Abstract
Iron homeostasis requires subtle control systems, as iron is both essential and toxic. In Aspergillus nidulans, iron represses iron acquisition via the GATA factor SreA, and induces iron-dependent pathways at the transcriptional level, by a so far unknown mechanism. Here, we demonstrate that iron-dependent pathways (e.g., heme biosynthesis) are repressed during iron-depleted conditions by physical interaction of HapX with the CCAAT-binding core complex (CBC). Proteome analysis identified putative HapX targets. Mutual transcriptional control between hapX and sreA and synthetic lethality resulting from deletion of both regulatory genes indicate a tight interplay of these control systems. Expression of genes encoding CBC subunits was not influenced by iron availability, and their deletion was deleterious during iron-depleted and iron-replete conditions. Expression of hapX was repressed by iron and its deletion was deleterious during iron-depleted conditions only. These data indicate that the CBC has a general role and that HapX function is confined to iron-depleted conditions. Remarkably, CBC-mediated regulation has an inverse impact on the expression of the same gene set in A. nidulans, compared with Saccharomyces cerevisae.
Keywords: CCAAT, Hap complex, iron, regulation, siderophore
Introduction
The cis-acting sequence CCAAT is present in approximately 30% of eukaryotic promoters (Bucher, 1990). An evolutionarily conserved protein complex able to bind to this motif has been found in all eukaryotes analyzed so far, ranging from yeast to mammals. It has been designated Hap complex in Saccharomyces cerevisiae (Pinkham and Guarente, 1985; McNabb et al, 1995), Kluyveromyces lactis (Mulder et al, 1994), and Arabidopsis thaliana (Edwards et al, 1998), Php in Schizosaccharomyces pombe (McNabb et al, 1997), AnCF in Aspergillus species (reviewed in Brakhage et al, 1999), CBF in Xenopus laevis (Li et al, 1998), and NF-Y in mammals (Hooft van Huijsduijnen et al, 1990; Maity et al, 1990), respectively. The S. cerevisiae Hap complex was the first CCAAT-binding complex to be identified. It comprises four subunits, Hap2p, Hap3p, Hap4p and Hap5p. Hap2/3/5p form the core CCAAT-binding complex (here termed CBC), which is responsible for DNA binding, while Hap4p is involved in transcriptional activation (McNabb et al, 1995). Orthologs of Hap2/3/5p are present in all eukaryotes. Moreover, S. cerevisiae Hap2p, A. nidulans HapB and human NF-YA are functionally interchangeable (Becker et al, 1991; Tuncher et al, 2005), which demonstrates high evolutionary conservation of the CBC. However, with the exception of the yeast species K. lactis and Hansenula polymorpha, clear evidence for a Hap4p ortholog in other organisms is inconclusive (Bourgarel et al, 1999; Sybirna et al, 2005). In S. cerevisiae, the Hap4p/CBC complex acts as an activator of genes involved in oxidative phosphorylation in response to growth on non-fermentable carbon sources (Pinkham and Guarente, 1985). The A. nidulans CBC, consisting of the Hap2/3/5p orthologs HapB/C/E, modulates the expression of numerous genes, including the anabolic penicillin biosynthesis genes acvA, ipnA and aatA (Litzka et al, 1996; Then Bergh et al, 1996) and the catabolic acetamidase encoding amdS (Littlejohn and Hynes, 1992). In this respect it has also been shown that the activation of gene expression by pathway-specific regulators can depend on the presence of a functional CBC (Steidl et al, 1999). At the same time, evidence for CBC-mediated repression of gene expression was found in A. nidulans for homoaconitase-encoding lysF and in the auto-regulation of hapB expression (Steidl et al, 2001; Weidner et al, 2001). Recently, the CBC was also found to act as a repressor of mitochondrial electron transport components in Candida albicans (Johnson et al, 2005). However, a clear picture of the CBC regulon in fungi is missing so far.
A yeast-two-hybrid screen suggested physical interaction of A. nidulans HapB with a protein of a yet unknown function, termed HapX (Tanaka et al, 2002). HapX displays no similarity to S. cerevisiae Hap4p, apart from an N-terminal 17 amino-acid-motif, which has been shown to be essential for interaction of Hap4p with the S. cerevisiae CBC (McNabb and Pinto, 2005). Deletion of hapX in A. nidulans did not result in a slow-growth and weak-conidiation phenotype, as caused by a deletion of any of the three CBC subunit-encoding genes (Steidl et al, 1999). Furthermore, expression of hapX in S. cerevisiae did not complement the deletion of Hap4. Therefore, a function of HapX in CBC-mediated gene expression appeared unlikely at that time.
We resumed the functional analysis of hapX, as we found that its expression is repressed by iron via the GATA factor SreA. As a cofactor for numerous enzymes and in electron transport systems, iron is indispensable for all eukaryotes. However, an excess of iron is toxic, due to its capacity to catalyze the production of cell damaging reactive oxygen species (Halliwell and Gutteridge, 1984). Therefore, subtle control systems are required to maintain iron homeostasis. In A. nidulans, iron represses siderophore-mediated iron uptake, which is the major iron acquisition system and is essential for fungal virulence (Schrettl et al, 2004; Oide et al, 2006), and induces iron-dependent pathways (Haas et al, 1999; Oberegger et al, 2001, 2002b). The repression of the siderophore system under iron-replete (+Fe) conditions is mediated, at least in part, by SreA. Here, we demonstrate that HapX mediates repression of iron-dependent pathways under iron-depleted (−Fe) conditions via interaction with the CBC. HapX target genes were identified by a proteomic approach. The importance of a tight interplay between HapX and SreA is demonstrated by mutual transcriptional control between hapX and sreA and synthetic lethality resulting from deletion of both regulatory genes.
Results
Deletion of hapX or of genes encoding CBC subunits impedes growth during −Fe conditions
A differential mRNA display screen for SreA target genes (Oberegger et al, 2001) suggested that the steady-state level of hapX mRNA is repressed by iron in the wild type and partially derepressed in an sreA deletion (Δ) strain (data not shown). This expression pattern was confirmed using Northern analysis (Figure 1A). Notably, deletion of sreA results in about 30% derepression, which is typical for SreA target genes, for example, siderophore transporter-encoding mirB (Haas et al, 2003).
Figure 1.
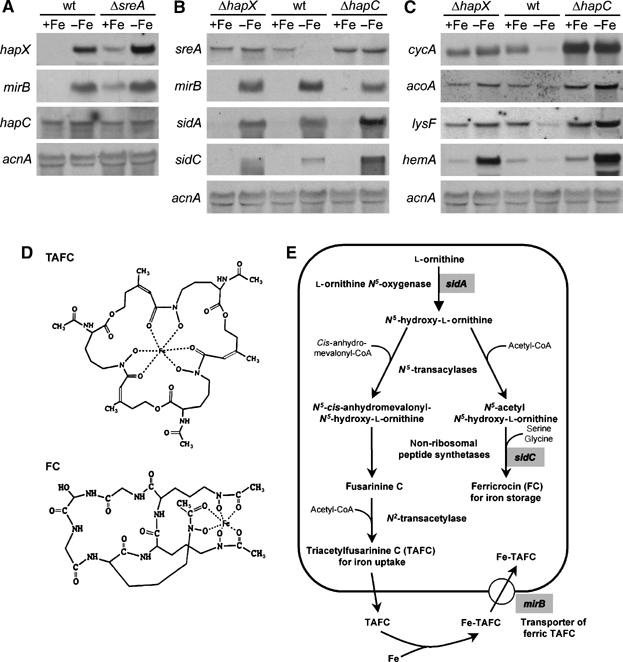
Iron-regulated gene expression in A. nidulans wild-type, ΔsreA, ΔhapX and ΔhapC strains. For Northern analysis, the total RNA was isolated from A. nidulans strains grown for 24 h under +Fe and −Fe conditions. As a control for loading and RNA quality, blots were hybridized with the γ-actin encoding acnA gene. (A) Expression of hapX but not hapC is partially controlled by SreA-mediated iron regulation. (B) Deletion of hapX or hapC causes derepression of sreA, during −Fe conditions but not the SreA regulon, during +Fe conditions. (C) Deletion of hapX or of hapC causes derepression of iron-dependent pathways during iron starvation. (D) A. nidulans siderophores. (E) A. nidulans siderophore metabolism. Known genes involved in siderophore biosynthesis and uptake are shaded in gray.
We compared the phenotypic consequences of hapX deletion during +Fe and −Fe conditions, because of the iron-dependent expression of hapX. Consistent with the previous analysis (Tanaka et al, 2002), during +Fe conditions, ΔhapX displayed no significant difference to a wild-type strain with respect to conidiation and growth rate on a solid medium (data not shown) and in a liquid medium (Table I). During −Fe conditions, however, the growth rate was significantly reduced in submerged cultures (Table I), and, on a solid medium, the mycelium was less dense and conidiation was severely impaired (data not shown). Ectopic integration of a functional hapX gene (strain hapXR) cured these and all other defects of ΔhapX (Table I; Figure 2), which demonstrates that the ΔhapX phenotype is a direct result of the loss of HapX activity.
