Abstract
The modification of an endogenous gene into a designed sequence by homologous recombination, termed gene targeting (GT), has broad implications for basic and applied research. Rice (Oryza sativa), with a sequenced genome of 389 Mb, is one of the most important crops and a model plant for cereals, and the single-copy gene Waxy on chromosome 6 has been modified with a frequency of 1% per surviving callus by GT using a strong positive-negative selection. Because the strategy is independent of gene-specific selection or screening, it is in principle applicable to any gene. However, a gene in the multigene family or a gene carrying repetitive sequences may preclude efficient homologous recombination-promoted GT due to the occurrence of ectopic recombination. Here, we describe an improved GT procedure whereby we obtained nine independent transformed calli having the alcohol dehydrogenase2 (Adh2) gene modified with a frequency of approximately 2% per surviving callus and subsequently isolated eight fertile transgenic plants without the concomitant occurrence of undesirable ectopic events, even though the rice genome carries four Adh genes, including a newly characterized Adh3 gene, and a copy of highly repetitive retroelements is present adjacent to the Adh2 gene. The results indicate that GT using a strong positive-negative selection can be widely applicable to functional genomics in rice and presumably in other higher plants.
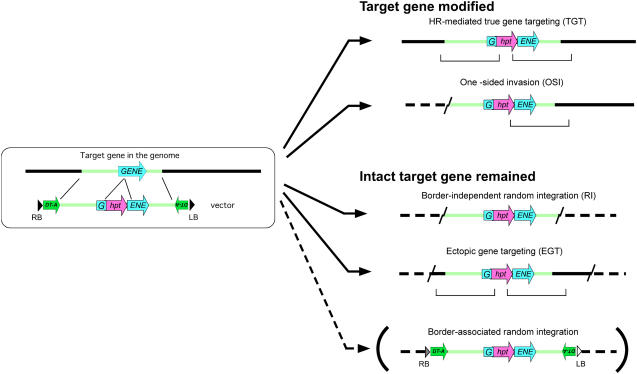
Although the modification of endogenous mouse genes by gene targeting (GT) via homologous recombination (HR) is common practice (Evans et al., 2001), only a few reproducible GT events of endogenous genes that resulted in generating fertile transgenic plants have been reported (Hanin et al., 2001; Terada et al., 2002; Shaked et al., 2005). While HR-mediated sequence-specific integration of a transgene in mouse embryogenic stem cells has been reported to be 1% or higher of the total integration events, most of which are random integration (RI) of the transgene by nonhomologous end joining (NHEJ; Jasin et al., 1996), the targeted integration of a transgene into the endogenous homologous sequence in higher plants has been regarded to be in the order of 0.01% to 0.1% compared with RI (Puchta, 2002; Hanin and Paszkowski, 2003; Reiss, 2003; Iida and Terada, 2004, 2005). In addition to RI, the concomitant occurrence of undesirable ectopic recombinations, such as one-sided invasion (OSI) and ectopic GT (EGT), has often been detected (Puchta, 2002; Hohn and Puchta, 2003; Iida and Terada, 2005; Endo et al., 2006b). OSI results from one homologous crossover and another NHEJ at the target locus, while EGT is thought to be generated by RI of a recombinant molecule produced by HR between the introduced transgene and a copy of the target sequence without altering the gene to be targeted (Fig. 1).
Figure 1.
Integration events of a transgene associated with homology-dependent GT with positive-negative selection. The hpt and DT-A genes were used as positive and negative selection markers, respectively (Terada et al., 2002). The large black arrowheads with RB and LB indicate the right and left borders of T-DNA, respectively. The anticipated TGT is generally regarded to occur via double crossovers at the hpt-flanking homologous regions on the vector, and the brackets under the maps indicate the junction fragments generated by the crossovers. One HR and another NHEJ at the target locus result in OSI. The majority of the surviving calli with our strong positive-negative selection were found to contain truncated T-DNA molecules mediated by border-independent RI, whereas the most efficient RI of T-DNA mediated by border-associated RI (shown in large parentheses), in which the integrated single T-DNA molecules contain the entire T-DNA segment with a well-conserved right border (dark gray arrowhead) and either conserved or slightly truncated left border (white arrowhead) sequences (Tinland and Hohn, 1995; Brunaud et al., 2002; Tzfira et al., 2004), would result in the killing of transformed calli by the expressed DT-A protein. These RI events in the transformed calli should be mediated by NHEJ. HR-promoted double crossovers between the transgene and a copy of the target sequence and subsequent RI of the resulting recombinant molecule by NHEJ generate EGT. It is noteworthy that the target gene remains intact in both RI and EGT. Zigzag lines represent breakpoints generated by NHEJ.
To detect true GT (TGT) efficiently among the overwhelming RI events, one approach is to apply either gene-specific selection or screening for the target genes; the Arabidopsis (Arabidopsis thaliana) protoporphyrinogen oxidase (PPO) gene was chosen for direct gene-specific selection, in which targeted plants acquired herbicide resistance (Hanin et al., 2001), and the Arabidopsis Cruciferin gene encoding a seed storage protein was employed for gene-specific screening, in which targeted integration of a promoterless GFP gene resulted in fluorescent seeds (Shaked et al., 2005). Another approach is to use a strong positive-negative selection for enriching targeted genes indirectly by reducing transformants with randomly integrated transgenes and to identify TGT subsequently by PCR screening; the mutants with the modified rice (Oryza sativa) Waxy gene have been obtained in this way (Terada et al., 2002; Iida and Terada, 2004, 2005). In Agrobacterium-mediated transformation, which has been used in all of the successful GT by HR in higher plants (Kempin et al., 1997; Hanin et al., 2001; Terada et al., 2002; Shaked et al., 2005; Endo et al., 2006b), T-DNA appears to integrate randomly throughout the plant genome as a single molecule or multiple sequences ligated to each other in various orientations (Tzfira et al., 2004). Because the majority of the integrated single-copy T-DNA molecules are known to contain the entire T-DNA segment with a well-conserved right border and a left-border sequence that is either conserved or truncated by a few to approximately 100 bp (Tinland and Hohn, 1995; Brunaud et al., 2002; Zhu et al., 2006), which we collectively termed here as border-associated RI, two inversely oriented diphtheria toxin-A (DT-A) genes encoding the DT-A fragments were placed as lethal negative selection markers at both ends of the T-DNA segment adjacent to its border sequences in the vector used (Terada et al., 2002, 2004) to efficiently remove transgenes integrated through border-associated RI (Fig. 1). There appears to be another RI with relatively large deletions at both ends of the T-DNA segment without the border proximal regions (Matsumoto et al., 1990), which we collectively termed as border-independent RI, although they seem to occur much less frequently than the border-associated RI. Because border-independent RI is likely to result in transgenes without an active DT-A gene, transformants with such transgenes must be recovered as those having randomly integrated transgenes. To identify transformants formed by TGT (Fig. 1), systematic PCR-based screening was used to detect the junction fragments generated by HR (Terada et al., 2002; Iida and Terada, 2004). In principle, this second approach is applicable to any genes to which gene-specific selection or screening is difficult to adapt directly. Wild-type plants were used for modifying the PPO and Waxy genes (Hanin et al., 2001; Terada et al., 2002), whereas a transgenic line overexpressing the yeast (Saccharomyces cerevisiae) RAD54 gene for an SWI2/SNF2 chromatin-remodeling factor was used for GT in the Cruciferin gene to increase the HR-promoted GT frequencies more than 10-fold (Shaked et al., 2005). Although transgenic Arabidopsis plants expressing the yeast RAD54 gene were reported to exhibit no altered phenotypes except for resistance to γ-irradiation, such overactive recombination-repair plants may hamper the functional characterization of certain endogenous genes to be modified.
