Abstract
Mixl1 is a member of the Mix/Bix family of paired-like homeodomain proteins and is required for proper axial mesendoderm morphogenesis and endoderm formation during mouse development. Mix/Bix proteins are transcription factors that function in Nodal-like signaling pathways and are themselves regulated by Nodal. Here, we show that Foxh1 forms a DNA-binding complex with Smads to regulate transforming growth factor β (TGFβ)/Nodal-dependent Mixl1 gene expression. Whereas Foxh1 is commonly described as a transcriptional activator, we observed that Foxh1-null embryos exhibit expanded and enhanced Mixl1 expression during gastrulation, indicating that Foxh1 negatively regulates expression of Mixl1 during early mouse embryogenesis. We demonstrate that Foxh1 associates with the homeodomain-containing protein Goosecoid (Gsc), which in turn recruits histone deacetylases to repress Mixl1 gene expression. Ectopic expression of Gsc in embryoid bodies represses endogenous Mixl1 expression and this effect is dependent on Foxh1. As Gsc is itself induced in a Foxh1-dependent manner, we propose that Foxh1 initiates positive and negative transcriptional circuits to refine cell fate decisions during gastrulation.
Keywords: Foxh1, gastrulation, Gsc, Mix1l, transcription
Introduction
During gastrulation, the three germ layers, ectoderm, mesoderm and endoderm, are generated. In mice, this is first apparent with the formation of the primitive streak at the posterior end of the epiblast near the embryonic/extraembryonic junction (Tam and Behringer, 1997). The primitive streak elongates towards the distal end of the epiblast and cells move through the streak to emerge as mesoderm and endoderm. The organizer or node, which is located at the anterior end of the primitive streak, is a specialized population of cells with axis-inducing abilities that also contributes to the formation of axial mesendoderm, prechordal mesoderm and notochord (Robb and Tam, 2004).
Numerous growth factors regulate gastrulation, including transforming growth factor β (TGFβ) superfamily members such as Nodal and Activin (Schier, 2003). Nodal, like other TGFβ ligands, initiates signaling through its interaction with a heteromeric complex of serine/threonine kinase receptors. It also requires the presence of the EGF-CFC co-receptors, Tdgf1 (Cripto) or Cfc1 (Cryptic) (Attisano and Wrana, 2002; Schier, 2003; Feng and Derynck, 2005). Following activation of the receptor complex, the signal is transmitted to Smad proteins, which translocate to the nucleus and interact with specific transcription factors to regulate the expression of target genes (Attisano and Wrana, 2002; Feng and Derynck, 2005). Loss- and gain-of-function studies in frog, fish and mice have demonstrated that several components of the Nodal signaling pathway are implicated in the establishment of mesoderm and endoderm as well as axial mesendoderm patterning (Schier, 2003).
Several transcription factors are targeted by Nodal signaling and function to regulate gastrulation. For example, Mix/Bix paired-like homeobox genes, including Xenopus Mix.2, zebrafish og9x/mezzo and mouse Mixl1 act downstream of Nodal-like signaling pathways to regulate both mesoderm and endoderm formation (Chen et al, 1997; Hart et al, 2002; Poulain and Lepage, 2002). In mice, Mixl1 is expressed in the primitive streak and emerging mesoderm at the onset of gastrulation and becomes restricted to the posterior primative streak at the early bud stage (Pearce and Evans, 1999; Robb et al, 2000). The important role played by Mixl1 during mouse development is revealed by the numerous defects displayed by Mixl1−/− mutant embryos, which include an enlarged primitive streak, abnormal anterior midline structures, absence of heart tube formation and defective gut morphogenesis.
Goosecoid (Gsc) is another paired-like homeodomain-containing transcription factor whose expression is regulated by Nodal-like signaling pathways that functions in gastrulation as well as in axial mesendoderm formation (Blum et al, 1992; Kinder et al, 2001). In mice, Gsc expression marks the onset of gastrulation and is first detected in the primitive streak. As gastrulation proceeds, Gsc expression is restricted to the anterior primitive streak and the anterior visceral endoderm (Blum et al, 1992; Belo et al, 1997). Although its role in early development remains unclear as Gsc-null mice present no overt gastrulation or axial midline defects (Rivera-Perez et al, 1995; Yamada et al, 1995), Gsc can function as a transcriptional repressor by directly binding to paired homeodomain-binding sites to repress transcription of its own promoter as well as those of other genes, including Xenopus brachyury (Xbra) and wnt8 (Xwnt8) (Latinkic et al, 1997; Danilov et al, 1998; Yao and Kessler, 2001). The N-terminus of Gsc comprises a conserved Goosecoid Engrailed Homology (GEH) domain, and in Drosophila, the domain has been shown to interact with Groucho and mediate the repressive activity of Gsc (Jimenez et al, 1999).
A key mediator of the Nodal signaling pathway is the forkhead/winged-helix transcription factor, Foxh1 (Attisano et al, 2001). Genetic ablation of Foxh1 in mice results in embryonic lethality and a range of defects, including a total lack of embryonic development, aberrant anterior primitive streak patterning, loss of anterior and midline structures, and failure to form definitive endoderm (Hoodless et al, 2001; Yamamoto et al, 2001). In Xenopus and zebrafish, loss of Foxh1 activity results in anterior and axial defects as well as aberrant mesoderm development (Schier, 2003). Thus, loss of Foxh1 activity mimics numerous phenotypes observed in mutants where components of the Nodal signaling pathway have been disrupted (Schier, 2003). In general, Foxh1 binds directly to DNA and cooperates with Smad2/4 complexes to activate Nodal-dependent expression of target genes such as the TGFβ family members, Nodal and Lefty2 and the homeobox factors, Mix.2, Gsc and Pitx2 (Chen et al, 1997; Labbé et al, 1998; Saijoh et al, 2000; Shiratori et al, 2001). However, Foxh1-dependent induction of the Mef2c gene in the anterior heart field also requires cooperation with Nkx2-5, a heart-specific homeodomain transcription factor (von Both et al, 2004). Here, we demonstrate that recruitment of Gsc to the Mixl1 promoter by Foxh1 represses Mixl1 expression. Thus, our work reveals that Foxh1 can function either positively or negatively to control target gene expression and we propose that this precise control of gene expression contributes to cell fate determination during gastrulation.
Results
Foxh1 and Smads mediate TGFβ/Activin-dependent transcription of Mixl1
Activin-dependent induction of the Xenopus Mix.2 gene, a Mix/Bix family member, occurs via Foxh1, a DNA-binding forkhead protein, in complex with activated Smads (Chen et al, 1997). As expression of Foxh1 and Mixl1 appears to overlap during early mouse embryogenesis (Weisberg et al, 1998; Hart et al, 2002), we sought to determine whether Foxh1 might mediate the TGFβ/Activin-dependent transcriptional regulation of the murine Mixl1 gene. Examination of human, mouse, rat and rhesus monkey Mixl1 promoters revealed the presence of a putative Foxh1-binding site (Supplementary Figure 1), suggesting a conserved role for Foxh1 in Mixl1 transcription. Thus, to examine whether Foxh1 regulates Mixl1 expression, a 248-bp fragment from the murine Mixl1 promoter, encompassing the putative Foxh1 site, was subcloned upstream of a luciferase reporter gene (Mixl1-luc) and the response to TGFβ was determined in the human hepatocarcinoma cell line, HepG2, which lacks Foxh1 activity (Labbé et al, 1998). Co-expression of Foxh1 with the Mixl1-luc reporter yielded strong TGFβ-dependent activation of the Mixl1-luc reporter (Figure 1A). Direct binding of Foxh1 to the Mixl1 promoter fragment was confirmed by electrophoretic mobility shift assays (EMSA) using bacterially expressed Foxh1 (Figure 1B). Moreover, Smad2 and Smad4 enhanced both the basal and TGFβ-induced Foxh1-dependent activation of the Mixl1-luc reporter (Figure 1C). Although expression of Smad3 and Smad4 also increased the basal Foxh1-mediated activation of the Mixl1 promoter, TGFβ-dependent responsiveness was lost (Figure 1C).
Figure 1.
