Abstract
Root-knot nematode (RKN; Meloidogyne spp.) is a major crop pathogen worldwide. Effective resistance exists for a few plant species, including that conditioned by Mi in tomato (Solanum lycopersicum). We interrogated the root transcriptome of the resistant (Mi+) and susceptible (Mi–) cultivars ‘Motelle’ and ‘Moneymaker,’ respectively, during a time-course infection by the Mi-susceptible RKN species Meloidogyne incognita and the Mi-resistant species Meloidogyne hapla. In the absence of RKN infection, only a single significantly regulated gene, encoding a glycosyltransferase, was detected. However, RKN infection influenced the expression of broad suites of genes; more than half of the probes on the array identified differential gene regulation between infected and uninfected root tissue at some stage of RKN infection. We discovered 217 genes regulated during the time of RKN infection corresponding to establishment of feeding sites, and 58 genes that exhibited differential regulation in resistant roots compared to uninfected roots, including the glycosyltransferase. Using virus-induced gene silencing to silence the expression of this gene restored susceptibility to M. incognita in ‘Motelle,’ indicating that this gene is necessary for resistance to RKN. Collectively, our data provide a picture of global gene expression changes in roots during compatible and incompatible associations with RKN, and point to candidates for further investigation.
Root-knot nematodes (RKN; Meloidogyne spp.) are obligate parasites of essentially all vascular plants and negatively impact production of most crops (Sasser, 1980). Central to the parasitic interaction is the ability of the nematode to reprogram root parenchyma cells to differentiate into highly specialized feeding cells called giant cells (GCs). Infective RKN juveniles (J2) hatch in the soil, mechanically penetrate the root, and migrate intercellularly to the stele. Migration is accompanied by secretion of cell wall-modifying enzymes from the nematode stylet (Bird et al., 1975; Wyss et al., 1992; Smant et al., 1998), and it has long been hypothesized that the primary inductive signal for GC formation also involves stylet secretions (Christie, 1936). Numerous candidate molecules have been proposed (Davis and Mitchum, 2005), and recent evidence points to a role for a molecule with functional similarities to rhizobial Nod factor as the initial RKN-plant signal (Weerasinghe et al., 2005). Each individual J2 induces up to 10 metabolically active GCs, which become strong nutrient sinks (Jones and Northcote, 1972; Bird, 1975; McClure, 1977) and serve as the sole food source for the developing nematode. Depending on the RKN species, cortical tissue surrounding the GCs exhibits hyperplasia and hypertrophy, leading to the stereotypic root-knot galls. Resistance to RKN has been identified in a number of plant species, and, in some cases, the responsible loci identified. The best understood of these is the tomato (Solanum lycopersicum) Mi gene (Watts, 1947; Milligan et al., 1998; Williamson and Kumar, 2006), which conditions resistance to Meloidogyne incognita and Meloidogyne javanica (but not to Meloidogyne hapla) and has been widely bred into commercial tomato varieties (Gilbert and McGuire, 1956).
Various approaches, including construction of subtractive cDNA libraries from individual GCs (Wilson et al., 1994), promoter-trapping strategies (Sijmons et al., 1994; Barthels et al., 1997; Favery et al., 2004), and in situ hybridizations (Lohar et al., 2004; Gal et al., 2006), have examined gene expression patterns during RKN feeding site initiation. Collectively, these experiments have revealed that genes regulating the cell cycle (Niebel et al., 1996; de Almeida Engler et al., 1999), cell wall synthesis (Niebel et al., 1993; Goellner et al., 2001; Vercauteren et al., 2002), and transcription regulation (Bird and Wilson, 1994) are up-regulated in GCs. Gheysen and Fenoll (2002) provide a detailed review of the approximately 50 genes known to be up-regulated and a few that are repressed. One emerging picture is that GCs share many features with rhizobial nodules, including coexpression of specific transcription regulators (Koltai and Bird, 2000), early nodulation genes (Bird, 1996; Koltai et al., 2001; Favery et al., 2002), and cytokinin-responsive genes (Lohar et al., 2004); genetic data reinforce these similarities (Lohar and Bird, 2003; Bird, 2004; Weerasinghe et al., 2005). Numerous putative defense genes also are up-regulated during RKN infection, including peroxidases, chitinases, extensins, and proteinase inhibitors, perhaps as a global response to pathogen invasion. Callose or lignin may also be deposited as a physical barrier along the cell wall (Balhadère and Evans, 1995).
Recently, two laboratories used microarrays to examine changes in Arabidopsis (Arabidopsis thaliana) gene expression responsive to RKN infection. Based on the hypothesis that GCs are transfer cells (Jones and Northcote, 1972), Hammes et al. (2005) examined expression of transporter genes in RKN-infected and healthy root tissue at weekly intervals and established that multiple transport processes are regulated, some in GCs and others in uninfected areas of the root. Another study (Jammes et al., 2005) interrogated GC-enriched root tissue, establishing that as many genes are repressed as are up-regulated upon nematode infection. They further substantiated that successful RKN infection is associated with suppression of a number of plant defenses.
Although there is a strong correlation between water uptake and RKN inoculum (Meon et al., 1978), suggesting that much of the yield loss caused by RKN can be attributed to compromised root function, it is clear that RKN infection broadly influences whole plant physiology (Myuge, 1956; Owens and Rubinstein, 1966; Loveys and Bird, 1973; Wallace, 1974). Various lines of evidence also have implicated auxin and cytokinin flux in the RKN-host interaction (Mathesius et al., 1998; Hutangura et al., 1999; Karczmarek et al., 2004; Lohar et al., 2004) as part of the regulation of root architecture. Thus, rather than focus on gene expression in GCs per se, we designed experiments to interrogate genes representative of the broader tomato root transcriptome during compatible and incompatible interactions with RKN. Time points were chosen to capture the host gene response to key aspects of the RKN life cycle in susceptible and resistant tomato iso-lines (‘Moneymaker’ and ‘Motelle,’ respectively) infected with either M. hapla or M. incognita.
Based on a mixed-model analysis (Wolfinger et al., 2001), we found that, in the absence of RKN, only one gene, encoding a glycosyltransferase, was found to be differentially regulated between the ‘Moneymaker’ and ‘Motelle’ transcriptomes. Experimental down-regulation of this gene via virus-induced gene silencing (VIGS) restores susceptibility to M. incognita in ‘Motelle,’ indicating that this function is necessary for Mi-mediated resistance. Glycosyltransferases have been implicated in carbohydrate biosynthesis and associated in plant stress and defense responses (Dixon, 2001; Qi et al., 2005; Vogt and Jones, 2000) and cell wall synthesis (Egelund et al., 2004; Lao et al., 2003); this is the first report, to our knowledge, of a role for a glycosyltransferase in nematode resistance.
RESULTS
cDNA Microarray Construction and Annotation
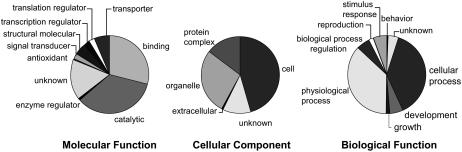
At the time we initiated this study, the most comprehensive source of tomato gene sequences was a collection of ESTs clustered into tentative consensus (TC) sequences (corresponding to gene predictions) by The Institute for Genomic Research (TIGR). Because RKN is a root pathogen, we selected ESTs obtained from root cDNA libraries for array construction. A complete list of the genes used, along with their identities, is given in Supplemental Table S1. For nomenclature and annotation uniformity, if a sequence had a match to a TC, that information was retained. Each clone without a match to a TC (e.g. a singleton) was named according to its GenBank accession number and individually hand annotated. For the small number of sequences for which some ambiguity remained, the clone name was retained. To further categorize the genes, we queried the TIGR and GenBank annotation files with a set of key words (Supplemental Table S2) related to various biological functions we hypothesize might be germane to the RKN-plant interaction (Table I). For example, all genes associated with hormone or hormone regulation were grouped into a category called Hormone, and each gene that fell into this category was then tagged with the letter H. Second, the Gene Ontology (GO; http://www.geneontology.org) identifier for each was traced to the identifier category immediately below the head ontology category in the hierarchy and tallied (Fig. 1). Protein motifs were identified by HMM and Interproscan (Zdobnov and Apweiler, 2001) queries (Supplemental Table S3). Motif names were manually curated to find groups of genes with functions corresponding to the list in Table I and correlated with genes that showed differential expression in the various array experiments (vide infra). Key findings are summarized in Table II.
Table I.