Table 1.
Deletion of hapX, hapB, hapC or hapE causes a reduced growth rate during −Fe conditions
| Strain | Growth condition | Ratio | |
|---|---|---|---|
| +Fe | −Fe | −Fe/+Fe | |
| Wild type | 100.0 | 66.9±8.4 | 0.67 |
| ΔhapX | 91.6±7.0 | 49.5±5.3 | 0.54 |
| hapXR | 114.3±5.8 | 74.2±6.4 | 0.65 |
| ΔhapB | 46.3±5.9 | 23.7±3.1 | 0.51 |
| ΔhapC | 61.6±7.6 | 28.3±4.2 | 0.46 |
| ΔhapE | 54.6±6.3 | 25.5±2.7 | 0.47 |
| Strains were grown for 24 h in –Fe and +Fe conditions. Dry weights were normalized to that of the wild type grown during iron-replete conditions, which was 0.58±0.04 g. | |||
Figure 2.
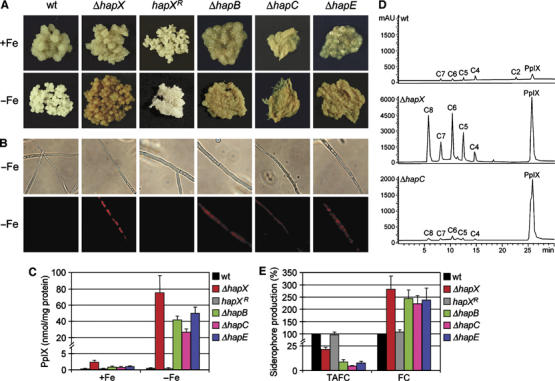
Deletion of hapX or genes encoding CBC subunits leads to cellular accumulation of PpIX, decreased TAFC synthesis and increased FC production, during −Fe conditions. (A) Mycelia of A. nidulans strains after growth for 24 h during +Fe and –Fe conditions. (B) Characteristic red auto-fluorescence caused by PpIX accumulation during −Fe conditions. During +Fe conditions, no auto-fluorescence was detectable in any strain (data not shown). (C) Quantification of PpIX accumulation. (D) Representative chromatograms of porphyrin analysis of wild-type, ΔhapX and ΔhapC strains after growth under −Fe conditions. C8, C7, C6, C5, C4 and C2 denote uroporphyrin, heptacarboxylporphyrin, hexacarboxylporphyrin, pentacarboxylporphyrin, coproporphyrin and protoporphyrin, respectively. (E) Quantification of siderophore production during −Fe conditions normalized to that of the wild type. The data represent the mean±s.d. of three simultaneously harvested flasks.
HapX was initially found to interact with the CBC subunit HapB. To ascertain a possible metabolic connection, we also compared the growth rates of strains lacking individual CBC subunits during +Fe and –Fe conditions. Deletion of hapB, hapC or hapE impairs growth during +Fe, as shown previously (Steidl et al, 1999), but in particular during −Fe conditions (Table I), which indicates a special function of the CBC during −Fe conditions consistent with a possible HapX/CBC interaction.
Deletion of hapX or of genes encoding CBC subunits causes accumulation of the iron-free heme precursor protoporphyrin IX (PpIX) during −Fe conditions
In contrast to the wild type, ΔhapX, ΔhapB, ΔhapC and ΔhapE mycelia displayed a red pigmentation concomitant with red auto-fluorescence during −Fe but not +Fe conditions (Figure 2A and B), which is typical for PpIX accumulation. Determination of the PpIX content revealed no significant difference between −Fe and +Fe conditions in the wild type (Figure 2C). In contrast, the PpIX content of ΔhapX increased slightly during +Fe conditions, and increased about 80-fold during −Fe conditions (Figure 2C and D). Moreover, the content of the PpIX precursors uroporphyrin, heptacarboxylporphyrin, hexacarboxylporphyrin, pentacarboxylporphyrin and coproporphyrin was significantly increased in ΔhapX during −Fe conditions (Figure 2D). Similarly, deletion of any of the CBC subunit-encoding genes caused a 20- to 50-fold increase in PpIX accumulation (Figure 2C).
Deletion of hapX or of genes encoding CBC subunits causes decreased extracellular and increased intracellular siderophore production
A. nidulans produces two major siderophores (Figure 1D), which are essential for growth (Eisendle et al, 2003); triacetylfusarinine C (TAFC) mobilizes extracellular iron and ferricrocin (FC) stores iron intracellularly (Eisendle et al, 2006). Because of the observed growth defect, we analyzed siderophore production in strains lacking HapX or CBC subunits (Figure 2E). During +Fe conditions, production of TAFC and FC was low and resembled the wild type in all strains (data not shown). During −Fe conditions, however, deletion of hapX or of any CBC subunit-encoding gene caused significantly decreased TAFC and increased FC production. Notably, the decrease in TAFC production was more pronounced in CBC subunit deletion mutants and FC accumulation was higher in a ΔhapX mutant.
Supplementation with iron-free TAFC to a final concentration of 20 μM only partially cured the growth defect of ΔhapX during −Fe conditions, whereas PpIX accumulation remained unaffected (data not shown). These data suggest that reduced TAFC production does not account for the full extent of growth reduction and PpIX accumulation resulting from hapX deletion.
HapX represses sreA expression without a general effect on the SreA regulon
To further investigate the altered siderophore production of ΔhapX and ΔhapC strains, we performed a Northern analysis of the known genes involved in siderophore metabolism (Haas et al, 2003): sreA (repressor of siderophore metabolism), mirB (transporter for uptake of ferric TAFC), sidA (ornithine monooxygenase catalyzing the common step for biosynthesis of TAFC and FC) and sidC (non-ribosomal peptide synthetase required for FC synthesis) (Figure 1B). The proposed siderophore biosynthetic pathway is provided in Figure 1E.
As shown previously (Oberegger et al, 2002b), expression of sreA is repressed during −Fe conditions, whereas that of the SreA target genes mirB, sidA, and sidC is repressed during +Fe conditions. Deletion of hapX or of hapC caused derepression of sreA during −Fe conditions (Figure 1B). In turn, as SreA represses siderophore biosynthesis and uptake, it was conceivable that SreA-regulated genes were repressed during −Fe conditions in these strains. However, regulation of mirB and sidA was unaffected in both ΔhapX and ΔhapC and, therefore, does not explain the reduced TAFC production. These data suggest that SreA-mediated repression requires either post-translational activation, and/or other additional factors. In contrast, the transcript levels of sidC were elevated in ΔhapX and ΔhapC during −Fe conditions, which agrees with the increased FC accumulation. Notably, sidC transcripts are approximately 15 kb in length and are therefore preferentially subject to physical degradation during RNA preparation. The upregulation of sidC expression in ΔhapX was also confirmed by dot blot analysis (data not shown).
HapX and the CBC are required to repress iron-dependent pathways during –Fe conditions
sreA belongs to a class of genes, which are downregulated during iron starvation. Most members of this class encode proteins requiring iron-containing cofactors, such as cycA, which encodes the heme-protein cytochrome c, as well as acoA and lysF, which encode the iron–sulfur cluster proteins aconitase and homoaconitase, respectively (Oberegger et al, 2002a). Another example is hemA, which codes for 5-aminolevulinate synthase (Bradshaw et al, 1993). HemA does not require iron by itself, but catalyzes the first committed step in heme biosynthesis. Deletion of hapX or of CBC subunit-encoding genes resulted in derepressed expression of all four genes during –Fe conditions (Figure 1C). In agreement with PpIX accumulation (Figure 2C), hemA expression was not only derepressed, but highly upregulated in both ΔhapX and ΔhapC (Figure 1C). A hypothetical explanation for the latter would be the lack of feedback inhibition of hemA expression by heme. Remarkably, hapC deletion resulted not only in derepression of iron-dependent pathways during −Fe conditions, but additionally in their upregulation during +Fe growth, in particular that of cycA (Figure 1C).
Notably, neither sreA (Figure 1A) nor hapX deletion affected the hapC transcript levels, and hapC deletion did not affect hapX expression (data not shown).
The A. nidulans wild-type BPU used is auxotrophic for uracil (pyrG89) and pyridoxamine (pyroA4). With respect to siderophore production and PpIX accumulation, BPU did not show any difference to A. nidulans strain TRAN, which is prototrophic for uracil and pyridoxamine, demonstrating that these auxotrophies do not influence iron metabolism (data not shown). Consistently, regulation of siderophore biosynthesis and iron-dependent pathways in BPU was as previously shown in TRAN (Oberegger et al, 2002a, 2002b).
Interaction of HapX with DNA-bound CBC is abolished by iron
To investigate putative HapX/CBC interaction in vivo, we applied bimolecular fluorescence complementation (BiFC) assays, previously shown to be valuable to define in vivo protein interaction (Hink et al, 2002; Hu and Kerppola, 2003; Hoff and Kück, 2005). BiFC was detected between enhanced-yellow-fluorescent-protein (eYFP) split fragments fused to HapX and HapB in strain yXB, under −Fe (Figure 3B) but not +Fe conditions (Figure 3A). BiFC could not be detected between HapX and HapB in the ΔhapC strain yXBΔC (Figure 3C), but was reconstituted by complementation with the hapC gene in strain yXBCc (Figure 3D). Northern analysis confirmed constitutive expression of the two eYFP split fragment-encoding genes in the used strains (Supplementary Figure 1). These data indicate that the entire CBC is required for in vivo interaction with HapX.