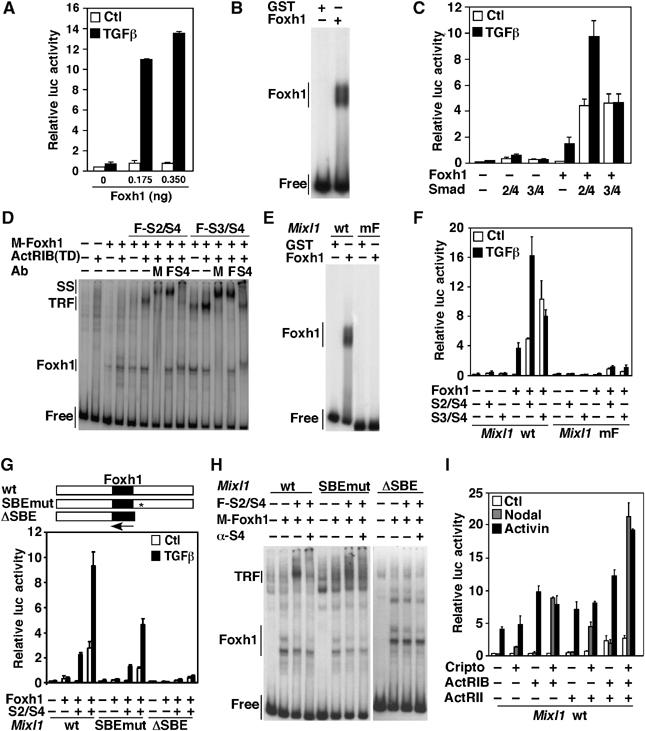
Foxh1 and Smads bind the Mixl1 promoter and mediate the TGFβ-dependent induction of Mixl1. (A, C, F, G, I) HepG2 cells were transiently transfected with the Mixl1-luc reporter, Foxh1 and Smads (S2, S3, S4), Cripto, ActRIB or ActRII, as indicated. Error bars represent standard deviation of the mean. (B, D, E, H) EMSA. A 248-bp Mixl1 promoter fragment containing a wild-type or mutated Foxh1-binding site was incubated with bacterially expressed proteins (B, E) or crude extracts from COS-1 cells transiently transfected with the indicated DNA (D, H). Protein–DNA complexes were visualized by autoradiography. For supershift assays (D, H), anti-myc (M), anti-Flag (F), and anti-Smad4 (S4) antibodies were added to the reactions.
We next examined whether Foxh1 and Smads cooperate to form a higher-order DNA-binding complex by EMSA. Comparison of DNA-binding complexes from mock-transfected COS-1 cells versus myc-Foxh1-expressing cells revealed the appearance of a slower migrating band in both the presence and absence of the activated Activin type I receptor, ActRIB(TD) (Figure 1D). Co-expression of Smad4 and either Smad2 or Smad3 with Foxh1 resulted in a further decrease in DNA migration, which was most evident in the presence of ActRIB(TD) (Figure 1D). Incubation with antibodies resulted in supershift or loss of DNA-binding complexes, demonstrating the presence Foxh1 and Smads in the TGFβ Responsive Factor (TRF) (Figure 1D). These observations indicate that Foxh1 can bind the Mixl1 promoter and, on activation of the signaling pathway, forms a DNA-binding complex with Smad2 or Smad3 and Smad4.
To confirm a requirement for Foxh1 binding, two point mutations that prevent Foxh1 binding (Labbé et al, 1998) were introduced in the putative Foxh1 site. These mutations abrogated Foxh1 binding to the Mixl1 promoter (Figure 1E) and abolished TGFβ-dependent signaling (Figure 1F). Smads bind to GC-rich sequences, and thus, to determine the Smad-binding requirements, we generated Mixl1 promoter constructs harboring either 8 GC to AT point mutations (SBEmut) or a complete deletion of a GC-rich region located downstream of the Foxh1 site (ΔSBE) (Figure 1G). The point mutations reduced, whereas complete deletion abolished, both TGFβ responsiveness and TRF formation on the promoter (Figure 1G and H). Thus, our results, in agreement with previous studies (Hart et al, 2005), show that Foxh1 and Smad DNA binding is required for maximal TGFβ-dependent activation of Mixl1.
Nodal induces transcriptional activation of Mixl1
Foxh1 expression is confined to early embryogenesis and is thought to be downstream of the Nodal signaling pathway (Schier, 2003). Nodal activates a TGFβ-like pathway by binding Activin receptors and the GPI-linked co-receptors, Cripto or Cryptic (Schier, 2003). To investigate whether Nodal signaling could induce Foxh1-dependent activation of the Mixl1 promoter, HepG2 cells were transiently transfected with the Mixl1-luc reporter and Foxh1 with various combinations of Activin type I and II receptors and the co-receptor, Cripto. Whereas co-transfection of Activin receptor type IB (ActRIB) alone or in the presence of Activin receptor type II (ActRII) mediated induction of the Mixl1 promoter on Activin treatment, Nodal-dependent activation of Mixl1 required co-expression of Cripto (Figure 1I), in agreement with a recent study (Hart et al, 2005). Taking into account that Nodal plays an essential role in the induction of mesendodermal cell fates (Schier, 2003) and that Mixl1 is implicated in axial mesendoderm morphogenesis and patterning (Hart et al, 2002), Foxh1-dependent induction of Mixl1 is most likely driven by Nodal signaling during embryogenesis.
Foxh1 negatively regulates Mixl1 expression during early mouse embryogenesis
To evaluate the contribution of Foxh1 to the regulation of Mixl1 transcription in vivo, we examined Mixl1 expression levels in wild-type and Foxh1 mutant (Foxh1−/−) embryos. RNA was extracted from genotyped pools of wild-type and Foxh1−/− embryos at embryonic day 7.5 (E7.5), and transcript levels were determined by quantitative PCR (QPCR) using primers specific for Mixl1 transcripts. In RNA extracted from a pool of Foxh1−/− embryos, Mixl1 expression was increased by almost twofold over levels expressed in wild-type embryos (Figure 2A). The level of Foxh1 expression and the absence of Foxh1 mRNA in our pool of wild-type and Foxh1−/− embryos, respectively, was confirmed by QPCR (Figure 2A).
Figure 2.
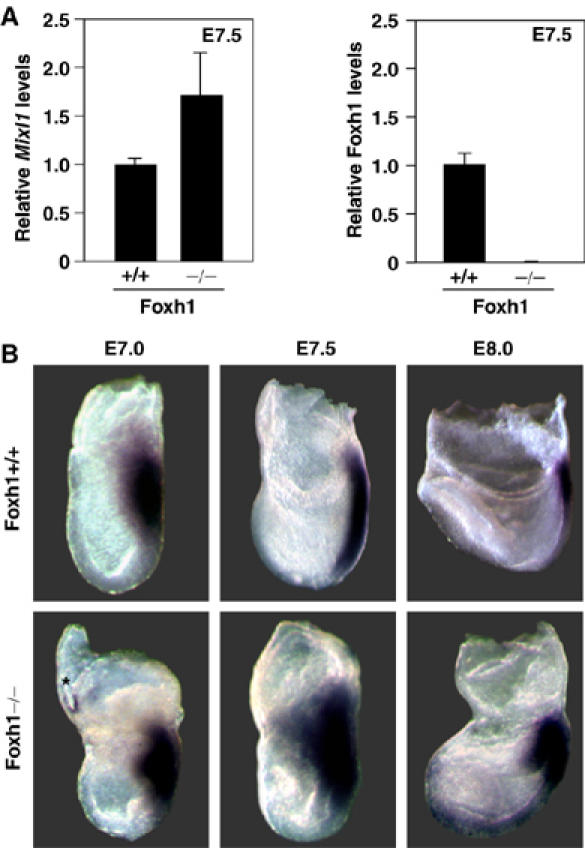
Mixl1 expression is upregulated in gastrulating Foxh1-null embryos. (A) RNA was extracted from E7.5 Foxh1+/+ and Foxh1−/− embryos. Mixl1 and Foxh1 levels were quantified by QPCR and normalized to Hprt expression. (B) At E7.0 and E7.5, Mixl1 expression is expanded and upregulated in Foxh1 mutant (−/−) embryos within the posterior primitive streak and the wings of the nascent mesoderm. At about E8.0, Mixl1 is ectopically expressed in the endoderm overlying the head process. Images taken from the left. Background staining (*) is noted.