Although plant genes affecting HR are known (Britt and May, 2003; Reiss, 2003; Schuermann et al., 2005; Emmanuel et al., 2006; Endo et al., 2006a; Kirik et al., 2006), we continue pursuing the second approach for GT using the wild-type rice to avoid potential side effects caused by altering plant genes involved in the recombination and/or repair systems, because we are attempting to develop a general reverse genetic method for characterizing an endogenous gene by modifying the gene of interest (Terada et al., 2002; Iida and Terada, 2004, 2005). While Waxy is a unique gene in the rice genome, the alcohol dehydrogenase (Adh) genes, which play a key function for survival in response to an anaerobic condition (Sachs et al., 1996), comprise a small multigene family in the rice genome (International Rice Genome Sequencing Project, 2005). As a model gene residing adjacent to a repetitive sequence and belonging to a small multigene family, which may preclude efficient HR-promoted GT due to the possible frequent occurrence of ectopic recombination at the repetitive and/or homologous sequences in the gene family, we chose the Adh2 gene, which comprises 10 exons, because no adh2 mutation in rice has been reported and because a highly repetitive Copia-like element is present 1.0 kb downstream of Adh2 (Tarchini et al., 2000). By an improved GT procedure described here, we were able to obtain eight fertile targeted transgenic plants without the concomitant occurrence of undesirable ectopic events among transformed calli having Adh2 modified with a frequency of approximately 2% per surviving callus.
The moss Physcomitrella patens, which serves as a model plant for studying evolutionary developmental genetics, exhibits highly frequent GT capability (Schaefer, 2002; Hohe and Reski, 2003); soon after the first report on the efficient integrative HR of introduced vectors by Schaefer and Zryd (1997) using the wild-type moss protoplasts, the reproducible GT of two genes, ftsZ and DES6, was reported to generate knockout mutants conferring distinctive traits (Girke et al., 1998; Strepp et al., 1998). Since then, various gene functions have been characterized by HR-promoted gene disruptions in P. patens (Cove, 2005), which can be regarded as a reference organism concerning GT in plants (Puchta, 1998; Reski, 1998; Reiss, 2003). Because the characteristic features of GT in the moss system and our rice system are considerably different, however, the differences are also discussed.
RESULTS AND DISCUSSION
The Adh Genes in the Rice Genome
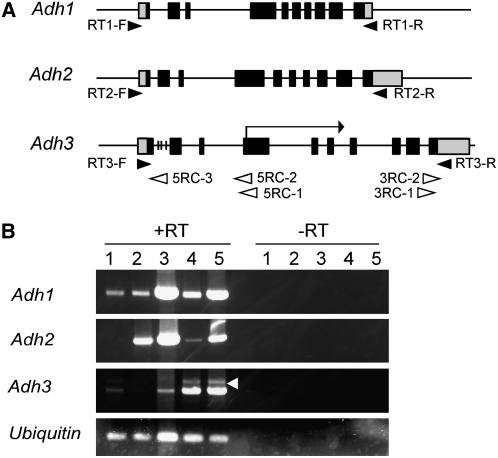
Available data indicate that the rice genome carries four copies of the Adh-related sequences (International Rice Genome Sequencing Project, 2005): Adh1 (X16296) and Adh2 (X16297) on chromosome 11 (Xie and Wu, 1989; Tarchini et al., 2000), Fdh1 (U77637) encoding glutathione-dependent formaldehyde dehydrogenase (class III ADH) on chromosome 2 (Dolferus et al., 1997), and another Adh-related gene on chromosome 11, which we designated here as Adh3. Because the full-length Adh3 cDNA reported (AK101288) lacks the ATG initiation codon, we reinvestigated the Adh3 transcripts (Fig. 2). The most upstream and two major transcriptional initiation sites of Adh3 were at −162, −81, and −53 bp from the ATG initiation codon, respectively, and the major and the longest 3′-untranslated region (UTR) were at 357 and 369 bp, respectively (AB267278). The Adh1, Adh2, and Adh3 genes all comprise 10 exons with identical intron positions and are clustered in the same orientation on chromosome 11 (Figs. 2A and 3A). Adh1 is expressed in leaves, roots, and coleoptiles, whereas Adh2 is mainly expressed in roots (Fig. 2B). Adh3 is chiefly expressed in coleoptiles, and additional larger Adh3 transcripts (AB267279) were found to contain the unspliced intron 1, which carries three in-frame stop codons (Fig. 2A). Among the four Adh genes, only a single adh1 mutant has been isolated in rice (Matsumura et al., 1998; Saika et al., 2006). Although an expressed gene (AK104537) on chromosome 10 was annotated to be Adh related (Tiger Rice Annotation Release 4, http://www.tigr.org), only partial homology to the four Adh genes could be found. We chose the Adh2 gene for GT in a small multigene family because: (1) no adh2 mutation in rice has been reported; (2) a highly repetitive Copia-like element is present 1.0 kb downstream of Adh2; (3) Adh2 resides in the middle of Adh1 and Adh3 in the genome; and (4) all three Adh genes are expressed significantly in rice calli (Fig. 2B), where somatic HR for GT occurs.
Figure 2.
Structure and expression of the Adh genes. A, Genomic structure of the Adh genes. The white and gray segments within exon boxes indicate the untranslated and coding regions, respectively. The black arrowheads indicate RT-PCR primers, and the white arrowheads indicate primers for 5′ or 3′ RACEs (Supplemental Table S1). The short vertical bars within Adh intron 1 indicate in-frame stop codons. The horizontal arrow with the vertical bar above the Adh3 map indicates the reported 5′ terminus of the full-length cDNA in the wild-type ‘Nipponbare’ (AK101288). B, Expression of the Adh genes detected by RT-PCR in various tissues of ‘Nipponbare’. The constitutively expressed Ubiquitin gene was used as an internal control, and −RT indicates RT-PCR amplification without SuperScript III reverse transcriptase. The white arrowhead in Adh3 points to RT-PCR-amplified fragments containing unspliced intron 1. Lane 1, Seedling leaves; lane 2, seedling roots; lane 3, calli; lane 4, coleoptiles; lane 5, submerged coleoptiles.
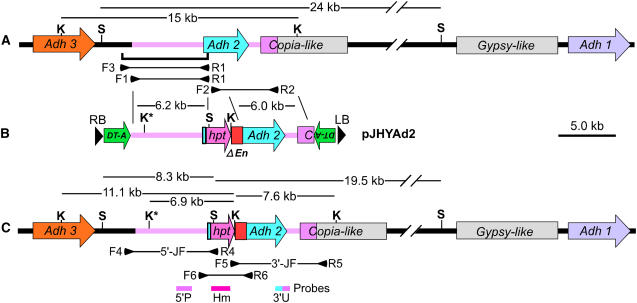
Figure 3.
