Abstract
The domestication of maize (Zea mays sp. mays) from its wild progenitors represents an opportunity to investigate the timing and genetic basis of morphological divergence resulting from artificial selection on target genes. We compared sequence diversity of 30 candidate selected and 15 reference loci between the three populations of wild teosintes, maize landraces, and maize inbred lines. We inferred an approximately equal ratio of genes selected during early domestication and genes selected during modern crop breeding. Using an expanded dataset of 48 candidate selected and 658 neutral reference loci, we tested the hypothesis that candidate selected genes in maize are more likely to have transcriptional functions than neutral reference genes, but there was no overrepresentation of regulatory genes in the selected gene dataset. Electronic northern analysis revealed that candidate genes are significantly overexpressed in the maize ear relative to vegetative tissues such as maize shoot, leaf, and root tissue. The maize ear underwent dramatic morphological alteration upon domestication and has been a continuing target of selection for maize yield. Therefore, we hypothesize that genes targeted by selection are more likely to be expressed in tissues that experienced high levels of morphological divergence during domestication and crop improvement.
Crop domestication has generated striking morphological differences between agricultural species and their wild relatives. For example, among the cereals (family Poaceae) artificial selection by early farmers produced larger grains, reduced dispersal, and alterations in plant architecture and flowering phenology (Hammer, 1984; Paterson et al., 1995). While many of these morphological changes occurred during the initial stages of domestication, the process of morphological change is an ongoing function of artificial selection. A prominent example of recent selection is the Green Revolution of the mid-20th century, which focused on altering plant morphology and biochemistry to improve yield in cereal crops such as maize (Zea mays sp. mays), rice (Oryza sativa), and wheat (Triticum aestivum; Khush, 2001; Evenson and Gollin, 2003). As a result of these and other modern breeding efforts, elite cultivars are often as diverged from native landraces (i.e. primitive forms of domesticated crops) as landraces are from their wild ancestors (Meyerowitz, 1994; Khush, 2001).
Maize, and specifically the maize female inflorescence or ear, is a particularly striking example of the morphological divergence between a crop species and its wild progenitor, teosinte (maize sp. parviglumis). The maize ear contains up to a 100-fold more seeds than the teosinte ear, and is composed of naked kernels firmly attached to the cob (Doebley, 2004). As a result, the maize plant can no longer disperse seeds at maturity, and seeds (lacking a protective fruitcase) are vulnerable to predators, leaving the plants dependent on human assistance for propagation. Differences in reproductive structures between maize and teosinte highlight the phenotypic shift resulting from artificial selection for domestication that was initiated between 5,000 and 9,000 years ago (Matsuoka et al., 2002; Sluyter and Dominguez, 2006). The question remains as to the genetic basis for these morphological shifts. Ultimately, the discovery and characterization of genes responsible for these morphological changes will not only improve our understanding of the molecular consequences of artificial selection, but also benefit modern crop breeding (McCouch, 2004).
Thus far, the most productive approach for identifying genes underlying phenotypes has been quantitative trait loci (QTL) mapping. Yet, few genes contributing to selected traits have been identified in crop species (Doebley et al., 2006), with the most successes coming from studies of maize. Among the genes that have been characterized in maize are three linked to the five major genomic regions predicted by Beadle (1939) and confirmed by Doebley and coworkers (Doebley et al., 1990; Doebley and Stec, 1991) to explain the major morphological differences between teosinte and maize. These genes include teosinte glume architecture1 (tga1), responsible for the reduced glume and exposed seeds in maize (Wang et al., 2005), and both barren stalk1 and teosinte branched1 (tb1), which modify patterns of lateral branching and the location of the male inflorescence or tassel (Doebley et al., 1997; Gallavotti et al., 2004). Despite the small number of loci isolated from domesticated species thus far, the genes contributing to major morphological shifts appear to be biased functionally in favor of transcription factors (Doebley et al., 2006), supporting the hypothesis that changes in gene expression patterns, rather than alterations in protein sequence, are the most likely molecular model for shifts in plant form (Doebley and Lukens, 1998).
Recently, molecular population genetics has been used as a complementary approach to identify loci that may contribute to domestication phenotypes. Unlike QTL mapping, which begins with a phenotype of interest, the molecular population genetic approach searches for the signature of artificial selection (or a selective sweep) in genetic polymorphism data to identify genes of historical importance. As a result, molecular population genetic methods serve as a bottom-up approach relative to the top-down methods of QTL mapping (Ross-Ibarra et al., 2007). For example, Wright et al. (2005) screened DNA polymorphism within 774 gene fragments among maize inbred lines and teosinte accessions to identify approximately 30 candidate selected genes, more than tripling the number of potential maize domestication genes discovered to date. Based on the proportion of candidate loci identified, the authors hypothesized as many as 2% to 4% (or approximately 1,200) of maize genes carry the signature of selection and are therefore candidates to contribute to agronomic traits. In a similar study, Yamasaki et al. (2005) screened 1,095 maize genes and identified at least 18 genes with evidence of a selective sweep, eight of which pass stringent and conservative statistical criteria.
Yamasaki et al. (2005) also determined the timing of selection on these genes by sequencing a stratified sample of teosintes, maize landraces, and maize inbred lines. These samples represent predomesticated, early domesticated, and highly improved germplasm, respectively. Yamasaki et al. (2005) reasoned that genes targeted by artificial selection during domestication should experience the hallmark features of selection (i.e. a sharp decline in genetic diversity, a marked shift in the frequency spectrum of polymorphisms, and increased linkage disequilibrium) in landraces relative to teosintes. In contrast, genes targeted by artificial selection more recently, during crop breeding, should exhibit these features between elite inbreds and maize landraces. Of the genes examined, roughly half appeared to have been under selection early during the process of domestication, with the remainder having been selected during more recent crop improvement. Ultimately, both gene categories represent the result of ongoing selection on important agronomic traits.
Taken together, the studies of Wright et al. (2005) and Yamasaki et al. (2005) have generated a number of loci that contain the signature of selection and are thus strong candidates to contribute to phenotypes of agronomic interests. However, the drawback of the molecular population genetic approach is that the phenotype to which the genes contribute is unknown (Ross-Ibarra et al., 2007). Once candidate genes are identified, an obvious next step is to examine them further with population genetic and bioinformatics methods to reveal more not only about the timing and pattern of selection but also insights about expression and function.
In this article, we perform these additional steps to address three topics of fundamental importance to understanding crop domestication. First, we study the population genetics of 30 candidate selected genes previously identified by Wright et al. (2005) with the goal of learning more about the timing and consequence of artificial selection. Were these genes under selection early in the process of domestication or later as a consequence of crop improvement? The question of timing is important because it may provide insights into the progression of phenotypic changes associated with domestication. Second, we examine the predicted functions of these genes in more detail. Is there any evidence that they are biased toward transcriptional regulation, and does that bias vary with the timing of selection? Finally, we conduct a bioinformatic analysis of tissue-specific expression, comparing selected genes to genes without a history of selection. Do selected genes have expression patterns consistent with a role in morphological divergence?