We next used whole-mount in situ hybridization to examine Mixl1 expression patterns in Foxh1−/− mutant embryos. At E7.0, Mixl1 expression is confined to the nascent primitive streak and emerging mesodermal wings in both wild-type (Figure 2B) (Pearce and Evans, 1999; Robb et al, 2000) and Foxh1−/−embryos (Figure 2B). However, in all Foxh1−/− mutant embryos examined, we observed an increase in the intensity of the staining relative to the wild-type embryos (Figure 2B). At E7.5, Mixl1 staining becomes limited to the primitive streak in wild-type embryos (Figure 2B) (Pearce and Evans, 1999; Robb et al, 2000), whereas in Foxh1−/− mutant embryos, Mixl1 expression was expanded anteriorly in the embryonic mesoderm and proximally in the embryonic/extra-embryonic border (Figure 2B). At E8.0, Mixl1, which becomes restricted to the posterior primitive streak and the base of the allantois in wild-type embryos (Figure 2B) (Pearce and Evans, 1999; Robb et al, 2000), exhibited an expanded expression domain in the caudal end of the primitive streak of Foxh1−/− embryos (Figure 2B). Ectopic Mixl1 staining was also observed anteriorly in mutant embryos (Figure 2B). Thus, increased Mixl1 transcript levels are due to both enhanced gene activity and spatial expansion of the Mixl1 expression domain. Although Foxh1 is best known as transcriptional activator downstream of the Activin/Nodal signaling pathway (Attisano et al, 2001), our data suggest that Foxh1 can also act as a negative regulator of target gene expression.
Goosecoid negatively regulates Foxh1-dependent activation of Mixl1
The repressive activity of Foxh1 on endogenous Mixl1 expression observed in mouse embryos was not recapitulated in HepG2 cells, where Foxh1 promotes activation of the Mixl1-luc reporter (Figure 1A). This suggested the possibility that a cofactor present in embryos, but absent in HepG2 cells, was required for the repressive effect. Forkhead and homeodomain-containing proteins have been shown to physically interact to negatively regulate target genes (Foucher et al, 2002; Guo et al, 2002; Rausa et al, 2003). As the expression of Foxh1, Mixl1 and the paired-like homeodomain gene Gsc partially overlap during gastrulation (Blum et al, 1992; Belo et al, 1998; Weisberg et al, 1998; Pearce and Evans, 1999; Robb et al, 2000) and Gsc expression is lost in Foxh1 mutant embryos (Hoodless et al, 2001; Yamamoto et al, 2001), we next determined whether Gsc might modulate Foxh1-dependent expression of Mixl1. For this, HepG2 cells were transfected with Foxh1 and increasing amounts of Gsc and TGFβ-dependent activation of the Mixl1-luc reporter was examined. Ectopic expression of Gsc strongly repressed TGFβ-dependent induction of luciferase activity mediated by Foxh1 alone (Figure 3A) or when Smad2 and Smad4 were co-expressed with Foxh1 (Figure 3B). Thus, Gsc functionally interacts with Foxh1 to repress Mixl1 transcription. To determine whether Gsc modulates the expression of Mixl1 in vivo, we next examined the expression of Mixl1 in E7.5 wild-type and Gsc-null embryos by QPCR. In RNA extracted from Gsc-null embryos, average Mixl1 expression was increased by almost twofold over wild-type levels (Figure 3C). Interestingly, the range of Mixl1 mRNA expression in Gsc-null embryos was wider as compared to wild-type embryos. As Gsc-null embryos do not display overt defects in early embryogenesis (Rivera-Perez et al, 1995; Yamada et al, 1995), we speculate that compensatory mechanisms overcome the increased expression of Mixl1 to allow gastrulation to proceed. As loss of Gsc enhances expression of Mixl1, these results suggest that Gsc can negatively regulate Mixl1 expression in vivo.
Figure 3.
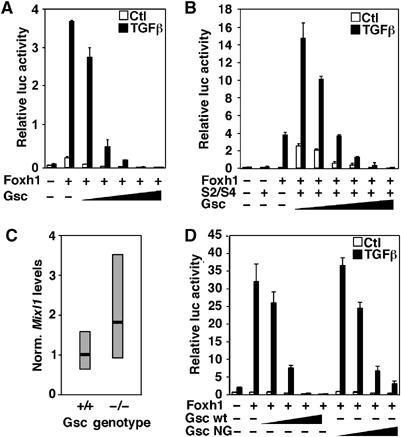
Goosecoid (Gsc) represses Foxh1-mediated induction of the Mixl1. (A, B, D) HepG2 cells were transiently transfected with the Mixl1-luc reporter with combinations of Foxh1, increasing amounts (0.25–250 ng) of Gsc or Gsc NG (N210G). Error bars represent standard deviation of the mean. (C) QPCR analysis of Mixl1 expression in wild-type (n=5) and Gsc-null (n=3) embryos. Expression was normalized to Gapdh. Black line represents average and gray box represents range of Mixl1 expression. Data represent the average of two QPCR experiments.
Physical interaction of Foxh1 with Goosecoid
Gsc can repress transcription of target genes such as Xbra and its own promoter by binding directly to DNA at paired homeodomain sites (Danilov et al, 1998; Latinkic and Smith, 1999). The Mixl1 promoter does not contain consensus paired homeodomain sites, and the Gsc DNA-binding mutant (Gsc NG; N210G) repressed Foxh1-mediated induction of the Mixl1 promoter (Figure 3D), indicating that Mixl1 repression by Gsc occurs independent of its DNA-binding activity.
We next examined whether Gsc might repress transcription by associating with Foxh1. Analysis of immunoprecipitates of cell lysates from COS-1 cells transfected with Flag-Foxh1 and T7-Gsc revealed an interaction between Foxh1 and Gsc (Figure 4A). To determine the region in Foxh1 that mediates the binding to Gsc, we evaluated the ability of two deletion constructs of Flag-Foxh1 to interact with Gsc expressed as a GST fusion. We observed that GST–Gsc bound full-length Flag-Foxh1 and a mutant (ΔC) containing only the forkhead domain, but not Foxh1 lacking the forkhead domain (ΔN, Figure 4B). A similar approach was used to identify the region of Gsc mediating the association with Foxh1. In this case, full-length GST–Gsc and GST–Gsc 1–219 bound Flag-Foxh1, whereas larger deletions lacking the homeodomain did not (Figure 4C). The Gsc homeodomain alone was not sufficient to interact with Flag-Foxh1 (Figure 4C), suggesting that in the context of full-length Gsc, other regions are also necessary for efficient binding to Foxh1. Gsc lacking the homeodomain (Gsc 1–143) did not block Foxh1-mediated induction of the Mixl1 promoter in luciferase reporter assays (Figure 4D). Thus, our data show that the interaction between Gsc and Foxh1 is mediated by their conserved DNA-binding regions, and that the Gsc homeodomain is required for the repressive activity of Gsc.
Figure 4.
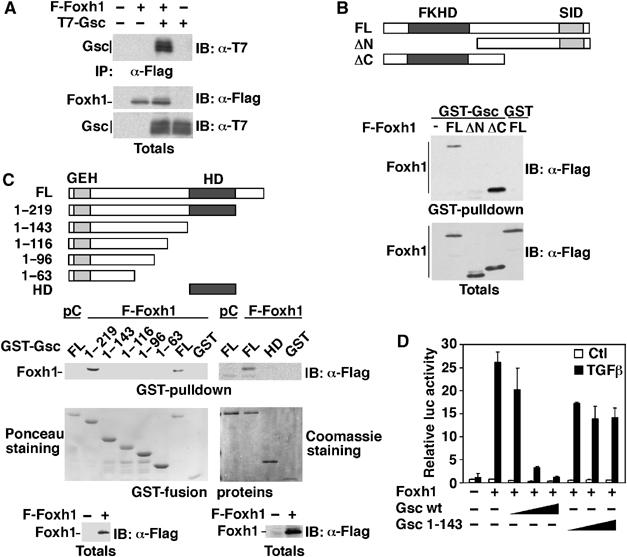
Gsc interacts with Foxh1 and represses Foxh1-mediated activation of the Mixl1 promoter. (A) COS-1 cells were co-transfected with T7-Gsc and Flag-Foxh1 or vector control. Cell lysates were immunoprecipitated (IP) with anti-Flag antibody, and immunoblotted (IB) with anti-T7 antibody. (B, C) GST pull-down assay was performed by incubating Gsc or Gsc deletion constructs expressed as GST-fusion proteins with lysates from COS-1 cells expressing full-length or deletion mutants of Flag-Foxh1. Bound proteins were detected by immunoblotting with anti-Flag antibody. Equal protein expression was confirmed by immunoblotting or by Ponceau red or Coomassie blue staining as indicated. (D) HepG2 cells were transiently transfected with the Mixl1-luc reporter, with Foxh1 and increasing amounts (0.25–250 ng) of Gsc or Gsc 1-143 (closed triangles). Error bars represent standard deviation of the mean.
