region of the insulin gene promoter. The 5′ end of the A2 element from the conserved RIPE3b region within the first intron of the CHOP gene is lacking the NFAT-binding site found within the A2C1 region of the insulin gene promoter, indicating that NFAT may not be required for CHOP intronic gene regulation. The other major point of divergence of the A2C1 region of the CHOP intron from the insulin promoter is a deletion of a single guanine nucleotide within the C1 region that is conserved in human and mouse insulin genes. This deletion lies within the overlapping region of the CEB-MARE sites. These consensus sequences are consistent with binding observed in ChIP experiments.
MafA Represses the CHOP Gene Promoter and Its MARE-CEB Site.
MafA bound to the 5′ flanking promoter of the CHOP gene with high affinity when human islets were in high glucose. We therefore sought to determine the effect of MafA on CHOP promoter activity. 293 cells, which do not express endogenous MafA, and Min6 cells were cotransfected with a MafA mammalian expression vector and a reporter linked to the first continuous 954 bp of the the 5′ flanking region of the CHOP gene promoter. Overexpressed MafA caused a 75% decrease in reporter activity from this region of the CHOP gene promoter in Min6 or 293 cells (data not shown; see Fig. 2B). C/EBP-β, ATF-2, c-Fos, and c-Jun alone had little effect on the activity from this reporter and could not relieve the inhibitory effect of MafA. The combined overexpression of c-Fos and c-Jun inhibited basal CHOP promoter activity by 50% but did not alter repression caused by MafA. These data indicate that MafA is a strong repressor of the CHOP gene promoter and that other bZIP factors cannot disrupt its ability to inhibit promoter activity. The functional role of MafA in regulating the identified MARE-CEB consensus sites of the CHOP gene promoter was explored by cotransfecting 293 and Min6 cells with a reporter containing the MARE-CEB site within −320 to −300 bp of the CHOP gene and a MafA mammalian expression vector. MafA inhibited basal activity of this MARE-CEB reporter (Fig. 2B) in a manner remarkably similar to its effects on the full-length CHOP gene promoter. C/EBP-β, ATF-2, c-Fos, and c-Jun alone did not affect reporter activity, and none relieved the inhibitory effect of MafA. Like the full-length CHOP promoter, the MARE-CEB site was inhibited by the combined expression of c-Fos and c-Jun. These data indicate that MafA is a strong repressor of the MARE-CEB site identified between −320 to−-300 bp and that other bZIP factors do not prevent its ability to inhibit promoter activity. Furthermore, the results suggest that the c-Fos/c-Jun (AP-1) complex does not inhibit the CHOP promoter via interaction with the isolated MARE-CEB composite site.
MafA Activates the RIPE3b-Like CEB-MARE Site (+2,624 to +2,643 bp) of the Human CHOP Intron.
Overexpression of MafA in 293 (data not shown) and Min6 cells (Fig. 2C) resulted in a 4-fold activation of a reporter coupled to the +2,624- to +2,643-bp CEB-MARE site of the first intron of the CHOP gene. C/EBP-β, ATF-2, c-Fos, and c-Jun alone did not affect reporter activity; however, ATF-2 slightly enhanced the effect of overexpressed MafA. Moreover, overexpressed c-Fos/c-Jun potently inhibited the CEB-MARE reporter but did not repress MafA-induced reporter activity. These results indicate that the CHOP intron contains DNA sequences capable of responding to the MafA transcription factor and of promoting transcription. These results combined with results from the 5′ flank CHOP promoter–reporters suggest that MafA has opposing roles in regulating CHOP gene transcription through its actions on distinct portions of the gene.
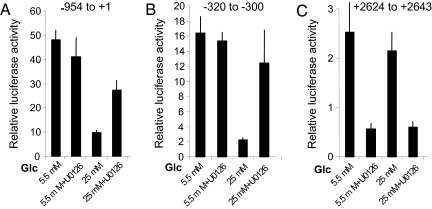
CHOP Promoter Activity in β-Cells Is Greater in 5.5 mM Glucose.
The studies above indicate that CHOP gene expression in human islets is highly responsive to changes in glucose concentration and is affected both positively and negatively by MafA. We evaluated changes in the sensitivity of the elements of the CHOP gene identified above as a function of the exposure to glucose to assess the potential role of MafA and ERK1/2 in regulating the expression of this gene in Min6 cells. Activity of the reporter linked to the 5′ promoter was ≈3-fold or more higher in 5.5 mM than in 25 mM glucose (Fig. 3A). In time courses, we found that the activity of the reporter in cells maintained in 5.5 mM glucose was rapidly suppressed by 25 mM glucose (data not shown). U0126 had little effect on CHOP promoter activity in cells in 5.5 mM glucose. Similar effects were observed using the region containing −320 to −300 bp to drive the reporter (Fig. 3B).
Fig. 3.
Glucose and ERK1/2 sensitivity of elements of the CHOP gene. Promoter–reporter assays of CHOP gene promoter regions −954 to +1 bp (A), 3× −320 to −300 bp (B), and +2,624 to + 2,643 bp (C) in Min6 cells maintained in 5.5 mM or 25 mM glucose in the presence or absence of U0126. Promoter assays were repeated a minimum of three times.
U0126 Enhances CHOP Promoter Activity in the Presence of 25 mM Glucose in β-Cells.