Strategy for the modification of the Adh locus. A, Genomic structure of the Adh locus containing Adh3, Adh2, Copia- and Gypsy-like retroelements, and Adh1 on chromosome 11. The bracket under the map indicates the 7.0-kb fragment cloned into pINA134 to produce pJHYAd2Ct5 for the authentic 5′-junction fragment in PCR analysis. B, Structure of the vector pJHYAd2. C, Structure of the modified adh2 gene having the hpt and ΔEn sequences inserted in front of its initiation codon. The pink regions represent the homologous sequences that correspond to the flanking Adh2 segments carried by pJHYAd2. Restriction sites S and K indicate SacI and KpnI, respectively, and K* represents the additional KpnI site generated during the pJHYAd2 construction. The horizontal lines and their flanking small arrowheads under the map represent PCR fragments and primers, respectively. The probes 5′P, Hm, and 3′U contain the 0.8-kb Adh2 promoter, the 0.9-kb hpt coding region, and a 0.6-kb segment containing the Adh2-specific 3′ UTR and its 3′ adjacent sequence, respectively. Other symbols are as in Figure 1.
Experimental Design for Targeting of the Adh2 Gene
We have basically adapted the same strategy used for Waxy, employing a large-scale Agrobacterium-mediated transformation with a strong positive-negative selection (Terada et al., 2002, 2004; Iida and Terada, 2005), and the targeted double crossovers by two consecutive HR events at the homologous segments were expected to lead to the integration of a transgene into the endogenous corresponding sequence (Fig. 3). The positive selection marker gene hpt for hygromycin B resistance (Hmr) in the targeting vector pJHYAd2 is flanked by the 6.2-kb Adh2 promoter sequence, including a 0.1-kb 5′ UTR, and the 4.0-kb Adh2 and its adjacent region together with the 2.0-kb 3′ part of a Copia-like retroelement, and DT-A is placed next to the border sequences at both ends of T-DNA to efficiently eliminate border-associated RI. The lengths of the two homologous segments in Adh2 used for somatic HR in rice calli are comparable to those in the case of Waxy (Table I). In the previous experiments for Waxy, vigorously growing calli obtained from about 350 mature seeds were used for Agrobacterium-mediated transformation, and, on average, one fertile transgenic rice line with targeted waxy was isolated per experiment (Terada et al., 2002). We have modified our transformation protocol in “Materials and Methods”; embryogenic rice calli for efficient and nonlaborious Agrobacterium-mediated transformation were prepared from approximately 1,000 to 2,000 mature seeds (Table I). We have also incorporated an additional step in the systematic PCR-based screening for the transformants having Adh2 modified by HR among the surviving calli with positive-negative selection; the junction fragments, which were generated by HR between the homologous segments in the introduced vector and the endogenous target gene (Fig. 3), were not only PCR-amplified but also sequenced at both ends to exclude ectopic recombination events, including OSI that might yield PCR-amplified fragments similar in size to the anticipated junction fragments (Fig. 1). In addition, Southern-blot hybridization analysis of genomic DNAs from homologous recombinant calli with the hpt sequence as a probe would reveal whether only one copy of the transgene was integrated into Adh2 or an additional copy of a randomly integrated hpt sequence was present in the genome. To distinguish TGT from EGT, plants having Adh2 modified in the homozygous condition were obtained among the selfed progeny of fertile transgenic plants that had been regenerated from the homologous recombinant calli, and the structure of the endogenous Adh2 gene in the T1 segregants was examined to determine whether it was properly modified by TGT or remained intact due to EGT (Fig. 1).
Table I.
Summary of the transformation experiments for targeting the Adh2 gene and comparison with the previous experiments for the Waxy gene
| Gene | Length of Homology for HR
|
Experiment(s) | Seeds (Callia) Used | Surviving Calli | Targeted Calli | Frequencyb | |
|---|---|---|---|---|---|---|---|
| 5′ | 3′ | ||||||
| Experiment 1 | 1,050 (95) | 236 | 6c | 2.5% | |||
| Adh2 | 6.2 kb | 6.0 kb | Experiment 2 | 1,900 (109) | 232 | 3d | 1.3% |
| Total | 2,950 (204) | 468 | 9 | 1.9% | |||
| Waxye | 6.3 kb | 6.8 kb | Total | 2,160 (194) | 638 | 6 | 0.94% |
The numerals in parentheses indicate the fresh weight of calli (grams) to be used for transformation.
The targeting frequency represents targeted calli per surviving callus.
One of the six calli regenerated slowly.
One of the three calli was accidentally lost.
The Waxy targeting represents a total of six independent experiments added with approximately 360 seeds in each experiment, and only the 3′-junction fragment of waxy can be amplified (Terada et al., 2002).
Isolation and Characterization of Homologous Recombinant Calli
From two experiments with the vector pJHYAd2, we were able to obtain the 468 surviving calli with positive-negative selection (Table I). To screen the homologous recombinant calli with properly modified adh2 by HR, we first examined by PCR analysis with primers F4 and R4 whether the 6.4-kb 5′-junction fragment containing the Adh2 promoter was detectable and then analyzed by PCR with primers F5 and R5 whether the 6.8-kb 3′-junction fragment containing the Adh2 coding region was also present in the calli that had yielded the 6.4-kb 5′-junction fragment (Fig. 3). All nine of the calli that produced the 5′-junction fragment were found to yield the 3′-junction fragment. To confirm that these 6.4- and 6.8-kb fragments corresponded to the anticipated 5′- and 3′-junction fragments, respectively, both ends of these PCR-amplified fragments were sequenced. All of the PCR-amplified fragments sequenced were shown to be the anticipated junction fragments, confirming that the expected HR had occurred in all nine calli. Thus, we estimated that the HR frequency was about 2% per surviving callus (Table I). We further analyzed by PCR screening whether the 3′-junction fragment could be detectable in the remaining 459 surviving calli, and none of them produced the 6.8-kb fragment. Therefore, no OSI events occurred in the 468 surviving calli examined, because none of them gave only one of the two junction fragments by PCR screening.
Calli with a Randomly Integrated hpt Gene
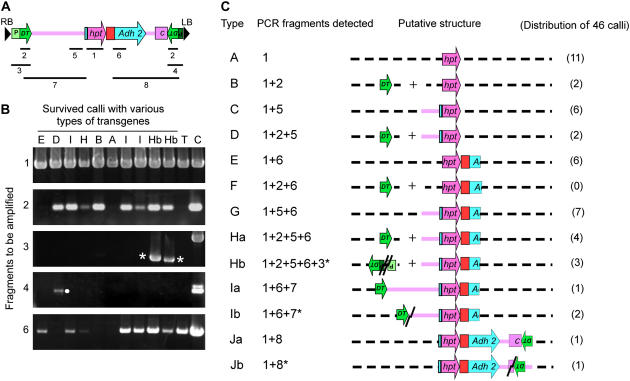
To gain information about surviving calli that did not produce the proper junction fragments, we randomly chose 46 such calli together with two homologous recombinant calli and examined whether they contained: (1) the hpt coding sequence; (2) the DT-A coding sequence; (3) the DT-A and Ubiquitin promoter sequence; (4) the DT-A and 35S promoter sequence; (5) at least 1.0-kb left-flanking sequence of the hpt-positive marker; (6) at least 1.0-kb right-flanking sequence of the hpt-positive marker; (7) the entire 6.2-kb left-flanking sequence of the hpt gene from the vector pJHYAd2; and (8) the entire 6.0-kb right-flanking sequence of the hpt gene from the vector pJHYAd2 (Fig. 4, A and B) by PCR analysis with appropriate primers (Supplemental Table S1). All the surviving calli examined contained the hpt sequence, and none of them carried the intact DT-A promoters originated from the vector pJHYAd2, even though about 30% of the calli examined were found to contain the DT-A coding sequence (Fig. 4C). Because the outmost primers to detect the DT-A with Ubiquitin and 35S promoter sequences were designed to be at 380 and 850 bp from the outmost right and left border sequences, respectively (Supplemental Table S1), the absence of the intact DT-A promoters implies that the integrated T-DNA molecules have more than the 380- and 850-bp distal regions deleted from the right and left border sequences, respectively. The results indicate that the integrated DT-A genes were not active and that none of them was an escapee from the positive-negative selection. It is noteworthy that none of the nine homologous recombinant calli contained the DT-A coding sequence (data not shown). Some surviving calli carried discrete segments (e.g. only the hpt and DT-A coding sequences) integrated in the genomes, suggesting that multiple RI events were likely to occur (Fig. 4C). Of 46 calli examined, 33 calli produced at least either one of the two 1.0-kb flanking sequences. Although 14 of these 33 calli also contained the DT-A coding sequence, only one callus (Ia) carried the whole 6.2-kb Adh2 promoter region with the DT-A coding sequence, and two other calli (Ib) appeared to bear deletions at the Adh2 promoter distal region (Fig. 4C) because they yielded shorter PCR fragments than the anticipated PCR fragment 7. Similarly, one callus (Ja) carried the hpt and entire 6.0-kb Adh2 genes and the DT-A coding region, and another (Jb) seemed to contain a deletion at the junction region between the 6.0-kb Adh2 gene and the DT-A coding region. Interestingly, three calli (Hb) were found to contain hpt and at least its 1.0-kb flanking sequences as well as a truncated Ubiquitin promoter fused with a truncated 35S promoter sequence, which must have been generated by fusing at least two T-DNA molecules in the tail-to-tail configuration with (Hb-2) and without (Hb-1 and Hb-3) filler sequences (Supplemental Fig. S1). The simplest explanation would be that these tail-to-tail fusions had occurred between two T-DNA intermediates, and the resultant fused molecules were integrated into the genome (Tzfira et al., 2004).