Gooseceoid is recruited to the Mixl1 promoter through Foxh1
As Gsc can bind Foxh1, we next examined whether Gsc could be recruited to the Mixl1 promoter through its interaction with Foxh1 by performing sequential protein–DNA immunoprecipitations. COS-1 cells were transiently transfected with the Mixl1-luc reporter together with cDNAs encoding Flag-Foxh1 and T7-Gsc. Protein–DNA complexes were crosslinked by formaldehyde treatment, cell lysates were sequentially immunoprecipitated with anti-Flag and anti-T7 antibodies (Figure 5A) and the amount of immunoprecipitated Mixl1-luc reporter was then assessed by QPCR. When Flag-Foxh1 and T7-Gsc were co-expressed, the Mixl1 promoter fragment was detected (Figure 5A), whereas only basal levels were observed when either protein was expressed alone. In contrast, Gsc 1–143, which does not interact with Foxh1 nor block Foxh1-dependent induction of Mixl1, did not bind to the Mixl1 promoter (Figure 5B). Thus, Gsc can be recruited to the Mixl1 promoter through its interaction with Foxh1.
Figure 5.
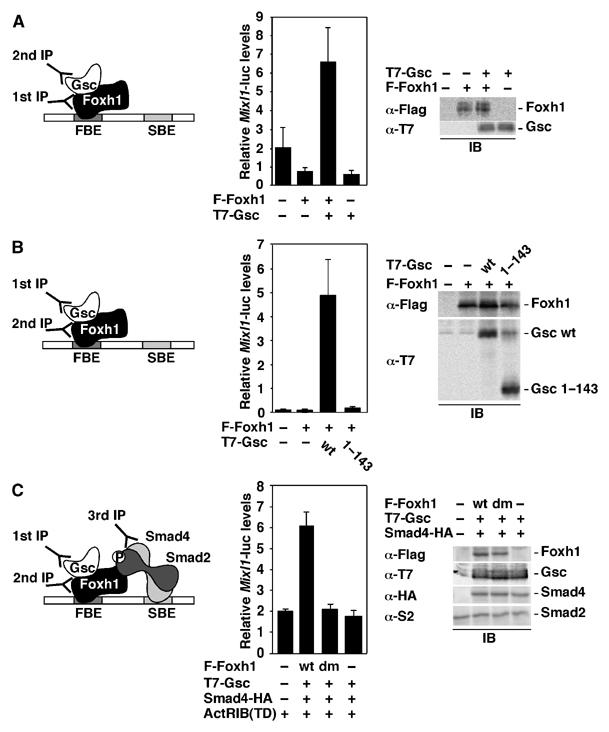
Gsc interacts with Foxh1 and Smads on the Mixl1 promoter. (A–C) COS-1 cells were transiently transfected with the Mixl1-luc reporter and the indicated cDNAs. Protein–DNA complexes were collected by sequential immunoprecipitations, followed by elution in 1% SDS, using anti-Flag, anti-T7 and anti-HA antibodies as indicated (left panels). Recovered DNA was analyzed by QPCR, and corrected for DNA inputs (middle panels). Protein expression was confirmed by immunoblotting (left panels).
We next evaluated whether Gsc could interact with Foxh1 within the context of a TRF complex by performing a three-step sequential protein–DNA immunoprecipitation. COS-1 cells were transiently transfected with the Mixl1-luc reporter along with cDNAs encoding Flag-Foxh1, Smad4-HA, T7-Gsc and ActRIB(TD). Cell lysates were subjected to immunoprecipitation with anti-T7 antibody, followed by elution with 1% SDS, immunoprecipitation of the eluate with anti-Flag antibody, a second elution with 1% SDS and a final immunoprecipitation with anti-HA antibody (Figure 5C). QPCR analysis demonstrated that T7-Gsc bound Flag-Foxh1 within a TRF complex (Figure 5C). In contrast, negligible amounts of Mixl1 were immunoprecipitated when T7-Gsc was co-expressed with a Foxh1 DNA-binding mutant (Foxh1 dm; R61H, K64N) or with a control plasmid (Figure 5C). Our data show that Foxh1, Smads and Gsc can coexist on the Mixl1 promoter and suggest that the transcriptional repression mediated by Gsc is unlikely to occur through disruption of the TRF.
The repressive activity of Goosecoid is mediated by histone deacetylases
We next examined whether the conserved GEH repressor domain present at the N-terminus of Gsc is required for the repressive activity. However, Gsc Δ14, which lacks the GEH domain, was able to repress Mixl1-luc reporter activity as well as wild-type Gsc (Figure 6A). To investigate whether Gsc might recruit transcriptional corepressors such as HDACs to abrogate transcriptional activation of Mixl1, we examined the effect of Trichostatin A (TSA), an inhibitor of HDAC activity on Mixl1-luc reporter activity. HepG2 cells, transfected with the Mixl1-luc reporter, Foxh1, in the presence or absence of Gsc were incubated with TGFβ and TSA. Treatment with increasing doses of TSA resulted in the enhancement of Mixl1-luc activity by up to threefold (Figure 6B), suggesting that recruitment of HDACs by Gsc contributes to the repression of Mixl1 promoter activity. The association of Gsc with HDACs was then examined using COS-1 cells transfected with T7-Gsc and various Flag-HDACs. Gsc interacted with Class I HDACs, HDAC1 and HDAC3 and the Class IV HDAC, HDAC11 but not with the others (Figure 6C). To determine the region in Gsc that mediates the interaction with HDACs, we tested the ability of Gsc N-terminal deletion mutants to interact with HDAC1 expressed as a GST-fusion protein (Figure 6D). HDAC1 bound full-length Gsc as well as deletion mutant Δ81 but not deletion mutants Δ111 and Δ143 (Figure 6D). The effect of these deletion mutants on Mixl1 promoter activation demonstrated that Gsc Δ81 repressed transcription, whereas the larger deletion mutants did not (Figure 6E). Thus, our data show that the interaction between Gsc and HDAC1 is mediated by a region located between residues 81 and 111 of Gsc, and that this region is required for Gsc-mediated repression of the Mixl1 promoter. Taken together, our biochemical analysis suggests a model (Figure 6F) in which Gsc associates with Foxh1 that is bound to the Mixl1 promoter and then functions to recruit histone deacetylases and thereby repress Mixl1 expression.
Figure 6.
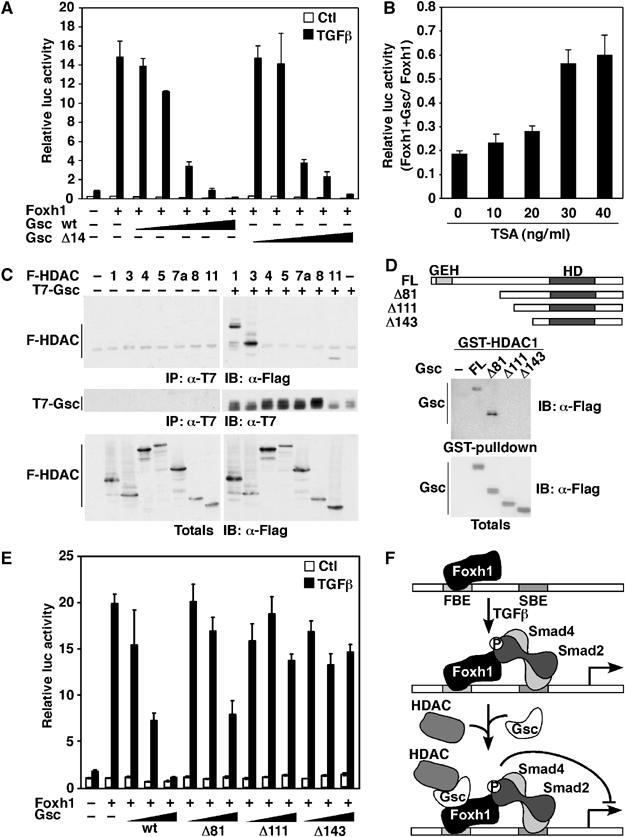
Gsc recruits histone deacetylases (HDACs) to repress Mixl1 promoter activity. (A, E) HepG2 cells were transiently transfected with the Mixl1-luc reporter, with Foxh1 and increasing amounts (0.25–250 ng) of full-length Gsc, or indicated deletion mutants (closed triangles). Error bars represent standard deviation of the mean. (B) HepG2 cells were transiently transfected with the Mixl1-luc reporter, Foxh1 and either a control or Gsc expression vector. Cells were incubated overnight with 100 pM TGFβ and the indicated amount of TSA. Luciferase activity from cell lysates was determined and presented as luciferase activity induced by TSA in cells transfected with Gsc relative to luciferase activity induced by TSA in cells transfected with the control plasmid. Error bars represent propagated error on the quotient calculated from standard errors of the mean of numerators and denominators. (C) Lysates of COS-1 cells co-transfected with T7-Gsc and the indicated Flag-HDAC constructs or vector control were immunoprecipitated with anti-T7 antibody and immunoblotted with anti-Flag antibody. Protein expression was verified by immunoblotting. (D) GST pull-down assay was performed by incubating GST-HDAC1 with lysates from COS-1 cells expressing full-length or deletion mutants of Flag-Gsc. Bound and total protein expression was detected by anti-Flag immunoblotting. (F) A model for Foxh1-dependent regulation of Mixl1. Foxh1 constitutively binds the Mixl1 promoter and on TGFβ signaling, Smads are recruited to Foxh1. Gsc binds Foxh1 and recruits HDACs to repress Mixl1 promoter activity.