In contrast to the behavior of this promoter in 5.5 mM glucose, inhibition of ERK1/2 activation with U0126 in cells in 25 mM glucose caused a 2.5-fold increase in reporter activity (Fig. 3A). Results were generally similar using the region containing −320 to −300 bp to drive the reporter (Fig. 3B). Additional time courses with U0126 further substantiate the effects noted above in that U0126 has its most significant effect on the 5′ promoter to increase activity in cells grown in elevated glucose (data not shown).
Comparable experiments examined the effect of glucose exposure and U0126 on reporter activity driven by the intronic element (Fig. 3C). Reporter activity driven by +2,624 to +2,643 bp was higher in cells maintained in 5.5 mM glucose and decreased slightly upon growth of cells in 25 mM glucose. In both cases, U0126 suppressed reporter activity driven by the intronic sequence. The inhibition by U0126 was most pronounced (up to 90%) under conditions with the highest reporter activity.
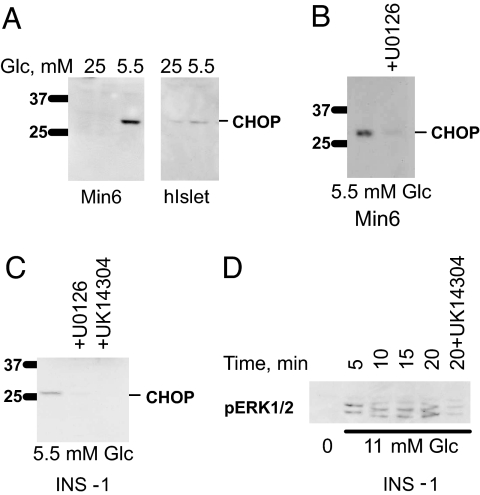
ERK1/2 Repress CHOP Protein Expression in β-Cells.
To investigate the relationship between the effects of inhibiting ERK1/2 on CHOP transcription and protein expression, CHOP protein was probed by immunoblotting. Little CHOP was detectable in Min6 cells or islets in high glucose (Fig. 4A). U0126 was sufficient to induce CHOP in the presence of 25 mM glucose (Fig. 5A). CHOP expression was readily detected in cells or islets maintained in 5.5 mM glucose. However, expression in 5.5 mM glucose was reduced if cells were exposed to U0126 to inhibit activation of ERK1/2 (Fig. 4 B and C). Likewise, α2-adrenergic agonists, which block ERK1/2 activation by glucose (Fig. 4D) (38), also inhibited CHOP expression in the INS-1 β-cell line (Fig. 4C).
Fig. 4.
Effects of U0126 on CHOP expression in β-cells and islets. (A–C) Expression of CHOP protein in Min6 cells and human islets in 5.5 mM glucose or 25 mM glucose (A); Min6 cells in 5.5 mM glucose in the presence or absence of U0126 (B); INS-1 cells in 5.5 mM glucose in the presence or absence of U0126 or UK14304 (C). (D) Activation of ERK1/2 in INS-1 cells by 11 mM glucose in the presence or absence of UK14304. Blots of islet proteins were repeated twice and, for cell lines, a minimum of three times. The time course of D was repeated twice.
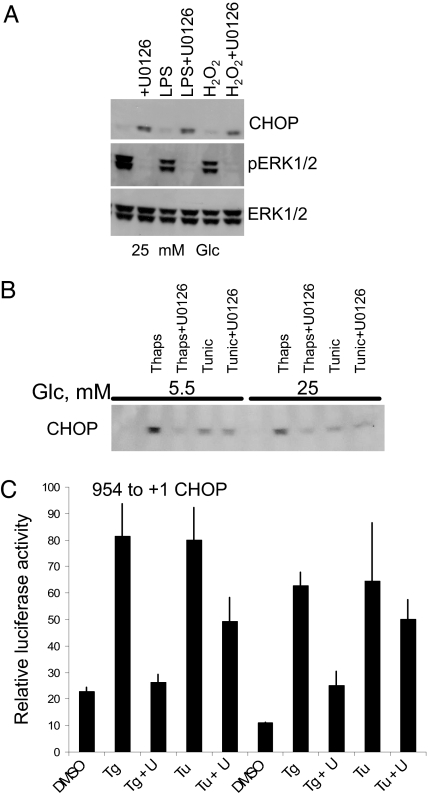
Fig. 5.
Effect of stresses on CHOP protein expression. (A) Min6 cells were maintained in 25 mM glucose then treated with or without LPS or hydrogen peroxide in the absence or presence of U0126. (B) Min6 cells in 25 mM or 5.5 mM glucose with or without thapsigargin (Thaps) or tunicamycin (Tunic) in the presence or absence of U0126. (C) Promoter–reporter assays of CHOP gene promoter −954 to +1 bp in Min6 maintained in 5.5 or 25 mM glucose then exposed to thapsigargin (Tg) or tunicamycin (Tu) for 4 h. Blots were repeated three times. Promoter assays were repeated a minimum of three times. U, U0126.
Tunicamycin and Thapsigargin Induce CHOP Protein Expression in β-Cells.