Figure 4.
PCR analysis of randomly integrated transgenes found in calli that survived with positive-negative selection. A, Strategy for the characterizing transgene segments in the surviving calli by PCR analysis. The thin lines with numerals below the map indicate the fragments to be amplified by PCR with appropriate primers (Supplemental Table S1). B, Typical examples of the PCR analysis. The fragments to be amplified by PCR correspond to those illustrated in A. The lanes T and C show the PCR-amplified fragments from the TGT calli and the control vector pJHYAd2, respectively, and other lanes show those from surviving calli containing various truncated transgene sequences. The fragments with the white asterisks were derived from truncated and fused promoters, whereas the one marked with the white circle was a nonspecific fragment, as confirmed by DNA sequencing analysis. C, Types of truncated transgene sequences carried by surviving calli deduced from the PCR analysis. The numerals in parentheses represent the occurrence of each type (A–J) illustrated, and those with asterisks (3*, 7*, and 8*) in the column of “PCR fragments detected” indicate that the fragments that appeared were shorter than the anticipated fragments. Zigzag lines represent breakpoints generated by NHEJ, and the junction sequences between two fused promoters in Hb are shown in Supplemental Figure S1. The Ubiquitin and 35S promoters for DT-A (Figs. 1 and 3) adjacent to the right and left border sequences are indicated by light- and dark-green boxes, respectively.
It is clear that the surviving calli have the truncated segments including the active hpt gene integrated into the genome by NHEJ processes independent of the T-DNA border-associated RI. Therefore, the HR frequency estimated as homologous recombinant calli per surviving calli with the positive-negative selection in Table I can be interpreted to be HR-promoted integration per NHEJ-mediated integration of the given vector, pJHYAd2, independent of the T-DNA border-associated integration events (Fig. 1). In the previous GT experiments for Waxy, the observed GT frequency as determined from the ratio of the number of homologous recombinant calli to the number of transformed calli resulting from the border-associated RI (transformants obtained by using another vector without containing the negative DT-A gene) was estimated to be 0.065%, whereas the GT frequency as determined from the ratio of the number of homologous recombinant calli to the number of surviving calli with positive-negative selection, the latter of which roughly corresponds to transformants generated via border-independent RI events (Fig. 1), was 0.94% (Terada et al., 2002). In this study, homologous recombinant calli were obtained with a frequency of 1.9% per surviving callus (Table I). Therefore, it is emphasized that the GT frequency in rice as determined by HR-promoted integration per NHEJ-mediated integration that is independent of the T-DNA-mediated integration processes must be approximately 1% or higher, which is comparable with the GT frequency in mouse embryogenic stem cells (Jasin et al., 1996). Assuming that most of the integration processes of transgenes that are independent of the T-DNA border-associated integrations are in common with those introduced by direct delivery methods with double-stranded DNA molecules (Tinland and Hohn, 1995; Somers and Makarevitch, 2004), because a significant portion of single-stranded T-DNA imported into the plant nucleus can become double-stranded in Agrobacterium-mediated transformation and because the double-stranded and tail-to-tail fused molecules characterized (Supplemental Fig. S1) are thought to be produced before integration (Tzfira et al., 2004), not only the GT frequencies but also RI processes of transgenes between mouse embryogenic stem cells and rice calli employing Agrobacterium-mediated transformation with a strong positive-negative selection may be similar to each other. Therefore, the previous concept that the overwhelming occurrence of RI of transgenes by NHEJ relative to targeted HR is the main obstacle to the development of an efficient GT system in higher plants (Reiss, 2003; Iida and Terada, 2004; Tzfira and White, 2005) may be applicable only to GT systems based on the Agrobacterium-mediated transformation without using our positive-negative selection (Terada et al., 2002), because Agrobacterium-mediated transformation via T-DNA border-associated integrations is a highly efficient NHEJ process (Tzfira et al., 2004; Tzfira and Citovsky, 2006; Fig. 1).
Isolation and Characterization of Transgenic Plants Having Adh2 Modified
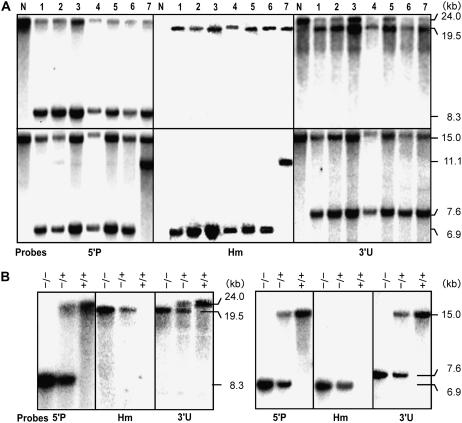
Of these nine homologous recombinant calli, one was accidentally lost during regeneration and seven calli gave normal fertile transgenic plants through multiple shoots (Table I). The remaining one callus regenerated slowly (about 2.5 times longer than the others), and only four regenerants were obtained. Three fertile plants showed a late flowering phenotype, and the remaining one was sterile, probably due to somaclonal variation, which refers to genetic and epigenetic changes induced by tissue culture (Larkin and Scowcroft, 1981; Kaeppler et al., 2000). Southern-blot analysis revealed that all seven normal fertile plant lines were heterozygotes, with only one copy of hpt integrated into the anticipated site of Adh2 by HR; no additional ectopic integration of hpt was detectable (Fig. 5A). We noticed that six out of seven plants tested carried an additional KpnI site at around 2 kb from the left end of the 6.2-kb Adh2 promoter segment provided by the vector pJHYAd2 (Figs. 3 and 5A). The original sequence, GGTATC, appeared to have converted into the KpnI site GGTACC during the construction of pJHYAd2, because pJHYAd2 recovered from Agrobacterium to be used for transformation was found to contain the KpnI site. The lost targeted callus and the late-flowering plants also produced patterns in Southern-blot hybridization that were identical to the six major fertile plant lines containing the additional KpnI site (data not shown). The results indicated that the left crossover occurred within the 2-kb distal region of the introduced vector sequence in eight out of nine homologous recombinants obtained.