Goosecoid-mediated repression of endogenous Mixl1 requires Foxh1
Analyses of Foxh1−/− mutant embryos suggested that Foxh1 negatively regulates Mixl1 expression in vivo and our biochemical studies showed that Gsc associates with Foxh1 to mediate the transcriptional repression of Mixl1. Thus, we sought to determine whether increased expression of Gsc could repress endogenous Mixl1 expression and whether this repressive effect of Gsc requires Foxh1. As Gsc is itself a Foxh1-regulated gene, the interpretation of in vivo data is particularly complex. Therefore, we utilized an in vitro assay in which mouse embryonic stem cells are induced to differentiate into embryoid bodies (EBs), a model system that recapitulates gastrulation events, including the generation of all three germ layers (Keller, 2005). For this, wild-type or Foxh1−/− mutant ES cell lines stably expressing murine Gsc or an empty vector control were generated. Individual lines were isolated and clones overexpressing Gsc were identified by QPCR using RNA extracted from ES cell colonies (Figure 7A). Using the hanging drop method (Spector et al, 1998), ES cell lines were induced to differentiate by the removal of leukemia-inhibiting factor (LIF) and EBs were collected for up to 7 days post-differentiation. Increased Gsc expression in the stable transfectants was maintained throughout the differentiation time course (Figure 7A). Consistent with previous reports, Mixl1 was transiently induced (Mossman et al, 2005; Ng et al, 2005; Willey et al, 2006), with maximal expression detected at day 5 in both wild type (Figure 7B, top panel) and Foxh1−/− control lines (Figure 7B, bottom panel). This concurs with our in vivo expression data, demonstrating that activation of Mixl1 expression occurs independent of Foxh1. However, in EBs derived from Foxh1, wild-type ES cells stably overexpressing Gsc, the enhanced expression of Mixl1 was abrogated (Figure 7B, top panel). In marked contrast, overexpression of Gsc in all four Foxh1−/− EBs, resulted in enhanced Mixl1 expression at either day 5 (for clones 3, 6) or with a short delay on day 6 (for clones 9, 21) (Figure 7B, bottom panel). Thus, in cells lacking Foxh1, Gsc is unable to repress Mixl1 expression.
Figure 7.
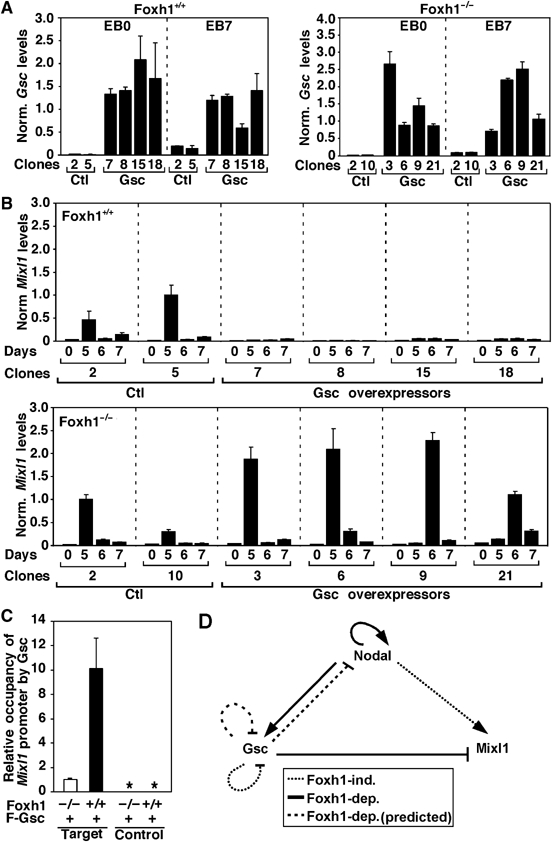
Mixl1 expression is downregulated in wild-type but Foxh1-null EBs overexpressing Gsc. Quantitative real–time RT–PCR analysis of Gsc (A) and Mixl1 (B) expression in control and Gsc-overexpressing undifferentiated ES cells (EB0) and EBs cultured for the indicated number of days. Expression was normalized to Gapdh. (C) EBs were differentiated for 5 days. Protein–DNA complexes were immunoprecipitated with control or anti-Flag antibodies and recovered DNA was analyzed by QPCR with control or target primer pairs. *No amplification detected. (D) A model representing positive and negative feedback loops regulating Mixl1 and Gsc expression through Foxh1.
To determine whether Gsc is recruited to the endogenous Mixl1 promoter, we next performed chromatin immunoprecipitation assays (ChIP) using cell lysates from differentiated EBs. Wild-type or Foxh1−/− ES cell lines overexpressing Flag-Gsc were induced to differentiate into EBs. After 5 days of differentiation, EBs were collected and protein–DNA complexes were fixed by formaldehyde treatment and immunoprecipitated with either control or anti-Flag antibody. Binding of Flag-Gsc to the region of the Mixl1 promoter encompassing the Foxh1 site was then assessed by QPCR. Efficient amplification of the target Mixl1 promoter fragment was detected in Foxh1 wild-type EBs as compared to EBs derived from Foxh1-null cells (Figure 7C). No amplification of a control DNA fragment, located 2.5 Kb away from the Foxh1-binding site, was detected, although efficient amplification of the DNA fragment was observed in total cell lysates (Figure 7C and data not shown). Thus, our results demonstrate that Gsc is recruited to the endogenous Mixl1 promoter and that this is dependent on Foxh1. Taken together, our results indicate that Gsc binds to the Mixl promoter to downregulate the expression of Mixl1, and that this binding and repressive activity requires Foxh1.
Discussion
Foxh1 negatively regulates Mixl1 expression in mouse embryos
The mouse Mix-like gene, Mixl1, is essential for normal gastrulation and for proper development of the node, notochord, axial mesendoderm, heart and gut (Hart et al, 2002). In Xenopus and zebrafish, the related Mix/Bix family members play pivotal roles in early development, functioning in Nodal-like signaling pathways to induce and specify mesoderm and endoderm (Chen et al, 1997; Hart et al, 2002; Poulain and Lepage, 2002). Whereas the requirements for induction of Mix/Bix genes in frogs and fish have been extensively studied, little is known of the molecular mechanism controlling the expression of the mouse Mixl1 gene. Our phylogenetic analysis of the Mixl1 promoter revealed the presence of a conserved DNA-binding site for the forkhead protein Foxh1, a positive transcriptional mediator of Nodal-like signals in early embryos (Attisano et al, 2001). However, rather than promoting gene expression, we show that Foxh1 functions as a negative regulator of Mixl1 transcription and that association of Foxh1 with the homeodomain protein Gsc is required for this repressive effect. Thus, we propose that positive and negative regulation of target gene transcription by Foxh1, a key mediator of Nodal signaling, contributes to refining cell fate decisions in the primitive streak.