To gain perspective on the effects of other stresses on CHOP expression, cells were treated with lipopolysaccharide or peroxide. Neither of these induced CHOP protein on its own (Fig. 5A). A major inducer of CHOP expression is ER stress (3–6). To determine whether ERK1/2 have an effect on CHOP expression in response to drugs that induce ER stress, we evaluated the effects of thapsigargin and tunicamycin that cause ER stress. Tunicamycin inhibits proper glycosylation required for biosynthesis of secreted proteins. Thapsigargin inhibits the sarco-ER Ca2+ ATPase (SERCA) that pumps calcium into the ER. Both agents induced CHOP expression in INS-1 (data not shown) and Min6 β-cell lines (Fig. 5B). Blockade of ERK1/2 activation by U0126 had relatively little effect on CHOP induction by tunicamycin in either cell line. U0126 inhibited CHOP expression caused by thapsigargin.
Activation of CHOP Promoter Activity by Tunicamycin and Thapsigargin Is ERK1/2-Dependent.
We also examined the effects of ER stress on the activity of the CHOP 5′ promoter. Both thapsigargin and tunicamycin stimulated reporter activity controlled by the 5′ promoter by at least 4-fold (Fig. 5C). U0126 markedly inhibited thapsigargin-stimulated reporter activity. Partial inhibition of tunicamycin-stimulated activity was noted, consistent with the modest effect of U0126 on CHOP protein expression induced by tunicamycin.
Discussion
Nutrient sensing permits β-cells to titrate the synthesis and release of insulin to maintain circulating glucose within a narrow concentration range. Postprandial glucose concentrations stimulate glucose-sensitive factors in β-cells to increase insulin gene expression, translation, processing, and secretion (39–43). Among the signaling proteins that mediate the effects of glucose on insulin production in β-cells are the MAPKs ERK1/2 (44, 45). ERK1/2 modulate the activities of several transcription factors that act on the insulin gene in a glucose-dependent manner, including MafA and C/EBP-β (29). MafA as well as other ERK1/2-sensitive factors also influence expression of the CHOP gene often associated with apoptosis (46).
The CHOP gene contains at least two functional sequences with overlapping MARE-CEB consensus sites. One of these is similar to the RIPE3b motif in the insulin gene promoter (47). MafA binds to both sites in an ERK1/2-dependent manner but affects the sites in an opposite manner. MafA represses the 5′ promoter but activates the intron element. In reconstitution assays, the effects of MafA are dominant over actions of the other factors tested, including several previously shown to regulate CHOP expression (12, 20, 24–26). The binding of MafA to the intron was apparently stronger than its binding to the 5′ promoter under conditions in which MafA expression is lower. Our data suggest that the intron sequence serves to restrict or permit the transcriptional response caused by the 5′ promoter, depending on the conditions to which β-cells are exposed. Intron sequences have been shown to regulate mRNA accumulation and mRNA splicing and to enhance transcript processing (48–50). The regulation of both the CHOP promoter and the intron by MafA, a factor with tissue-restricted expression, suggests that this mechanism for regulation of CHOP expression may be limited to β-cells. The similarity of the CHOP intronic sequence to a glucose-sensitive region of the insulin gene promoter further suggests the specificity of regulation of this portion of the CHOP gene in β-cells.
The effects of ERK1/2 on CHOP protein expression in low and high glucose track reasonably well with their effects on transcriptional reporter activity. Maintenance in fasting glucose stimulates the transcriptional reporter activity of the 5′ CHOP promoter comparable to what is observed for expression of the protein. U0126 suppresses protein expression under this condition but has little effect on the 5′ promoter. However, at 5.5 mM glucose, U0126 has the most pronounced effect to inhibit transcription driven by the intron sequence. In the presence of chronically high glucose, inhibition of ERK1/2 is associated with increased CHOP protein expression. A comparable effect of U0126 to increase transcription is also observed on the 5′ promoter, although U0126 inhibits transcription directed by the intron element in high glucose. At this time we cannot attribute ERK1/2-dependent changes in CHOP protein expression entirely to effects on these elements. The specific contributions of individual elements in the gene to net transcription are not known. We found, using quantitative PCR, that the mRNA encoding CHOP was increased in β-cells in low glucose if ERK1/2 activities were inhibited (data not shown). We cannot currently reconcile this observation with the findings on transcription and protein expression. We suggest that mRNA stability and other transcriptional and posttranscriptional regulation must also occur.
The demand for insulin biosynthesis places β-cells at risk of ER stress. During the pathogenesis of diabetes, the profile of β-cell gene expression is altered initially to enable the cell to adapt to metabolically induced stresses (51–54). In the early stages, β-cells proliferate to keep up with the increasing insulin requirement that accompanies insulin-resistance syndromes. In later stages, chronic hyperglycemia unduly stresses the β-cell and may cause cell failure (55–57). The effects of ER stresses on CHOP expression parallel the effects of the stressors and U0126 on the 5′ promoter. Thapsigargin itself has a short-term effect to interfere with ERK1/2 activation by glucose (58). ERK1/2 activation by glucose depends on calcium signaling both from the external milieu and release from the internal storage compartment in the ER. Thus, the greater relative effect of U0126 on thapsigargin compared with tunicamycin further supports a mechanistic relationship between ER regulation of calcium signaling and ERK1/2 activity in the β-cell.