Figure 5.
Southern-blot analysis of the adh2 region. A, Normally grown fertile T0 transgenic plants. N and numerals indicate ‘Nipponbare’ and seven targeted plants, respectively. B, T1 segregants of plant number 5. The top and bottom segments in A and the left and right segments in B show the SacI and KpnI digests, respectively.
To confirm whether the modified adh2 genes disrupted by hpt were transmitted into the next generation in a Mendelian fashion, we chose three plants, numbers 3, 5, and 7, and the genotypes of their selfed progeny were scored by PCR analysis with the primers F6 and R6 (Fig. 3); the ratios of T1 plants with the homozygous Adh2/Adh2 alleles, the heterozygous Adh2/adh2∷hpt (adh2 disrupted by the hpt integration) alleles, and the homozygous adh2∷hpt/adh2∷hpt alleles were approximately 1:2:1 (no. 3, 5:13:6; no. 5, 9:24:7; no. 7, 7:13:6). Southern-blot analysis of the T1 segregants of plant number 5 (Fig. 5B) and two other plants, numbers 3 and 7 (data not shown), further confirmed the conclusion that HR-mediated, targeted disruptants had been obtained without the concomitant occurrence of ectopic recombination events; no segment corresponding to the wild-type Adh2 band was present in the targeted homozygotes with the adh2∷hpt/adh2∷hpt alleles, excluding the possibility of EGT (Fig. 1). The results clearly indicate that the GT frequency of Adh2, as determined by TGT per surviving callus with positive-negative selection (NHEJ-mediated integration, which is independent of the T-DNA-mediated integration processes), is approximately 2%, which is comparable with the GT frequency in mouse embryogenic stem cells (Jasin et al., 1996); furthermore, they show that no ectopic events, such as OSI, EGT, or simultaneous RI of the positive selection marker, can be detected. Although the molecular mechanisms generating a copy of the target sequence in EGT are largely unknown, an Adh2 sequence of more than 12.2 kb, which corresponds to the two junction fragments together, must be copied for generating EGT. Thus, the longer the homologous segments used for a targeting vector, the less frequently EGT can occur, because the generation of a longer copy of the target gene is likely to become less probable. In this respect, EGT has usually been observed in GT leading to base changes (Hanin et al., 2001; Iida and Terada, 2005; Endo et al., 2006b).
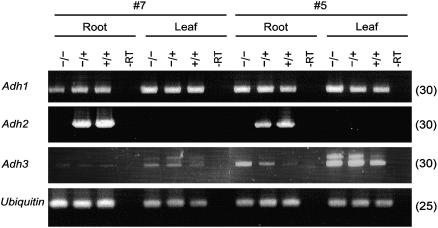
No Adh2 expression was observed in these adh2-disrupted mutants, not even in roots (Fig. 6), where the Adh2 transcripts are known to accumulate in wild-type plants (Xie and Wu, 1989; Matsumura et al., 1998). While an adh1 mutant in rice was reported to be vulnerable to submergence stress (Matsumura et al., 1998; Saika et al., 2006), no apparent phenotypic alterations could be detected in the adh2 mutants (data not shown).
Figure 6.
Expression of the Adh genes in fertile transgenic plants. RT-PCR analysis of the Adh transcripts in T1 plant numbers 5 and 7. The symbols −/−, −/+, and +/+ indicate the targeted homozygote (adh2∷hpt/adh2∷hpt), the heterozygote (adh2∷hpt/Adh2), and the wild type (Adh2/Adh2), respectively, and other symbols are as described in Figure 2B.
Characteristic Features of GT in Rice
We demonstrated here that eight independently obtained fertile plants, regenerated from nine targeted recombinant calli isolated out of 468 surviving calli of the positive-negative selection with the vector pJHYAd2, have Adh2 modified by HR without the concomitant occurrence of ectopic recombination events (Table I; Fig. 3). Even though the Adh genes comprise a small multigene family in the rice genome and a highly repetitive Copia-like element is present 1.0 kb downstream of Adh2, no apparent preclusion by homologous and repetitive sequences was observed. As described in “Materials and Methods,” the main improvements over the previous version of the GT procedure for the single-copy gene Waxy (Terada et al., 2002) are as follows: (1) the scale of calli used for one transformation experiment is increased, from calli prepared from about 350 seeds to those prepared from approximately 2,000 seeds; (2) end sequencing of the PCR-amplified junction fragments obtained by PCR screening for homologous recombinant calli is incorporated; and (3) Southern-blot hybridization of genomic DNA from the homologous recombinant calli with an hpt probe is also introduced to examine whether the homologous recombinant calli carry an additional copy of the hpt sequence by RI. With the improved protocol, we were able to enrich the targeted calli and identify the TGT events efficiently by combining PCR screening with end sequencing of the junction fragments. The calli with OSI and other ectopic recombinations, which might produce PCR-amplified fragments similar in size to the anticipated junction fragments, could be excluded unambiguously by end sequencing. Subsequent Southern-blot analysis confirmed that the transformed calli or their regenerated T0 plants were heterozygotes with TGT alleles without containing an ectopically inserted additional transgene, and the analysis of their homozygous T1 segregants clearly showed no occurrence of EGT. The method described here is, in principle, applicable to any rice gene. Using the same strategy, we were able to obtain transgenic rice plants with the homozygously knocked-out adh1 gene by HR and targeted T0 plants with heterozygously modified ddm1a and ddm1b genes encoding SWI2/SNF chromatin remodeling proteins (Jeddeloh et al., 1999; International Rice Genome Sequencing Project, 2005), although we still need to remove the remote possibility that EGT might have occurred in some of these T0 plants (Y. Johzuka-Hisatomi, R. Terada, K. Yamaguchi, and S. Iida, unpublished data). Thus, targeted plants could be obtained in all five of the endogenous rice genes tested, Waxy, Adh1, Adh2, DDM1a, and DDM1b, by our strong positive-negative procedure. Because Agrobacterium-mediated transformation of rice calli derived from about 2,000 seeds in one experiment is routinely feasible, we can conclude that our GT procedure is widely applicable to modifying various endogenous genes in rice. The present GT protocol with positive-negative selection must also be applicable to other higher plants when calli are used for transformation mediated by Agrobacterium. As the floral dip procedure for Agrobacterium-mediated transformation is routinely employed in Arabidopsis and the resulting transformants can be selected in their progeny seedlings (Clough and Bent, 1998), it remains to be seen whether GT with the floral dip procedure is as effective as the GT protocol with embryogenic rice calli described here.