Figure 3.
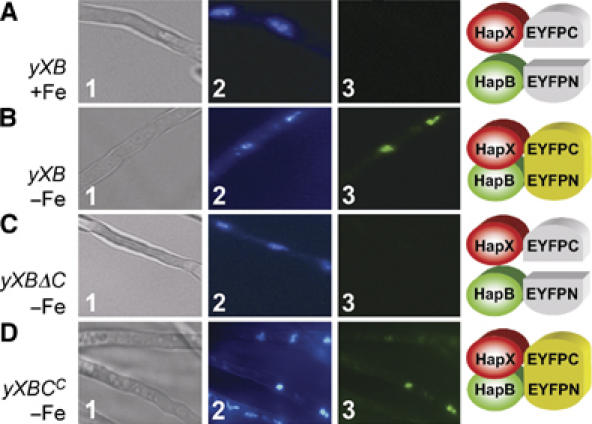
HapX and HapB interact in vivo. The interaction was observed after 24 h of growth, using BiFC in A. nidulans strains producing HapX and HapB fused with the C-terminal and N-terminal split fragments of eYFP, respectively. Panels 1, light microscopy; panels 2 and 3, fluorescence microscopy of DAPI-stained nuclei and BiFC, respectively. HapX and HapB interact during −Fe (B) but not +Fe (A) conditions in strain yXB. (C) HapX/HapB interaction is abolished by deletion of hapC in strain yXBΔC and is (D) reconstituted after complementation of yXBΔC with hapC in yXBCc.
HapE contains two evolutionary conserved regions (Figure 4A). Domain B is conserved among all HapE orthologs and is essential for the assembly of the CBC, as shown for S. cerevisiae Hap5p (McNabb et al, 1997). In contrast, domain A is only conserved among fungal HapE orthologs and has been shown to be required for the recruitment of Hap4p in S. cerevisiae. Recently, Tanoue et al (2006) reported that only the B-domain is required for CBC-mediated transcriptional activation in A. nidulans. Here, we found that deletion of the non-conserved N-terminal region and the A-domain phenocopies hapX deletion, that is, wild type-like growth during +Fe conditions, but decreased growth rate, reduced TAFC production, and increased accumulation of FC and PpIX during −Fe conditions (Figure 4B). In contrast, truncation of the non-conserved C-terminal domain of HapE had no effect (Figure 4B). These data suggest that the region encompassing the N-terminus and the A-domain of HapE is involved in interaction with HapX in vivo.
Figure 4.
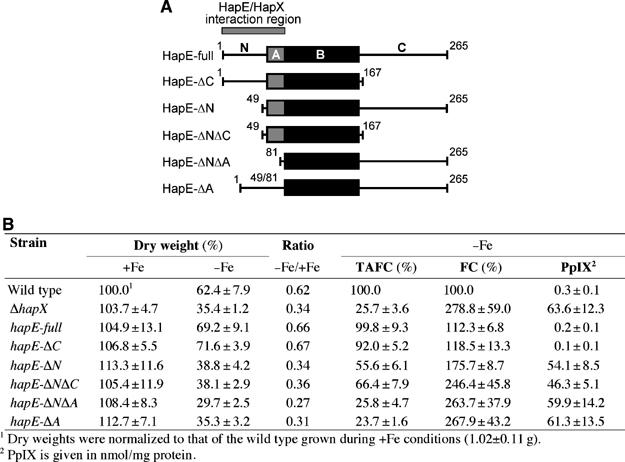
Deletion of the non-conserved N-terminal region or the conserved A-domain of HapE phenocopies hapX deletion. (A) Schematic representation of the HapE versions investigated. (B) Growth rates and production of TAFC, FC and PpIX after growth for 48 h during+Fe and −Fe conditions. For induction of amylase promoter-driven genes (Tanoue et al, 2006), strains were grown in medium containing starch as the sole carbon source. During +Fe conditions, production of TAFC, FC and PpIX was wild type-like in all strains (data not shown).
The 5′-upstream regions of the putative HapX/CBC target genes cycA, acoA, lysF, hemA and sreA all contain CCAAT boxes (Supplementary Figure 2). To study interaction of the CBC with the CCAAT sequences from promoter regions of iron-induced genes, we overproduced recombinant HapB, HapC, HapE, HapE-ΔNΔA and HapX in Escherichia coli and purified the proteins to homogeneity (Figure 5A). The CBC was reconstituted by mixing equimolar amounts of the CBC subunits. Real-time protein–DNA interaction analysis was performed with two 50-bp DNA duplexes immobilized on flow cells of a surface plasmon resonance (SPR) biosensor. The two sequences used covered the CCAAT box at position –1235 of the sreA promoter, which perfectly matches the consensus CBC-binding sequence (Mantovani, 1998), and the CCAAT box at position –181 of the lysF promoter, respectively. The apparent dissociation constants of the CBC were 1.8 and 4.6 nM for the CCAAT boxes from sreA and lysF, respectively, indicating specific and high-affinity binding. In contrast, HapX alone bound non-specifically, and with low affinity to sensor-bound DNA. (Figure 5B).
Figure 5.
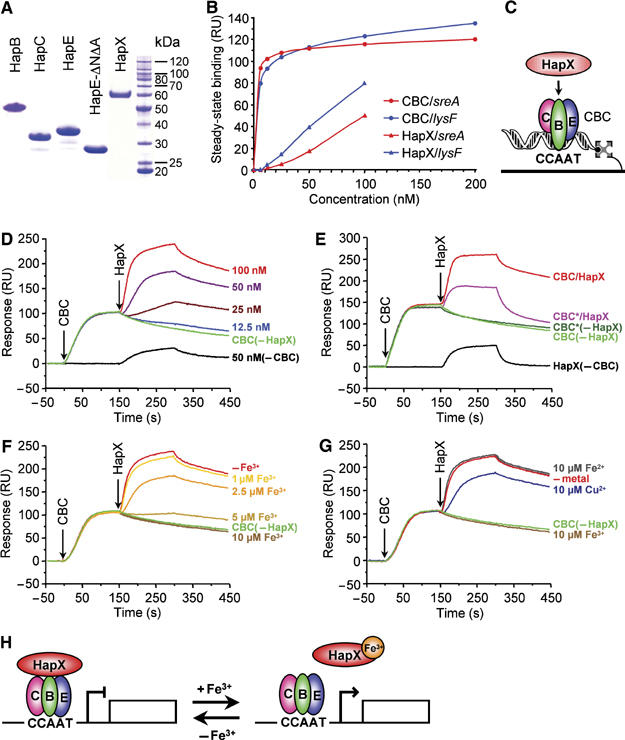
SPR analysis of iron-regulated HapX binding to DNA-bound CBC. (A) SDS–PAGE analysis of 1.5 μg of purified HapB, HapC, HapE, HapE-ΔNΔA and HapX proteins. (B) Concentration-dependent, steady-state binding of the CBC to biosensor-bound CCAAT boxes derived from the 5′-upstream regions of sreA and lysF, respectively. (C) Schematic representation of the SPR analysis of HapX/CBC interaction. HapX was injected onto preformed CBC/DNA complexes after reaching the steady-state level (D) Concentration-dependent association of HapX (12.5, 25, 50 and 100 nM, respectively) to the CBC (6.25 nM) bound to the biosensor-linked sreA CCAAT box. ‘CBC(−HapX)' shows the steady-state association of the CBC to the CCAAT box without application of HapX. ‘(−CBC)' shows the unspecific interaction of HapX (50 nM) with sensor-bound DNA. (E) Comparison of the interaction of HapX (100 nM) with the CBC and with the CBC containing HapE-ΔNΔA (CBC*). Note that 12.5 nM CBC* was necessary to reach an equilibrium response equivalent to 6.25 nM CBC. (F) Interaction of HapX (100 nM) after preincubation with iron (1, 2.5, 5 and 10 μM FeCl3, respectively) or without iron (−Fe), with the DNA-bound CBC (6.25 nM). (G) Interaction of HapX (100 nM) after preincubation with 10 μM FeCl3, 10 μM (NH4)2Fe(SO4)2, 10 μM CuCl2, or without any metal (−metal), with DNA-bound CBC (6.25 nM). (H) Proposed model for HapX/CBC-mediated regulation of iron-dependent pathways and sreA.