RESULTS
Sequence Diversity and Tests of Selection
We gathered DNA polymorphism data from 30 candidate selected loci and 15 reference (nonselected) loci from a panel of maize landrace individuals (see “Materials and Methods”), with the purpose of determining whether selection acted early or late during the process of domestication and crop improvement. The landrace data were aligned to published sequence data from samples of teosinte and elite inbred lines (Wright et al., 2005), providing an approximate timeline spanning predomestication (teosintes) to early domestication (landraces) to recent cultivation (elite inbreds). The alignments of the 30 candidate selected loci and 15 reference loci represented a total of 33,293 and 26,109 bp, respectively, when pooled across populations of inbreds, landraces, and teosintes. The mean length of sequence alignments was 636.9 bp with a range of 357 to 1,197 bp, and there was an average of 11 sequences per gene per population. The loci were designated as candidate or reference loci based on the selection analyses of Wright et al. (2005).
The stratified samples of teosintes, landraces, and elite inbreds exhibited four characteristics expected from previous studies of maize sequence diversity (Table I). First, the average level of diversity for reference genes for the landrace and inbred samples was similar to that previously reported for maize (Tenaillon et al., 2001). Second, diversity levels varied markedly between the candidate selected genes and reference genes. This was not surprising because genes were initially classified as candidate or reference in large part on the basis of diversity in inbred lines (Wright et al., 2005). Nonetheless, this contrast was also evident for the new diversity data from the landrace samples, suggesting that some diversity differences between selected and reference genes took place early in domestication, such that differences are evident in landrace samples. Third, Tajima's D, which is a measure of the frequency of polymorphisms, increased from teosintes to landraces for reference genes, as detected previously (Tenaillon et al., 2001).
Table I.
Average sequence diversity for 30 candidate selected genes and 15 neutral reference genes among maize inbred lines, landraces, and teosintes
Parameters include the average number of sequences (n), average pairwise nucleotide diversity (π), average Watterson's estimator of diversity (θ), and Tajima's D. ses are in parentheses.
| Diversity Statistics | Maize Inbreds | Maize Landraces | Teosintes |
|---|---|---|---|
| Candidate genes | |||
| n | 9.8 | 11.7 | 11.6 |
| π | 0.0005 (0.0002) | 0.0041 (0.0037) | 0.0099 (0.0010) |
| θ | 0.0005 (0.0002) | 0.0050 (0.0007) | 0.0116 (0.0011) |
| D | −0.150 (0.282) | −0.840 (0.131) | −0.608 (0.129) |
| Reference genes | |||
| n | 11.9 | 11.7 | 12.9 |
| π | 0.0084 (0.0011) | 0.0087 (0.0011) | 0.0115 (0.0013) |
| θ | 0.0080 (0.0001) | 0.0093 (0.0010) | 0.0125 (0.0013) |
| D | 0.200 (0.192) | −0.224 (0.281) | −0.367 (0.133) |
Finally, the level of genetic diversity, as measured by the standard diversity statistics π and θ, decreased from the teosinte sample to the maize landrace sample to elite inbreds for both candidate and reference genes. This pattern is consistent with a bottleneck process winnowing genetic diversity (Eyre-Walker et al., 1998). A decline in sequence diversity from teosinte to maize inbred lines was also apparent for the number of segregating sites and the number of haplotypes (Supplemental Table S1). However, the decrease in π among maize inbred lines, landraces, and teosintes was not statistically significant for the 15 neutral genes (Kruskal-Wallis; P = 0.1601). The decrease was more prominent among samples for candidate selected loci (Kruskal-Wallis; P < 0.0001), with t tests also detecting significant declines in sequence diversity for all pairwise comparisons (inbreds < landraces < teosintes, P < 0.0001; Fig. 1). The exaggerated effect in candidate loci relative to reference loci likely reflects the action of artificial selection.
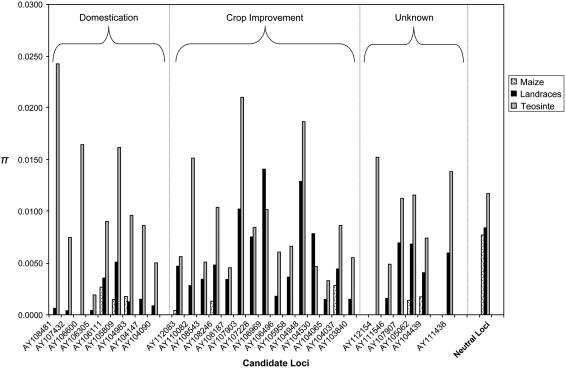
Figure 1.
Graph of pairwise nucleotide diversity (π) for the 30 candidate selected loci and 15 pooled neutral reference loci in populations of maize inbreds, landraces, and teosintes. Loci are sorted into domestication and crop improvement categories based on MLHKA results. Nucleotide diversity for the 15 neutral reference genes are pooled within populations.
Our main purpose with regard to diversity statistics was to determine whether selection occurred during early domestication or later crop improvement. To address this issue, we conducted maximum likelihood Hudson, Kreitman, Aguadé tests (MLHKA; Wright and Charlesworth, 2004) for 29 of the 30 candidate genes among maize inbreds, landraces, and teosintes. Genes were included in analyses based on the availability of a Sorghum bicolor outgroup sequence (see “Materials and Methods”). Using MLHKA, we compared individual candidate genes to the 15 reference loci for each population. Following Yamasaki et al. (2005), if a candidate locus was significant for selection in both maize inbreds and landraces, we concluded that the selection occurred early in the process of domestication (i.e. before the geographic scattering of landraces) and accordingly classified the gene as an early domestication gene. In contrast, if a candidate gene was significant for selection solely in maize inbreds, we classified it as a crop improvement gene based on the observation that the gene did not appear to be a target of selection prior to formation of landraces.
Because there was a priori evidence for selection on candidate genes, we used a lenient criterion of P < 0.10 for MLHKA significance to place genes into categories (Table II). Even with this lenient criterion, two of the 29 candidate loci (AY105062 and AY107907) did not produce a significant MLHKA test and thus could not be categorized as either domestication or improvement genes. In addition, MLHKA tests for three candidate loci (AY104439, AY111546, and AY112154) deviated from neutrality for the teosinte sample. In these cases, we could not reliably assign the genes to the domestication or improvement because selection may have occurred in the wild, prior to domestication. However, we were able to assign the remaining 24 genes as nine domestication genes and 15 improvement genes based on the MLHKA results (Table II).
Table II.