Number of genes categorized according to known or suspected involvement in feeding site and gall formation
All Array Genes and All Significant Genes are all genes used on tomato root gene array and all genes statistically determined to be regulated (at q ≤ 0.05) in tissue comparisons, respectively. Statistical comparisons of Mixed-Stage Infection, Onset of RKN Reproduction, and Resistant Infected are made to uninfected tissue. Mixed-stage roots are nonsynchronous infected roots (grown in a greenhouse). Categories are based as follows: 1, all genes on the array; 2, hormones (Hutangura et al., 1999; Bird and Koltai, 2000); 3, pathogenesis-related genes (Bowles, 1990); 4, transport (Hammes et al., 2005); 5, nucleic acid binding; 6, (I) shikimate (Doyle and Lambert, 2002); 7, (Y) cell cycle (Niebel et al., 1996; de Almeida Engler et al., 1999); 8, aquaporin, water channel-related genes (Opperman et al., 1994); 9, (X) peroxidase; 10, cell wall (Bird,1974); 11, GC genes (Wilson et al., 1994); 12, (S) ribosome-related genes; and 13, genes with unknown function. Annotations were searched with keywords (Supplemental Table S2) to identify genes to categorize.
| Array Probes | All1 | H2 | P3 | T4 | B5 | I6 | Y7 | A8 | X9 | W10 | GC11 | S12 | U13 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| All Array Genes | 1,547 | 20 | 13 | 33 | 22 | 4 | 9 | 8 | 8 | 14 | 181 | 75 | 318 |
| All Significant Genes | 753 | 8 | 7 | 19 | 8 | 4 | 5 | 7 | 6 | 4 | 96 | 59 | 146 |
| Mixed-Stage Infection | 258 | 5 | 2 | 7 | 2 | 3 | 4 | 2 | 3 | 2 | 25 | 7 | 50 |
| Onset of RKN Reproduction | 264 | 4 | 4 | 9 | 3 | 1 | 1 | 5 | 5 | 1 | 34 | 15 | 41 |
| All Infected (between 12 and 36 hpi) | 354 | 3 | 4 | 8 | 3 | 1 | 2 | 2 | 1 | 2 | 53 | 51 | 64 |
| Susceptible Infected (between 12 and 36 hpi) | 217 | 3 | 2 | 6 | 1 | 0 | 1 | 2 | 1 | 1 | 31 | 43 | 38 |
| Resistant Infected | 58 | 1 | 0 | 3 | 0 | 0 | 0 | 0 | 1 | 0 | 6 | 4 | 12 |
Figure 1.
GO classification of all tomato genes used to construct microarray, expressed as a percentage of head ontologies. Each GO annotation (downloaded from TIGR) from all array tomato root genes is mapped to the subcategory, and subcategories are tallied and are represented as a percentage of head ontology.
Table II.
Predicted protein motifs found in tomato array root genes (a portion of Supplemental Table S3)
Tomato gene sequences were analyzed using protein motif prediction programs. Gene Groups and Protein Motif Names were manually curated to find groups of genes with functions corresponding to the list in Table I. I, P, and X identify motifs related to the shikimate pathway, pathogen response, and peroxidases, respectively; protein motifs were found with prediction programs (we found 1,040 motifs in all; see Supplemental Table S3); predicted protein motifs were tallied for all genes and results tabulated in All Motifs; only predicted motifs in genes found to be significantly up-regulated (up-reg.) and repressed (repress.) in different comparisons were tabulated and can be compared to determine how the same motif is “regulated” in other treatment comparisons. Mixed Roots, Roots nonsynchronously infected with RKN; REL, infected roots at nematodes' egg-laying stage; s12–s36, infected susceptible tissue 12 hpi compared to same at 36 hpi; Resist., resistant tissue compared to uninfected tissue. No entry indicates a motif was not regulated in the direction given by the column heading.
| Gene Groups | Protein Motif Names | All Motifs | Mixed Roots, up-reg. | Mixed Roots, repress. | REL up-reg. | REL repress. | s12–s36 up-reg. at 36 hpi | s12–s36 repress. at 36 hpi | Resist. up-reg. | Resist. repress. |
|---|---|---|---|---|---|---|---|---|---|---|
| I | 3-Dehydroquinate synthase | 1 | 1 | |||||||
| I | DAHP synthetase, class II | 1 | 1 | |||||||
| I | Dehydroquinase class I | 2 | 2 | 1 | ||||||
| I | EPSP synthase (3-phosphoshikimate 1-carboxyvinyltransferase) | 1 | ||||||||
| I | Quinate/shikimate 5-dehydrogenase | 1 | 1 | 1 | ||||||
| I | Shikimate/quinate 5-dehydrogenase | 1 | 1 | 1 | ||||||
| Late embryogenesis abundant protein 2 | 1 | 1 | 1 | 1 | ||||||
| Late embryogenesis abundant protein 3 | 2 | 1 | 1 | 1 | 2 | |||||
| P | Glutathione S-transferase, C terminal | 4 | 1 | 1 | 1 | 1 | ||||
| P | Glutathione S-transferase, C terminal-like | 7 | 2 | 1 | 1 | 1 | 1 | |||
| P | Glutathione S-transferase, N terminal | 5 | 1 | 1 | 1 | 1 | ||||
| X | Haem peroxidase | 7 | 2 | 4 | 1 | 1 | ||||
| X | Haem peroxidase, plant/fungal/bacterial | 10 | 2 | 4 | 1 | 1 | 2 | |||
| X | Plant peroxidase | 7 | 2 | 4 | 1 | 1 |
Resistant and Compatible Tomato Roots Have Near Equivalent Transcriptomes in the Absence of RKN
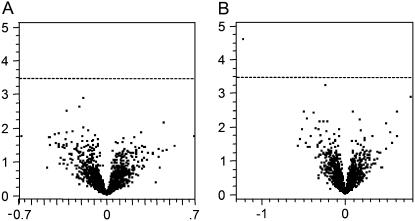
‘Moneymaker’ and ‘Motelle’ differ for practical purposes by the presence of Mi in the latter. Because other genetic differences between the cultivars may lead to different transcription profiles, possibly confounding analysis of gene expression changes in response to RKN, we compared the transcriptome of each of the cultivars in the absence of nematodes. Similarly, because the life cycle of RKN takes 4 weeks to complete, we examined temporal changes in the transcriptome of mature tomato roots over that time span. The experimental loop design with four replications per sample to simultaneously test these differences is shown in Supplemental Figure S1. As indicated (Fig. 2), none of the genes exhibited a significant age-dependent difference and only one gene, a glycosyltransferase, exhibited a significant cultivar-dependent difference in gene expression at q ≤ 0.05 (Storey, 2003).
Figure 2.
Gene significance results for cultivar and age comparison in tomatoes. Volcano plots depict the log2 fold change (horizontal axes) of normalized and averaged intensity values for each gene (each point) plotted against the −log10(P value) (vertical axes) of tomato age (A) and cultivar (B) comparisons. Any spots above the dashed line (representing an FDR of 1 in 20) are considered to be significantly expressed. No significant difference in gene expression was detected between 4- and 8-week-old tomato plants. One gene was considered differentially expressed between ‘Motelle’ and ‘Moneymaker.’
RKN Infection Causes Substantial Changes in Root Expression Profiles
Although genes for array construction were chosen based on their presence in root cDNA libraries, suggesting that their transcripts were sufficiently abundant to be sampled, we wanted to establish that we could detect changes in the root transcriptome during nematode infection. We used our array to interrogate the transcriptome of greenhouse-grown tomato roots either uninfected or nonsynchronously infected with M. incognita. Using an experimental design with seven direct comparison replicates (Supplemental Fig. S2), we detected many significant gene expression changes in roots infected with RKN; Supplemental Table S1 provides a complete list. Approximately 17% of the genes interrogated were differently regulated following RKN infection. Slightly fewer than half the genes were up-regulated, and, accordingly, slightly more than half were found to be repressed in infected tissue. Notably, 25% of the genes annotated as Hormone were modulated by RKN. Nearly half of the genes annotated as Cell Cycle were regulated, and three of the four shikimate pathway-related genes responded to M. incognita infection (Table I).
The Root Transcriptome at RKN Egg Laying
Having established that we could detect differential regulation of many root genes following nonsynchronous infection by RKN, we wanted to distinguish tomato responses throughout the parasitic life cycle. Maximal development of GCs is coincidental with egg laying, so we interrogated the transcriptome of tomato roots 4 weeks postinfection. Secondary galling also is greatest at this time, and we compared roots infected by M. incognita (induces large galls) with those infected by M. hapla (elicits small galls). Because Mi is not effective against M. hapla, we exploited ‘Moneymaker’ and ‘Motelle’ to compare tissues from susceptible responses, resistant responses, and uninfected roots; the experiment design is shown in Supplemental Figure S3.
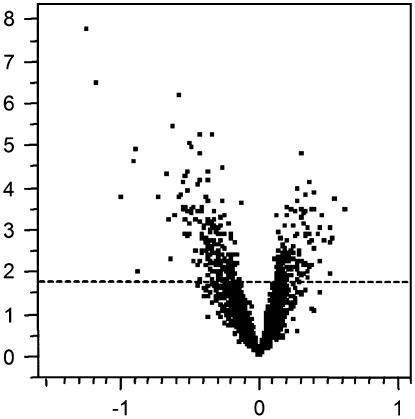
No significant differences in gene expression were observed between ‘Motelle’ roots that had been infected 4 weeks previously by M. incognita and uninfected ‘Motelle’ roots. Because Mi-mediated resistance is effected within a narrow time window after RKN invasion (Dropkin, 1969), this result was not surprising. We also found no differences in gene expression between roots infected with M. hapla compared to ‘Moneymaker’ infected with M. incognita, despite the morphological differences in gall appearance. All susceptible observations were then pooled and compared to all observations from uninfected roots. We found that 17% of the genes were significantly differentially expressed (q ≤ 0.05) between all infected roots and uninfected roots. Exactly one-third of the significant genes were up-regulated and two-thirds were repressed in infected roots compared to uninfected roots (Fig. 3). Over half the tomato genes grouped as peroxidases and aquaporins were regulated by nematodes at the egg-laying stage compared to uninfected roots. Only 25% of the transporter genes were observed to be significantly regulated (Table I), and most of those were repressed. Supplemental Table S1 provides a complete list of genes regulated, and in which direction.
Figure 3.
Gene significance results for roots infected with nematode at the onset of reproduction compared to uninfected tissue. Volcano plots depict log2 fold change (horizontal axes) of normalized and averaged intensity values for each gene (each point) plotted against the −log10(P value) (vertical axes) of infected roots at onset of nematode reproduction compared to uninfected tissue. Spots above the dashed line are considered to be significantly expressed at q ≤ 0.05. Points to the right of center above the dashed line represent genes that are up-regulated, and points to the left and above the dashed line are genes that are repressed in infected tissue.