Our initial studies (this paper) and those of others (Hart et al, 2005) using mammalian tissue culture cells, have shown that Foxh1 binds to the Mixl1 promoter and can co-operate with Smads to mediate Nodal-dependent signaling. This is consistent with reports defining a requirement for Nodal-like signaling for the induction of Mix/Bix family members such as Xenopus Mix.2, and Mix.3/Mixer (Chen et al, 1997; Henry and Melton, 1998) as well as zebrafish ogx9/mezzo and bonnie and clyde (bon) (Poulain and Lepage, 2002) and more specifically via Foxh1 for Mix.2 (Chen et al, 1997). In contrast, our in vivo analysis of embryos lacking Foxh1 revealed prominent expression of Mixl1, clearly indicating that in mice Foxh1 is dispensable for induction of Mixl1 expression. Interestingly, use of morpholinos to knock down maternally expressed FAST1 (i.e., Foxh1) in Xenopus oocytes before the onset of zygotic gene expression did not significantly affect Mix.2 or Mix.3/Mixer expression (Kofron et al, 2004). Furthermore, reduction of both FAST1 and the Xenopus-specific Foxh1-related gene, FAST3, in animal caps dissected from morpholino-injected early gastrulae, similarly revealed minimal effects on Mix.1/Mix.2 expression (Howell et al, 2002). Thus, elimination of Foxh1 in either frogs or mice reveals that Foxh1 is not required for the initial induction of Mix-like gene expression. Instead, our data indicate that Foxh1 acts to downregulate established Mixl1 expression. Intriguingly, in the late Xenopus blastula, maternal Foxh1 is required to prevent ectopic expression of Xnr5 and Xnr6 but not Xnr3 in the ventral vegetal area (Kofron et al, 2004). The mechanism for the inhibitory Foxh1 activity was not determined, but the authors speculate that perhaps these promoters might harbor Foxh1-binding sites that function to negatively regulate their expression. Recently, a role for Foxh1 in negatively regulating flk1 expression in zebrafish was also reported (Choi et al, 2007). These observations, together with our data, suggest that Foxh1 may function as a negative regulator of transcription for diverse genes.
Gsc is recruited to Foxh1 and represses Mixl1 expression via histone deacetylases
Negative regulation of certain Foxh1 target genes can occur when the TRF is comprised of Smad3/Smad4 heteromers rather than the positively acting Smad2/Smad4 complex (Labbé et al, 1998). In addition, disruption of Foxh1 DNA binding via DRAP1, a corepressor of basal RNA polymerase II transcription, has been reported to prevent Foxh1 target gene expression (Iratni et al, 2002). In contrast, our analysis has revealed a distinct mechanism whereby, the homeobox transcription factor Gsc associates with DNA-bound Foxh1 to repress Mixl1 gene expression. Gsc has been shown to negatively regulate gene expression by binding directly to DNA at specific sites (Danilov et al, 1998; Latinkic and Smith, 1999) and by recruiting the corepressor protein Groucho via the N-terminal GEH domain (Jimenez et al, 1999). In the case of Mixl, the repressive activity of Gsc is mediated by recruitment of HDACs. Foxh1 can also recruit the homeodomain-containing protein Nkx2-5 (von Both et al, 2004) but, unlike Gsc, recruitment of Nkx2-5 mediates heart-specific induction of Mef2c during cardiogenesis (von Both et al, 2004). The ability of homeodomain proteins to act as either co-activators or corepressors of forkhead transcription factors has been previously reported (Foucher et al, 2002; Kim et al, 2003) and may, thus, be a general property of this class of transcription factors.
Nodal regulated gene expression during early development
During early mouse development, Nodal expression is highly dynamic and its activity is tightly regulated. It is first expressed throughout the epiblast and in the visceral endoderm and later is confined to the posterior proximal epiblast, where it marks the site of primitive streak induction (Schier, 2003). Data suggest that at the onset of gastrulation Nodal induces expression of Gsc in the primitive streak via Foxh1 (Labbé et al, 1998; Hoodless et al, 2001; Yamamoto et al, 2001), whereas Mixl1 expression is induced via a Foxh1-independent mechanism (Figure 7D). As gastrulation proceeds, Gsc accumulates in cells fated for the anterior region of the streak, where it is recruited through Foxh1 to the Mixl1 promoter to repress transcription, thus restricting Mixl1 expression to the posterior region. Accordingly, in Foxh1 mutant embryos, we observed anterior expansion of Mixl1 expression. Consistent with this, in Gsc-null mice, Mixl expression is increased. Thus, Foxh1 functions in a transcriptional regulatory loop that acts through Gsc to negatively regulate Mixl1. This mechanism likely reflects part of a much larger transcriptional network that acts to refine gene expression patterning and cell fate decisions in the primitive streak. For instance, the expression of Gsc itself requires Foxh1 (Labbé et al, 1998), and thus downregulation of Gsc expression could occur via an autoregulatory loop either by direct interaction with Foxh1 or through direct DNA binding of Gsc to its own promoter as has been previously reported (Danilov et al, 1998). Furthermore, Foxh1-dependent enhancer elements present in the first intron of the Nodal gene are required for the maintenance and amplification of Nodal expression (Saijoh et al, 2000). Thus, it will be interesting to investigate whether Foxh1 can also function to negatively regulate Nodal expression through the recruitment of Gsc. In Xenopus, Nodal-dependent induction has also been reported to occur either through Mixer (Germain et al, 2000), the Xenopus homolog of the human gene Williams–Beuren syndrome critical region 11 (XWBSCR11) (Ring et al, 2002) or p53 (Cordenonsi et al, 2003), although the role of these modulators has not been examined during early mouse development. The exact mechanisms controlling transcriptional events occurring during mouse gastrulation are likely to have specific spatial, temporal and promoter context requirements and will require further investigation. However, our results indicate that Foxh1, functioning within positive and negative feedback loops may serve to spatially and temporally refine Gsc and Mixl1 activities to ensure appropriate cell fate choices during gastrulation.
Materials and methods
Reporter constructs and transcriptional reporter assays
The Mixl1 promoter (position −390 to −143 from the translational start site) was amplified by PCR from a mouse Mixl1 genomic DNA clone (kindly provided by Jonathan J Pearce) and subcloned into the SacI/BglII sites of a modified pGL2-promoter vector (Promega), as described previously (Labbé et al, 1998). All mutant Mixl1-luc reporters were generated by overlap PCR mutagenesis using primers indicated in Supplementary Table 1. For luciferase assays, HepG2 cells were transiently transfected using the calcium phosphate DNA precipitation method, as described previously (Labbé et al, 1998). Unless otherwise indicated, transfections contained 0.083 μg of Mixl1-luc reporter, 0.0035 μg of Foxh1, 0.035 μg of each Smad, 0.25–250 ng of Gsc or its derived mutants, 0.1 μg of pCMV-βgal and pCMV5 empty vector to a total of 1 μg per well in a 24-well dish. Trichostatin A (Sigma) in 100% ethanol was added as indicated 18 h before lysis.
Construction of mammalian and bacterial expression vectors
The mature ligand region of Nodal was amplified by PCR and subcloned into a pCMV5 vector containing the Activin pro-region. C-terminal Flag-tagged mouse Cripto was generated by PCR using NIA clone H3029H05 as template. Epitope-tagged Gsc full-length and deletion mutants in pCMV5B were generated by PCR using mouse Gsc cDNA and subcloned in pGEX4T1 for bacterial expression. Flag-Foxh1 ΔN where the first 183 amino acids were deleted was generated by PCR. GST-HDAC1 was generated by PCR using HDAC1 cDNA and subcloned pGEX4T1 for bacterial expression. All primers are listed in Supplementary Table 1.
Gel-shifts, immunoprecipitation, GST pull-downs and immunoblotting
EMSAs were performed as described previously (Labbé et al, 1998) except that cell extracts or bacterial fusion proteins were incubated with 30 000 cpm [32P]-labeled DNA before nondenaturing electrophoresis.
For immunoprecipitations and GST pull-downs, COS-1 cells were transfected using the polyethylenimine (PEI; Sigma-Aldrich catalog no. 408727) method. Briefly, for a 100 mm dish, 25 μl of 2 mg/ml of PEI stock was added to 10 μg of DNA made up to 750 μl in serum-free medium. The DNA/PEI solution was vortexed, incubated at room temperature for 5 min and added to cells containing 10 ml of growth medium. Immunoprecipitations, GST pull-downs and immunoblotting were carried out as reported (Labbé et al, 1998) using M2 anti-Flag (Sigma) or anti-T7 (Novagen) monoclonal antibodies.