Results of MLHKA tests to compare sequence diversity of each of 30 candidate agronomic loci in maize inbred lines, landraces, and teosintes with 15 neutral reference loci
Candidate genes are defined as domestication (D) genes and crop improvement (CI) genes based on interpretation of MLHKA P values. *, AY111438 had no S. bicolor outgroup at the date of analyses. **, Values for the 15 reference loci are reported for HKA results (see “Materials and Methods”). NA, Not applicable.
| Gene | MLHKA P Values
|
Category | ||
|---|---|---|---|---|
| Inbreds | Landraces | Teosintes | ||
| AY104090 | 0.009 | 0.019 | 0.510 | D |
| AY104147 | 0.010 | 0.053 | 0.603 | D |
| AY104983 | 0.024 | 0.079 | 0.752 | D |
| AY105809 | 0.004 | 0.083 | 0.545 | D |
| AY106111 | 0.268 | 0.044 | 0.584 | D |
| AY106305 | 0.025 | 0.076 | 0.135 | D |
| AY106600 | 0.004 | 0.033 | 0.445 | D |
| AY107432 | 0.017 | 0.096 | 0.665 | D |
| AY108481 | 0.003 | 0.003 | 0.201 | D |
| AY103840 | 0.008 | 0.412 | 0.365 | CI |
| AY104037 | 0.005 | 0.497 | 0.468 | CI |
| AY104065 | 0.043 | 0.387 | 0.421 | CI |
| AY104530 | 0.033 | 0.247 | 0.390 | CI |
| AY104948 | 0.056 | 0.658 | 0.305 | CI |
| AY105958 | 0.003 | 0.536 | 0.749 | CI |
| AY106496 | 0.100 | 0.781 | 0.677 | CI |
| AY106970 | 0.028 | 0.804 | 0.279 | CI |
| AY107228 | 0.003 | 0.754 | 0.690 | CI |
| AY107903 | 0.082 | 0.553 | 0.371 | CI |
| AY108187 | 0.003 | 0.216 | 0.115 | CI |
| AY108246 | 0.081 | 0.514 | 0.473 | CI |
| AY108543 | 0.031 | 0.580 | 0.708 | CI |
| AY110082 | 0.003 | 0.114 | 0.551 | CI |
| AY112083 | 0.001 | 0.181 | 0.382 | CI |
| AY104439 | 0.010 | 0.013 | 0.015 | Unknown |
| AY105062 | 0.469 | 0.811 | 0.701 | Unknown |
| AY107907 | 0.105 | 0.927 | 0.145 | Unknown |
| AY111546 | 0.024 | 0.010 | 0.054 | Unknown |
| AY112154 | 0.026 | 0.011 | 0.015 | Unknown |
| AY111438* | NA | NA | NA | |
| 15 reference loci** | 0.914 | 0.862 | 0.901 | |
Although these designations are admittedly approximate (see “Discussion”), patterns of genetic diversity in the two classes roughly match expectations (Fig. 1). For example, most improvement genes retain a relatively large proportion of genetic diversity in the landrace sample, averaging 63% of diversity, as measured by π, compared to the teosinte sample. In the inbred sample, this number decreases dramatically to 3%, consistent with the most marked declines in diversity occurring during the process of crop improvement. In contrast, domestication genes exhibit extensive losses of diversity between the teosinte and the landrace sample, retaining only 6% of diversity on average in the landraces relative to teosinte. The forces acting to reduce diversity in domestication genes did so by the time of formation of primitive landraces.
Gene Ontology Annotations of Selected and Neutral Genes
A prominent idea in the evolution of plant form is that major phenotypic changes are driven by changes in transcriptional regulators, with the thought that these genes act as switches between phenotypic states (Doebley and Lukens, 1998). Thus far, this notion seems to be consistent with phenotypic changes in crop plants, in that genes associated with major phenotypic changes during domestication are enriched for transcription functions (Doebley et al., 2006). However, the number of genes studied carefully to date is small. Molecular population genetics approaches have greatly increased the number of genes of potential importance during domestication or crop improvement, providing the opportunity to assess whether this class of genes is also enriched for transcription functions. Critically, the maize studies have also provided a set of nonselected, reference genes for comparison.
To examine the hypothesis that domestication and crop improvement loci are biased in favor of transcriptional regulators, we compared gene ontology (GO) assignments between selected and reference genes. Our set of 48 selected genes included all of those identified by molecular population genetic approaches (Wright et al., 2005; Yamasaki et al., 2005) as well as 10 genes identified by functional analyses, such as tb1 (Doebley et al., 1995), tga1 (Wang et al., 2005), c1 (Hanson et al., 1996), and others (Supplemental Table S3). The comparison set consisted of 658 genes deemed to be nonselected by Wright et al. (2005) (also see “Materials and Methods”). Using the InterPro database, we functionally annotated genes in the selected and reference sets, and made GO assignments. InterPro searches resulted in the assignment of GO terms for 54% of the 658 neutral reference genes and 51% of the 48 candidate selected genes, reducing our dataset for GO analysis by half.
We first tested for overrepresentation of GO functional categories between the selected and reference genes using GeneMerge (Castillo-Davis and Hartl, 2003). No significant differences were identified in any functional category between the selected and neutral gene datasets (data not shown). To simplify the comparison, we also compared the proportion of genes identified with a transcription-related GO function between the selected and reference sets. The selected set had 4% of genes with transcription-related functions, and the reference set had 12% (selected versus reference; χ2 = 1.54, P = 0.214). Thus, with this expanded data set, there is no obvious trend toward transcription-related functions for genes with an adaptive history.
It is possible that transcription factors are particularly important in the initial steps of domestication, during the initiation of major phenotypic changes. We thus hypothesized that domestication genes could be more biased for transcription factors than improvement genes. We compared the proportion of transcription-related GO functions among domestication genes, improvement genes, and reference genes. None of the pairwise contrasts between these three gene classes were significant (data not shown), but it must also be noted that statistical power was low due to a small number of observations in individual categories.
Electronic Northern
Selected genes did not have biased GO functions relative to reference genes, but do selected genes differ in expression profile relative to reference genes? To answer this question, we performed an electronic northern (e-northern) analysis. Expression was based on a database of 679,266 maize EST sequences produced from 171 cDNA libraries. We also examined a subset of 87 libraries confirmed to be nonnormalized (Supplemental Table S2), because nonnormalized libraries should provide a more quantitative measure of gene expression.