Successful Initiation of Feeding Sites Elicits Major Shifts in Gene Expression
In broad terms, establishment of the parasitic interaction by RKN involves three phases: (1) migration through root tissues, (2) initiation of GCs in the stele, and (3) onset of sustained feeding with concomitant development of the GCs and surrounding gall. We hypothesized that we could capture snapshots of the changes in gene expression associated with those stages following synchronous infection of tomato by RKN at 12 h postinfection (hpi), 36 hpi, and 72 hpi, respectively. Using an interconnected loop design (Supplemental Fig. S3), we compared root tissue of ‘Moneymaker’ and ‘Motelle’ infected both by M. incognita and M. hapla at these three time points. Uninfected tissue (0) was harvested at the same time points and pooled.
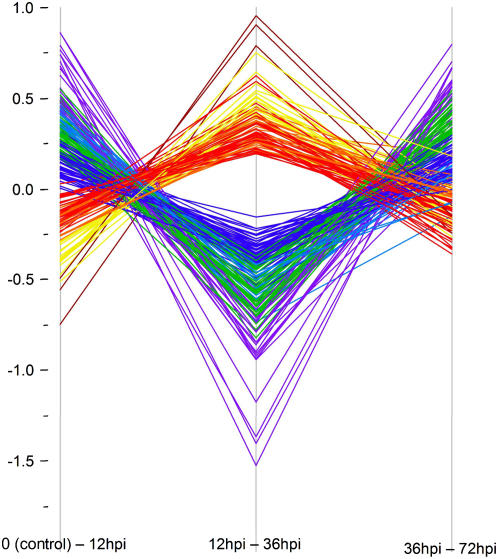
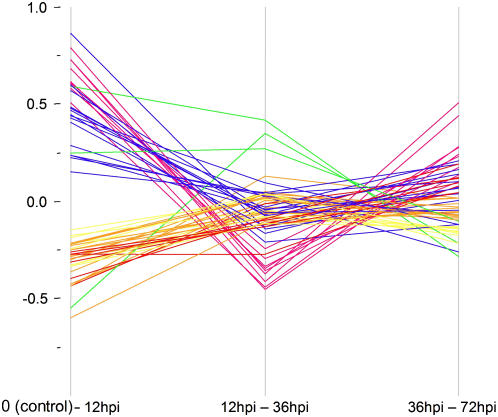
To observe gene expression changes following successful initiation of feeding sites (compatible interactions), observations from ‘Moneymaker’ plants infected with both RKN species were pooled with observations from ‘Motelle’ infected by M. hapla and statistically compared to uninfected controls for each time-course (0–12–36–72 hpi) point. We found 217 (14%) of the arrayed genes to be significantly differentially expressed between 12 and 36 hpi. To examine expression changes in these genes over time, differences in expression data for each time point comparison were plotted (Fig. 4). Each of the 217 lines was color coded to enhance visual interpretation using a hierarchical clustering algorithm (Ward, 1963). Clusters were assigned colors based on how differences of expression changed over time, with the warm colors (red, orange, yellow, and brown) representing genes with expression significantly repressed in a susceptible interaction between the 12- and 36-hpi time points, following a generally smaller increase in gene expression between 0 and 12 hpi. The slope of the lines reflects the magnitude of change in gene expression; genes represented by the brown lines, for example, have the biggest change of expression (Fig. 4). The three genes represented by the brown lines are TC164156 and L24025, which encode unknown functions, and gene TC155176, annotated as “similar to peroxidase.” Those genes annotated as Hormone and Aquaporin (Table I) that we found to be significantly regulated were repressed in this time period.
Figure 4.
Gene expression differences in susceptible tissue over four time periods postinfection. Each colored line represents one of 217 genes exhibiting significant differential regulation. Differences in gene expressions were plotted over time-course comparisons. Hierarchical clustering groups genes into eight classes of broadly similar expression pattern, indicated by color coding. The vertical axis represents intensity differences in log2, normalized values for each gene from each statistical comparison (horizontal axis). Significant genes were determined by using a q value ≤0.05.
The cool colors (purple, green, blue, and teal) in Figure 4 represent genes whose expression is significantly up-regulated in susceptible responses between 12 and 36 hpi after a generally smaller repression of gene expression between 0 and 12 hpi. Genes represented by purple lines have the steepest slopes, indicating the greatest change in difference in gene expression. Approximately 72% of the genes in the hierarchical cluster colored purple encode ribosomal proteins, as do more than 30% of the genes represented by the green lines. In all, nearly two-thirds of the ribosomal protein genes are differentially expressed in this comparison. This is in striking contrast to the warm-color genes, where only one such gene shows differential expression (it is repressed). Other genes up-regulated include a gene annotated into the Pathogenesis category (an Erwinia-induced gene), a gene involved in cell cycle regulation, and 10 genes classified as having an unknown function (Table I). It is clear from Figure 4 that overall gene expression comparing 12- and 36-hpi time points changes in the opposite direction from that seen when comparing uninfected (0) with the 12-hpi samples. Most (99%) of those genes that were significantly repressed in the susceptible reaction between 12 and 36 hpi were up-regulated in the 0 to 12 hpi comparison (and vice versa). In most cases (94%), the expression of all genes again changed between 36 and 72 hpi.
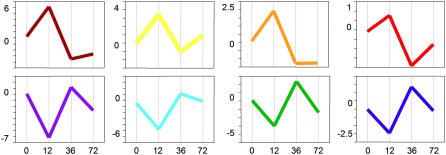
Figure 4 graphically illustrates how the differences between tissue comparisons change over time. To see how individual gene expression changes, the normalized, log2 averaged expression of each gene was plotted over time. Gene expression patterns consistent within each hierarchically (color) coded gene set and individual genes, chosen to represent each cluster, are depicted in Figure 5. Thus, an example in Figure 5 shows a red line depicting typical expression pattern of a gene hierarchically clustered into the red category of Figure 4. Although nearly all of the genes found to exhibit significant regulation between 12 and 36 hpi have a predicable expression profile over the entire early time course, none of these genes is considered significant in the 0 to 12 hpi or the 36 to 72 hpi statistical comparisons. This is consistent with the plotted expression profiles in Figure 5 that indicate gene expression is less robustly manipulated by RKN between these time points or that changes in gene expression are very small. Careful examination of the plots also reveals that by 72 hpi, gene expression is returning to expression levels of the uninfected (control) tissue (0). In Figure 5, the steeper the slope, the higher the fold change in gene expression. The expression plots of the hierarchically clustered genes reveal that the largest amount of fold change in gene expression occurs between 12 and 36 hpi. It is also apparent that the slopes between uninfected tissue (0) and 12 hpi are quite steep, although in this comparison absolute changes in gene expression are less (i.e. there is less of a fold change). Variation between the plots is most noticeable between 36 and 72 hpi. In some gene clusters, such as those represented by the orange, brown, and teal graphs, the slope of the line between 36 and 72 hpi is modest, compared to the slopes represented by the red, yellow, green, blue, and purple graphs.
Figure 5.
Graphical representation of expression changes of each of the eight classes of genes represented in Figure 4. The x axis is time points postinfection; y axis is arbitrary units of expression level change. Note differences in scale.
Gene Expression in Resistant Plants
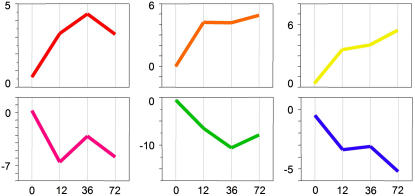
One consequence of our loop design is that it yields only one-third the observations of the transcriptome in resistant plants compared to those we have for the susceptible responses, thus reducing the statistical power. To redress this, we pooled all observations from ‘Motelle’ plants over the early time points and compared them to uninfected tissue. This analysis identified 58 genes that were significantly differentially expressed between resistant tissue infected with M. incognita and uninfected tissue. We plotted expression differences to show how these genes behave over time (Fig. 6). Each gene is represented by a line and is color coded by hierarchical clusters based on differences of expression. It is clear from this plot that genes involved in the resistant response have very different expression profiles from those that are differentially regulated in the susceptible response. These genes tend to change in one absolute direction over time, as opposed to being systematically switched up and down (Fig. 4). To further dissect the patterns of expression, we plotted both the normalized, log2 averaged expression of the genes over time and a typical gene chosen to represent the expression profile of the cluster (Fig. 7). The warm colors represent 31 genes that are consistently up-regulated over all three early infection time points in relation to gene expression level in uninfected tissue. Up-regulated genes include those encoding a glycosyltransferase, a peroxidase, and an ethylene-responsive gene. Some of the genes represented by the orange and yellow profiles (Fig. 7) are also regulated in roots at the nematode egg-laying stage, but are not regulated in any other comparison. Four of these genes were first discovered based on differential expression in GCs (Bird and Wilson, 1994; Wilson et al., 1994); eight are of unknown function. None of the up-regulated genes was found to be regulated in the susceptible tissue comparison at 12 to 36 hpi.
Figure 6.
Gene expression differences in resistant tissue over four time periods postinfection. Each colored line represents one of 58 genes exhibiting significant differential regulation. Differences in gene expressions from statistical comparisons were plotted over time-course comparisons. Hierarchical clustering groups genes into six classes of broadly similar expression pattern, indicated by color coding. The vertical axis represents intensity differences in log2, normalized values for each gene from each statistical comparison (horizontal axis). Significant genes were determined by using a q value ≤0.05.