ERK1/2 are essential for glucose-regulated transcription of the insulin gene and additionally contribute to reduced insulin gene transcription in cells exposed to chronically elevated glucose (29, 59). ERK1/2 appear to play equally complex roles in modulating the expression of CHOP under nonideal conditions, repressing it during extended periods in high glucose and enhancing it during extended fasting. Chronic exposure of β-cells to high glucose can lead to glucose toxicity and CHOP-induced apoptosis (23). Our data suggest that ERK1/2 may protect β-cells from glucose toxicity by suppressing CHOP expression during extended high-glucose conditions. Less is known about long-term effects of fasting glucose on β-cells and whether or not apoptosis occurs in this state. Because CHOP is a transcription factor also known to regulate growth arrest and differentiation (4, 60), it may also have a role other than apoptosis in β-cells.
Experimental Procedures
Materials.
Early passages of Min6 cells (p16) were obtained from Gene Webb (University of Chicago, Chicago, IL). The Dual-Luciferase Reporter Assay System and passive lysis buffer were purchased from Promega (Madison, WI). The ERK1/2 antibody was Y691 (61), and the anti-pERK1/2 antibody was from Sigma (St. Louis, MO). The following antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA): GADD153 (B-3), c-Fos (4), c-Jun (H-79), ATF-2 (C-19), C/EBP-β (C-19), c-Maf (M-153), PDX-1 (N-18), NeuroD1 (N-19), and NFATc3 (F-1).
Cell Culture.
Min6 and INS-1 cells were maintained in DMEM and RPMI 1640 media (Invitrogen, Carlsbad, CA), respectively, supplemented with 10% FBS, 10 mM Hepes, pH 7.4, 10.2 mM l-glutamine, 50 mM sodium pyruvate, 2.5 mM β-mercaptoethanol, 0.1 mg/ml streptomycin, and 100 units/ml penicillin at 37°C in 10% CO2. Each cell line was maintained in two glucose concentrations: a fasting glucose concentration of 5.5 mM and a high glucose concentration of 25 mM (standard for Min6) or 11 mM (standard for INS-1) cells. U0126 was used from 10 to 25 μM in all experiments.
Isolation of Human Pancreatic Islets.
Islets were isolated from human cadaveric pancreata according to previously described methods (62–64) at the cGMP Islet Cell Processing Laboratory, Baylor University Medical Center. After procurement, the tissue was preserved using the two-layer method before islet isolation (65). The pancreatic duct was cannulated and initially perfused with cold liberase enzyme (Roche, Indianapolis, IN). The pancreatic tissue was then digested in a specialized chamber [“Ricordi Chamber” (62)] with enhanced mechanical agitation at 37°C. The resulting pancreatic digest was subjected to continuous density gradient centrifugation by using a COBE2991 cell processor to separate the islets from the exocrine tissue. The purity and quantity of islets were determined by staining with diphenylthiocarbazone. Islets were cultured in RPMI medium 1640 or Krebs–Ringer bicarbonate-Hepes (KRBH) medium in low glucose (5.5 mM) or high glucose (16–25 mM).
DNA Constructs.
The pcDNA3 plasmid expression vector encoding ATF-2 was kindly provided by Phyllis LuValle (University of Florida, Gainesville, FL). RSV-c-fos was obtained from Michael Karin (University of California at San Diego, La Jolla, CA). The pGL3-Luc reporter construct containing the −954 bp 5′ flank of the CHOP gene transcriptional start site was a generous gift from Pierre Fafournoux (National Institute of Agronomic Research, Theix, France). Complementary oligonucleotides from regions −320 to −300 bp and from +2,624 to +2,643 bp of the CHOP gene were synthesized (Integrated DNA Technologies, Coralville, IA) and blunt-end ligated as 3× repeats into the SmaI site of the pGL3-Luc reporter (Promega). The pCMV5-MafA and MSV-C/EBP-β constructs were described in ref. 29.
ChIP.