Comparison of GT between the Moss and Rice Systems
The observed GT frequencies of Waxy and Adh2, as determined by TGT per surviving callus with positive-negative selection, are approximately 1% and 2%, respectively, and no ectopic events have ever been observed (Terada et al., 2002; this study), indicating that our GT procedure is a relatively efficient method. As the moss P. patens exhibits high frequencies of HR-dependent GT (Schaefer, 2002; Cove, 2005), we compare here the characteristics of GT between the moss system and our rice system. Because the GT terminology used in higher plants and moss is slightly different, it is necessary to clarify similar and/or equivalent terminology used in both systems for discussion. TGT in higher plants appears to roughly correspond to allele replacement (Schaefer, 2002; Cove, 2005; Kamisugi et al., 2005), resulting from two HR events at both ends, but allele replacement may include the integration of multiple copies of a transgene into the target locus. Kamisugi et al. (2005) have introduced a new term, targeted gene replacement (TGR), for single copy and single insertion at the target locus, which is synonymous to TGT in higher plants. Similarly, OSI in higher plants appears to be related to insertional gene disruption (Schaefer, 2002) or targeted insertion (Kamisugi et al., 2006), which resulted from one HR and another NHEJ event and usually brought about the integration of concatenated multiple copies of transgenes at the target loci. There appears to be no term in moss corresponding to EGT in higher plants, probably because HR events are so efficient that no one seems to have ever looked specifically for EGT in moss. Ectopic insertion in moss, termed by Kamisugi et al. (2005), appears to roughly correspond to RI in higher plants, whereas RI was also used in moss by Schaefer (2002). As GT in higher plants is rare compared with RI, the focus has been on TGT as well as concomitantly occurring undesirable OSI and EGT. However, HR activity in moss is so high that only TGR (TGT) without additional ectopic events remains a relatively minor fraction among allele replacements (Reiss, 2003; Kamisugi et al., 2005), and almost all targeted insertions at target loci carry tandemly repeated transgenes in moss (Kamisugi et al., 2006).
While transformation for GT in moss is usually achieved by polyethylene glycol-mediated direct gene transfer of double-stranded DNA into protoplasts (Schaefer, 2002; Hohe and Reski, 2003; Cove, 2005), Agrobacterium-mediated transformation is commonly used for GT in higher plants, including rice (Iida and Terada, 2005). Transforming DNA containing homologous sequences becomes integrated into its target locus by HR in moss (Schaefer, 2002; Hohe and Reski, 2003), whereas T-DNA becomes integrated randomly throughout the higher plant genome as a single molecule or multiple sequences ligated to each other in various orientations (Tzfira et al., 2004) even though T-DNA carries homologous sequences to target sequences (Puchta, 2002; Reiss, 2003; Iida and Terada, 2005). It is known that a single-stranded T-DNA molecule is imported into the plant nucleus and then becomes double stranded in Agrobacterium-mediated transformation (Tzfira and Citovsky, 2006). The exact molecular mechanism for T-DNA integration processes and single T-DNA integration processes in particular is largely unknown; multiple T-DNA sequences are thought to occur before integration by ligating double-stranded T-DNA molecules in various orientations (Tzfira et al., 2004). In our rice GT system, the DT-A gene as a cell-autonomous, nonconditional, and lethal negative selection marker (Iida and Terada, 2005) was placed at both ends of the T-DNA segment adjacent to its border sequences of the vector used (Figs. 1 and 3). Because the transient expression of the double-stranded DT-A gene before integration is thought to kill host plant cells (Morton and Hooykaas, 1995), it is assumed that, in the calli that lead to successful GT in our rice system, the entire DT-A gene regions with their promoters in the introduced vector would neither become double stranded nor express transiently before integration (Iida and Terada, 2004, 2005). Although we do not know whether an intermediate T-DNA molecule for successful GT processes is single stranded or double stranded, we speculate that two DT-A genes placed at both T-DNA ends act effectively to suppress undesirable ectopic events, including OSI, EGT, or simultaneous RI of the positive selection marker and ensure the promotion of an accurate and clean TGT event. Because new virulent Agrobacterium strains were recently found to promote moss transformation successfully (D.G. Schaefer, unpublished data [cited by Cove, 2005]), it would be intriguing to know whether Agrobacterium-mediated transformation of P. patens with positive-negative selection using a cell-autonomous, nonconditional, and lethal negative selection marker leads to the exclusive generation of TGR (TGT) in the moss GT system.
MATERIALS AND METHODS
Nucleic Acid Procedures
General nucleic acid procedures, including plasmid preparation, plant DNA and RNA preparation, PCR and reverse transcription (RT)-PCR amplification, and Southern-blot and DNA sequencing analyses were performed as described before (Terada et al., 2002, 2004; Morita et al., 2005). For Southern-blot hybridization, the probes 5′P, Hm, and 3′U were prepared by PCR amplification with the primer sets 5′P-F/5′P-R, Hm-F/Hm-R, and 3′U-F/3′U-R, respectively (Fig. 3). To determine the entire sequence of the Adh3(AB267278) transcripts, we employed the 5′ and 3′ RACEs and the RT-PCR analyses (Fig. 2). The 5′ and 3′ RACEs were performed with a Gene RACER kit (Invitrogen) with two appropriate primers consecutively for nested PCR amplification. To detect approximately 1.5-kb transcripts of the Adh genes bearing the entire coding regions, first-strand cDNAs were synthesized using SuperScript III reverse transcriptase (Invitrogen), and Ex Taq polymerase (TaKaRa Biomedicals) was used for subsequent PCR amplification in Adh1 (X16296), Adh2 (X16297), and Adh3 with the primer sets RT1-F/RT1-R, RT2-F/RT2-R, and RT3-F/RT3-R, respectively, which also correspond to their unique 5′- and 3′-untranslated sequences. The constitutively expressed gene for Ubiquitin (D12629) was used as an internal control for RT-PCR analysis with the primer sets Ubiq-F/Ubiq-R. The sequences of primers used for PCR and RT-PCR amplification are listed in Supplemental Table S1.
Vector Construction
To construct the vector pJHYAd2 for targeting Adh2 (Fig. 3B), the 6.2-kb fragment containing the Adh2 promoter was prepared from the ‘Nipponbare’ genome by PCR amplification (Terada et al., 2002; Morita et al., 2005) using LA Taq polymerase (TaKaRa Biomedicals) with the primers F1 and R1: initial denaturation (94°C for 1 min), 16 cycles of denaturation (98°C for 10 s), annealing and extension (58°C for 15 min), then 19 cycles of denaturation (98°C for 10 s), annealing and extension (58°C for 15 min with the autoextension feature to add 15 s/cycle), and final extension (72°C for 10 min). Similarly, the 6.0-kb fragment containing the Adh2 coding region was also amplified by PCR with the primers F2 and R2: initial denaturation (94°C for 1 min), 16 cycles of denaturation (98°C for 10 s), annealing and extension (62°C for 15 min), then 19 cycles of denaturation (98°C for 10 s), annealing and extension (62°C for 15 min with the autoextension feature to add 15 s/cycle), and final extension (72°C for 10 min). The resultant fragments were first cloned individually into the vector pCR-XL-TOPO (Invitrogen) and subsequently recloned into the targeting backbone vector pINA134 (Terada et al., 2002), yielding pJHYAd2. We also amplified the 7.0-kb fragment containing the Adh2 promoter (Fig. 3A) using LA Taq polymerase with the primers F3 and R1: initial denaturation (94°C for 1 min), 16 cycles of denaturation (98°C for 10 s), annealing and extension (68.1°C for 15 min), then 19 cycles of denaturation (98°C for 10 s), annealing and extension (68.1°C for 15 min with the autoextension feature to add 15 s/cycle), and final extension (72°C for 10 min). The fragment was cloned into pCR-XL-TOPO and then recloned into pINA134 to produce pJHYAd2Ct5, which was used for producing the authentic 5′-junction fragment in PCR screening with the primers F4 and R4 (Fig. 3C).