Figure 7.
Graphical representation of expression changes of each of the six classes of genes represented in Figure 6. The x axis is time points postinfection; y axis is arbitrary units of expression level change. Note differences in scale.
The cool colors represent genes with expression generally repressed over time in resistant reactions. The pink profiles represent genes whose expressions shift somewhat over the time points (Fig. 7) but follow a general trend of down-regulation. Interestingly, all genes that follow this profile are regulated in other comparisons (Supplemental Table S1) with the exception of gene AI637343, a gene with unknown function shown to be expressed in GCs (Bird and Wilson, 1994; Wilson et al., 1994). In all, half of the 27 repressed genes (cool colors: Table I and Supplemental Table S2) are represented by genes considered significant in other comparisons.
Susceptible Roots Infected by M. hapla versus M. incognita Have Similar Transcriptomes
Because the morphology of the galls induced on susceptible tomato roots by M. incognita is visually distinguishable from those induced by M. hapla and may reflect underlying transcriptional differences, we interrogated the transcriptome of ‘Moneymaker’ infected with each RKN species. Although the smaller number of replications limited the statistical power, 16 genes were found to be differentially regulated in the combined observations of 12 and 36 hpi. Predicted functions include a calcium-binding protein (Cab39) and ethylene response factor number 5. Other genes potentially regulated between these interactions can be found in Supplemental Table S4.
Comparison of the RKN-Responsive Transcriptome across Treatments
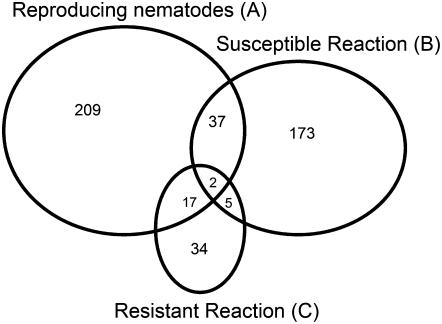
Comparing the expression data from all treatments (Supplemental Table S1) revealed that not many of the genes are regulated in every treatment (Fig. 8). This is particularly evident when comparing the susceptible and resistant responses, where only five genes exhibit a pattern of differential regulation in both treatments. All five genes are repressed in resistant roots and up-regulated between 12 and 36 hpi in susceptible tissue. One of these genes encodes a nuclear transporter factor, three encode ribosomal proteins, and the other encodes a protein of unknown function. Although nearly one-third of the genes regulated in the resistant response are also regulated in roots at the egg-laying stage, the overall number of genes is small (Fig. 8). It is worth noting, however, that each gene in common is regulated in the same direction.
Figure 8.
Venn diagram comparing genes identified as being significantly regulated in roots at the onset of nematode reproduction (A), genes significantly regulated during a susceptible response (B), and genes significantly regulated during a resistant response (C).
Many comparisons can be made about the behavior of individual genes. For example, in the nonsynchronous, mixed-stage greenhouse experiment (Mixed), we found that five of the seven genes containing shikimate pathway protein motifs are repressed compared to genes containing these motifs in uninfected material. Some shikimate genes are also regulated in infected tomato root at the egg-laying stage (REL), although it should be noted that none of the shikimate pathway genes appears to be regulated in the susceptible or resistant reactions during early infection time points. Another noteworthy group is the pathogen response-related motifs that include the glutathione S-transferase domains; one-third of genes encoding these domains are repressed or up-regulated in Mixed and susceptible infected root tissue, but completely repressed in REL. Genes encoding Leu-rich repeats, a protein motif common in resistance genes, are more regulated (both up and down) in susceptible reactions in Mixed than they are in the resistance comparison. The two genes encoding late embryogenesis abundant motifs are regulated in all experiment comparisons except in the resistance reaction, consistent with previous findings that such genes are globally up-regulated following RKN infection (van der Eycken et al., 1996; Lambert et al., 1999). Of the genes encoding domains identified as peroxidase motifs that were regulated in resistant roots, all were up-regulated. By contrast, all but one of these genes were repressed in the susceptible interaction with RKN.
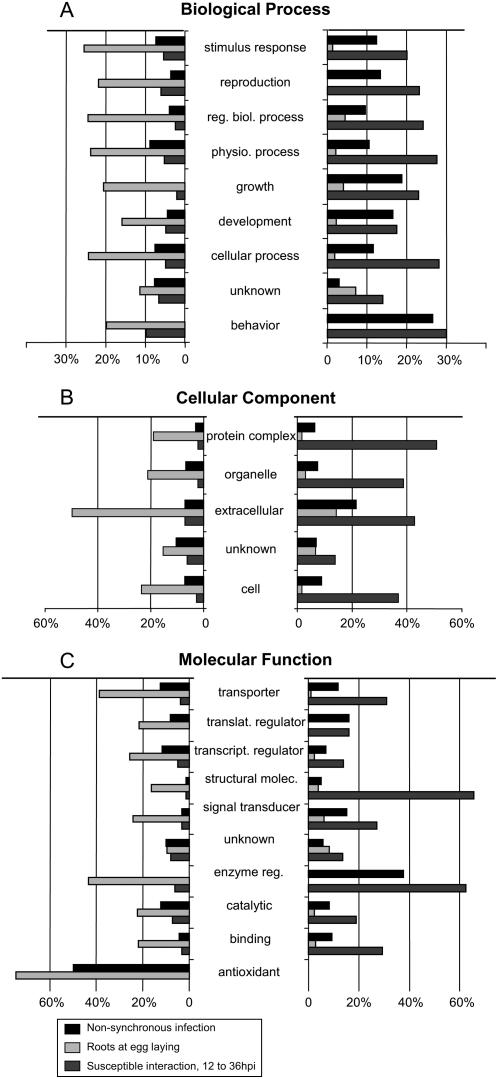
To further categorize plant processes that respond to RKN infection, we plotted the percentages of genes that showed a significant change based on their GO subcategory (Fig. 9). Comparing the Molecular Function category genes in Mixed and REL against uninfected tissue (0) experiments revealed that all “antioxidants” (in this case, all peroxidases) that are regulated are repressed in infective tissue and all “regulatory enzymes” are up-regulated in Mixed, but they are all repressed in the REL comparison. REL genes classified to the Biological Process “growth” that exhibit significant differential expression are repressed. Nearly all “transporter” genes considered significant between 12 and 36 hpi are up-regulated, whereas in the REL comparison “transporter” genes considered significant are repressed. More than 25% of the REL genes with the GO annotation “response to stimulus” were down-regulated, whereas fewer than 5% of genes thus classified were up-regulated. Nearly one-half of the GO tags in the subcategory “extracellular” were repressed; this category also has a higher proportion of up-regulation compared to all other subcategories. The second highest proportion of REL up-regulation throughout the subcategories is in the “unknown” categories in each of the three major GO subdivisions.
Figure 9.
Summery of expression responses of categories of genes differentially expressed in the nematode plant interaction. A, Biological Process; B, Cellular Component; C, Molecular Function. Genes were categorized based on GO annotation. Percentage indicates number of array genes in the given category that exhibit significant differential regulation. Figure design based on figure 4 in Jammes et al. (2005).
Validation of Microarray Results and Functional Analysis of a Candidate Gene in the Resistance Response to RKN
We selected eight RKN-regulated genes for verification by quantitative PCR (Tables III and IV). Except for the glycosyltransferase gene (TC166108), these genes were arbitrarily selected from the list of differentially expressed genes. In each case, these experiments confirmed the results obtained from the array experiments.
Table III.
Genes selected for verification of statistical analysis using RT-PCR
TIGR LeGI gene ID and annotations were downloaded from the LeGI Web site. (AI63743 is a singleton and therefore retains its GenBank accession number.) ▴ indicates up-regulation of expression, and ▾ indicates repression of gene expression when REL, roots infected with egg-laying nematodes, are compared to uninfected tissue; s12–s36, susceptible roots infected 36 hpi compared to 12 hpi; and Resist, resistant roots compared to uninfected root tissue. Gene Group identifies category of genes identified in Table I: U = gene with unknown function; P = gene related to pathogen response. GC No. corresponds to clone DB number in GC library (Wilson et al., 1994).
| TIGR Gene ID | REL | s12–s36 | Resist | Gene Group | GC No. | TIGR Annotation |
|---|---|---|---|---|---|---|
| TC166108 | ▴ | Glycogenin glucosyltransferase | ||||
| TC166934 | ▴ | ▾ | U | Arabidopsis expressed protein | ||
| TC163802 | ▴ | Fibrillarin 2 (FIB2) | ||||
| AI637343 | ▾ | U | 361 | Unknown | ||
| TC156238 | ▴ | P | Homolog to UP|Q84XG6 (Q84XG6) | |||
| TC165974 | ▴ | U | 197 | Lycopersicon esculentum (DB197) Meloidogyne-induced GC protein | ||
| TC155133 | ▾ | 12-Oxophytodienoic acid 10,11-reductase | ||||
| TC161840 | ▾ | CBL-interacting protein kinase 23 (CIPK23) |
Table IV.