For DNA immunoprecipitations, COS-1 cells were transfected with the appropriate cDNA and Mixl1-luc reporter using LipofectAMINE (Invitrogen), and immunoprecipitations were performed as described previously (von Both et al, 2004) with the following modifications. For double DNA immunoprecipitation, protein–DNA complexes were immunoprecipitated with 2 μg of Flag antibody or T7 antibody, followed by incubation with protein G Sepharose for 1 h. Protein–DNA complexes were eluted by incubation with 1% SDS, rediluted to a final concentration of 0.1% SDS, and subjected to immunoprecipitation with 2 μg of T7 or Flag antibody overnight. For triple DNA immunoprecipitations, lysates were sequentially immunoprecipitated with 2 μg of T7 antibody for 3 h, followed by an overnight immunoprecipitation with 2 μg of Flag antibody, and a 3-h immunoprecipitation with 2 μg of Y11 rabbit anti-HA antibody (Santa Cruz). Protein–DNA complexes were eluted each time with 1% SDS. For endogenous ChIP, EB cultures were established as described previously (von Both et al, 2004) except that ES cells were suspended in 0.05% trypsin and differentiated in bacterial grade Petri-dishes. After 5 days of differentiation, EBs were collected and ChIPs were carried out as described previously (von Both et al, 2004), except that EBs were fixed in 1% formaldehyde for 20 min at room temperature and Protein G sepharose beads were blocked with 0.5 mg/ml BSA and 0.2 mg/ml salmon sperm DNA. Levels of Mixl1 promoter precipitated DNA were analyzed by QPCR using SYBR Green PCR Master Mix (Applied Biosystems) and primers as indicated in Supplementary Table 1. For DNA immunoprecipitation, Ct values were normalized with corresponding DNA input Ct values. Net Ct values were plotted using Comparative Ct method (docs.appliedbiosystems.com/pebiodocs/04303859.pdf). For ChIPs, net Ct values for the anti-Flag immunoprecipitation were corrected with net Ct values of control anti-T7 immunoprecipitation and plotted using the Comparative Ct method.
ES cells and embryoid body differentiation
Flag-tagged Gsc cDNA was subcloned in the episomal expression vector pCAGIP, and this vector was linearized with PvuI and electroporated into the previously described Foxh1+/+ (no. 9b) and Foxh1−/− (no. 3) ES cell lines (Hoodless et al, 2001). Individual puromycin-resistant colonies were selected, and Gsc mRNA expression was confirmed by quantitative RT–PCR. For in vitro differentiation assays, ES cells grown in ES medium (Nagy, 2003) were passaged twice on 0.1% gelatin-coated plates and grown to about 70% confluency. ES cells were trypsinized in 0.05% trypsin, harvested and resuspended in EB medium (ES medium minus LIF). Hanging drop cultures were established as described previously (Spector et al, 1998). EBs from 2-day hanging drop cultures were transferred to 24-well ultralow attachement plates at a density of 50 EBs/well. Three days later, EBs were transferred to gelatin-coated six-wells dishes and maintained in EB media until harvesting for RNA extraction.
Quantitative RT–PCR and in situ hybridization analysis of Mixl1 in mouse embryos
Embryonic RNA was isolated using TRIzol reagent (Invitrogen) according to standard procedures from pools of E7.5 embryos genotyped for Foxh1 as described previously (Hoodless et al, 2001). Gsc mutant embryos were genotyped by PCR as described previously (Belo et al, 1998) with DNA extracted from the extraembryonic region of E7.5 embryos. Embryonic RNA from wild-type and Gsc-null embryos were isolated using the Nanoprep kit (Stratagene) according to manufacturer's instructions. ES cell and EB RNA were isolated using the RNeasy Mini Kit (Qiagen). RNA samples were treated with DNaseI (Fermentas), primed with oligo p(dT)20 (ACGT Corp.) and reverse transcribed using RevertAid H Minus M-MuLV Reverse Transcriptase (Fermentas). QPCR on embryonic samples was performed using SYBR Green PCR Master Mix (Applied Biosystems) and Mixl1 and Hprt primers as indicated in Supplementary Table 1. QPCR on ES and EB samples and wild-type and Gsc-null embryos was performed with TaqMan Universal PCR Master Mix (Applied Biosystems) and pre-developed Taqman Gene expression assays (Applied Biosystems) for mouse Mixl1, Gsc and Gapdh using the ABI Prism 7000 or 7900 sequence detection system (Applied Biosystems). Analysis was performed using the Comparative Ct method. Whole-mount in situ hybridization analysis of Mixl1 in mouse embryos was performed as described previously (von Both et al, 2004) using a Mixl1 probe (Pearce and Evans, 1999).
Supplementary Material
Supplementary Figure and Table
Acknowledgments
We thank Edward M De Robertis for generously providing the Gsc-null embryos, Bryan W Miller for the Flag-HDAC constructs and Jonathan J Pearce for the murine Mixl1 genomic clone and probe. This work was supported by grants to LA from the National Cancer Institute of Canada, with funds from the Canadian Cancer Society, and to JLW from the Canadian Institutes of Health Research (CIHR). LI was supported by a CIHR studentship and Heart and Stroke Foundation of Canada doctoral research award and CS and EL by CIHR doctoral studentships. LZ is an Associate of the Howard Hughes Medical Institute (HHMI). LA and JLW hold Canadian Research Chairs, and JLW is an International Scholar of the HHMI.
References
- Attisano L, Silvestri C, Izzi L, Labbé E (2001) The transcriptional role of Smads and FAST (FoxH1) in TGFβ and activin signalling. Mol Cell Endocrinol 180: 3–11 [DOI] [PubMed] [Google Scholar]
- Attisano L, Wrana JL (2002) Signal transduction by the TGF-β superfamily. Science 296: 1646–1647 [DOI] [PubMed] [Google Scholar]
- Belo JA, Bouwmeester T, Leyns L, Kertesz N, Gallo M, Follettie M, De Robertis EM (1997) Cerberus-like is a secreted factor with neutralizing activity expressed in the anterior primitive endoderm of the mouse gastrula. Mech Dev 68: 45–57 [DOI] [PubMed] [Google Scholar]
- Belo JA, Leyns L, Yamada G, De Robertis EM (1998) The prechordal midline of the chondrocranium is defective in Goosecoid-1 mouse mutants. Mech Dev 72: 15–25 [DOI] [PubMed] [Google Scholar]
- Blum M, Gaunt SJ, Cho KWY, Steinbeisser H, Blumberg B, Bittner D, DeRobertis EM (1992) Gastrulation in the mouse: the role of the homeobox gene goosecoid. Cell 69: 1097–1106 [DOI] [PubMed] [Google Scholar]
- Chen X, Weisberg E, Fridmacher V, Watanabe M, Naco G, Whitman M (1997) Smad4 and FAST-1 in the assembly of activin-responsive factor. Nature 389: 85–89 [DOI] [PubMed] [Google Scholar]
- Choi J, Dong L, Ahn J, Dao D, Hammerschmidt M, Chen JN (2007) FoxH1 negatively modulates flk1 gene expression and vascular formation in zebrafish. Dev Biol 304: 735–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordenonsi M, Dupont S, Maretto S, Insinga A, Imbriano C, Piccolo S (2003) Links between tumor suppressors: p53 is required for TGF-beta gene responses by cooperating with Smads. Cell 113: 301–314 [DOI] [PubMed] [Google Scholar]
- Danilov V, Blum M, Schweickert A, Campione M, Steinbeisser H (1998) Negative autoregulation of the organizer-specific homeobox gene goosecoid. J Biol Chem 273: 627–635 [DOI] [PubMed] [Google Scholar]
- Feng XH, Derynck R (2005) Specificity and versatility in tgf-beta signaling through Smads. Annu Rev Cell Dev Biol 21: 659–693 [DOI] [PubMed] [Google Scholar]
- Foucher I, Volovitch M, Frain M, Kim JJ, Souberbielle JC, Gan L, Unterman TG, Prochiantz A, Trembleau A (2002) Hoxa5 overexpression correlates with IGFBP1 upregulation and postnatal dwarfism: evidence for an interaction between Hoxa5 and Forkhead box transcription factors. Development 129: 4065–4074 [DOI] [PubMed] [Google Scholar]
- Germain S, Howell M, Esslemont GM, Hill CS (2000) Homeodomain and winged-helix transcription factors recruit activated Smads to distinct promoter elements via a common Smad interaction motif. Genes Dev 14: 435–451 [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Costa R, Ramsey H, Starnes T, Vance G, Robertson K, Kelley M, Reinbold R, Scholer H, Hromas R (2002) The embryonic stem cell transcription factors Oct-4 and FoxD3 interact to regulate endodermal-specific promoter expression. Proc Natl Acad Sci USA 99: 3663–3667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart AH, Hartley L, Sourris K, Stadler ES, Li R, Stanley EG, Tam PP, Elefanty AG, Robb L (2002) Mixl1 is required for axial mesendoderm morphogenesis and patterning in the murine embryo. Development 129: 3597–3608 [DOI] [PubMed] [Google Scholar]