Chromatin from Min6 or human pancreatic islets was subjected to DNA–protein cross-linking and sonic fragmentation. DNA–protein complexes were immunoprecipitated as described in ref. 29 with the modifications that follow. Treated cells were fixed with 1% formaldehyde in medium and collected in ChIP lysis buffer (50 mM Tris, pH 8.0/10 mM EDTA/1% SDS). The chromatin lysates were sonicated by a Fisher Scientific (Hanover Park, IL) Sonic Dismembrator 500. Sonication was optimized to produce ≈200- to 300-bp DNA fragments with a microtip probe setting at 40% amplitude. Each sample was treated with four rounds of 15 pulses of 1.5-sec duration, with a 1-sec pause between pulses. Samples were placed on ice for 2–3 min between rounds. DNA–protein complexes were immunoprecipitated with antibodies immobilized on protein A-Sepharose beads. The immunoprecipitates were adjusted to 0.2 M NaCl and incubated at 65°C for 5 h to reverse cross-links. DNA was purified with phenol/CHCl3, and ethanol was precipitated. Total precipitated DNA was resuspended in TE (10 mM Tris/1 mM EDTA). Ten-fold serial dilutions of the precipitated DNA were used in PCR reactions with the following primers for the targeted CHOP gene DNA regions: 5′-GGTGAAACGTAGTCTCGCTCT-3′and 5′-AGCGGATCACTTGAGGTCAG-3′ (−954 to −725); 5′-AAGTGATCCGCTCTCCTCAG-3′ and 5′-TAGTCGGTCGTGAGCCTCTT-3′ (−736 to −453); 5′-AAGAGGCTCACGACCGACTA-3′ and 5′-GGAGGAGGTGGGTGAGTCAT-3′ (−472 to −226); 5′-CATGACTCACCCACCTCCTC-3′ and 5′-AGCGCCTGGCTGTAATCTT-3′ (−246 to −10); 5′-GCTCCCGAGGTCAGAGACTT-3′ and 5′-GGACTCTCCCCATTCCTCTC-3′ (−12 to +223); 5′-GTCCCTTATTCTGGGGTGGT-3′ and 5′-GCGATGGTCACCCAGTCTTA-3′ (+220 to +456); 5′-TCTAAGACTGGGTGACCATCG-3′ and 5′-CCCAAATGGCCTCCTACAC-3′ (+435 to +730); 5′-CTCCCCAGGGTATTGTCCTT-3′ and 5′-TATAGGGAAGGGGCAGAAGC-3′ (+733 to +1,016); 5′-TTCTGCCCCTTCCCTATAAGA-3′ and 5′-TGTCATGGAAGTCACTGAAGG-3′ (+999 to +1,297); 5′-GGAAGTCATTGGAGGGTTTG-3′ and 5′-CTCCCAAAGTGCTGGGATTA-3′ (+1,305 to + 1,597); 5′-ACTTTGGGAGACCAGAGGTG-3′ and 5′-ACGGGTCTCACTCTTTTTGC-3′ (+1,588 to +1,830); 5′-TGGGCAAAAAGAGTGAGACC-3′ and 5′-TGGACCTCTTCCAGAACTTCA-3′ (+1,808 to +2,083); 5′-TGAAGTTCTGGAAGAGGTCCA-3′ and 5′-CATTTCGAAGGGCAGAAGAG-3′ (+2,063 to +2,311); 5′-GAAATGGCTACAGGGACCAA-3′ and 5′-CGCCCAGCTAATTTTTGTGT-3′ (+2,306 to +2,529); 5′-AAAAATTAGCTGGGCGTGGT-3′ and 5′-TGCTTTCAGGTGTGGTGATG-3′ (+2,514 to +2,805); 5′-CCACACCTGAAAGCAGGTAAA-3′ and 5′-CTGCAGGATAATGGGGAGTG-3′; (+2,791 to +3,073); 5′-CCCCATTATCCTGCAGATGT-3′ and 5′-CATTTCCAGGAGGTGAAACA-3′ (+3,058 to +3,238).
cDNA Synthesis.
Treated cells were harvested with TRI Reagent (Ambion, Austin, TX) for isolation of total RNA. For each treatment, 10 μg of total RNA was converted to cDNA by reverse transcriptase by High-Capacity cDNA Archive kit (Applied Biosystems, Foster City, CA) using random hexamers.
Real-Time Quantitative PCR.
TaqMan probe-based PCR was performed on the ABI 7500 DNA Sequence Detection System with standard fluorescent chemistries and thermal cycling conditions specified by the manufacturer: 50°C for 2 min, 95°C for 10 min for one cycle, an additional 40 cycles at 95°C for 15 sec, and ramp to 60°C for 1 min. TaqMan Gene Expression Assays were purchased from Applied Biosystems, including the CHOP gene expression assay DDIT3 (Mn00492097_m1) and custom assay that targets the +2,626 bp CHOP intronic sequence with primers 5′-GGAGGCAGAGGTTGCAGTGA-3′ and 5′-GCTCTGTTGCCAGGCTGAAG-3′ and TaqMan probe 5′-CGAGATCGCACCACTG-3′. As an internal expression control, 18S rRNA was used and was amplified using primer and probe sequences from the Ribosomal RNA Control Reagents kit (Applied Biosystems).
Statistical Analyses.
Results are expressed as means ± SEM determined from at least three independent experiments, unless otherwise stated. Statistical significance was calculated by one-tailed unpaired Student's t test.
Transfections and Reporter Assays.
Cells were cultured for 2 days in 5.5 or 25 mM glucose with or without U0126 in six-well dishes and transfected with plasmids by using Lipofectamine 2000 reagent in antibiotic-free medium overnight. Cells were harvested in passive lysis buffer supplemented with 100 mM β-glycerophosphate, 2 mM Na3VO4, and 100 mM NaF. The lysates were vortexed for 30 sec, and the supernatants were collected after centrifugation for 30 min at 16,000 × g at 4°C in a microcentrifuge. The supernatants were stored at −80°C. Reporter assays were performed using a TD-20/20 bioluminometer (Turner Designs, Sunnyvale, CA). Data were normalized to Renilla luciferase driven by an SV40 promoter.
Immunoblot Analysis.
Extracts were prepared from treated cells harvested in passive lysis buffer supplemented with 100 mM β-glycerophosphate, 2 mM Na3VO4, and 100 mM NaF. Samples containing 20 μg of protein were subjected to SDS/polyacrylamide gel electrophoresis. Proteins were electrotransferred to nitrocellulose membranes (Millipore, Billerica, MA) and blocked with 1% BSA and 1% nonfat milk powder in TBS buffer containing 0.1% Tween-20 for 4–18 h at 4°C and incubated with a primary antibody for 2 h at 4°C, followed by a horseradish peroxidase-conjugated secondary antibody diluted 1:3,000 in blocking buffer for 30 min at room temperature. Proteins were detected by enhanced chemiluminescence.