Plant Transformation
The Agrobacterium-mediated rice (Oryza sativa) transformation procedure was based upon the previously described method (Terada et al., 2002, 2004) with the following modifications. Embryogenic calli for the transformation were prepared from approximately 1,000 to 2,000 mature seeds of rice sp. japonica ‘Nipponbare’ (Table I). After cocultivation with Agrobacterium, vancomycin (200 mg/L) was employed for washing to remove Agrobacterium (Sallaud et al., 2003). Cefatoxim (400 mg/L) and vancomycin (100 mg/L) were continuously used in the selection 2N6-CH medium containing hygromycin B (50 mg/L; Terada et al., 2002, 2004).
To detect the 6.4-kb 5′-junction fragment containing the Adh2 promoter, PCR amplification was performed using LA Taq polymerase with the primers F4 and R4, and an equimolar mixture of pJHYAd2Ct5 and ‘Nipponbare’ DNA mimicking the heterozygous state was used as a control DNA sample. The cycles of the PCR reactions were as follows: initial denaturation (94°C for 1 min), 15 cycles of denaturation (98°C for 10 s), annealing and extension (63.5°C for 15 min), then 19 cycles of denaturation (98°C for 10 s), annealing and extension (63.5°C for 15 min with the autoextension feature to add 15 s/cycle), and final extension (72°C for 10 min). To detect the 6.8-kb 3′-junction fragment containing the Adh2 coding region, PCR analysis was performed with the primers F5 and R5: initial denaturation (94°C for 1 min), 16 cycles of denaturation (98°C for 10 s), annealing and extension (66.5°C for 15 min), then 19 cycles of denaturation (98°C for 10 s), annealing and extension (66.5°C for 15 min with the autoextension feature to add 15 s/cycle), and final extension (72°C for 10 min). To confirm that these 6.4- and 6.8-kb fragments corresponded to the anticipated 5′- and 3′-junction fragments, respectively, both ends of these PCR-amplified fragments were sequenced directly.
An MSRE (Murashige and Skoog for regeneration) medium, which comprises the basic salts and vitamins of the Murashige and Skoog medium (Murashige and Skoog, 1962), Suc (30 g/L), sorbitol (30 g/L), naphthylacetic acid (1 mg/L), benzylaminopurine (2 mg/L), and 0.8% agarose Type I (Sigma), was routinely used for regenerating multiple shoots from calli (Terada et al., 2002). For slowly regenerating calli, a modified MSRE medium containing reduced inorganic salts and vitamins (one-quarter of the normal MSRE medium) and additional indole-3-acetic acid (1 mg/L) and zeatin (0.5 mg/L) was employed. From a targeted callus, more than 20 transgenic plants were routinely regenerated through multiple shoots (Terada et al., 2002) and subjected to the PCR analysis for detecting the 5′- and 3′-junction fragments to confirm that the regenerants contained the adh2∷hpt allele. As expected, most of the regenerants usually carried the adh2∷hpt allele, and vigorously growing transgenic plants were chosen for further analysis. To examine the genotype of Adh2 in the T1 segregants, the appearance of the 1.3- and 5.0-kb fragments for the Adh2 and adh2∷hpt alleles, respectively, was examined by PCR amplification with the primers F6 and R6.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AB267278 and AB267279.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. A possible process to generate a truncated Ubiquitin promoter fused with a truncated 35S promoter found in a surviving callus with positive-negative selection.
Supplemental Table S1. List of primers used for PCR and RT-PCR amplification.
Supplementary Material
Acknowledgments
We thank Miwako Matsumoto and Miki Shimamoto for technical assistance, Kazuo Tsugane for discussions about rice functional genomics, Mitsuyasu Hasebe for enlightening us about various aspects of the moss GT, and Mikio Nakazono for sharing information on rice Adh genes.
This work was supported by the Ministry of Agriculture, Forestry, and Fisheries of Japan (Green Technology Project grant no. IP1007), by the Ministry of Education, Culture, Sports, Science, and Technology in Japan, by the Bio-oriented Technology Research Advancement Institution (PROBRAIN), and by the Japan Society for the Promotion of Science for Young Scientists (fellowship to Y.J.-H.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Shigeru Iida (shigiida@nibb.ac.jp).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Britt AB, May GD (2003) Re-engineering plant gene targeting. Trends Plant Sci 8 90–95 [DOI] [PubMed] [Google Scholar]
- Brunaud V, Balzergue S, Dubreucq B, Aubourg S, Samson F, Chauvin S, Bechtold N, Cruaud C, DeRose R, Pelletier G, et al (2002) T-DNA integration into the Arabidopsis genome depends on sequences of pre-insertion sites. EMBO Rep 3 1152–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Cove D (2005) The moss Physcomitrella patens. Annu Rev Genet 39 339–358 [DOI] [PubMed] [Google Scholar]
- Dolferus R, Osterman JC, Peacock WJ, Dennis ES (1997) Cloning of the Arabidopsis and rice formaldehyde dehydrogenase genes: implications for the origin of plant ADH enzymes. Genetics 146 1131–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmanuel E, Yehuda E, Melamed-Bessudo C, Avivi-Ragolsky N, Levy AA (2006) The role of AtMSH2 in homologous recombination in Arabidopsis thaliana. EMBO Rep 7 100–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo M, Ishikawa Y, Osakabe K, Nakayama S, Kaya H, Araki T, Shibahara K, Abe K, Ichikawa H, Valentine L, et al (2006. a) Increased frequency of homologous recombination and T-DNA integration in Arabidopsis CAF-1 mutants. EMBO J 25 5579–5590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo M, Osakabe K, Ichikawa H, Toki S (2006. b) Molecular characterization of true and ectopic gene targeting events at the acetolactate synthase gene in Arabidopsis. Plant Cell Physiol 47 372–379 [DOI] [PubMed] [Google Scholar]
- Evans MJ, Smithies O, Capecchi MR (2001) Mouse gene targeting. Nat Med 7 1081–109011590418 [Google Scholar]
- Girke T, Schmidt H, Zähringer U, Reski R, Heinz E (1998) Identification of a novel Δ6-acyl-group desaturase by targeted gene disruption in Physcomitrella patens. Plant J 15 39–48 [DOI] [PubMed] [Google Scholar]
- Hanin M, Paszkowski J (2003) Plant genome modification by homologous recombination. Curr Opin Plant Biol 6 157–162 [DOI] [PubMed] [Google Scholar]
- Hanin M, Volrath S, Bogucki A, Briker M, Ward E, Paszkowski J (2001) Gene targeting in Arabidopsis. Plant J 28 671–677 [DOI] [PubMed] [Google Scholar]
- Hohe A, Reski R (2003) A tool for understanding homologous recombination in plants. Plant Cell Rep 21 1135–1142 [DOI] [PubMed] [Google Scholar]
- Hohn B, Puchta H (2003) Some like it sticky: targeting of the rice gene Waxy. Trends Plant Sci 8 51–53 [DOI] [PubMed] [Google Scholar]
- Iida S, Terada R (2004) A tale of two integrations, transgene and T-DNA: gene targeting by homologous recombination in rice. Curr Opin Biotechnol 15 132–138 [DOI] [PubMed] [Google Scholar]
