Abstract
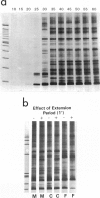
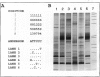
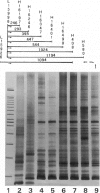
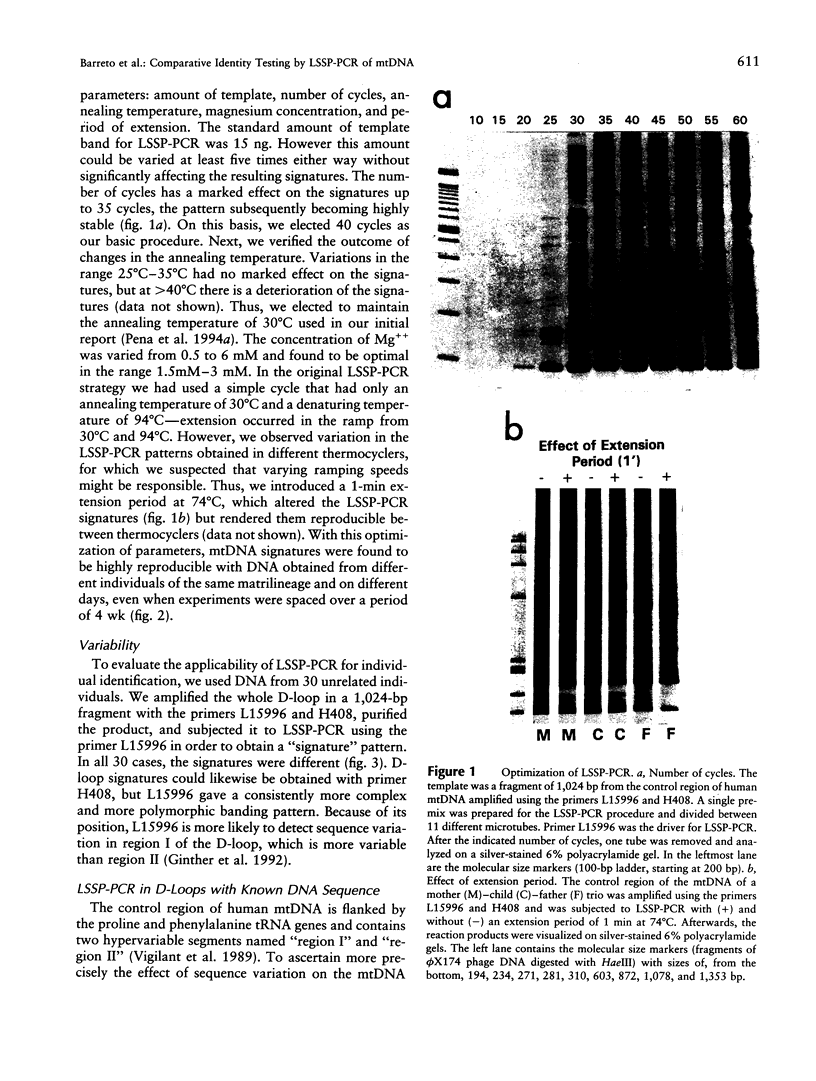
We have developed a technique called "LSSP-PCR" (low-stringency single specific primer PCR) that detects single or multiple mutations in DNA. A purified DNA fragment is submitted to PCR by using a single primer specific for one of the extremities of the fragment, under conditions of very low stringency. The primer hybridizes specifically to its complementary extremity and nonspecifically to multiple sites within the fragment, in a sequence-dependent manner. A complex set of reaction products is thus created that, when separated by electrophoresis, constitutes a unique "gene signature." We here report the application of LSSP-PCR to the detection of sequence variation in the control (D-loop) region of human mtDNA, which is known to differ significantly between unrelated individuals. We prepared human DNA samples from blood and amplified a 1024-bp portion of the mtDNA control region, using primers L15996 and H408. The amplified mtDNA fragments were then reamplified under LSSP-PCR conditions by using L15996 or H408 as drivers to produce complex signatures that always differed between unrelated individuals and yet were highly reproducible. In contrast, all mother-child pairs tested were identical, as expected from the matrilineal inheritance of mtDNA. Thus, the use of LSSP-PCR to produce D-loop signatures constitutes a powerful new technique for mtDNA-based comparative identity testing.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson S., Bankier A. T., Barrell B. G., de Bruijn M. H., Coulson A. R., Drouin J., Eperon I. C., Nierlich D. P., Roe B. A., Sanger F. Sequence and organization of the human mitochondrial genome. Nature. 1981 Apr 9;290(5806):457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- Di Rienzo A., Wilson A. C. Branching pattern in the evolutionary tree for human mitochondrial DNA. Proc Natl Acad Sci U S A. 1991 Mar 1;88(5):1597–1601. doi: 10.1073/pnas.88.5.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giles R. E., Blanc H., Cann H. M., Wallace D. C. Maternal inheritance of human mitochondrial DNA. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6715–6719. doi: 10.1073/pnas.77.11.6715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginther C., Issel-Tarver L., King M. C. Identifying individuals by sequencing mitochondrial DNA from teeth. Nat Genet. 1992 Oct;2(2):135–138. doi: 10.1038/ng1092-135. [DOI] [PubMed] [Google Scholar]