Fold changes of transcript abundance for indicated genes determined using quantitative PCR
Fold change is relative to uninfected tissue. Interaction: R = resistant interaction; S = susceptible interaction. Postinfection: in hours (h) and weeks (w).
| Gene | Interaction | Postinfection | Fold Change |
|---|---|---|---|
| TC166108 | R | 36 h | 5.73 |
| AI637343 | R | 12 h | 0.44 |
| TC156238 | R | 12 h | 0.80 |
| TC166934 | S | 36 h | 1.79 |
| TC163802 | S | 36 h | 2.61 |
| TC165974 | S | 4 w | 1.60 |
| TC155133 | S | 4 w | 0.27 |
| TC161840 | S | 4 w | 0.44 |
To further investigate the possibility that the glycosyltransferase gene plays a functional role in Mi-mediated resistance, we used VIGS (Liu et al., 2002a, 2002b; Ryu et al., 2004) to down-regulate its expression in susceptible (‘Moneymaker’) and resistant (‘Motelle’) tomato plants that were subsequently challenged with M. incognita. Because Mi is known to function only during a narrow temporal window corresponding to the period of GC initiation in a susceptible plant (Dropkin, 1969), we performed a calibration experiment to gauge when effective silencing in roots was occurring. Tomato plants (‘Rutgers Large Red’) that had been stably transformed with CaMV35S∷GFP were treated with a GFP-VIGS silencing construct and GFP fluorescence in whole roots was assessed. Silencing of GFP was taken as evidence for a VIGS effect and defined the optimal period for RKN infection of VIGS roots. During this window, ‘Moneymaker’ and ‘Motelle’ plants that had been agroinfected with the TC166108-VIGS construct, in parallel with those agroinfected with the GFP-VIGS construct, were inoculated with M. incognita J2 and scored 1 week later for the presence of galls. To further ensure that the VIGS process per se did not influence the resistant or susceptible response to RKN, nontransgenic ‘Moneymaker’ and ‘Motelle’ plants were agroinfected with the GFP-VIGS construct and infected with M. incognita J2.
At 1 week postinfection, all ‘Moneymaker’ plants exhibited an average of 20 symptomatic galls, and, like the uninfected controls, none of the nonagroinfected or GFP-VIGS agroinfected ‘Motelle’ plants exhibited galling. However, half (4/8) of the ‘Motelle’ plants agroinfected with the TC166108-VIGS construct exhibited galling indistinguishable from ‘Moneymaker’ and control plants; the remainder (4/8) had no galls. A preliminary histological analysis of hand-sectioned roots from the TC166108-VIGS-containing ‘Motelle’ plants (data not shown) indicated that the galls and GCs appeared indistinguishable from those in ‘Moneymaker’ plants. Down-regulation of the specific glycosyltransferase target was confirmed by quantitative reverse transcription (RT)-PCR, and the presence in ‘Motelle’ of the resistance-conferring allele at the Mi locus was confirmed by PCR and restriction analysis (Williamson et al., 1994a). Unrelated to nematode infection, all VIGS-treated plants exhibited additional symptoms, including stem lesions (Supplemental Fig. S5), consistent with effective agroinfection. Collectively, these results point to a functional role for the glycosyltransferase in the Mi-mediated resistance response; a more detailed description will be presented elsewhere.
DISCUSSION
Identifying the broad transcriptional events associated with successful parasitism of plants by RKN or the successful defense by resistant hosts is a prerequisite to understanding the biology of the host-parasite interaction. Further, understanding the response of individual plant genes to RKN invasion may suggest new strategies for development of nematode control in crop plants. Our approach differs from previous transcriptome analysis of plants infected with RKN in several ways, including choice of plant species. Tomato not only is a robust host for RKN, but also encodes robust resistance via the Mi locus. The existence of the resistance-breaking RKN species M. hapla has permitted a comprehensive comparison of the response to RKN invasion of the resistant and susceptible tomato root transcriptomes. It also included a comprehensive examination of the host transcriptome in the first few days post RKN infection.
Using a microarray approach, we found that nearly half the plant genes queried were significantly regulated in one or more treatment comparisons. Included in this list are genes previously reported to be regulated during RKN pathogenesis, including Aquaporin (Opperman et al., 1994) and “transport” (Hammes et al., 2005) genes. We also examined the response of genes encoding host biochemical pathways that have been implicated in the response to RKN. Doyle and Lambert (2002), for example, proposed that RKN-encoded Chorismate Mutase, a key enzyme of the shikimate pathway, is injected into plant cells to modulate the local balance of auxin in roots as a means to initiate GC formation and establish feeding sites. Careful examination of the shikimate pathway genes showed that none of those tested exhibited significant changes in regulation during the 72-hpi temporal window spanning GC induction, thus failing to provide evidence that would either refute or bolster the Doyle-Lambert hypothesis. We did, however, observe significant differential regulation of shikimate pathway genes later in the infection cycle (i.e. in roots with established feeding sites), consistent with the notion that global alterations of auxin balance accompany RKN infection in general (Hutangura et al., 1999).
More than 70% of the genes encoding ribosomal proteins were found to be regulated during some aspect of successful infection of RKN, suggesting that protein production is substantially altered in infected roots, likely associated with the substantial morphological remodeling that occurs in the GC and surrounding tissue. More than half the ribosomal protein-related genes are up-regulated in the compatible interaction between 12 and 36 h, and these genes follow a pattern of repression (0 to 12 hpi comparison) before a significant increase of expression (12 to 36 hpi). The expression of most of these genes is down-regulated between 36 and 72 hpi. Substantial fluctuation around baseline (noninfected) levels of gene expression is not limited to ribosomal protein-related genes; genes found significantly regulated during nematode pathogenesis are expressed in this manner. This pattern suggests that substantial changes in root gene expression have occurred by 12 hpi, a time point prior to the appearance of recognizable GCs. During this period, RKN J2 penetrate the root and migrate into the stele (Gheysen and Fenoll, 2002). It seems reasonable that host genes are responding to the migrating nematodes, and, indeed, previous work has revealed that nonspecific defense responses are detected by 12 hpi (Williamson et al., 1994b; Williamson and Hussey, 1996). However, it is an intriguing possibility that our analysis also captured gene expression in the proto-GC. Later changes in expression (12 to 36 hpi) likely indicate development of the GC initials into recognizable feeding cells. This represents the first time (to our knowledge) that global gene expression has been examined so soon after infection. Key to this was the use of a bulk infection system designed such that those nematodes that failed to penetrate the host within 2 to 3 hpi became desiccated and died.
It is worth noting that after establishment of feeding sites (roots during nematode reproduction and in mixed-stage infected roots) there are substantially more genes repressed than up-regulated. A similar conclusion was made by Jammes et al. (2005), who examined differential gene expression in RKN-infected Arabidopsis roots at weekly intervals postinfection. Collectively, this suggests that the nematode might manipulate multiple pathways to coordinate feeding site formation, protect from host defense responses, and maintain GCs. Fluctuation in gene expression patterns may also indicate that GC formation requires multiple events to occur in the proper order for successful formation in a susceptible root. It seems reasonable that the nematode needs to both induce certain pathways and interrupt others to initiate feeding sites, and these pathways are not necessarily those restricted to normal root development. Elucidating the sequence of events may be key to understanding the induction of GCs.
In contrast to the response of host genes in a susceptible plant, those genes regulated in a resistant response are either up-regulated or are repressed over time and do not fluctuate; only one-third of these genes show any evidence that their expression levels had returned to basal levels by 72 h. This suggests that activation of the resistant response persists over the first few days after nematode infection, consistent with Dropkin's (1969) observations. Not surprisingly, by 4 weeks after infection, gene expression in an infected resistant host was indistinguishable from that in uninfected plant roots; resistant plants do not sustain nematodes after 4 weeks and, hence, are essentially uninfected.
Because those genes regulated during a resistant reaction define candidates that may play a role in the resistance response, we were particularly interested a glycosyltransferase that we observed to be up-regulated nearly 6 times more in resistant roots infected with M. incognita than in uninfected roots. Using a VIGS approach, we confirmed that expression of this gene is necessary for expression of the resistance phenotype. Not surprisingly, not all plants exhibited loss of resistance, presumably reflecting incomplete silencing. Intriguingly, some plants exhibited normal (and complete) resistance, whereas others developed numbers of galls indistinguishable from the control plants (‘Moneymaker’ challenged with M. incognita), suggesting that glycosyltransferase acts in an all-or-nothing manner. Although this might be a coincidence of all-or-nothing silencing, it might also point to this enzyme functioning (or not) to effect resistance via a threshold effect, perhaps as some sort of switch.
How repression of gene expression of a specific glycosyltransferase interferes with the resistance of ‘Motelle’ to M. incognita remains to established, but the variety and nature of roles of the ubiquitous glycosyltransferase family members (Lim and Bowles, 2004; Langlois-Meurinne et al., 2005) provide many possible avenues for plant defense against RKN. Low-Mr secondary metabolites (including phytoalexins) produced from transcriptional activation of response genes (Hammond-Kosack and Jones, 1996) are often stabilized by conjugation to sugars through the action of glycotransferases (Vogt and Jones, 2000). Recent reports suggest that glycosyltransferases act in roles related to defense and stress response from plants (Vogt and Jones, 2000; Dixon, 2001; Qi et al., 2005). Langlois-Meurinne et al. (2005) found that expression of specific glycosyltransferases was necessary for resistance to Pseudomonas syringae in Arabidopsis, and suggested that up-regulation in response to pathogens and during senescence points toward a role in the cell death process and possibly in the hypersensitive response. We also found a glycosyltransferase that is necessary for resistance of RKN in tomato, an interaction that typically results in a hypersensitive response. This suggests that glycosyltransferase may act across plant species as a defense to very different pathogens. Glycosyltransferases also play a role in cell wall synthesis (Lao et al., 2003; Egelund et al., 2004) and may suggest a role in defense to RKN via this function. Research to better understand the role of this enzyme in RKN resistance is currently in progress.