To investigate HapX/CBC interaction in vitro, HapX was injected onto the preformed CBC/DNA complex after reaching the steady-state level, as represented schematically in Figure 5C. Apart from its unspecific DNA binding, HapX bound with a remarkably high affinity to the CBC bound to the CCAAT box from the sreA promoter (Figure 5D). However, HapX did not bind to a CBC containing a HapE version that lacks the N-terminal region and the A-domain (HapE-ΔNΔA), that is, the measured response did not exceed the unspecific interaction of HapX with CBC-free DNA (Figure 5E). The latter agrees with the in vivo requirement for the N- and A-domains of HapE for repression of iron-dependent pathways during −Fe conditions (Figure 4B). Consistent with the observed lack of interaction between HapX and the CBC during +Fe conditions in vivo, the in vitro interaction was abolished upon preincubation of HapX with Fe3+ (Figure 5F). Notably, the interaction of HapX with the CBC was not abolished by preincubation with the same concentration of Fe2+ or Cu2+, indicating that the Fe3+ effect is specific (Figure 5G).
Identification of putative HapX targets
To identify other possible targets for HapX-dependent regulation, we examined the global effect of hapX deletion at the proteomic level. For this purpose, protein extracts of wild-type and ΔhapX strains grown for 19 h during −Fe conditions were compared by 2D-PAGE (Supplementary Figure 3). Reproducibly, 19 proteins displayed an increase and 23 proteins a decrease in their levels of more than 1.8-fold in the ΔhapX mutant. Some proteins appeared in gels as more than one spot with the same apparent molecular mass, but with different pI values and abundance, presumably due to post-translational modifications or isoenzyme variation, for example, 3-isopropylmalate dehydratase. Therefore, the 42 spots identified represented only 30 different proteins (Table II). Increased levels of 5-aminolevulinic acid synthase (HemA) and aconitase (AcoA) in ΔhapX agreed with the transcriptional upregulation of the encoding genes (Figure 1C). Further iron-related proteins with increased levels in ΔhapX during −Fe conditions were iron–sulfur cluster containing 3-isopropylmalate dehydratase, iron-containing 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase, the mitochondrial processing peptidase, the respiratory protein ATP synthase subunit D, and the respiratory heme-proteins cytochrome c peroxidase, and ubiquinol–cytochrome c reductase complex core protein 2. This finding suggests that the encoding genes are further targets for HapX-mediated repression. Moreover, deletion of hapX also resulted in decreased levels of some proteins, for example, several dehydrogenases, oxidoreductases and proteins involved in amino-acid metabolism.
Table 2.
Comparison of the proteome of the wild type and ΔhapX during −Fe conditions
| Putative function | Locus | pI/MW | Spot | Fold changes | Sequence coverage (%) | Identified peptides | Mascot score |
|---|---|---|---|---|---|---|---|
| (A) Proteins with higher levels in ΔhapX versus wild type | |||||||
| 3-Isopropylmalate dehydratase | AN5886.2 | 5.5/84 | 1 | +2.9 | 20 | 8 | 109 |
| 2 | +2.6 | 14 | 6 | 89.5 | |||
| 3 | +1.7 | 35 | 12 | 166 | |||
| 4 | +3.5 | 44 | 14 | 211 | |||
| 5 | +3.4 | 35 | 12 | 171 | |||
| 6 | +2.3 | 37 | 12 | 124 | |||
| Aconitate hydratase, mitochondrial | AN5525.2 | 5.9/83 | 7 | +2.1 | 17 | 8 | 95 |
| 8 | +2.0 | 39 | 17 | 177 | |||
| 9 | +1.7 | 26 | 12 | 95.7 | |||
| Cytochrome c peroxidase, mitochondrial precursor | AN1630 | 9.4/40 | 12 | +1.9 | 34 | 8 | 80.7 |
| Ubiquinol–cytochrome c reductase complex core protein 2 | AN8273.2 | 9.4/48 | 13 | +1.8 | 16 | 5 | 58.3 |
| 5-Aminolevulinic acid synthase | AN2284.2 | 9.5/69 | 14 | +19.2 | 21 | 8 | 118 |
| 9.5/69 | 15 | +2.1 | 30 | 10 | 160 | ||
| 9.5/69 | 16 | +2.5 | 34 | 8 | 188 | ||
| Phe-inhibited 3-deoxy-D-arabino-heptulosonate- 7-phosphate synthase (DAHP) | AAG36950 | 6.6/40 | 18 | +1.7 | 28 | 6 | 88.6 |
| ATP synthase D chain, mitochondrial | AN6631.2 | 7.2/20 | 22 | +3.1 | 49 | 4 | 98.3 |
| Cobalamin-independent methionine synthase | AAF82115 | 6.4/87 | 27 | +13.7 | 31 | 15 | 194 |
| Phosphoribosylaminoimidazolecarboxamide for myltransferase/IMP cyclohydrolase | AN4464.2 | 6.2/66 | 29 | +5.1 | 31 | 9 | 185 |
| Predicted RNA-binding protein | AN1044.2 | 4.3/66 | 30 | +3.1 | 21 | 4 | 82 |
| Mitochondrial processing peptidase β subunit | AN0747.2 | 5.5/53 | 31 | +2.3 | 33 | 8 | 105 |
| RNA recognition motif | AN6004.2 | 5.6/29 | 32 | +6.3 | 40 | 6 | 136 |
| (B) Proteins with lower levels in ΔhapX versus wild type | |||||||
| NADH-ubiquinone oxidoreductase 75 kDa subunit | AN9411.2 | 6.0/81 | 10 | −2.1 | 31 | 9 | 181 |
| Fe-containing alcohol dehydrogenase | AN1868.2 | 6.9/53 | 11 | −2.6 | 19 | 6 | 63.6 |
| 3-Hydroxy-3-methylglutaryl CoA synthase | AN4923.2 | 5.8/51 | 17 | −2.0 | 32 | 7 | 101 |
| Aspartic protease | AN2903.2 | 4.7/43 | 19 | −3.0 | 26 | 5 | 76.4 |
| Aminotransferase, classes I and II | AN5591.2 | 5.7/54 | 20 | −2.1 | 18 | 6 | 89.6 |
| Woronin body major protein | AN4695.2 | 6.8/25 | 21 | −3.2 | 51 | 3 | 101 |
| Hydantoinase/oxoprolinase | AN5652.2 | 5.2/141 | 23 | −3.5 | 29 | 17 | 260 |
| 24 | −2.2 | — | — | ||||
| 25 | −3.3 | 34 | 17 | 329 | |||
| 26 | −2.7 | 40 | 20 | 351 | |||
| Proteasome alpha subunit | AN4869.2 | 6.7/28 | 28 | −2.0 | 52 | 8 | 150 |
| GMC oxidoreductase | AN3206.2 | 5.4/63 | 33 | −2.7 | 16 | 6 | 91.3 |
| UDP-glucose pyrophosphorylase | AAW49005 | 7.9/58 | 34 | −2.3 | 39 | 12 | 157 |
| Alcohol dehydrogenase, zinc containing | AN8406.2 | 6.0/38 | 35 | −2.2 | 53 | 7 | 169 |
| Alcohol dehydrogenase, zinc containing | AN0443.2 | 6.2/40 | 36 | −2.0 | 56 | 10 | 189 |
| Haloacid dehalogenase-like hydrolase | AN4572.2 | 5.7/36 | 37 | −3.9 | 30 | 5 | 81 |
| Carboxyphosphonoenolpyruvate phosphonomutase | AN3805.2 | 5.6/32 | 38 | −6.5 | 49 | 8 | 133 |
| L-Xylulose reductase | AN7590.2 | 6.4/28 | 39 | −3.4 | 56 | 8 | 120 |
| Oxidoreductase, short-chain dehydrogenase/ reductase family | AN3161.2 | 9.6/37 | 40 | −4.1 | 29 | 7 | 91.4 |
| Transcription initiation factor subunit TAF30 | AN0083.2 | 8.7/35 | 41 | −2.6 | 25 | 5 | 82.6 |
| Hypothetical protein | AN1152.2 | 5.4/19 | 42 | −2.8 | 50 | 3 | 72.8 |
Deletion of sreA in combination with deletion of hapX or of hapB is lethal
To address the role of HapX in iron metabolism, we attempted to generate a mutant lacking both hapX and sreA, via a meiotic cross of strains ΔhapX and ΔsreA. With arginine supplementation, 26% of the progeny were hapX+/sreA+ recombinants, 40% hapX+/ΔsreA and 34% ΔhapX/sreA+; however, ΔhapX/ΔsreA recombinants were not recovered (Supplementary Table 1). Without arginine supplementation, hapX+/sreA+ recombinants cannot grow, due to the argB2 mutation of both parental strains. Because hapX and sreA are located on different chromosomes (II and VIII, respectively), 33% of the progeny was expected to be ΔhapX/ΔsreA double mutants. Among the 50 randomly chosen progeny screened by PCR (Supplementary Table 2), 27 had a ΔhapX/sreA+ genotype and 23 had a hapX+/ΔsreA genotype, but again ΔhapX/ΔsreA double mutants could not be recovered. The probability of not recovering the ΔhapX/ΔsreA double mutant without arginine supplementation is (1–0.33)50, or 2.0 × 10−9. In both cases, the three additional genetic markers (pyroA4, pyrG89 and wA3) segregated according to the Mendelian rules (Supplementary Table 1). These results strongly suggest that the deletion of both hapX and sreA is lethal.