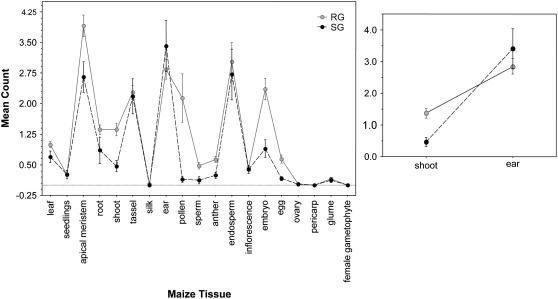
We BLASTn queried our set of 48 selected and 658 reference maize loci to EST data. Based on a BLAST e value < 10−30, we identified 38,747 hits for the screen of all libraries and 19,888 hits for the screen of nonnormalized libraries. These hits provided count data for each gene query, pooled across cDNA libraries representing approximately 20 maize tissues or tissue combinations (Supplemental Table S3). The average count for all tissues was 1.19 for the 658 reference genes (range of 0–10.375) and 0.844 (0.08–3.24) for candidate selected loci. A large subset of these hits occurred in libraries created with mixed maize tissues, and thus they could not be incorporated into tissue-specific analyses. However, comparison of e-northern counts from 19 distinct (nonmixed) maize tissues revealed that maize candidate selected loci were consistently underrepresented, on average, compared to neutral reference loci in all but two tissues: the ear and the pericarp (Fig. 2). This general pattern was also observed when analyses were limited to nonnormalized libraries, but note that tissue-specific (i.e. nonmixed) cDNA samples were not available from pericarp tissue, which prohibited inclusion of pericarp in analyses based solely on nonnormalized libraries (Fig. 3).
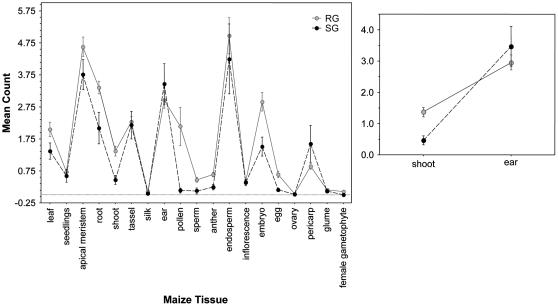
Figure 2.
Tissue expression patterns for results of e-northern screens of all cDNA libraries, including both normalized and nonnormalized libraries, for neutral reference loci (RG) and candidate selected loci (SG). Expression patterns for ear and shoot tissues are illustrated separately. Error bars represent ±1 se.
Figure 3.
Tissue expression patterns for results of e-northern screens of nonnormalized cDNA libraries for neutral reference loci (RG) and candidate selected loci (SG). Expression patterns for ear and shoot tissues are illustrated separately. Error bars represent ±1 se.
Principal components analysis (PCA) reduced the set of 19 maize tissues to eight factors, which accounted for approximately 75% of the variance in the nonnormalized count data (Table III). The first principal component explained the largest proportion (23%) of the variance and predominantly represented the maize shoot. The second two principal components explained >10% of the variance and represented male reproductive tissues (pollen and anther) and the maize ear, respectively. In comparison, PCA analyses of the e-northern data for all cDNA libraries resulted in similar tissue combinations but shifted factor loadings so that the first principal component corresponded to the maize ear and subsequent components corresponded to male reproductive tissues, the female gametophyte, and the maize shoot, respectively.
Table III.
Oblique factor loadings for the first four principal components for a subset of maize tissues in both the nonnormalized and complete e-northern datasets
Only factor loadings greater than 0.25 are shown.
| Maize Tissue | Nonnormalized Libraries
|
All Libraries
|
||||||
|---|---|---|---|---|---|---|---|---|
| PC1 | PC2 | PC3 | PC4 | PC1 | PC2 | PC3 | PC4 | |
| Shoot | 0.927 | – | – | – | – | – | – | 0.793 |
| Root | – | – | – | – | 0.258 | – | – | – |
| Leaf | 0.334 | – | – | – | – | – | – | 0.805 |
| Inflorescence | – | 0.639 | 0.362 | – | 0.484 | – | 0.603 | – |
| Pollen | – | 0.846 | – | – | – | – | 0.849 | – |
| Anther | – | 0.809 | – | – | – | – | 0.824 | – |
| Ear | – | – | 0.892 | – | 0.861 | – | – | – |
| Female gametophyte | – | – | – | – | – | 0.856 | – | – |
| Ovary | – | – | – | 0.862 | – | – | – | – |
| Cumulative proportion of variance explained | 0.211 | 0.317 | 0.413 | 0.480 | 0.223 | 0.321 | 0.415 | 0.482 |
We used log-linear analysis to examine the effects of tissue (maize ear or shoot), gene status (selected or neutral), and the tissue × status interaction on patterns of gene expression resulting from e-northern screens of all libraries and the nonnormalized library subset. Shoot tissue was selected for comparison with ear tissue based on principal component scores for individual tissue variables. In both normalized and nonnormalized libraries, we found a significant effect of tissue and the tissue × status interaction (Table IV), and near significance of the effect of gene status. Namely, candidate selected loci were significantly overexpressed in maize ear tissue relative to maize shoot tissue. We also ran the same model to compare ear count data with a broader vegetative category of combined leaf, root, and shoot tissues and found a similar interactive effect of the expression patterns for selected and reference genes (tissue × status; P = 0.0164). To sum, candidate selected loci are overexpressed in maize ear tissue relative both to reference genes and to other tissues. These results were robust for analyses of both normalized and nonnormalized libraries and also for the alternate BLAST values used to detect homology (data not shown).
Table IV.
Summary of log-linear analysis designed to test the effects of maize tissue (ear versus shoot) and gene status (selected versus neutral) on e-northern gene expression data
Results are presented for e-northern analyses representing all libraries and nonnormalized libraries for the e-value criterion of 10−30. Degrees of freedom are the same for both tests of nonnormalized and complete libraries. Num DF, Numerator degrees of freedom. Den DF, Denominator degrees of freedom.
| Effect | Num DF | Den DF | Nonnormalized Libraries
|
All Libraries
|
||
|---|---|---|---|---|---|---|
| χ2 | P | χ2 | P | |||
| Tissue | 1 | 1,410 | 43.51 | <0.0001 | 45.65 | <0.0001 |
| Status | 1 | 1,410 | 3.53 | 0.0603 | 3.70 | 0.0543 |
| Tissue × status | 1 | 1,410 | 7.43 | 0.0064 | 7.13 | 0.0076 |
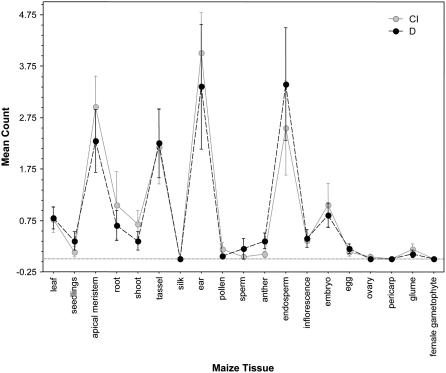
As a final step, we examined e-northern expression patterns between the early domestication and crop improvement subcategories using the full candidate gene set excluding the six genes unassigned by MLHKA analyses. Overall, tissue expression patterns were highly similar for early domestication and crop improvement loci (Fig. 4). We noted a qualitative increase in expression of crop improvement genes relative to domestication genes in several tissues (e.g. meristem and ear tissue), but detected no statistical difference in tissue expression patterns between the two gene classes.