MATERIALS AND METHODS
cDNA Array Preparation
We queried the Tomato Gene Index database for ESTs identified in root cDNA libraries, established by TIGR, via their Web portal (http://www.tigr.org/) and identified approximately 4,300 genes. Clone representatives with the longest sequence (>200 bp) from each of the selected TIGR TC for each gene were purchased from the Clemson University Genomics Institute (CUGI) as a set of 202 microtiter plates, and consolidated using a QBOT robot (Genepix). Plasmids were isolated by alkaline lysis in 96-well format using 96-well Whatman filter and collection plates. Clone inserts were amplified using universal M13 forward and reverse primers, and PCR products were purified by ethanol precipitation and resuspended in filtered, distilled water. Aliquots of these products were then sequenced. A substantial number of the 4,300 wells of interest from CUGI were not turbid (i.e. not viable), and many were significantly contaminated. BLAST analysis revealed that approximately 10% had significant matches to a TC in the TIGR database other than the desired clone and 20% of the sequences did not have a significant match to any clone (including EST singletons) in the TIGR tomato (Solanum lycopersicum) database. Ultimately, we were able to recover and consolidate 1,547 root-expressed genes, including 186 genes previously characterized from a GC-specific library (Bird and Wilson, 1994; Wilson et al., 1994).
Amplified insert DNA was dissolved in 50% DMSO at a final concentration of 150 ng/μL, and genes were arrayed in an arbitrary order (some genes were duplicated) on Corning UltraGaps II slides using Affymetrix GMS 417 pin and ring arrayer. Stored slides were later rehydrated and spots “set” in four cycles of 10 s of steam (slides were suspended face down over steaming water) and 1 min on a 65°C hotplate and then fixed by 250 mJ of UV irradiation. Slides were prehybridized and washed according to TIGR standard operating protocol M005.
Synchronous RKN Infection of Root Tips
Tomato ‘Moneymaker’ and ‘Motelle’ seeds were surface sterilized and planted three to a growth pouch (Mega International; Supplemental Fig. S6) dampened with sterile water and grown in an environmentally controlled chamber (16 h light/8 h dark, 250 μE m−2 s−1, 26°C). Sprouted seedlings were fertilized with a 0.5× Hoagland (Sigma) solution one or two times per week and watered with filter-sterilized tap water between fertilizing. Three- to 4-week-old seedlings were inoculated with 2,000 freshly hatched J2 in 1 mL of water per plant, and pouches were left to dry in the dark in a lateral position overnight.
Collection of Plant Tissue, RNA Preparation, and Dye Ester Coupling
Tissue Collected to 72 hpi
Pouches were hydrated and placed upright under light at least 2 h before the 12-h collection period to ensure dryness and light cycle differences did not affect gene expression between the early time points. Plants exhibiting wilting were rejected. By 36 h, small galls could be seen forming in susceptible roots. Tissue was collected at 12, 36, and 72 hpi by excising root tips extending approximately 3 cm from cap and flash freezing in 1.5-mL tubes set in a metal block on dry ice. RNA was isolated from excised root tips using the RNeasy kit (Qiagen) according to the manufacturer's instructions and amplified using Low RNA Input Fluorescent Linear Amplification kit (Agilent Technologies). cRNA amplification was conducted according to the manufacturer's instructions with the following modifications: Instead of directly incorporating labeled CTP, we indirectly incorporated 4.5 μL of a 50 mm solution 5-(3-aminoallyl)-UTP (Ambion) and 9 μL of a 25 mm mix of RNA nucleotides A, C, and G into each reaction. Amplified cRNA was cleaned using the RNeasy kit. Typically, starting with 0.5 μg of total RNA yielded 5 to 10 μg cRNA. A total of 1.5 μg cRNA was coupled to Cy3 or Cy5 dye ester (GE Healthcare) using TIGR standard operating protocol M005. Coupled RNA for each direct comparison was combined and cleaned using the RNeasy kit and stored at −80°C. Half of each combined reaction was used to hybridize microarrays.
Tissue Collected at Onset of Nematode Reproduction, Age, and Variety Controls
Whole, noninfected root systems were grown as described above and collected from both ‘Moneymaker’ and ‘Motelle’ plants at 4 and 8 weeks. Collection of infected root material during nematode reproduction was conducted approximately 4 weeks after inoculation, when eggs were first visible on the surface of the roots (approximate plant age 8 weeks). Whole root systems were collected and flash frozen in liquid nitrogen. RNA was isolated using the RNeasy kit and indirectly labeled (without amplification), purified, coupled to the dye ester, and stored as above.
Mixed-Stage Infected Tissue
‘Moneymaker’ plants were grown in a glasshouse in spring and summer months with no additional light source. Temperature was regulated to below 30°C. Two-week-old seedlings were planted in a mixture of half soil, half river sand, and contained slow-release fertilizer granules. Tomato plants with four expanded leaves were inoculated with 2 × 104 Meloidogyne incognita eggs. Uninfected roots were grown the same but were not inoculated with RKN. Whole roots were washed and flash frozen in liquid nitrogen, and RNA was isolated using the RNeasy kit and indirectly labeled (without amplification), purified, coupled to the dye ester, and stored as above.
Hybridizations and Image Collection
Coupled samples were dried down and resuspended in a hybridization buffer [33% formamide, 5× SSC, 0.1% SDS, 5 μg poly(A) DNA] heated to 90°C and snap chilled. Slides were hybridized for 14 h and washed according to TIGR standard protocol M005. Each slide was scanned twice using ScanArray Express software (Perkin Elmer) and a ScanArray 4000 scanner (Packard BioChip Technologies) at low (approximately 60% laser power) and medium (approximately 75% laser power) intensity settings. The fluorescent intensity for each spot was captured and quantified using Spot v3 (CSIRO) using the GOGAC setting. Combined images from both Cy3 and Cy5 cRNA channels, with spot locations marked, were examined manually, and any scratches or artifacts that caused the program to incorrectly identify spots were removed using R, the parent program of Spot v3. It was determined that spot intensities greater than 33,000 indicated some pixels were saturated; consequently, the lower-intensity scan was used (across all arrays) for genes where the medium-level scan gave an intensity reading greater than 33,000 for 20 or more measurements.
Data Analysis
We adopted a two-stage approach (Wolfinger et al., 2001) for normalization and analysis. To normalize the data, we used mixed-model ANOVA to remove random effects (global effects of the arrays and pins) and fixed effects (dyes and scan intensity levels). All two-way interactions also were fitted. Interactions that included arrays and pins were considered random effects; all other interactions were fixed effects. Analyses were performed using SAS Proc Mixed (SAS Institute). For genes printed in duplicate, averages were taken of the post-normalized spot data. Residual values from the normalization step were used for the subsequent gene model analyses, the second stage of the Wolfinger et al. (2001) approach. It is in this stage that individual genes were tested for differential expression patterns, by fitting an appropriate mixed-model ANOVA. In addition to the effects of interest described in “Results,” terms for slide (random) and dye (fixed) were also fitted in the gene models to adjust for possible gene-specific dye and array effects, as well as to control for locational array effects that may not have been addressed by the global normalization procedure.
The specific hypotheses of interest were tested through the use of appropriate “estimate” statements in SAS Proc Mixed (SAS Institute). To verify the distributional assumptions of this procedure, we performed permutation tests for a subset of the hypotheses examined, and compared the resulting P values to those derived from the SAS procedure. A PHP script was used to write and automatically run SAS code to permute the data. For each permutation thus produced, the mixed-model ANOVA was run and the per-gene F-statistic values generated by the model were stored. The same model was run on the original, nonpermuted data and the F statistics from this run compared with the collection of all F values from the permuted runs to generate a P value for each gene. To preserve the within-array correlation structure, we used only those permutations that maintained the pairing of samples on the arrays. The level of consistency between the permutation-based results and the pure mixed-model approach was concordant, indicating that the results obtained by Proc Mixed were reliable (as well as much more computationally efficient). Results reported here were based on those calculated by the SAS procedure.
To adjust for the multiple tests performed for the microarray experiments, we employed the techniques of Storey and Tibshirani (2003) and Storey (2003) for controlling false discovery rate (FDR). Their method determines an empirical significance threshold based on maintaining the FDR at a prespecified level. We prespecified that level at 5%, meaning that an estimated 5% of the genes identified as significantly regulated are presumed to be false positives. P values, q values, and direction changes for each comparison are presented in Supplemental Table S4.
Real-Time PCR
Quantitative PCR analysis was performed using the ABI Prism 7900 HT detection system with SDS2.1 software (Applied Biosystems) according to the manufacturer's suggestions. Primers for target genes were designed using DNAStar 6 Primer Select program according to Applied Biosystems primer design suggestions for RT-PCR and are listed in Supplemental Table S5. cDNA was synthesized from RQ1 Dnased (Fisher Scientific) total RNA using Taqman RT reagents (Applied Biosystems) and the 3′ gene-specific primers. RT-PCR was performed using SYBR green PCR master mix and gene-specific primers for 40 cycles at 95°C for 15 s, 54°C for 30 s, and 60°C for 1 min. A dissociation curve (95°C for 15 s, 60°C for 15 s, 95°C for 15 s) was generated after the final PCR cycle. Fluorescent signals were detected in the 7900 HT detection system. Transcript level comparisons were determined using the comparative Ct method (ΔΔCt method) for each gene during exponential amplification. Dissociation curves were confirmed to have single dominant peaks for each gene. Average ΔCt values and sd for each tissue-gene and control combination for each replicate can be viewed in Supplemental Table S6. The control gene TC163896 showed no change in any of the experimental treatments. The reproducibility of expression analysis results was confirmed in two independent experiments.