Acknowledgments
We thank Gray Pearson (Salk Institute, La Jolla, CA) for helpful suggestions; Malavika Raman, Chunli Shao, Kyle Wedin, Lisa Lenertz, and other members of the H.H.C. Laboratory for comments about the data and manuscript; and Dionne Ware for administrative assistance. This work was supported by National Institutes of Health Grant DK55310 (to M.H.C.). M.C.L. was supported by a mentor-based postdoctoral fellowship from the American Diabetes Foundation.
Abbreviations
- ATF
activating transcription factor
- CEB
CAAT enhancer binding
- CRE
cyclic AMP response element
- ER
endoplasmic reticulum
- MARE
MafA response elements.
Footnotes
The authors declare no conflict of interest.
References
- 1.Fornace AJ, Jr, Alamo I, Jr, Hollander MC. Proc Natl Acad Sci USA. 1988;85:8800–8804. doi: 10.1073/pnas.85.23.8800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ron D, Habener JF. Genes Dev. 1992;6:439–453. doi: 10.1101/gad.6.3.439. [DOI] [PubMed] [Google Scholar]
- 3.Yang L, Carlson SG, McBurney D, Horton WE., Jr J Biol Chem. 2005;280:31156–31165. doi: 10.1074/jbc.M501069200. [DOI] [PubMed] [Google Scholar]
- 4.Tang QQ, Lane MD. Proc Natl Acad Sci USA. 2000;97:12446–12450. doi: 10.1073/pnas.220425597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yoshida H, Okada T, Haze K, Yanagi H, Yura T, Negishi M, Mori K. Mol Cell Biol. 2000;20:6755–6767. doi: 10.1128/mcb.20.18.6755-6767.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oyadomari S, Takeda K, Takiguchi M, Gotoh T, Matsumoto M, Wada I, Akira S, Araki E, Mori M. Proc Natl Acad Sci USA. 2001;98:10845–10850. doi: 10.1073/pnas.191207498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guyton KZ, Xu Q, Holbrook NJ. Biochem J. 1996;314:547–554. doi: 10.1042/bj3140547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McCullough KD, Martindale JL, Klotz LO, Aw TY, Holbrook NJ. Mol Cell Biol. 2001;21:1249–1259. doi: 10.1128/MCB.21.4.1249-1259.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tang JR, Nakamura M, Okura T, Takata Y, Watanabe S, Yang ZH, Liu J, Kitami Y, Hiwada K. Biochem Biophys Res Commun. 2002;290:1255–1259. doi: 10.1006/bbrc.2002.6336. [DOI] [PubMed] [Google Scholar]
- 10.Carlson SG, Fawcett TW, Bartlett JD, Bernier M, Holbrook NJ. Mol Cell Biol. 1993;13:4736–4744. doi: 10.1128/mcb.13.8.4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bruhat A, Jousse C, Wang XZ, Ron D, Ferrara M, Fafournoux P. J Biol Chem. 1997;272:17588–17593. doi: 10.1074/jbc.272.28.17588. [DOI] [PubMed] [Google Scholar]
- 12.Averous J, Bruhat A, Jousse C, Carraro V, Thiel G, Fafournoux P. J Biol Chem. 2004;279:5288–5297. doi: 10.1074/jbc.M311862200. [DOI] [PubMed] [Google Scholar]
- 13.Friedman AD. Cancer Res. 1996;56:3250–3256. [PubMed] [Google Scholar]
- 14.Zinszner H, Kuroda M, Wang X, Batchvarova N, Lightfoot RT, Remotti H, Stevens JL, Ron D. Genes Dev. 1998;12:982–995. doi: 10.1101/gad.12.7.982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brenner B, Koppenhoefer U, Weinstock C, Linderkamp O, Lang F, Gulbins E. J Biol Chem. 1997;272:22173–22181. doi: 10.1074/jbc.272.35.22173. [DOI] [PubMed] [Google Scholar]
- 16.Marciniak SJ, Yun CY, Oyadomari S, Novoa I, Zhang Y, Jungreis R, Nagata K, Harding HP, Ron D. Genes Dev. 2004;18:3066–3077. doi: 10.1101/gad.1250704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maytin EV, Ubeda M, Lin JC, Habener JF. Exp Cell Res. 2001;267:193–204. doi: 10.1006/excr.2001.5248. [DOI] [PubMed] [Google Scholar]
- 18.Carriere A, Carmona MC, Fernandez Y, Rigoulet M, Wenger RH, Penicaud L, Casteilla L. J Biol Chem. 2004;279:40462–40469. doi: 10.1074/jbc.M407258200. [DOI] [PubMed] [Google Scholar]
- 19.Huang H, Lane MD, Tang QQ. Biochem Biophys Res Commun. 2005;338:1185–1188. doi: 10.1016/j.bbrc.2005.10.057. [DOI] [PubMed] [Google Scholar]