- Hart AH, Willson TA, Wong M, Parker K, Robb L (2005) Transcriptional regulation of the homeobox gene Mixl1 by TGF-beta and FoxH1. Biochem Biophys Res Commun 333: 1361–1369 [DOI] [PubMed] [Google Scholar]
- Henry GL, Melton DA (1998) Mixer, a homeobox gene required for endoderm development. Science 281: 91–96 [DOI] [PubMed] [Google Scholar]
- Hoodless PA, Pye M, Chazaud C, Labbé E, Attisano L, Rossant J, Wrana JL (2001) FoxH1 (Fast) functions to specify the anterior primitive streak in the mouse. Genes Dev 15: 1257–12371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell M, Inman GJ, Hill CS (2002) A novel Xenopus Smad-interacting forkhead transcription factor (XFast-3) cooperates with XFast-1 in regulating gastrulation movements. Development 129: 2823–2834 [DOI] [PubMed] [Google Scholar]
- Iratni R, Yan YT, Chen C, Ding J, Zhang Y, Price SM, Reinberg D, Shen MM (2002) Inhibition of excess nodal signaling during mouse gastrulation by the transcriptional corepressor DRAP1. Science 298: 1996–1999 [DOI] [PubMed] [Google Scholar]
- Jimenez G, Verrijzer CP, Ish-Horowicz D (1999) A conserved motif in goosecoid mediates groucho-dependent repression in Drosophila embryos. Mol Cell Biol 19: 2080–2087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller G (2005) Embryonic stem cell differentiation: emergence of a new era in biology and medicine. Genes Dev 19: 1129–1155 [DOI] [PubMed] [Google Scholar]
- Kim JJ, Taylor HS, Akbas GE, Foucher I, Trembleau A, Jaffe RC, Fazleabas AT, Unterman TG (2003) Regulation of insulin-like growth factor binding protein-1 promoter activity by FKHR and HOXA10 in primate endometrial cells. Biol Reprod 68: 24–30 [DOI] [PubMed] [Google Scholar]
- Kinder SJ, Tsang TE, Wakamiya M, Sasaki H, Behringer RR, Nagy A, Tam PP (2001) The organizer of the mouse gastrula is composed of a dynamic population of progenitor cells for the axial mesoderm. Development 128: 3623–3634 [DOI] [PubMed] [Google Scholar]
- Kofron M, Puck H, Standley H, Wylie C, Old R, Whitman M, Heasman J (2004) New roles for FoxH1 in patterning the early embryo. Development 131: 5065–5078 [DOI] [PubMed] [Google Scholar]
- Labbé E, Silvestri C, Hoodless PA, Wrana JL, Attisano L (1998) Smad2 and Smad3 positively and negatively regulate TGFβ-dependent transcription through the forkhead DNA binding protein, FAST2. Mol Cell 2: 109–120 [DOI] [PubMed] [Google Scholar]
- Latinkic BV, Smith JC (1999) Goosecoid and mix.1 repress Brachyury expression and are required for head formation in Xenopus. Development 126: 1769–1779 [DOI] [PubMed] [Google Scholar]
- Latinkic BV, Umbhauer M, Neal KA, Lerchner W, Smith JC, Cunliffe V (1997) The Xenopus Brachyury promoter is activated by FGF and low concentrations of activin and suppressed by high concentrations of activin and by paired-type homeodomain proteins. Genes Dev 11: 3265–3276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossman AK, Sourris K, Ng E, Stanley EG, Elefanty AG (2005) Mixl1 and oct4 proteins are transiently co-expressed in differentiating mouse and human embryonic stem cells. Stem Cells Dev 14: 656–663 [DOI] [PubMed] [Google Scholar]
- Nagy A (2003) Manipulating the Mouse Embryo: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Ng ES, Azzola L, Sourris K, Robb L, Stanley EG, Elefanty AG (2005) The primitive streak gene Mixl1 is required for efficient haematopoiesis and BMP4-induced ventral mesoderm patterning in differentiating ES cells. Development 132: 873–884 [DOI] [PubMed] [Google Scholar]
- Pearce JJH, Evans MJ (1999) Mml, a mouse Mix-like gene expressed in the primitive streak. Mech Dev 87: 189–192 [DOI] [PubMed] [Google Scholar]
- Poulain M, Lepage T (2002) Mezzo, a paired-like homeobox protein is an immediate target of Nodal signalling and regulates endoderm specification in zebrafish. Development 129: 4901–4914 [DOI] [PubMed] [Google Scholar]
- Rausa FM, Tan Y, Costa RH (2003) Association between hepatocyte nuclear factor 6 (HNF-6) and FoxA2 DNA binding domains stimulates FoxA2 transcriptional activity but inhibits HNF-6 DNA binding. Mol Cell Biol 23: 437–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ring C, Ogata S, Meek L, Song J, Ohta T, Miyazono K, Cho KW (2002) The role of a Williams–Beuren syndrome-associated helix-loop-helix domain-containing transcription factor in activin/nodal signaling. Genes Dev 16: 820–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Perez JA, Mallo M, Gendron-Maguire M, Gridley T, Behringer RR (1995) Goosecoid is not an essential component of the mouse gastrula organizer but is required for craniofacial and rib development. Development 121: 3005–3012 [DOI] [PubMed] [Google Scholar]
- Robb L, Hartley L, Begley CG, Brodnicki TC, Copeland NG, Gilbert DJ, Jenkins NA, Elefanty AG (2000) Cloning, expression analysis, and chromosomal localization of murine and human homologues of a Xenopus mix gene. Dev Dyn 219: 497–504 [DOI] [PubMed] [Google Scholar]
- Robb L, Tam PP (2004) Gastrula organiser and embryonic patterning in the mouse. Semin Cell Dev Biol 15: 543–554 [DOI] [PubMed] [Google Scholar]
- Saijoh Y, Adachi H, Sakuma R, Yeo CY, Yashiro K, Watanabe M, Hashiguchi H, Mochida K, Ohishi S, Kawabata M, Miyazono K, Whitman M, Hamada H (2000) Left-right asymmetric expression of lefty2 and nodal is induced by a signaling pathway that includes the transcription factor FAST2. Mol Cell 5: 35–47 [DOI] [PubMed] [Google Scholar]
- Schier AF (2003) Nodal signaling in vertebrate development. Annu Rev Cell Dev Biol 19: 589–621 [DOI] [PubMed] [Google Scholar]
- Shiratori H, Sakuma R, Watanabe M, Hashiguchi H, Mochida K, Sakai Y, Nishino J, Saijoh Y, Whitman M, Hamada H (2001) Two-step regulation of left-right asymmetric expression of Pitx2: initiation by nodal signaling and maintenance by Nkx2. Mol Cell 7: 137–149 [DOI] [PubMed] [Google Scholar]
- Spector DL, Goldman RD, Leinwand LA (1998) Culture and in vitro differentiation of mouse embryonic stem cells. In Cells A Laboratory Manual. Volume 1: Culture and biochemical analysis of cells, pp 8.1–8.22. New York: Cold Spring Harbor Laboratory Press [Google Scholar]
- Tam PP, Behringer RR (1997) Mouse gastrulation: the formation of a mammalian body plan. Mech Dev 68: 3–25 [DOI] [PubMed] [Google Scholar]
- von Both I, Silvestri C, Erdemir T, Lickert H, Walls JR, Henkelman RM, Rossant J, Harvey RP, Attisano L, Wrana JL (2004) Foxh1 is essential for development of the anterior heart field. Dev Cell 7: 331–345 [DOI] [PubMed] [Google Scholar]
- Weisberg E, Winnier GE, Chen X, Farnsworth CL, Hogan BLH, Whitman M (1998) A mouse homologue of FAST-1 transduces TGFβ superfamily signals and is expressed during early embryogenesis. Mech Dev 79: 17–27 [DOI] [PubMed] [Google Scholar]
- Willey S, Ayuso-Sacido A, Zhang H, Fraser ST, Sahr KE, Adlam MJ, Kyba M, Daley GQ, Keller G, Baron MH (2006) Acceleration of mesoderm development and expansion of hematopoietic progenitors in differentiating ES cells by the mouse Mix-like homeodomain transcription factor. Blood 107: 3122–3130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada G, Mansouri A, Torres M, Stuart ET, Blum M, Schultz M, De Robertis EM, Gruss P (1995) Targeted mutation of the murine goosecoid gene results in craniofacial defects and neonatal death. Development 121: 2917–2922 [DOI] [PubMed] [Google Scholar]
- Yamamoto M, Meno C, Sakai Y, Shiratori H, Mochida K, Ikawa Y, Saijoh Y, Hamada H (2001) The transcription factor FoxH1 (FAST) mediates nodal signaling during anterior-posterior patterning and node formation in the mouse. Genes Dev 15: 1242–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J, Kessler DS (2001) Goosecoid promotes head organizer activity by direct repression of Xwnt8 in Spemann's organizer. Development 128: 2975–2987 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure and Table