Plasmid Construction and VIGS
pTRV1 and pTRV2 VIGS vectors (Liu et al., 2002a) were obtained from Dr. Dinesh-Kumar (Yale University). A 266-bp GFP fragment was amplified from a CaMV35S∷GFP expression vector (Lohar et al., 2004) using primers 5′-ggggacaagtttgtacaaaaaagcaggctTATCATTATCCTCGGCCGAA-3′ and 5′-ggggaccactttgtacaagaaagctgggtGTCGTGCCGCTTCATATGAT-3′. A fragment spanning bps 433 through 1,035 of the glycosyltransferase gene amplified from the 1,063-bp TIGR LeGI clone cLEW19P6 (the same clone printed on the microarrays) was cloned using tomato-EST, GATEWAY ready primers (Liu et al., 2002a). PCR-amplified gene products were introduced into the GATEWAY ready pTRV2 (Liu et al., 2002a) using the GATEWAY cloning system according to manufacturer's specifications (Invitrogen). Plasmids were electroporated into Agrobacterium tumefaciens GV2260 and transformants were verified by restriction digestion and sequencing.
Seeds (‘Moneymaker,’ ‘Motelle,’ and ‘Rutgers Large Red’ stably transformed with CaMV35S∷GFP) were surface sterilized and germinated in potting soil in a growth chamber on a 16- h/8-h daylight/night cycle at 26°C. Approximately 2 weeks after germination, CaMV35S∷GFP transformants (verified for GFP expression) were planted into 1:1 soil:sand. Seedlings were agroinoculated by leaf infiltration and agrodrench (Ryu et al., 2004) with appropriate VIGS constructs. Two and 3 weeks after agroinoculation, when GFP fluorescence was no longer detected in ‘Rutgers Large Red’ CaMV35S∷GFP roots, all plants were inoculated with 500 to 1,000 M. incognita J2. Roots were assayed for galls 7 d postinfection. The presence of the Mi gene in ‘Motelle’ was verified by PCR and restriction digest (Williamson et al., 1994a).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figures S1 to S4. Experimental designs for age and variety comparisons, nonsynchronously infected roots compared to uninfected roots, roots at onset of nematode reproduction compared to uninfected roots, and tissue comparisons made over the first 72 hpi in susceptible and resistant roots compared to uninfected tissue.
Supplemental Figure S5. Photograph depicting typical symptoms of TRV infection on tomato, an indication that VIGS infection is successful.
Supplemental Figure S6. Photograph of tomato seedlings grown in growth pouches, ready for inoculation with RKN.
Supplemental Table S1. Gene IDs and annotations, and direction change of significant genes.
Supplemental Table S2. Keywords used to identify gene groups.
Supplemental Table S3. Interproscan (ftp://ftp.ebi.ac.uk/pub/software/unix/iprscan) was used to locate predicted protein motifs in tomato array gene sequences.
Supplemental Table S4. Gene IDs and annotations, direction change of significant genes, and P and q values associated with each gene for each statistical comparison. Annotations are as described above.
Supplemental Table S5. RT-PCR primers for tomato genes used in quantitative PCR analysis.
Supplemental Table S6. RT-PCR ΔCts for genes used in quantitative PCR analysis.
Supplementary Material
Acknowledgments
We thank Sandy Gove for technical assistance in the lab, and Mark Burke, Sam Kalat, Jacob Frelinger, and D. Eric Windham for computational support and advice.
This work was supported by the National Research Initiative of the U.S. Department of Agriculture Cooperative State Research, Education, and Extension Service (grant no. 2006–35604–1673).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: David McK. Bird (david_bird@ncsu.edu).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Balhadère P, Evans AAF (1995) Histopathogenesis of susceptible and resistant responses of wheat, barley and wild grasses to Meloidogyne naasi. Fundam Appl Nematol 18 531–538 [Google Scholar]
- Barthels N, van der Lee FM, Klap J, Goddijn OJ, Karimi M, Puzio P, Grundler FM, Ohl SA, Lindsey K, Roberson L, et al (1997) Regulatory sequences of Arabidopsis drive reporter gene expression in nematode feeding structures. Plant Cell 9 –2134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird AF (1974) Plant response to root-knot nematode. Annu Rev Phytopathol 12 69–85 [Google Scholar]
- Bird AF (1975) Symbiotic relationships between nematodes and plants. In Twenty Ninth Symposium of the Society for Experimental Biology: Symbiosis, Ed 1, Vol 29. Cambridge University Press, Cambridge, UK, pp 351–371 [PubMed]
- Bird AF, Downton WJS, Hawker JS (1975) Cellulase secretion by second stage larvae of the root-knot nematode (Meloidogyne javanica). Marcellia 38 165–169 [Google Scholar]
- Bird DM (1996) Manipulation of host gene expression by root-knot nematodes. J Parasitol 82 881–888 [PubMed] [Google Scholar]
- Bird DM (2004) Signaling between nematodes and plants. Curr Opin Plant Biol 7 372–376 [DOI] [PubMed] [Google Scholar]
- Bird DM, Koltai H (2000) Plant parasitic nematodes: habitats, hormones, and horizontally-acquired genes. J Plant Growth Regul 19 183–194 [DOI] [PubMed] [Google Scholar]
- Bird DM, Wilson MA (1994) DNA sequence and expression analysis of root-knot nematode-elicited giant cell transcripts. Mol Plant Microbe Interact 7 419–424 [DOI] [PubMed] [Google Scholar]
- Bowles DJ (1990) Defense-related proteins in higher plants. Annu Rev Biochem 59 873–907 [DOI] [PubMed] [Google Scholar]
- Christie JR (1936) The development of root-knot nematode galls. Phytopathology 26 1–22 [Google Scholar]
- Davis EL, Mitchum MG (2005) Nematodes. Sophisticated parasites of legumes. Plant Physiol 137 1182–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Almeida Engler J, De Vleesschauwer V, Burssens S, Celenza JL Jr, Inzé D, Van Montagu M, Engler G, Gheysen G (1999) Molecular markers and cell cycle inhibitors show the importance of cell cycle progression in nematode-induced galls and syncytia. Plant Cell 11 793–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon RA (2001) Natural products and plant disease resistance. Nature 411 843–847 [DOI] [PubMed] [Google Scholar]
- Doyle EA, Lambert KN (2002) Meloidogyne javanica chorismate mutase 1 alters plant cell development. Mol Plant Microbe Interact 16 123–131 [DOI] [PubMed] [Google Scholar]
- Dropkin VH (1969) The necrotic reaction of tomatoes and other hosts resistant to Meloidogyne: reversal by temperature. Phytopathology 59 1632–1637 [Google Scholar]
- Egelund J, Skjøt M, Geshi M, Ulvskov P, Peterson BL (2004) A complementary bioinformatics approach to identify potential plant cell wall glycosyltransferase-encoding genes. Plant Physiol 136 2609–2620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favery B, Chelysheva LA, Lebris M, Jammes F, Marmagne A, de Almeida-Engler J, Lecomte P, Vaury C, Arkowitz RA, Abad P (2004) Arabidopsis formin AtFH6 is a plasma membrane-associated protein upregulated in giant cells induced by parasitic nematodes. Plant Cell 16 2529–2540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favery B, Complainville A, Vinardell JM, Lecomte P, Vaubert D, Mergaert P, Kondorosi A, Crespi M, Abad P (2002) The endosymbiosis-induced genes ENOD40 and CCS52a are involved in endoparasitic nematode interactions in Medicago truncatula. Mol Plant Microbe Interact 15 1008–1013 [DOI] [PubMed] [Google Scholar]
- Gal TZ, Kapulnik Y, Koltai H, Aussenburg ER, Burdman S (2006) Expression of a plant expansin is involved in the establishment of root knot nematode parasitism in tomato. Planta 224 155–162 [DOI] [PubMed] [Google Scholar]
- Gheysen G, Fenoll C (2002) Gene expression in nematode feeding sites. Annu Rev Phytopathol 40 191–219 [DOI] [PubMed] [Google Scholar]
- Gilbert JC, McGuire DC (1956) Inheritance of resistance to severe root knot from Meloidogyne incognita in commercial type tomatoes. Proc Amer Soc Hort Sci 68 437–442 [Google Scholar]
- Goellner M, Wang X, Davis EL (2001) Endo-β-1,4-glucanse expression in compatible plant-nematode interactions. Plant Cell 13 2241–2255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammes UZ, Schachtman DP, Berg RH, Nielson E, Koch W, McIntyre LM, Taylor CG (2005) Nematode-induced changes of transporter gene expression in Arabidopsis roots. Mol Plant Microbe Interact 18 1247–1257 [DOI] [PubMed] [Google Scholar]
- Hammond-Kosack KE, Jones JD (1996) Resistance gene-dependant plant defense responses. Plant Cell 8 1773–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutangura P, Mathesius U, Jones MGK, Rolfe BG (1999) Auxin induction is a trigger for root gall formation caused by root-knot nematodes in white clover and is associated with the activation of the flavonoid pathway. Aust J Plant Physiol 26 221–231 [Google Scholar]
- Jammes F, Lecomte P, de Almeida-Engler J, Bitton F, Martin-Magniette M, Renou JP, Abad P, Favery B (2005) Genome-wide expression profiling of the host response to root-knot nematode infection in Arabidopsis. Plant J 44 447–458 [DOI] [PubMed] [Google Scholar]