- 20.Sylvester SL, ap Rhys CM, Luethy-Martindale JD, Holbrook NJ. J Biol Chem. 1994;269:20119–20125. [PubMed] [Google Scholar]
- 21.Cardozo AK, Ortis F, Storling J, Feng YM, Rasschaert J, Tonnesen M, Van Eylen F, Mandrup-Poulsen T, Herchuelz A, Eizirik DL. Diabetes. 2005;54:452–461. doi: 10.2337/diabetes.54.2.452. [DOI] [PubMed] [Google Scholar]
- 22.Cnop M, Welsh N, Jonas JC, Jorns A, Lenzen S, Eizirik DL. Diabetes. 2005;54(Suppl 2):S97–S107. doi: 10.2337/diabetes.54.suppl_2.s97. [DOI] [PubMed] [Google Scholar]
- 23.Oyadomari S, Koizumi A, Takeda K, Gotoh T, Akira S, Araki E, Mori M. J Clin Invest. 2002;109:525–532. doi: 10.1172/JCI14550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fawcett TW, Martindale JL, Guyton KZ, Hai T, Holbrook NJ. Biochem J. 1999;339:135–141. [PMC free article] [PubMed] [Google Scholar]
- 25.Wolfgang CD, Chen BP, Martindale JL, Holbrook NJ, Hai T. Mol Cell Biol. 1997;17:6700–6707. doi: 10.1128/mcb.17.11.6700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bruhat A, Jousse C, Carraro V, Reimold AM, Ferrara M, Fafournoux P. Mol Cell Biol. 2000;20:7192–7204. doi: 10.1128/mcb.20.19.7192-7204.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kataoka K, Han SI, Shioda S, Hirai M, Nishizawa M, Handa H. J Biol Chem. 2002;277:49903–49910. doi: 10.1074/jbc.M206796200. [DOI] [PubMed] [Google Scholar]
- 28.Hartsough MT, Morrison DK, Salerno M, Palmieri D, Ouatas T, Mair M, Patrick J, Steeg PS. J Biol Chem. 2002;277:32389–32399. doi: 10.1074/jbc.M203115200. [DOI] [PubMed] [Google Scholar]
- 29.Lawrence MC, McGlynn K, Park BH, Cobb MH. J Biol Chem. 2005;280:26751–26759. doi: 10.1074/jbc.M503158200. [DOI] [PubMed] [Google Scholar]
- 30.Schmitt-Ney M, Habener JF. J Biol Chem. 2000;275:40839–40845. doi: 10.1074/jbc.M007440200. [DOI] [PubMed] [Google Scholar]
- 31.Ogino H, Yasuda K. Science. 1998;280:115–118. doi: 10.1126/science.280.5360.115. [DOI] [PubMed] [Google Scholar]
- 32.Kataoka K, Shioda S, Ando K, Sakagami K, Handa H, Yasuda K. J Mol Endocrinol. 2004;32:9–20. doi: 10.1677/jme.0.0320009. [DOI] [PubMed] [Google Scholar]
- 33.Olbrot M, Rud J, Moss LG, Sharma A. Proc Natl Acad Sci USA. 2002;99:6737–6742. doi: 10.1073/pnas.102168499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matsuoka TA, Artner I, Henderson E, Means A, Sander M, Stein R. Proc Natl Acad Sci USA. 2004;101:2930–2933. doi: 10.1073/pnas.0306233101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu M, Seufert J, Habener JF. J Biol Chem. 1997;272:28349–28359. doi: 10.1074/jbc.272.45.28349. [DOI] [PubMed] [Google Scholar]
- 36.Sharma A, Olson LK, Robertson RP, Stein R. Mol Endocrinol. 1995;9:1127–1134. doi: 10.1210/mend.9.9.7491105. [DOI] [PubMed] [Google Scholar]
- 37.Harmon JS, Stein R, Robertson RP. J Biol Chem. 2005;280:11107–11113. doi: 10.1074/jbc.M410345200. [DOI] [PubMed] [Google Scholar]
- 38.Gibson TB, Lawrence MC, Gibson C, Vanderbilt CA, McGlynn K, Arnette D, Chen W, Collins J, Naziruddin B, Levy MF, et al. Diabetes. 2006;55:1066–1073. doi: 10.2337/diabetes.55.04.06.db05-1266. [DOI] [PubMed] [Google Scholar]
- 39.Ohneda K, Ee H, German M. Semin Cell Dev Biol. 2000;11:227–233. doi: 10.1006/scdb.2000.0171. [DOI] [PubMed] [Google Scholar]
- 40.Wang J, Shen L, Najafi H, Kolberg J, Matschinsky FM, Urdea M, German M. Proc Natl Acad Sci USA. 1997;94:4360–4365. doi: 10.1073/pnas.94.9.4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Henquin JC. Diabetes. 2000;49:1751–1760. doi: 10.2337/diabetes.49.11.1751. [DOI] [PubMed] [Google Scholar]
- 42.Welsh M, Scherberg N, Gilmore R, Steiner DF. Biochem J. 1986;235:459–467. doi: 10.1042/bj2350459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wicksteed B, Uchizono Y, Alarcon C, McCuaig JF, Shalev A, Rhodes CJ. Cell Metab. 2007;5:221–227. doi: 10.1016/j.cmet.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 44.Frödin M, Sekine N, Roche E, Filloux C, Prentki M, Wollheim CB, Van Obberghen E. J Biol Chem. 1995;270:7882–7889. doi: 10.1074/jbc.270.14.7882. [DOI] [PubMed] [Google Scholar]