To test the genetic interaction between SreA and the CBC, we generated a strain, designated ΔsreA/ΔhapB/hapBc, that contained deletions of the sreA and hapB loci along with an ectopic copy of the hapB gene controlled by the alcA promoter, by meiotically crossing strains ΔsreA and ΔhapB/hapBc (Steidl et al, 2004). Under inducing conditions, ΔsreA/ΔhapB/hapBc showed growth comparable to ΔsreA. In contrast, no growth was observed under conditions that cause repression of the alcA promoter, supporting the synthetic lethal interaction also between SreA and the CBC (data not shown).
Discussion
All organisms face constantly changing availability of the essential metal, iron. To avoid deleterious consequences caused by iron overload, as well as iron deficiency, organisms have evolved mechanisms that maintain iron homeostasis. During iron deprivation, A. nidulans cells undergo a transcriptional remodeling, leading to the inverse regulation of two major sets of genes. On one hand, iron acquisition, for example, siderophore-mediated iron uptake, is upregulated (Haas, 2003). On the other hand, many iron-dependent pathways, including proteins involved in the tricarboxylic acid cycle, respiration and heme biosynthesis, are downregulated (Oberegger et al, 2002a). The latter pathways represent oxidative metabolism, which largely depends on iron.
We show that various iron-dependent pathways are repressed during −Fe conditions, by the interaction of HapX with the CBC. The lines of evidence are: (i) hapX and CBC subunit-encoding genes are required for repression of genes involved in oxidative metabolism during −Fe conditions and their deletion causes PpIX accumulation; (ii) HapX physically interacts with the CBC in vitro and in vivo only under –Fe conditions; (iii) HapX/CBC binds to the CCAAT boxes of at least two HapX/CBC target genes, sreA and lysF; (iv) the non-conserved N-terminal region and the A-domain of HapE are required for in vitro interaction of the CBC with HapX. In agreement, strains carrying HapE versions with N-terminal deletions phenocopy hapX deletion in vivo.
Proteome analysis confirmed some of the HapX/CBC targets detected by transcriptional analysis, identified some more obviously iron-related targets, and revealed putative HapX targets not previously shown to be affected by iron metabolism. Moreover, we found that HapX/CBC formally acts as a repressor of TAFC production, but not via transcriptional control of known siderophore biosynthesis genes, and as a repressor of FC production, which agrees with increased transcript levels of FC-biosynthetic sidC in ΔhapX and ΔhapC mutants.
Deletion of hapX is deleterious only during −Fe conditions. In contrast, deletion of genes encoding CBC subunits has deleterious consequences during both +Fe and −Fe conditions. Consistently, expression of hapX is confined to −Fe conditions, but that of the CBC subunit-encoding genes is constitutive, which is achieved by negative auto-regulation of the hapB gene (Steidl et al, 2001). Furthermore, hapB deletion is epistatic to hapX deletion, that is, a ΔhapX/ΔhapB double mutant displayed a phenotype identical to ΔhapB (data not shown). HapX-independent functioning of the CBC is also indicated by the CBC-independent nuclear localization of HapX (Goda et al, 2005), that is, HapC and HapE, but not HapX, depend on a HapB-mediated ‘piggy-back' nuclear transport (Steidl et al, 2004). Moreover, deletion of hapC, but not of hapX, caused upregulation of iron-dependent pathways during +Fe conditions. Together, these data show that the CBC has a role independent of the iron status, whereas HapX/CBC function appears to be confined to −Fe conditions. The more general effect of the CBC on gene regulation, compared to HapX/CBC, is probably also the explanation for the different extent of deregulation of TAFC, FC and PpIX production in hapX and CBC subunit-encoding gene deletion strains.
In C. albicans, CBC-mediated repression of respiration was reported (Johnson et al, 2005), and a putative ortholog of HapX was found to be subject to iron regulation mediated by Sfu1, the ortholog of A. nidulans SreA (Lan et al, 2004). Recently, mutual transcriptional control of the SreA ortholog Fep1 and Php4, which displays similarity to the S. cerevisiae Hap4p in the N-terminus, was found in S. pombe (Mercier et al, 2006). Moreover, Php4 and the CBC were shown to be required for repression of some iron-dependent pathways during −Fe conditions, but, in contrast to A. nidulans, the CBC is also required for transcriptional upregulation of the respective genes during +Fe conditions in this yeast. Nevertheless, these data indicate evolutionary conservation of HapX/CBC-mediated repression of oxidative metabolism in other fungal species. In striking contrast, in S. cerevisiae, the Hap4p/CBC complex acts as an inducer of oxidative metabolism (Pinkham and Guarente, 1985). In particular, the genes encoding cytochrome c, aconitase and 5-aminolevulinate synthase, which are repressed by the CBC in A. nidulans, are positively influenced in S. cerevisiae (Keng and Guarente, 1987; Pinkham et al, 1987; Liu and Butow, 1999). The downregulation of iron-dependent pathways in S. cerevisiae occurs by acceleration of the rate of mRNA decay, which is mediated by the RNA-binding protein Cth2p/Tis11p (Puig et al, 2005). Orthologs of Cth2p/Tis11p appear to be missing in the genomes of A. nidulans and all other filamentous fungal species. Taken together, these results suggest fundamental differences in the regulation of oxidative metabolism between S. cerevisiae and other fungi.
Intimate coupling of the regulatory mechanisms mediating the inverse regulation of iron-dependent pathways and iron acquisition systems is suggested by the synthetic lethality of double deletion of sreA and hapX, as well as of sreA and hapB. The requirement of at least one of these regulatory circuits suggests mutual compensation of essential functions. A tight interplay is further indicated by mutual transcriptional control between SreA and HapX. Here, we have shown that in vitro HapX/CBC binds to the sreA promoter, and SreA is able to bind to GATA sites of the hapX promoter (data not shown). However, the deregulation of hapX, as well as of sreA, had no impact on the regulation of the respective target genes. In agreement with these results, we have previously shown that constitutive expression of sreA does not repress siderophore biosynthesis (Haas et al, 1999), and the same is true for HapX targets in a strain constitutively expressing hapX (data not shown). These data suggest that the repressor function of both SreA and HapX/CBC requires additional factors and/or post-transcriptional modification. SreA might sense the iron status by an evolutionarily conserved cysteine-rich region, as suggested for other fungal SreA orthologs (Harrison and Marzluf, 2002; Pelletier et al, 2005). Remarkably, the putative HapX orthologs of Neurospora crassa, C. albicans and Cryptococcus neoformans also contain evolutionarily conserved cysteine-rich regions (although of different architecture as in SreA), which might be involved in sensing of iron (Supplementary Figure 4). Consistently, we found that in vitro and in vivo interaction of HapX with the CBC is abolished by iron. Taken together, these data suggest the following model (Figure 5H), in which HapX/CBC interaction is regulated at both transcriptional and post-translational levels. Iron starvation causes expression of hapX. Subsequent binding of HapX to the CBC results in transcriptional repression of iron-dependent pathways. During +Fe conditions, hapX is repressed and, therefore, iron-dependent pathways are derepressed. Moreover, HapX/CBC interaction is inhibited by increased iron concentrations. This post-translational mechanism allows rapid adjustment to iron availability by disruption of the HapX/CBC complex. In this respect it is interesting to note that the iron content of A. nidulans increases about 86-fold to approximately 20 μmol g−1 fungal dry weight during a shift from −Fe to +Fe conditions (Eisendle et al, 2006). Mutual transcriptional control of hapX and sreA coordinates iron acquisition and iron-dependent pathways, thereby serving for both iron supply and prevention of iron toxicity.
It will be interesting to analyze how iron directly influences the binding affinity of HapX to the CBC. As mentioned above, HapX contains three cysteine-rich domains, which are potential ligands for this metal. In comparison, S. cerevisiae Hap4p displays similarity to HapX in the N-terminal region, which interacts with the CBC, but lacks the bZip domain and cysteine-rich motifs (Tanaka et al, 2002).
In contrast to oxidative metabolism, utilization of secondary carbon and nitrogen sources (e.g., acetamide, formamide, γ-aminobutyrate and starch), as well as the production of the secondary metabolite penicillin are positively regulated by the CBC in Aspergillus (Brakhage et al, 1999; Kato, 2005). This might indicate that CBC-mediated regulation coordinates oxidative metabolism, utilization of secondary carbon and nitrogen sources, as well as secondary metabolism.
Materials and methods
Strains, oligonucleotides, media and growth conditions
The A. nidulans strains and oligonucleotides used in this study are listed in Table III and Supplementary Table 2, respectively. Generally, fungal strains were grown at 37°C in Aspergillus minimal medium (AMM), according to Pontecorvo et al (1953), containing 1% (w/v) glucose as the carbon source, 20 mM glutamine as the nitrogen source, 10 μM FeSO4 as the iron source and the respective supplements (+Fe conditions). Addition of iron was omitted for creating −Fe conditions. BiFC analyses were performed after growth of fungi on coverslips submerged in liquid medium. Under these conditions, biomass production is low and therefore additional treatment with 1 mM deferoxamine mesylate salt for 1 h was essential to cause iron starvation. The amylase promoter of hapE versions and the alcA promoter were induced using AMM with 1% (w/v) starch and with 3% (w/v) lactose plus 10 mM threonine as the carbon source, respectively. For repression of the alcA promoter, 1% (w/v) glucose was used. Shake flask culture (180 r.p.m.) included inoculation of 108 conidia in 200 ml of medium, in 1.0 l Erlenmeyer flasks.