Figure 4.
Tissue expression patterns for results of e-northern screens of nonnormalized libraries for 48 candidate crop improvement (CI) and domestication (D) genes. Error bars represent ±1 se.
DISCUSSION
The Timing of Selection
Plant domestication fundamentally altered the course of human history, and humans still rely on crops that were domesticated approximately 7,000 to 12,000 years ago (Harlan, 1992). These crops have been subjected to selection continuously since their domestication, resulting in dramatic morphological shifts. In the case of maize, early domestication pressures contributed to the domestication syndrome (Hammer, 1984), including the striking alteration of the maize ear. More recently, modern crops have sustained greater yields and altered traits such as leaf angle, starch content, root lodging, and tassel weight (Duvick and Cassman, 1999). Morphological shifts resulting from domestication are important examples of the evolutionary process, but they also have broad economic and societal consequences. The isolation of the genes that contribute to phenotypic changes may facilitate further trait manipulation through modified breeding strategies (McCouch, 2004).
In this article, we build on the philosophical paradigm that understanding the process, targets, and the outcome of artificial selection is an important prerequisite for identifying and characterizing genes that contribute to agronomic phenotypes (Ross-Ibarra et al., 2007). We began with genes that had already been characterized as candidate selection genes through population genetic approaches (Wright et al., 2005; Yamasaki et al., 2005). Our first goal was to identify whether the loci were targeted during early domestication or more recently during the process of crop improvement. We compared sequence diversity of 30 candidate and 15 reference loci between the three populations of teosintes, maize landraces, and maize inbred lines, detecting an overall decline in genetic variation ranging from 27% in reference loci to 95% in candidate selected loci. Tajima's D statistics were higher, on average, in maize populations relative to teosintes, consistent with expectations of the loss of low frequency variants resulting from a domestication bottleneck (Tenaillon et al., 2004).
By comparing polymorphism among the three populations, we inferred that nine genes were targeted by selection relatively early in the domestication process, with approximately 15 genes showing evidence of more recent selection (Table II). The ratio of domestication to improvement genes is thus roughly 3 to 5 for our sample of genes. Yamasaki et al. (2005) also found a roughly equal ratio between domesticated and improvement genes, based on a different set of loci and test criteria. We should note, however, three limitations to our classification of genes as either domestication or improvement. First, we could not classify five loci either because the MLHKA test was significant for the teosinte sample or because the test was not significant with any Zea sample. These latter genes may represent false positives from the original analyses of Wright et al. (2005), but the discrepancy could also reflect differences in statistical tests. The MLHKA test is based on the ratio of outgroup divergence to polymorphism and, while relatively robust, does not explicitly consider demographic history, such as a population bottleneck (Hudson et al., 1987; Wright and Gaut, 2005). In contrast, Wright et al. (2005) explicitly considered demography but did not include outgroup information.
Second, we performed multiple MLHKA tests, potentially leading to a high experiment-wide error rate that favors the detection of selection. However, multiple test corrections such as the Bonferroni can be unduly conservative (Ryman and Jorde, 2001; Storey and Tibshirani, 2003). For example, using the Bonferroni correction, the MLHKA tests required a significance cutoff of P < 0.0006, and under this criterion none of the genes exhibited any evidence of selection. This strict interpretation is not merited, both because the genes were inferred to be under selection in a prior study that used different statistical approaches and also because we are interested in the result from each gene as opposed to only experiment-wide inferences. We were also lenient with respect to the interpretation of P values for significance, using a cutoff of P < 0.10. A more stringent P value would have altered the ratio of domestication to improvement genes to be 4:16 (Table II).
The third limitation to our designation of domestication and improvement genes is that these two classes are at best approximate. The classification scheme artificially assumed the historical process leading to elite maize germplasm is bimodal, where in fact it has probably been continual and ongoing. If the effects of selection are cumulative over this process, then there is more statistical power to detect episodes of crop improvement, and we may have underestimated the ratio of domestication to improvement genes. In addition, our study assumed that the landrace and teosinte data represent historical samples, when both are in fact present-day samples. Nonetheless, it is unmistakable that some selection events occurred early in the history of maize. For example, over 4,000 years ago selection for maize alleles was complete at two loci responsible for major morphological differences between maize and teosinte (Jaenicke-Despres et al., 2003). Here we extend those results to suggest that roughly half of the genes selected in the maize genome may have been targeted relatively early in crop history, prior to the geographic dispersal of maize landraces and preceding modern methods of improvement.
It is interesting to consider the ratio of domestication to improvement genes in light of the original discovery of five major genomic regions responsible for the morphological differences between teosinte and maize (Beadle, 1939; Doebley et al., 1990). Doebley et al. (1990) postulated that key traits are represented by a few genes of large effect, but also that there are likely many genes of small effect that also contribute to morphological change. Traditional QTL studies to date have isolated only genes of major effect (Doebley et al., 2006), based on a priori knowledge of phenotypic divergence. However, genomic screens have estimated that approximately 1,200 maize genes harbor the signature of selection. Molecular population genetic approaches may thus be more likely than QTL analysis to identify small-effect genes. Our results, coupled with previous results (Yamasaki et al., 2005), imply that several hundred genes were under selection early during the process of domestication, prior to the geographic dispersal of maize landraces.
Functional Biases and Transcription Factors
Major genes that contribute to morphological differences between crops and their wild ancestors are enriched for transcription factor functions, but the sample of available genes is small, consisting of about 30 genes over several crops (Doebley et al., 2006). In maize, a series of population genetic studies have identified at least 48 genes that have a high probability of selection and presumably contribute to agronomic phenotypes. This expanded set provided the opportunity to assess whether selected genes also exhibit a bias toward transcription function, based on gene ontologies. Comparison of the 48 selected loci against 658 neutral loci did not detect evidence that transcriptional regulators were overrepresented among selected genes. Instead, our candidate gene dataset represented a broad range of molecular functions. However, we were unable to assign GO terms to 25 of 48 candidate genes and 355 of 658 neutral reference genes, limiting statistical power to detect differences. Further, InterPro searches failed to identify two selected genes as transcription factors, despite a priori evidence for the regulatory status of those genes.