- Jones MGK, Northcote DH (1972) Multinucleate transfer cells induced in Coleus roots by the root-knot nematode, Meloidogyne arenaria. Protoplasma 75 381–395 [Google Scholar]
- Karczmarek A, Overmars H, Helder J, Goverse A (2004) Feeding cell development by cyst and root-knot nematodes involves a similar early, local and transient activation of a specific auxin-inducible promoter element. Mol Plant Pathol 5 343–346 [DOI] [PubMed] [Google Scholar]
- Koltai H, Bird DM (2000) Epistatic repression of PHANTASTICA and class I KNOTTED genes is uncoupled in tomato. Plant J 22 455–459 [DOI] [PubMed] [Google Scholar]
- Koltai H, Dhandaydham M, Opperman C, Thomas J, Bird D (2001) Overlapping plant signal transduction pathways induced by a parasitic nematode and a rhizobial endosymbiont. Mol Plant Microbe Interact 10 1168–1177 [DOI] [PubMed] [Google Scholar]
- Lambert KN, Ferrie BJ, Nombela G, Brenner ED, Williamson VM (1999) Identification of genes whose transcripts accumulate rapidly in tomato after root-knot nematode infection. Physiol Mol Plant Pathol 55 341–348 [Google Scholar]
- Langlois-Meurinne M, Gachon CMM, Saindrenan P (2005) Pathogen-responsive expression of glycosyltransferase genes UGT73B3 and UGT73B5 is necessary for resistance to Pseudomonas syringae pv tomato in Arabidopsis. Plant Physiol 139 1890–1901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lao NT, Long D, Kiang S, Coupland G, Shoue DA, Carpita NC, Kavanagh TA (2003) Mutation of a family 8 glycosyltransferase gene alters cell wall carbohydrate composition and causes a humidity-sensitive semi-sterile dwarf phenotype in Arabidopsis. Plant Mol Biol 53 687–701 [DOI] [PubMed] [Google Scholar]
- Lim EK, Bowles DJ (2004) A class of plant glycosyltransferases involved in cellular homeostasis. EMBO J 23 2915–2922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Dinesh-Kumar SP (2002. a) Virus-induced gene silencing in tomato. Plant J 31 777–786 [DOI] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Marathe R, Dinesh-Kumar SP (2002. b) Tobacco Rar1, EDS1 and NPR/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J 30 415–429 [DOI] [PubMed] [Google Scholar]
- Lohar DP, Bird DM (2003) Lotus japonicus: a new model to study root-parasitic nematodes. Plant Cell Physiol 44 1176–1184 [DOI] [PubMed] [Google Scholar]
- Lohar DP, Schaff JE, Laskey JG, Kieber JJ, Bilyeu KD, Bird DM (2004) Cytokinins play opposite roles in lateral root formation, and nematode and rhizobial symbioses. Plant J 38 203–214 [DOI] [PubMed] [Google Scholar]
- Loveys RR, Bird AF (1973) The influence of nematodes on photosynthesis in tomato plants. Physiol Plant Pathol 3 525–529 [Google Scholar]
- Mathesius U, Schlaman HRM, Spaink HP, Suatter C, Rolfe BG, Djordjevic MA (1998) Auxin transport inhibition precedes nodule formation in white clover roots and is regulated by flavonoids and derivatives of chitin oligosaccharides. Plant J 14 23–24 [DOI] [PubMed] [Google Scholar]
- McClure MA (1977) Meloidogyne incognita: a metabolic sink. J Nematol 9 88–90 [PMC free article] [PubMed] [Google Scholar]
- Meon S, Wallace HR, Fisher JM (1978) Water relations of tomato (Lycopersicon esculentum Mill. cv. Early Dwarf Red) infected with Meloidogyne javanica (Treub), Chitwood. Physiol Plant Pathol 13 275–281 [Google Scholar]
- Milligan SB, Bodeau J, Yaghoobi J, Kaloshian I, Zabel P, Williamson VM (1998) The root knot nematode resistance gene Mi from tomato is a member of the leucine zipper, nucleotide binding, leucine-rich repeat family of plant genes. Plant Cell 10 1307–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myuge SG (1956) The nutritional physiology of the gall nematodes (Meloidogyne incognita). Dokl Akad Nauk SSSR 108 164–165 [Google Scholar]
- Niebel A, de Almeida Engler J, Hemerly A, Ferriera P, Inzé D, Van Montagu M, Gheysen G (1996) Induction of cdc2A and cyc1At expression in Arabidopsis thaliana during early phases of nematode-induced feeding cell formation. Plant J 10 1037–1043 [DOI] [PubMed] [Google Scholar]
- Niebel A, de Almeida-Englar J, Tire C, Engler G, Van Montagu M, Gheysen G (1993) Induction patterns of an extension gene in tobacco upon nematode infection. Plant Cell 5 1697–1710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opperman CH, Taylor CG, Conkling MA (1994) Root-knot nematode directed expression of a plant root-specific gene. Science 263 221–223 [DOI] [PubMed] [Google Scholar]
- Owens RG, Rubinstein JH (1966) Metabolic changes induced by root-knot nematodes in host tissues. Contrib Boyce Thompson Inst 23 199–214 [Google Scholar]
- Qi Y, Kawano N, Yamauchi Y, Ling J, Li D, Tanaka K (2005) Identification and cloning of a submergence-induced gene OsGGT (glycogenin glucosyltransferase) from rice (Oryza sativa L.) by suppression subtractive hybridization. Planta 221 437–445 [DOI] [PubMed] [Google Scholar]
- Ryu C, Anand A, Kang L, Mysore KS (2004) Agrodrench: a novel and effective agroinoculation method for virus-induced gene silencing in roots and diverse Solanaceous species. Plant J 40 322–331 [DOI] [PubMed] [Google Scholar]
- Sasser JN (1980) Root-knot nematodes: a global menace to crop production. Plant Dis 64 36–41 [Google Scholar]
- Sijmons PC, Cardol EF, Goddijn OJM (1994) Gene activities in nematode-induced feeding structures. In MJ Daniels, JA Downie, AE Osbourn, eds, Advances in Molecular Genetics of Plant-Microbe Interactions. Kluwer, Dordrecht, The Netherlands, pp 333–338
- Smant G, Stokkermans JPWG, Yan Y, de Boer JM, Baum TJ, Wang X, Hussey RS, Gommers FJ, Henrissat B, Davis EL, et al (1998) Endogenous cellulases in animals: isolation of β-1,4-endoglucanase genes from two species of plant-parasitic cyst nematodes. Proc Natl Acad Sci USA 95 4906–4911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD (2003) The positive false discovery rate: a Bayesian interpretation and the q-value. Ann Statist 31 2013–2035 [Google Scholar]
- Storey JD, Tibshirani R (2003) Statistical significance for genome-wide experiments. Proc Natl Acad Sci USA 100 9440–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Eycken W, de Almeida Engler J, Inzé D, Van Montagu M, Gheysen G (1996) A molecular study of root-knot nematode-induced feeding sites. Plant J 9 45–54 [DOI] [PubMed] [Google Scholar]
- Vercauteren I, De Groodt R, de Almeida Engler J, Gheysen G (2002) An Arabidopsis thaliana pectin acetylesterase gene is upregulated in nematode feeding sites induced by root-knot and cyst nematodes. Mol Plant Microbe Interact 15 404–407 [DOI] [PubMed] [Google Scholar]
- Vogt T, Jones P (2000) Glycosyltransferases in plant natural product synthesis: characterization of a supergene family. Trends Plant Sci 5 380–386 [DOI] [PubMed] [Google Scholar]
- Wallace HR (1974) The influence of root-knot nematode, Meloidogyne javanica, on photosynthesis and nutrient demand by roots of tomato plants. Nematologica 20 27–33 [Google Scholar]
- Ward JH (1963) Hierarchical grouping to optimize an objective function. J Am Stat Assoc 58 236–244 [Google Scholar]
- Watts VM (1947) The use of Lycopersicon peruvianum as a source of nematode resistance in tomatoes. Proc Amer Soc Hort Sci 49 233 [Google Scholar]
- Weerasinghe RR, Bird DM, Allen NS (2005) Root-knot nematodes and bacterial Nod factors elicit common signal transduction events in Lotus japonicus. Proc Natl Acad Sci USA 102 3147–3152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson VM, Ho JY, Wu FF, Miller N, Kaloshian I (1994. a) A PCR-based marker tightly linked to the nematode resistance gene, Mi, in tomato. Theor Appl Genet 87 757–763 [DOI] [PubMed] [Google Scholar]
- Williamson VM, Hussey RS (1996) Nematode pathogenesis and resistance in plants. Plant Cell 8 1735–1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson VM, Kumar A (2006) Nematode resistance in plants: the battle underground. Trends Genet 22 396–403 [DOI] [PubMed] [Google Scholar]
- Williamson VM, Lambert KN, Kaloshian I (1994. b) Molecular biology of nematode resistance in tomato. In F Lamberti, C De Giorgi, DM Bird, eds, Advances in Molecular Plant Nematology. Plenum, New York, pp 211–219
- Wilson MA, Bird DM, van der Knaap E (1994) A comprehensive subtractive cDNA cloning approach to identify nematode induced transcripts in tomato. Phytopathology 84 299–303 [Google Scholar]
- Wolfinger RD, Gibson G, Wolfinger ED, Bennet L, Hamadeh H, Bushel Afshari C, Paules RS (2001) Assessing gene significance from cDNA microarray expression data via mixed models. J Comput Biol 8 625–637 [DOI] [PubMed] [Google Scholar]
- Wyss U, Grundler FMW, Münch A (1992) The parasitic behaviour of 2nd-stage juveniles of Meloidogyne incognita in roots of Arabidopsis thaliana. Nematologica 38 98–111 [Google Scholar]
- Zdobnov EM, Apweiler R (2001) InterProScan—an integration platform for the signature-recognition methods in InterPro. Bioinformatics 17 847–848 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.