Table 3.
Fungal strains used in this study
| Strain designation in text | Strain | Genotype | Reference |
|---|---|---|---|
| Gene regulation, synthetic lethality, epistasis | |||
| Wild type | BPU1 | pyrG89; biA; wA3; argB2; pyroA4; pAR5 (argB); ArgB+ | Tanaka et al (2002) |
| Wild type | TRAN | argB2; bgA0; biA1; argB2∷pTRAN; ArgB+ | Haas et al (2003) |
| ΔsreA | SRKO1 | argB2; bgA0; biA; sreAΔ∷argB; ArgB+ | Haas et al (1999) |
| ΔhapB | ΔB-HapEegfp | pyrG89; pabaA1; fwA1; hapBΔ∷argB; pHapE-GFP; PyrG+ | Steidl et al (2004) |
| ΔhapC | Nat24 | pyrG89; pabaA1; riboB; hapCΔ∷riboB; RiboB+ | Steidl et al (2004) |
| ΔhapE | pyrG89; biA; wA3; argB2; pyroA4; pabaA1; hapEΔ∷argB; pTG-Taa; PyrG+; ArgB+ | Tanoue et al (2006) | |
| ΔhapX | BPUΔX1 | pyrG89; biA; wA3; argB2; pyroA4; hapXΔ∷argB; ArgB+ | Tanaka et al (2002) |
| ΔhapB/hapBc | ΔB-HapEegfp-pALB | pyrG89; pabaA1; fwA1; hapBΔ∷argB; pHapE-GFP; pAL4H6HapB (alcAp-hapB); ArgB+; PyrG+ | Steidl et al (2004) |
| ΔsreA/ΔhapB/ hapBc | pyrG89; pabaA1; biA; wA3; argB2; sreAΔ∷argB; hapBΔ∷argB; pHapE-GFP; PyrG+; pAL4H6HapB (alcAp-hapB) | This study | |
| ΔhapX/ΔhapB | pyrG89; yA; argB2; pyroA4; hapXΔ∷argB; hapBΔ∷argB; ArgB+ | This study | |
| hapXR | pyrG89; biA; wA3; argB2; pyroA4; hapXΔ∷argB; ArgB+; pCAME703-AoHapX-full; PyrG+; pSK275 (PtrA+) | This study | |
| BiFC analysis | |||
| AXB4A2 | pyrG89, pabaA1; argB2; fwA1; bga0; argB2∷pAXB4A (acvAp-uidA; ipnAp-lacZ); ArgB+ | Weidner et al (1998) | |
| yXB | yHapX-HapB | pyrG89, pabaA1; argB2; fwA1; bga0; argB2∷pAXB4A (acvAp-uidA, ipnAp-lacZ); ArgB+; pHapX-YC-pyr4; PyrG+; pHapB-YN | This study |
| yXBΔC | yHapX-HapB-ΔC | pyrG89; pabaA1; riboB; hapCΔ∷riboB; RiboB+; pHapX-YC-pyr4; PyrG+; pHapB-YN | This study |
| yXBCC | yHapX-HapB-HapCC | pyrG89; pabaA1; riboB; hapCΔ∷riboB; RiboB+; pyr-4∷pHapX-YC-pyr4; PyrG+; pHapB-YN; hapC∷pAlcA-HapC; HapC+; pabaA1∷PabaAnid; PabaA+ | This study |
| hapE truncation mutants | |||
| hapE-full | pyrG89; biA; wA3; argB2; pyroA4; pabaA1; hapEΔ∷argB; pTG-Taa; PyrG+; ArgB+; pCAME3M-AohapE-Full | Tanoue et al (2006) | |
| hapE-ΔC | pyrG89; biA; wA3; argB2; pyroA4; pabaA1; hapEΔ∷argB; pTG-Taa; PyrG+; ArgB+; pCAME3M-AohapE-ΔC | Tanoue et al (2006) | |
| hapE-ΔN | pyrG89; biA; wA3; argB2; pyroA4; pabaA1; hapEΔ∷argB; pTG-Taa; PyrG+; ArgB+; pCAME3M-AohapE-ΔN | Tanoue et al (2006) | |
| hapE-ΔNΔC | pyrG89; biA; wA3; argB2; pyroA4; pabaA1; hapEΔ∷argB; pTG-Taa; PyrG+; ArgB+; pCAME3M-AohapE-ΔNΔC | Tanoue et al (2006) | |
| hapE-ΔNΔA | pyrG89; biA; wA3; argB2; pyroA4; pabaA1; hapEΔ∷argB; pTG-Taa; PyrG+; ArgB+; pCAME3M-AohapE-ΔNΔA | Tanoue et al (2006) | |
| hapE-ΔA | pyrG89; biA; wA3; argB2; pyroA4; pabaA1; hapEΔ∷argB; pTG-Taa; PyrG+; ArgB+; pCAME3M-AohapE-ΔA | Tanoue et al (2006) | |
Complementation of ΔhapX
A. nidulans hapXR was generated by ectopic integration of a single copy of the hapX-containing plasmid pCAME703-AoHapX-full (Goda et al, 2005), via co-transformation with plasmid pSK275, which carries the pyrithiamine resistance gene ptrA. The screening was performed by selection for pyrithiamine resistance and hapX integration by PCR (primer: o1_ hapX/o3_ hapX and o3_ hapX/o2_ argB) as well as Southern analysis.
Northern analysis, siderophores analysis and sexual crosses
Northern analysis as well as purification and analysis of TAFC and FC were carried out as previously described (Oberegger et al, 2001). Sexual crosses were performed according to Pontecorvo et al (1953).
Fluorescence analysis, PpIX analysis
Red auto-fluorescence was visualized with a Zeiss Axioplan fluorescence microscope with appropriate filters (excitation/emission at 546/590 nm). A digital Zeiss Axiocam MRc camera (Carl Zeiss AG) was used for documentation. Porphyrins were quantified by HPLC with UV and fluorescence detection, according to Bonkovsky et al (1986), and normalized to the sample protein content.
For all other fluorescence microscopic studies, a Leica DM4500 B digital fluorescence microscope (Leica Microsystems) was employed, using filtercubes A, GFP and YFP for nuclear staining, GFP localization studies and BiFC analysis, respectively. Nuclei were stained with 4′,6-diamidino-2-phenylindol (DAPI) for 1 min. For documentation, a Leica DFC480 digital camera (Leica Microsystems) was used. Photographs were processed with Photoshop 5.5 (Adobe Systems).
Proteome analysis
Proteome analysis of A. nidulans, including sample preparation, 2D-chromatography, protein visualization, quantification and MS identification of proteins, was essentially carried out as previously described for A. fumigatus (Kniemeyer et al, 2006). Gel images (n=12) were analyzed using the software Delta 2D 3.3 (Decodon). Afterwards, background subtraction spot volumes were normalized against total spot volume and total spot area. Spot values were logarithmically transformed and regarded as differentially regulated with a t-test value P<0.05. The MALDI-TOF data were used to search the NCBI database, using the Mascot algorithm (Matrix Science), with the following parameters: Cys as S-carbamidomethyl derivative and Met in oxidized form (variable), one missed cleavage site, peptide mass tolerance of 200 ppm.
Purification of HapB, HapC, HapE, HapE-ΔNΔA and HapX
Genes encoding N-terminally (His)6-tagged versions of full-length HapX, HapB, HapE and a HapE version with a deletion of the first 80 amino acids (HapE-ΔNΔA) were expressed using vector pET-43.1a (Novagen). A gene encoding full-length HapC with an extended N-terminus including maltose-binding protein (MBP), a (His)6 tag and a cleavage site for tobacco etch virus (TEV) protease was expressed using vector pMAL-c2X (New England Biolabs). All recombinant proteins were produced by auto-induction in E. coli Rosetta 2 (DE3) cells grown on Overnight Express Instant TB Medium (Novagen), and were subsequently purified to homogeneity, as described in detail in the Supplementary data. The MBP domain was removed from HapC by TEV protease cleavage.
SPR analysis
Real-time analysis was performed on a Biacore 2000 system at 25°C. Data were processed with the BIAevaluation software version 4.1 (Biacore). The running buffer used for DNA immobilization and the SPR analysis was 10 mM HEPES pH 7.4, 0.15 M NaCl, 1 mM DTT, 0.005% (v/v) surfactant P20. Refractive index errors due to bulk solvent effects were corrected by subtracting responses from the non-coated flow cell 1. Sample injection and dissociation time was set to 2.5 min at a flow rate of 30 μl/min.