Results of GO analyses did not support the hypothesis that candidate selected genes are biased in favor of transcriptional regulators. Even if our GO result is robust, it is possible that molecular population genetic screens identify genes with small effects that are difficult to detect using traditional QTL and association mapping approaches (Ross-Ibarra et al., 2007). In this case, selected genes may represent a mix of major genes, such as the transcriptional regulators tb1 and tga1, and minor genes without obvious morphological effects. Consequently, the Doebley-Lukens model (Doebley and Lukens, 1998) may be correct for genes that contribute to the modification of plant form, but perhaps needs to be revisited for selected genes with small phenotypic effects.
Tissue-Specific Expression in the Ear: Cause or Effect?
Analyses of gene expression profiles for 706 loci (48 selected and 658 reference) revealed that candidate selected genes were significantly overexpressed in maize ear tissue relative to vegetative tissues (Figs. 2 and 3; Table IV). The maize ear is one of the most prominent examples of morphological change documented in crop domestication (Doebley, 2004). These results indicate that our sample of selected genes exhibit significant differences in expression patterns in tissues that experienced high levels of morphological divergence during domestication.
Similar to MLHKA analyses, our e-northern results must be viewed in light of several weaknesses (Peri et al., 2001). First, we were limited to available cDNA libraries, and e-northern screens likely missed significant expression patterns in tissues excluded from the EST database (such as the maize pericarp that was present only in normalized or subtracted libraries). Second, many cDNA libraries were constructed from combinations of maize tissues and were therefore uninformative for tissue-specific analyses. Third, it is possible that expression of paralogous genes, particularly nearly identical paralogs (Emrich et al., 2007), complicate our results. Nonetheless, the key observation that selected genes are expressed at high levels in the maize ear was robust for several treatments, including variations in the BLAST selection criterion (e-value ≤ 10−30 or ≤10−50) and the use of normalized and nonnormalized libraries.
There remains the question of the underlying mechanism for our observation that selected genes are overexpressed in ear tissue. Were candidate loci selected for up-regulation in the maize ear? Altered expression levels of selected genes have been documented in several crop species (Doebley et al., 2006). For example, the domestication genes tb1 in maize and Q in wheat appear to have been selected for overexpression of gene products, and altered expression levels are sufficient to explain the domesticated phenotype (Doebley et al., 1997; Simons et al., 2006). Similarly, the domestication gene fw2.2 in tomato (Solanum lycopersicum) was selected for repression or underexpression of gene products (Nesbitt and Tanksley, 2001; Cong et al., 2002). Given precedence for shifts in expression under selection, it is possible that the expression of many candidate genes was increased in response to selection.
There is, however, an intriguing alternative. It is possible that selected loci were already expressed at high levels in the protoear tissues of teosinte, and thus were convenient targets for artificial selection on ear-related traits. Further research is needed to distinguish between this cause and effect, with appropriate controls for genetic background. One possibility will be to measure cis-allelic expression in F1 hybrids between maize and teosinte for a series of selected loci (e.g. Clark et al., 2006; de Meaux et al., 2006); this approach controls for background genetic effects and could distinguish whether selection resulted in gene expression changes. More generally, relatively little is known about the targets, mechanisms, and effects of selection at the level of genes and gene expression, and thus additional focus on this question will have broad significance. The outstanding question remains as to whether high expression of genes facilitated selection, or instead, selection led to up-regulation of gene expression.
MATERIALS AND METHODS
The names of products are necessary to report factually on available data; however, neither the U.S. Department of Agriculture nor the University of California, Irvine guarantees or warrants the standard of the product and the use of the name does not imply the approval of the product to the exclusion of others that may also be suitable.
Plant Materials and Sequence Polymorphism
For a sample of 16 maize (Zea mays sp. mays) landraces described by Tenaillon et al. (2001), we sequenced 30 of the original 32 loci ranked by Wright et al. (2005) as most likely to have experienced selection according to their posterior probability measure (Supplemental Table S1). In addition, we chose 15 of the original 774 genes with low (<0.05) selection probabilities as neutral reference loci. Sequences were generated by protocols described in Yamasaki et al. (2005). Briefly, both forward and reverse primers were used in touchdown PCR to amplify each locus. PCR products were sequenced using the Big Dye Terminator sequencing kit (Applied Biosystems) and analyzed with an ABI 3100 sequencer.
Sequences were aligned by both ClustalW (Thompson et al., 1994) and by manual editing with BioEdit version 7.0.5.2 (Hall, 1999). Base calling and quality checks were conducted using PHRED, and sequence assembly was accomplished with PHRAP (Ewing and Green, 1998; Ewing et al., 1998). DNA sequences for maize landraces were removed from the dataset in cases where PHRAP quality scores averaged <30, or sequences represented <70% of the average sequence length of corresponding maize inbred lines. Also, single nucleotide polymorphisms were only considered reliable if both variants had quality scores >30. These criteria resulted in an average of 11 sequences per locus for the 16 maize landraces. The landrace sequences have been deposited in GenBank (BV722945–BV723470). DNA sequences for maize inbred lines and a sample of teosintes were available from the study of Wright et al. (2005).
Population Genetic Analyses and Tests for Selection
Population genetic parameters were measured for each locus and population (maize inbred lines, landraces, and teosintes) using DNAsp software version 4.10.3 (Rozas et al., 2003). We calculated the number of segregating sites, pairwise nucleotide diversity (π), the number of haplotypes, Watterson's estimator (θ) of diversity, and Tajima's D for both candidate selected genes and reference loci. Comparisons of genetic parameters among populations were made using Kruskal-Wallis and Student's t tests.
Candidate agronomic loci were tested for evidence of selection during periods of early domestication or later crop improvement using a maximum likelihood version of the Hudson, Kreitman, Aguadé (HKA) test (Hudson et al., 1987) developed by Wright and Charlesworth (2004) and available from S.I. Wright (http://www.yorku.ca/stephenw/). Similar to the HKA test, the MLHKA compares sequence polymorphism within species to rates of divergence between species to test for deviation from the neutral model. However, while HKA tests do not identify which loci deviate from expectations, MLHKA is capable of directly testing specific loci for evidence of selection.
We conducted MLHKA tests for each locus and each population of maize inbreds, landraces, and teosintes using Sorghum bicolor as the outgroup taxon. Outgroup sequences were identified by BLAST comparisons of the longest maize inbred sequences to the S. bicolor GSS and EST databases at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov), and selected using an e-value BLAST criterion <10−7. No outgroup was available for one of 30 candidate loci (GenBank accession no. AY111438) and it was subsequently excluded from MLHKA analyses. To conduct the tests, we ran 100,000 simulations in MLHKA to compare the fit of a neutral model to a model specifying each selected gene in each population. We used the 15 neutral genes as reference loci for comparison with candidate genes. As a group these 15 loci did not deviate from neutral expectations based on the standard HKA test (Table II), as implemented in SITES (http://lifesci.rutgers.edu/∼heylab/ProgramsandData/Programs/SITES). Following Yamasaki et al. (2005), we designated candidate loci to be domestication genes if MLHKA was significant for both maize inbreds and landraces. Similarly, we designated candidate loci to be crop improvement genes if significant declines in diversity were only apparent in maize inbreds.
e-Northern
We downloaded the April 20, 2006 release of maize mRNA ESTs (dbEST) from the PlantGDB Web site (http://www.plantgdb.org/). The download consisted of 679,266 EST sequences. We sorted 171 cDNA libraries into 20 categories of specific maize tissues or tissue combinations following the National Center for Biotechnology Information-listed tissue types or communications with library authors. At the same time, we determined if libraries represented normalized data. Sequences were excluded from the dataset if information regarding library normalization was unavailable (Supplemental Table S2).