- Handt O., Richards M., Trommsdorff M., Kilger C., Simanainen J., Georgiev O., Bauer K., Stone A., Hedges R., Schaffner W. Molecular genetic analyses of the Tyrolean Ice Man. Science. 1994 Jun 17;264(5166):1775–1778. doi: 10.1126/science.8209259. [DOI] [PubMed] [Google Scholar]
- Lundstrom R., Tavaré S., Ward R. H. Estimating substitution rates from molecular data using the coalescent. Proc Natl Acad Sci U S A. 1992 Jul 1;89(13):5961–5965. doi: 10.1073/pnas.89.13.5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monnat R. J., Jr, Loeb L. A. Nucleotide sequence preservation of human mitochondrial DNA. Proc Natl Acad Sci U S A. 1985 May;82(9):2895–2899. doi: 10.1073/pnas.82.9.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena S. D., Barreto G., Vago A. R., De Marco L., Reinach F. C., Dias Neto E., Simpson A. J. Sequence-specific "gene signatures" can be obtained by PCR with single specific primers at low stringency. Proc Natl Acad Sci U S A. 1994 Mar 1;91(5):1946–1949. doi: 10.1073/pnas.91.5.1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena S. D., Macedo A. M., Gontijo N. F., Medeiros A. M., Ribeiro J. C. DNA bioprints: simple nonisotopic DNA fingerprints with biotinylated probes. Electrophoresis. 1991 Feb-Mar;12(2-3):146–152. doi: 10.1002/elps.1150120209. [DOI] [PubMed] [Google Scholar]
- Santos F. R., Pena S. D., Epplen J. T. Genetic and population study of a Y-linked tetranucleotide repeat DNA polymorphism with a simple non-isotopic technique. Hum Genet. 1993 Feb;90(6):655–656. doi: 10.1007/BF00202486. [DOI] [PubMed] [Google Scholar]
- Stoneking M., Hedgecock D., Higuchi R. G., Vigilant L., Erlich H. A. Population variation of human mtDNA control region sequences detected by enzymatic amplification and sequence-specific oligonucleotide probes. Am J Hum Genet. 1991 Feb;48(2):370–382. [PMC free article] [PubMed] [Google Scholar]
- Vigilant L., Pennington R., Harpending H., Kocher T. D., Wilson A. C. Mitochondrial DNA sequences in single hairs from a southern African population. Proc Natl Acad Sci U S A. 1989 Dec;86(23):9350–9354. doi: 10.1073/pnas.86.23.9350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa L. L., Caballero O. L., Levi J. E., Pena S. D., Simpson A. J. An approach to human papillomavirus identification using low stringency single specific primer PCR. Mol Cell Probes. 1995 Feb;9(1):45–48. doi: 10.1016/s0890-8508(95)90992-3. [DOI] [PubMed] [Google Scholar]
- Walsh P. S., Metzger D. A., Higuchi R. Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. Biotechniques. 1991 Apr;10(4):506–513. [PubMed] [Google Scholar]
- Ward R. H., Frazier B. L., Dew-Jager K., Päbo S. Extensive mitochondrial diversity within a single Amerindian tribe. Proc Natl Acad Sci U S A. 1991 Oct 1;88(19):8720–8724. doi: 10.1073/pnas.88.19.8720. [DOI] [PMC free article] [PubMed] [Google Scholar]