DNA duplexes containing the CCAAT boxes from the sreA (−1257 to −1208) and lysF (−200 to −151) promoter regions were generated on an SA sensor chip on flow cells 2 and 3 by injection of 5′-biotinylated 50-bp oligonucleotides B-SREAC1i and B-LYSFC1i (5 nM), at a flow rate of 10 μl/min until 20 RU had been bound. This was followed by injection of oligonucleotides SREAC1 and LYSFC1 (1 μM) until formation of a total of 40 RU DNA duplexes. The CBCs were preformed from the single HapC, HapE (or HapE-ΔNΔA) and HapB proteins by mixing 0.1 mM solutions of each subunit. Samples for SPR analysis were generated by 500-fold dilution of this stock solution in running buffer, followed by serial two-fold dilution.
To avoid possible oxidation reactions caused by DTT-mediated reduction of Fe3+ in HEPES buffer (Spear and Aust, 1998), the running buffer for analyses of CBC/HapX interaction in the presence of metal salts contained 10 mM phosphate buffer pH 7.4, 2.7 mM KCl, 137 mM NaCl, 1 mM DTT and 0.005%(v/v) surfactant P20. For metal treatment of HapX, freshly prepared stock solutions of FeCl3, (NH4)2Fe(SO4)2 or CuCl2 were used.
Regeneration of sensor chips was achieved by treatment with the respective running buffer containing additionally 0.5 M NaCl and 0.01% (w/v) SDS for 1 min. Dissociation constants were calculated from the concentration-dependent steady-state binding of the CBCs, using the 1:1 steady-state affinity model.
BiFC analysis
The hapB gene was amplified by PCR using primers HapB 5′NcoI and HapB 3′NotI, which also inserted NcoI and NotI sites at the 5′ and 3′ ends, respectively. The NcoI/NotI-digested PCR product was inserted into plasmid pEYFPN (Hoff and Kück, 2005), yielding plasmid pYN-HapB, which encodes a C-terminal fusion of HapB with the N-terminal split fragment of eYFP. The hapX ORF was amplified by PCR using primers HapX 5′NcoI and HapX 3′NotI, using plasmid pCR2.1-HapX as the template. The NcoI/NotI-digested PCR product was inserted into plasmid pEYFPC to yield plasmid pYC-HapX, which encodes a C-terminal fusion of HapX with the C-terminal split fragment of eYFP.
Co-transformation of pYC-HapX and pYN-HapB was carried out in both the wild-type strain AXB4A2 and the ΔhapC strain. The resulting strains were designated yXB and yXBΔC, respectively. Strain yXBCC was generated by ectopical integration of plasmid pAlcA-HapC, which contains hapC under the control of the alcA promoter, into strain yXBΔC. Plasmid pAlcA-HapC was generated by integration of the hapC-containing BamHI/XbaI fragment from plasmid pHapC-Topo in plasmid pAL4 (Steidl et al, 2001).
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Table 1
Supplementary Table 2
Supplementary data
Acknowledgments
We thank Peter Mayser for helpful discussions and Petra Loitzl for technical assistance in the PpIX determination. Olaf Scheibner is greatfully acknowledged for identification of proteins using mass spectrometry. We thank Birgit Hoff and Ulrich Kück for the gift of BiFC plasmid pEYFPN and pEYFPC, as well as Gary Sawers und Kenton E Barnes for advice regarding the manuscript. This work was supported by Austrian Science Foundation grants FWF P-15959-B11 and FWF P-18606-B11 (HH) and FWF P-19764 (ERW), the Deutsche Forschungsgemeinschaft Priority Program 1152 (AAB), and the Grant-in-Aid for Scientific Research from the Japan Society for the Promotion of Science 16580057 (MK).
References
- Becker DM, Fikes JD, Guarente L (1991) A cDNA encoding a human CCAAT-binding protein cloned by functional complementation in yeast. Proc Natl Acad Sci USA 88: 1968–1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonkovsky HL, Wood SG, Howell SK, Sinclair PR, Lincoln B, Healey JF, Sinclair JF (1986) High-performance liquid chromatographic separation and quantitation of tetrapyrroles from biological materials. Anal Biochem 155: 56–64 [DOI] [PubMed] [Google Scholar]
- Bourgarel D, Nguyen CC, Bolotin-Fukuhara M (1999) HAP4, the glucose-repressed regulated subunit of the HAP transcriptional complex involved in the fermentation-respiration shift, has a functional homologue in the respiratory yeast Kluyveromyces lactis. Mol Microbiol 31: 1205–1215 [DOI] [PubMed] [Google Scholar]
- Bradshaw RE, Dixon SW, Raitt DC, Pillar TM (1993) Isolation and nucleotide sequence of the 5-aminolevulinate synthase gene from Aspergillus nidulans. Curr Genet 23: 501–507 [DOI] [PubMed] [Google Scholar]
- Brakhage AA, Andrianopoulos A, Kato M, Steidl S, Davis MA, Tsukagoshi N, Hynes MJ (1999) HAP-like CCAAT-binding complexes in filamentous fungi: implications for biotechnology. Fungal Genet Biol 27: 243–252 [DOI] [PubMed] [Google Scholar]
- Bucher P (1990) Weight matrix descriptions of four eukaryotic RNA polymerase II promoter elements derived from 502 unrelated promoter sequences. J Mol Biol 212: 563–578 [DOI] [PubMed] [Google Scholar]
- Edwards D, Murray JA, Smith AG (1998) Multiple genes encoding the conserved CCAAT-box transcription factor complex are expressed in Arabidopsis. Plant Physiol 117: 1015–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisendle M, Oberegger H, Zadra I, Haas H (2003) The siderophore system is essential for viability of Aspergillus nidulans: functional analysis of two genes encoding l-ornithine N 5-monooxygenase (sidA) and a non-ribosomal peptide synthetase (sidC). Mol Microbiol 49: 359–375 [DOI] [PubMed] [Google Scholar]
- Eisendle M, Schrettl M, Kragl C, Muller D, Illmer P, Haas H (2006) The intracellular siderophore ferricrocin is involved in iron storage, oxidative-stress resistance, germination, and sexual development in Aspergillus nidulans. Eukaryot Cell 5: 1596–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goda H, Nagase T, Tanoue S, Sugiyama J, Steidl S, Tuncher A, Kobayashi T, Tsukagoshi N, Brakhage AA, Kato M (2005) Nuclear translocation of the heterotrimeric CCAAT binding factor of Aspergillus oryzae is dependent on two redundant localising signals in a single subunit. Arch Microbiol 184: 93–100 [DOI] [PubMed] [Google Scholar]
- Haas H (2003) Molecular genetics of fungal siderophore biosynthesis and uptake: the role of siderophores in iron uptake and storage. Appl Microbiol Biotechnol 62: 316–330 [DOI] [PubMed] [Google Scholar]
- Haas H, Schoeser M, Lesuisse E, Ernst JF, Parson W, Abt B, Winkelmann G, Oberegger H (2003) Characterization of the Aspergillus nidulans transporters for the siderophores enterobactin and triacetylfusarinine C. Biochem J 371: 505–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas H, Zadra I, Stoffler G, Angermayr K (1999) The Aspergillus nidulans GATA factor SREA is involved in regulation of siderophore biosynthesis and control of iron uptake. J Biol Chem 274: 4613–4619 [DOI] [PubMed] [Google Scholar]
- Halliwell B, Gutteridge JM (1984) Oxygen toxicity, oxygen radicals, transition metals and disease. Biochem J 219: 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison KA, Marzluf GA (2002) Characterization of DNA binding and the cysteine rich region of SRE, a GATA factor in Neurospora crassa involved in siderophore synthesis. Biochemistry 41: 15288–15295 [DOI] [PubMed] [Google Scholar]
- Hink MA, Bisselin T, Visser AJ (2002) Imaging protein–protein interactions in living cells. Plant Mol Biol 50: 871–883 [DOI] [PubMed] [Google Scholar]
- Hoff B, Kück U (2005) Use of bimolecular fluorescence complementation to demonstrate transcription factor interaction in nuclei of living cells from the filamentous fungus Acremonium chrysogenum. Curr Genet 47: 132–138 [DOI] [PubMed] [Google Scholar]
- Hooft van Huijsduijnen R, Li XY, Black D, Matthes H, Benoist C, Mathis D (1990) Co-evolution from yeast to mouse: cDNA cloning of the two NF-Y (CP-1/CBF) subunits. EMBO J 9: 3119–3127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu CD, Kerppola TK (2003) Simultaneous visualiation of multiple protein interaction in living cells using multicolor fluorescence complementation analysis. Nat Biotechnol 21: 539–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DC, Cano KE, Kroger EC, McNabb DS (2005) Novel regulatory function for the CCAAT-binding factor in Candida albicans. Eukaryot Cell 4: 1662–1676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M (2005) An overview of the CCAAT-box binding factor in filamentous fungi: assembly, nuclear translocation, and transcriptional enhancement. Biosci Biotechnol Biochem 69: 663–672 [DOI] [PubMed] [Google Scholar]