To conduct the e-northern, the dbEST was formatted and parsed using Perl to extract the accession number and corresponding tissue for all ESTs matching query sequences. Queries of the dbEST included full-length GenBank maize sequences representing the 30 candidate agronomic loci, an expanded set of 658 neutral loci, and an additional 18 selected genes from the maize literature. The neutral dataset consisted of all genes with posterior probabilities of selection less than 0.05 based on the analyses of Wright et al. (2005), and included the 15 neutral genes used in MLHKA comparisons. Newly added candidate genes consisted of eight loci discovered by Yamasaki et al. (2005), an additional gene from Wright et al. (2005), and nine genes known to be selected in maize based on published research (Supplemental Table S3). Prior to e-northern analyses, the supplementary candidate genes were sorted into domestication or crop improvement categories based on published evidence for ancestral or recent selection.
Given that the number of EST matches only corresponded to quantitative tissue expression patterns if libraries were nonnormalized (Peri et al., 2001), we performed the e-northern for both a reduced database of 87 nonnormalized libraries as well as the full database of 171 libraries. Alignment criteria for matching sequences or hits were established for expected values (e-values) of ≤10−30 or less to ensure an exact match for each gene query. For comparison, we repeated the e-northern using more stringent criteria for e-values of ≤10−50. e-northern results did not differ qualitatively between e-values and we only report results for analyses using the ≤10−30 criteria.
The outcome of e-northern analyses represented tissue counts for the two categories of candidate selected loci and neutral loci in maize. We next performed PCA to identify and extract patterns of gene expression among maize tissue categories. PCA was performed with oblique rotation of eigenvectors because the resulting factor variables better represented the biological organization of maize tissues (e.g. reproductive and vegetative categories). The eigenvectors were examined to determine which tissues contributed to each factor and we assumed that variables with high factor loadings best represented the variation in the dataset (Dunteman, 1989). Based on PCA analysis and a priori hypotheses that tissue expression patterns would differ in the maize ear, we selected data representing vegetative (shoot) and reproductive (ear) tissues to investigate differences in expression between the two gene categories (selected or neutral). Log-linear analysis to test the interaction of tissue type on gene status was conducted using PROC GENMOD in SAS version 9.1 with a loglink Poisson distribution and DSCALE option to allow for overdispersion of the data. We fit the model for both the full e-northern dataset as well as the reduced dataset of nonnormalized libraries.
GO
Functional characterization of the selected gene dataset and the complete (658) neutral gene dataset was performed using InterPro (Mulder et al., 2005). Six frame translations of the sequences were generated from which all open reading frames of two or more amino acids were selected. Amino acid sequences were compared to InterPro Release 12.0 using InterProScan (Quevillon et al., 2005) version 4.2, utilizing the option for mapping to GO terms (Ashburner et al., 2000). For each initial maize sequence, GO terms that mapped to any of its derived amino acid sequences were extracted. We subsequently ran GeneMerge software (Castillo-Davis and Hartl, 2003) to test for significant differences between the assigned GO categories of selected and neutral genes. As a last step, we tested the candidate gene dataset for enrichment in 123 GO terms specifying transcription factors using χ2 comparisons of selected and neutral genes (Sokal and Rholf, 1995). χ2 analyses were repeated for the subset of candidate selected loci to test whether the number of transcription factors differed between genes selected during early domestication and genes selected during later crop improvement.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers BV722945 to BV723470.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. Descriptive statistics and population genetic parameters for 30 maize candidate agronomic loci and 15 reference loci in maize inbred lines, landraces, and teosintes.
Supplemental Table S2. PlantGDB cDNA libraries, normalization status (N, Normalized; UN, nonnormalized), and corresponding maize tissues used in e-northern screens of maize candidate neutral and selected loci.
Supplemental Table S3. e-northern results based on comparisons of 658 neutral loci and 48 selected loci in maize with the PlantGDB mRNA EST database as of April 20, 2006.
Supplementary Material
Acknowledgments
The authors thank Ms. Katherine Houchins for technical assistance.
This work was supported by the National Science Foundation (grant no. DBI 0321467).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Brandon S. Gaut (bgaut@uci.edu).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beadle GW (1939) Teosinte and the origin of maize. J Hered 30 245–247 [Google Scholar]
- Castillo-Davis CI, Hartl DL (2003) GeneMerge—post-genomic analysis, data mining, and hypothesis testing. Bioinformatics 19 891–892 [DOI] [PubMed] [Google Scholar]
- Clark RM, Wagler TN, Quijada P, Doebley J (2006) A distant upstream enhancer at the maize domestication gene tb1 has pleiotropic effects on plant and inflorescent architecture. Nat Genet 38 594–597 [DOI] [PubMed] [Google Scholar]
- Cong B, Liu J, Tanksley SD (2002) Natural alleles at a tomato fruit size quantitative trait locus differ by heterochronic regulatory mutations. Proc Natl Acad Sci USA 99 13606–13611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Meaux J, Pop A, Mitchell-Olds T (2006) Cis-regulatory evolution of chalcone-synthase expression in the genus Arabidopsis. Genetics 174 2181–2202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley J (2004) The genetics of maize evolution. Annu Rev Genet 38 37–59 [DOI] [PubMed] [Google Scholar]
- Doebley J, Gaut BS, Smith BD (2006) The molecular genetics of crop domestication. Cell 127 1309–1321 [DOI] [PubMed] [Google Scholar]
- Doebley J, Lukens L (1998) Transcriptional regulators and the evolution of plant form. Plant Cell 10 1075–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley J, Stec A (1991) Genetic-analysis of the morphological differences between maize and teosinte. Genetics 129 285–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley J, Stec A, Gustus C (1995) Teosinte branched1 and the origin of maize: evidence for epistasis and the evolution of dominance. Genetics 141 333–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley J, Stec A, Hubbard L (1997) The evolution of apical dominance in maize. Nature 386 485–488 [DOI] [PubMed] [Google Scholar]