- 45.Khoo S, Cobb MH. Proc Natl Acad Sci USA. 1997;94:5599–5604. doi: 10.1073/pnas.94.11.5599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Laybutt DR, Preston AM, Akerfeldt MC, Kench JG, Busch AK, Biankin AV, Biden TJ. Diabetologia. 2007;50:752–763. doi: 10.1007/s00125-006-0590-z. [DOI] [PubMed] [Google Scholar]
- 47.Shieh SY, Tsai MJ. J Biol Chem. 1991;266:16708–16714. [PubMed] [Google Scholar]
- 48.Himes SR, Tagoh H, Goonetilleke N, Sasmono T, Oceandy D, Clark R, Bonifer C, Hume DA. J Leukoc Biol. 2001;70:812–820. [PubMed] [Google Scholar]
- 49.Hwang ES, Choi A, Ho IC. J Immunol. 2002;169:248–253. doi: 10.4049/jimmunol.169.1.248. [DOI] [PubMed] [Google Scholar]
- 50.Lee JG, Dahi S, Mahimkar R, Tulloch NL, Alfonso-Jaume MA, Lovett DH, Sarkar R. Proc Natl Acad Sci USA. 2005;102:16345–16350. doi: 10.1073/pnas.0508085102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schuit F, Flamez D, De Vos A, Pipeleers D. Diabetes. 2002;51(Suppl 3):S326–S332. doi: 10.2337/diabetes.51.2007.s326. [DOI] [PubMed] [Google Scholar]
- 52.Hoorens A, Van de CM, Kloppel G, Pipeleers D. J Clin Invest. 1996;98:1568–1574. doi: 10.1172/JCI118950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Laybutt DR, Kaneto H, Hasenkamp W, Grey S, Jonas JC, Sgroi DC, Groff A, Ferran C, Bonner-Weir S, Sharma A, et al. Diabetes. 2002;51:413–423. doi: 10.2337/diabetes.51.2.413. [DOI] [PubMed] [Google Scholar]
- 54.Brun T, Franklin I, St Onge L, Biason-Lauber A, Schoenle EJ, Wollheim CB, Gauthier BR. J Cell Biol. 2004;167:1123–1135. doi: 10.1083/jcb.200405148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Laybutt DR, Sharma A, Sgroi DC, Gaudet J, Bonner-Weir S, Weir GC. J Biol Chem. 2002;277:10912–10921. doi: 10.1074/jbc.M111751200. [DOI] [PubMed] [Google Scholar]
- 56.Weir GC, Bonner-Weir S. Diabetes. 2004;53(Suppl 3):S16–S21. doi: 10.2337/diabetes.53.suppl_3.s16. [DOI] [PubMed] [Google Scholar]
- 57.Jonas JC, Sharma A, Hasenkamp W, Ilkova H, Patane G, Laybutt R, Bonner-Weir S, Weir GC. J Biol Chem. 1999;274:14112–14121. doi: 10.1074/jbc.274.20.14112. [DOI] [PubMed] [Google Scholar]
- 58.Arnette D, Gibson TB, Lawrence MC, January B, Khoo S, McGlynn K, Vanderbilt CA, Cobb MH. J Biol Chem. 2003;278:32517–32525. doi: 10.1074/jbc.M301174200. [DOI] [PubMed] [Google Scholar]
- 59.Khoo S, Griffen SC, Xia Y, Baer R, German MS, Cobb MH. J Biol Chem. 2003;278:32969–32977. doi: 10.1074/jbc.M301198200. [DOI] [PubMed] [Google Scholar]
- 60.Shirakawa K, Maeda S, Gotoh T, Hayashi M, Shinomiya K, Ehata S, Nishimura R, Mori M, Onozaki K, Hayashi H, et al. Mol Cell Biol. 2006;26:6105–6116. doi: 10.1128/MCB.02429-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Boulton TG, Nye SH, Robbins DJ, Ip NY, Radziejewska E, Morgenbesser SD, DePinho RA, Panayotatos N, Cobb MH, Yancopoulos GD. Cell. 1991;65:663–675. doi: 10.1016/0092-8674(91)90098-j. [DOI] [PubMed] [Google Scholar]
- 62.Ricordi C, Lacy PE, Scharp DW. Diabetes. 1989;38(Suppl 1):140–142. doi: 10.2337/diab.38.1.s140. [DOI] [PubMed] [Google Scholar]
- 63.Linetsky E, Bottino R, Lehmann R, Alejandro R, Inverardi L, Ricordi C. Diabetes. 1997;46:1120–1123. doi: 10.2337/diab.46.7.1120. [DOI] [PubMed] [Google Scholar]
- 64.Lakey JR, Warnock GL, Shapiro AM, Korbutt GS, Ao Z, Kneteman NM, Rajotte RV. Cell Transplant. 1999;8:285–292. doi: 10.1177/096368979900800309. [DOI] [PubMed] [Google Scholar]
- 65.Tsujimura T, Kuroda Y, Avila JG, Kin T, Oberholzer J, Shapiro AM, Lakey JR. Transplantation. 2004;78:96–100. doi: 10.1097/01.tp.0000133515.37892.d5. [DOI] [PubMed] [Google Scholar]