- Iida S, Terada R (2005) Modification of endogenous natural genes by gene targeting in rice and other higher plants. Plant Mol Biol 59 205–219 [DOI] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project (2005) The map-based sequence of the rice genome. Nature 436 793–800 [DOI] [PubMed] [Google Scholar]
- Jasin M, Moynahan ME, Richardson C (1996) Targeted transgenesis. Proc Natl Acad Sci USA 93 8804–8808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeddeloh JA, Stokes TL, Richards EJ (1999) Maintenance of genomic methylation requires a SWI2/SNF2-like protein. Nat Genet 22 94–97 [DOI] [PubMed] [Google Scholar]
- Kaeppler SM, Kaeppler HF, Rhee Y (2000) Epigenetic aspects of somaclonal variation in plants. Plant Mol Biol 43 179–188 [DOI] [PubMed] [Google Scholar]
- Kamisugi Y, Cuming AC, Cove DJ (2005) Parameters determining the efficiency of gene targeting in the moss Physcomitrella patens. Nucleic Acids Res 33 e173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamisugi Y, Schlink K, Rensing SA, Schween G, von Stackelberg M, Cuming AC, Reski R, Cove DJ (2006) The mechanism of gene targeting in Physcomitrella patens: homologous recombination, concatenation and multiple integration. Nucleic Acids Res 34 6205–6214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempin SA, Liljegren SJ, Block LM, Rounsley SD, Yanofsky MF, Lam E (1997) Targeted disruption in Arabidopsis. Nature 389 802–803 [DOI] [PubMed] [Google Scholar]
- Kirik A, Pecinka A, Wendeler E, Reiss B (2006) The chromatin assembly factor subunit FASCIATA1 is involved in homologous recombination in plants. Plant Cell 18 2431–2442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin PJ, Scowcroft WR (1981) Somaclonal variation: a novel source of variability from cell cultures for plant improvement. Theor Appl Genet 60 197–214 [DOI] [PubMed] [Google Scholar]
- Matsumoto S, Ito Y, Hosoi T, Takahashi Y, Machida Y (1990) Integration of Agrobacterium T-DNA into a tobacco chromosome: possible involvement of DNA homology between T-DNA and plant DNA. Mol Gen Genet 224 309–316 [DOI] [PubMed] [Google Scholar]
- Matsumura H, Takano T, Takeda G, Uchimiya H (1998) Adh1 is transcriptionally active but its translational product is reduced in a rad mutant of rice (Oryza sativa L.), which is vulnerable to submergence stress. Theor Appl Genet 97 1197–1203 [Google Scholar]
- Morita Y, Hoshino A, Kikuchi Y, Okuhara H, Ono E, Tanaka Y, Fukui Y, Saito N, Nitasaka E, Noguchi H, et al (2005) Japanese morning glory dusky mutants displaying reddish-brown or purplish-gray flowers are deficient in a novel glycosylation enzyme for anthocyanin biosynthesis, UDP-glucose:anthocyanidin 3-O-glucoside-2″-O-glucosyltransferase, due to 4-bp insertions in the gene. Plant J 42 353–363 [DOI] [PubMed] [Google Scholar]
- Morton R, Hooykaas PJJ (1995) Gene replacement. Mol Breed 1 123–132 [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15 473–497 [Google Scholar]
- Puchta H (1998) Towards targeted transformation in plants. Trends Plant Sci 3 77–78 [Google Scholar]
- Puchta H (2002) Gene replacement by homologous recombination in plants. Plant Mol Biol 48 173–182 [PubMed] [Google Scholar]
- Reiss B (2003) Homologous recombination and gene targeting in plant cells. Int Rev Cytol 228 85–139 [DOI] [PubMed] [Google Scholar]
- Reski R (1998) Physcomitrella and Arabidopsis: the David and Goliath of reverse genetics. Trends Plant Sci 3 209–210 [Google Scholar]
- Sachs MM, Subbaiah CC, Saab IN (1996) Anaerobic gene expression and flooding tolerance in maize. J Exp Bot 47 1–15 [Google Scholar]
- Saika H, Matsumura H, Takano T, Tsutsumi N, Nakazono M (2006) A point mutation of Adh1 gene is involved in the repression of coleoptile elongation under submergence in rice. Breed Sci 56 69–74 [Google Scholar]
- Sallaud C, Meynard D, van Boxtel J, Gay C, Bes M, Brizard JP, Larmande P, Ortega D, Raynal M, Portefaix M, et al (2003) Highly efficient production and characterization of T-DNA plants for rice (Oryza sativa L.) functional genomics. Theor Appl Genet 106 1396–1408 [DOI] [PubMed] [Google Scholar]
- Schaefer DG (2002) A new moss genetics: targeted mutagenesis in Physcomitrella patens. Annu Rev Plant Biol 53 477–501 [DOI] [PubMed] [Google Scholar]
- Schaefer DG, Zryd J-P (1997) Efficient gene targeting in the moss Physcomitrella patens. Plant J 11 1195–1206 [DOI] [PubMed] [Google Scholar]
- Schuermann D, Molinier J, Fritsch O, Hohn B (2005) The dual nature of homologous recombination in plants. Trends Genet 21 172–181 [DOI] [PubMed] [Google Scholar]
- Shaked H, Melamed-Bessudo C, Levy AA (2005) High-frequency gene targeting in Arabidopsis plants expressing the yeast RAD54 gene. Proc Natl Acad Sci USA 102 12265–12269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers DA, Makarevitch I (2004) Transgene integration in plants: poking or patching holes in promiscuous genomes? Curr Opin Biotechnol 15 126–131 [DOI] [PubMed] [Google Scholar]
- Strepp R, Scholz S, Kruse S, Speth V, Reski R (1998) Plant nuclear gene knockout reveals a role in plastid division for the homolog of the bacterial cell division protein FtsZ, an ancestral tubulin. Proc Natl Acad Sci USA 95 4368–4373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarchini R, Biddle P, Wineland R, Tingey S, Rafalski A (2000) The complete sequence of 340 kb of DNA around the rice Adh1-Adh2 region reveals interrupted colinearity with maize chromosome 4. Plant Cell 12 381–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terada R, Asao H, Iida S (2004) A large-scale Agrobacterium-mediated transformation procedure with a strong positive-negative selection for gene targeting in rice (Oryza sativa L.). Plant Cell Rep 22 653–659 [DOI] [PubMed] [Google Scholar]
- Terada R, Urawa H, Inagaki Y, Tsugane K, Iida S (2002) Efficient gene targeting by homologous recombination in rice. Nat Biotechnol 20 1030–1034 [DOI] [PubMed] [Google Scholar]
- Tinland B, Hohn B (1995) Recombination between prokaryotic and eukaryotic DNA: integration of Agrobacterium tumefaciens T-DNA into the plant genome. Genet Eng 17 209–229 [PubMed] [Google Scholar]
- Tzfira T, Citovsky V (2006) Agrobacterium-mediated genetic transformation of plants: biology and biotechnology. Curr Opin Biotechnol 17 147–154 [DOI] [PubMed] [Google Scholar]
- Tzfira T, Li J, Lacroix B, Citovsky V (2004) Agrobacterium T-DNA integration: molecules and models. Trends Genet 20 375–383 [DOI] [PubMed] [Google Scholar]
- Tzfira T, White C (2005) Towards targeted mutagenesis and gene replacement in plants. Trends Biotechnol 23 567–569 [DOI] [PubMed] [Google Scholar]
- Xie Y, Wu R (1989) Rice alcohol dehydrogenase genes: anaerobic induction, organ specific expression and characterization of cDNA clones. Plant Mol Biol 13 53–68 [DOI] [PubMed] [Google Scholar]
- Zhu Q-H, Ramm K, Eamens AL, Dennis ES, Upadhyaya NM (2006) Transgene structures suggest that multiple mechanisms are involved in T-DNA integration in plants. Plant Sci 171 308–322 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.