- Doebley J, Stec A, Wendel J, Edwards M (1990) Genetic and morphological analysis of a maize teosinte F2 population—implications for the origin of maize. Proc Natl Acad Sci USA 87 9888–9892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunteman GH (1989) Principal Components Analysis. Sage University Paper Series on Quantitative Applications in the Social Sciences, 07-069. Sage Publications, Newbury Park, CA
- Duvick DN, Cassman KG (1999) Post-green revolution trends in yield potential of temperate maize in the north-central United States. Crop Sci 39 1622–1630 [Google Scholar]
- Emrich SJ, Li L, Wen TJ, Yandeau-Nelson MD, Fu Y, Guo L, Chou HH, Aluru S, Ashlock DA, Schnable PS (2007) Nearly identical paralogs: implications for maize (Zea mays L.) genome evolution. Genetics 175 429–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evenson RE, Gollin D (2003) Assessing the impact of the green revolution, 1960 to 2000. Science 300 758–762 [DOI] [PubMed] [Google Scholar]
- Ewing B, Green P (1998) Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res 8 186–194 [PubMed] [Google Scholar]
- Ewing B, Hillier L, Wendl MC, Green P (1998) Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res 8 175–185 [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A, Gaut RL, Hilton H, Feldman DL, Gaut BS (1998) Investigation of the bottleneck leading to the domestication of maize. Proc Natl Acad Sci USA 95 4441–4446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallavotti A, Zhao Q, Kyozuka J, Meeley RB, Ritter M, Doebley JF, Pe ME, Schmidt RJ (2004) The role of barren stalk1 in the architecture of maize. Nature 432 630–635 [DOI] [PubMed] [Google Scholar]
- Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41 95–98 [Google Scholar]
- Hammer K (1984) Das domestikationssyndrom. Kulturpflanze 32 11–34 [Google Scholar]
- Hanson MA, Gaut BS, Stec AO, Fuerstenberg SI, Goodman MM, Coe EH, Doebley JF (1996) Evolution of anthocyanin biosynthesis in maize kernels: the role of regulatory and enzymatic loci. Genetics 143 1395–1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlan JR (1992) Crops and Man, Ed 2. American Society of Agronomy, Madison, WI
- Hudson RR, Kreitman M, Aguade M (1987) A test of neutral molecular evolution based on nucleotide data. Genetics 116 153–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenicke-Despres V, Buckler ES, Smith BD, Gilbert MT, Cooper A, Doebley J, Paabo S (2003) Early allelic selection in maize as revealed by ancient DNA. Science 302 1206–1208 [DOI] [PubMed] [Google Scholar]
- Khush GS (2001) Green revolution: the way forward. Nat Rev Genet 2 815–822 [DOI] [PubMed] [Google Scholar]
- Matsuoka Y, Mitchell SE, Kresovich S, Goodman M, Doebley J (2002) Microsatellites in Zea—variability, patterns of mutations, and use for evolutionary studies. Theor Appl Genet 104 436–450 [DOI] [PubMed] [Google Scholar]
- McCouch S (2004) Diversifying selection in plant breeding. PLoS Biol 2 e347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerowitz EM (1994) Plant evolution: de-evolution and re-evolution of maize. Curr Biol 4 127–130 [DOI] [PubMed] [Google Scholar]
- Mulder NJ, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bradley P, Bork P, Bucher P, Cerutti L, et al (2005) InterPro, progress and status in 2005. Nucleic Acids Res 33 D201–D205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesbitt TC, Tanksley SD (2001) fw2.2 directly affects the size of developing tomato fruit, with secondary effects on fruit number and photosynthate distribution. Plant Physiol 127 575–583 [PMC free article] [PubMed] [Google Scholar]
- Paterson AH, Lin YR, Li Z, Schertz KF, Doebley JF, Pinson SRM, Liu SC, Stansel JW, Irvine JE (1995) Convergent domestication of cereal crops by independent mutations at corresponding genetic loci. Science 269 1714–1718 [DOI] [PubMed] [Google Scholar]
- Peri S, Ibarrola N, Blagoev B, Mann M, Pandey A (2001) Common pitfalls in bioinformatics-based analyses: look before you leap. Trends Genet 17 541–545 [DOI] [PubMed] [Google Scholar]
- Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, Apweiler R, Lopez R (2005) InterProScan: protein domains identifier. Nucleic Acids Res 33 W116–W120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Ibarra J, Morrell PL, Gaut BS (2007) Plant domestication, a unique opportunity to identify the genetic basis of adaptation. Proc Natl Acad Sci USA 104 8641–8648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R (2003) DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19 2496–2497 [DOI] [PubMed] [Google Scholar]
- Ryman N, Jorde PE (2001) Statistical power when testing for genetic differentiation. Mol Ecol 10 2361–2373 [DOI] [PubMed] [Google Scholar]
- Simons KJ, Fellers JP, Trick HN, Zhang Z, Tai YS, Gill BS, Faris JD (2006) Molecular characterization of the major wheat domestication gene Q. Genetics 172 547–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sluyter A, Dominguez G (2006) Early maize (Zea mays L.) cultivation in Mexico: dating sedimentary pollen records and its implications. Proc Natl Acad Sci USA 103 1147–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokal RR, Rholf FJ (1995) Biometry. W.H. Freeman, New York
- Storey JD, Tibshirani R (2003) Statistical significance for genomewide studies. Proc Natl Acad Sci USA 100 9440–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenaillon M, Sawkins MC, Long AD, Gaut RL, Doebley JF, Gaut BS (2001) Patterns of DNA sequence polymorphism along chromosome 1 of maize (Zea mays ssp. mays L.). Proc Natl Acad Sci USA 98 9161–9166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenaillon MI, U'Ren J, Tenaillon O, Gaut BS (2004) Selection versus demography: a multilocus investigation of the domestication process in maize. Mol Biol Evol 21 1214–1225 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) Clustal W—improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Nussbaum-Wagler T, Li B, Zhao Q, Vigouroux Y, Faller M, Bomblies K, Lukens L, Doebley JF (2005) The origin of the naked grains of maize. Nature 436 714–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright SI, Bi IV, Schroeder SG, Yamasaki M, Doebley JF, McMullen MD, Gaut BS (2005) The effects of artificial selection of the maize genome. Science 308 1310–1314 [DOI] [PubMed] [Google Scholar]
- Wright SI, Charlesworth B (2004) The HKA test revisited: a maximum-likelihood-ratio test of the standard neutral model. Genetics 168 1071–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright SI, Gaut BS (2005) Molecular population genetics and the search for adaptive evolution in plants. Mol Biol Evol 22 506–519 [DOI] [PubMed] [Google Scholar]
- Yamasaki M, Tenaillon MI, Bi IV, Schroeder SG, Sanchez-Villeda H, Doebley JF, Gaut BS, McMullen MD (2005) A large-scale screen for artificial selection in maize identifies candidate agronomic loci for domestication and crop improvement. Plant Cell 17 2859–2872 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.