- Keng T, Guarente L (1987) Constitutive expression of the yeast HEM1 gene is actually a composite of activation and repression. Proc Natl Acad Sci USA 84: 9113–9117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kniemeyer O, Lessing F, Scheibner O, Hertweck C, Brakhage AA (2006) Optimisation of a 2-D gel electrophoresis protocol for the human-pathogenic fungus Aspergillus fumigatus. Curr Genet 49: 178–189 [DOI] [PubMed] [Google Scholar]
- Lan CY, Rodarte G, Murillo LA, Jones T, Davis RW, Dungan J, Newport G, Agabian N (2004) Regulatory networks affected by iron availability in Candida albicans. Mol Microbiol 53: 1451–1469 [DOI] [PubMed] [Google Scholar]
- Li Q, Herrler M, Landsberger N, Kaludov N, Ogryzko VV, Nakatani Y, Wolffe AP (1998) Xenopus NF-Y pre-sets chromatin to potentiate p300 and acetylation-responsive transcription from the Xenopus hsp70 promoter in vivo. EMBO J 17: 6300–6315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littlejohn TG, Hynes MJ (1992) Analysis of the site of action of the amdR product for regulation of the amdS gene of Aspergillus nidulans. Mol Gen Genet 235: 81–88 [DOI] [PubMed] [Google Scholar]
- Litzka O, Then Bergh K, Brakhage AA (1996) The Aspergillus nidulans penicillin-biosynthesis gene aat (penDE) is controlled by a CCAAT-containing DNA element. Eur J Biochem 238: 675–682 [DOI] [PubMed] [Google Scholar]
- Liu Z, Butow RA (1999) A transcriptional switch in the expression of yeast tricarboxylic acid cycle genes in response to a reduction or loss of respiratory function. Mol Cell Biol 19: 6720–6728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maity SN, Vuorio T, de Crombrugghe B (1990) The B subunit of a rat heteromeric CCAAT-binding transcription factor shows a striking sequence identity with the yeast Hap2 transcription factor. Proc Natl Acad Sci USA 87: 5378–5382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani R (1998) A survey of 178 NF-Y binding CCAAT boxes. Nucleic Acids Res 26: 1135–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNabb DS, Pinto I (2005) Assembly of the Hap2p/Hap3p/Hap4p/Hap5p–DNA complex in Saccharomyces cerevisiae. Eukaryot Cell 4: 1829–1839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNabb DS, Tseng KA, Guarente L (1997) The Saccharomyces cerevisiae Hap5p homolog from fission yeast reveals two conserved domains that are essential for assembly of heterotetrameric CCAAT-binding factor. Mol Cell Biol 17: 7008–7018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNabb DS, Xing Y, Guarente L (1995) Cloning of yeast HAP5: a novel subunit of a heterotrimeric complex required for CCAAT binding. Genes Dev 9: 47–58 [DOI] [PubMed] [Google Scholar]
- Mercier A, Pelletier B, Labbe S (2006) A transcription factor cascade involving Fep1 and the CCAAT-binding factor Php4 regulates gene expression in response to iron deficiency in the fission yeast Schizosaccharomyces pombe. Eukaryot Cell 5: 1866–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder W, Scholten IH, de Boer RW, Grivell LA (1994) Sequence of the HAP3 transcription factor of Kluyveromyces lactis predicts the presence of a novel 4-cysteine zinc-finger motif. Mol Gen Genet 245: 96–106 [DOI] [PubMed] [Google Scholar]
- Oberegger H, Schoeser M, Zadra I, Abt B, Haas H (2001) SREA is involved in regulation of siderophore biosynthesis, utilization and uptake in Aspergillus nidulans. Mol Microbiol 41: 1077–1089 [DOI] [PubMed] [Google Scholar]
- Oberegger H, Schoeser M, Zadra I, Schrettl M, Parson W, Haas H (2002a) Regulation of freA, acoA, lysF, and cycA expression by iron availability in Aspergillus nidulans. Appl Environ Microbiol 68: 5769–5772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberegger H, Zadra I, Schoeser M, Abt B, Parson W, Haas H (2002b) Identification of members of the Aspergillus nidulans SREA regulon: genes involved in siderophore biosynthesis and utilization. Biochem Soc Trans 30: 781–783 [DOI] [PubMed] [Google Scholar]
- Oide S, Moeder W, Haas H, Krasnoff S, Gibson D, Yoshioka K, Turgeon BG (2006) NPS6, encoding a non-ribosomal peptide synthetase involved in siderophore-mediated iron metabolism, is a conserved virulence determinant of plant pathogenic ascomycetes. Plant Cell 18: 2836–2853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier B, Trott A, Morano KA, Labbe S (2005) Functional characterization of the iron-regulatory transcription factor Fep1 from Schizosaccharomyces pombe. J Biol Chem 280: 25146–25161 [DOI] [PubMed] [Google Scholar]
- Pinkham JL, Guarente L (1985) Cloning and molecular analysis of the HAP2 locus: a global regulator of respiratory genes in Saccharomyces cerevisiae. Mol Cell Biol 5: 3410–3416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkham JL, Olesen JT, Guarente LP (1987) Sequence and nuclear localization of the Saccharomyces cerevisiae HAP2 protein, a transcriptional activator. Mol Cell Biol 7: 578–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontecorvo G, Roper JA, Hemmons LM, Macdonald KD, Bufton AW (1953) The genetics of Aspergillus nidulans. Adv Genet 5: 141–238 [DOI] [PubMed] [Google Scholar]
- Puig S, Askeland E, Thiele DJ (2005) Coordinated remodeling of cellular metabolism during iron deficiency through targeted mRNA degradation. Cell 120: 99–110 [DOI] [PubMed] [Google Scholar]
- Schrettl M, Bignell E, Kragl C, Joechl C, Rogers T, Arst HN Jr, Haynes K, Haas H (2004) Siderophore biosynthesis but not reductive iron assimilation is essential for Aspergillus fumigatus virulence. J Exp Med 200: 1213–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spear N, Aust D (1998) The effects of different buffers on the oxidation of DNA by thiols and ferric ion. J Biochem Mol Toxicol 12: 125–132 [DOI] [PubMed] [Google Scholar]
- Steidl S, Hynes MJ, Brakhage AA (2001) The Aspergillus nidulans multimeric CCAAT binding complex AnCF is negatively autoregulated via its hapB subunit gene. J Mol Biol 306: 643–653 [DOI] [PubMed] [Google Scholar]
- Steidl S, Papagiannopoulos P, Litzka O, Andrianopoulos A, Davis MA, Brakhage AA, Hynes MJ (1999) AnCF, the CCAAT binding complex of Aspergillus nidulans, contains products of the hapB, hapC, and hapE genes and is required for activation by the pathway-specific regulatory gene amdR. Mol Cell Biol 19: 99–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steidl S, Tuncher A, Goda H, Guder C, Papadopoulou N, Kobayashi T, Tsukagoshi N, Kato M, Brakhage AA (2004) A single subunit of a heterotrimeric CCAAT-binding complex carries a nuclear localization signal: piggy back transport of the pre-assembled complex to the nucleus. J Mol Biol 342: 515–524 [DOI] [PubMed] [Google Scholar]
- Sybirna K, Guiard B, Li YF, Bao WG, Bolotin-Fukuhara M, Delahodde A (2005) A new Hansenula polymorpha HAP4 homologue which contains only the N-terminal conserved domain of the protein is fully functional in Saccharomyces cerevisiae. Curr Genet 47: 172–181 [DOI] [PubMed] [Google Scholar]
- Tanaka A, Kato M, Nagase T, Kobayashi T, Tsukagoshi N (2002) Isolation of genes encoding novel transcription factors which interact with the Hap complex from Aspergillus species. Biochim Biophys Acta 1576: 176–182 [DOI] [PubMed] [Google Scholar]
- Tanoue S, Kamei K, Goda H, Tanaka A, Kobayashi T, Tsukagoshi N, Kato M (2006) The region in a subunit of the Aspergillus CCAAT-binding protein similar to the HAP4p-recruiting domain of Saccharomyces cerevisiae Hap5p is not essential for transcriptional enhancement. Biosci Biotechnol Biochem 70: 782–787 [DOI] [PubMed] [Google Scholar]
- Then Bergh KT, Litzka O, Brakhage AA (1996) Identification of a major cis-acting DNA element controlling the bidirectionally transcribed penicillin biosynthesis genes acvA (pcbAB) and ipnA (pcbC) of Aspergillus nidulans. J Bacteriol 178: 3908–3916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuncher A, Sprote P, Gehrke A, Brakhage AA (2005) The CCAAT-binding complex of eukaryotes: evolution of a second NLS in the HapB subunit of the filamentous fungus Aspergillus nidulans despite functional conservation at the molecular level between yeast, A. nidulans and human. J Mol Biol 352: 517–533 [DOI] [PubMed] [Google Scholar]
- Weidner G, d'Enfert C, Koch A, Mol PC, Brakhage AA (1998) Development of a homologous transformation system for the human pathogenic fungus Aspergillus fumigatus based on the pyrG gene encoding orotidine 5′-monophosphate decarboxylase. Curr Genet 33: 378–385 [DOI] [PubMed] [Google Scholar]
- Weidner G, Steidl S, Brakhage AA (2001) The Aspergillus nidulans homoaconitase gene lysF is negatively regulated by the multimeric CCAAT-binding complex AnCF and positively regulated by GATA sites. Arch Microbiol 175: 122–132 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Table 1
Supplementary Table 2
Supplementary data


