Abstract
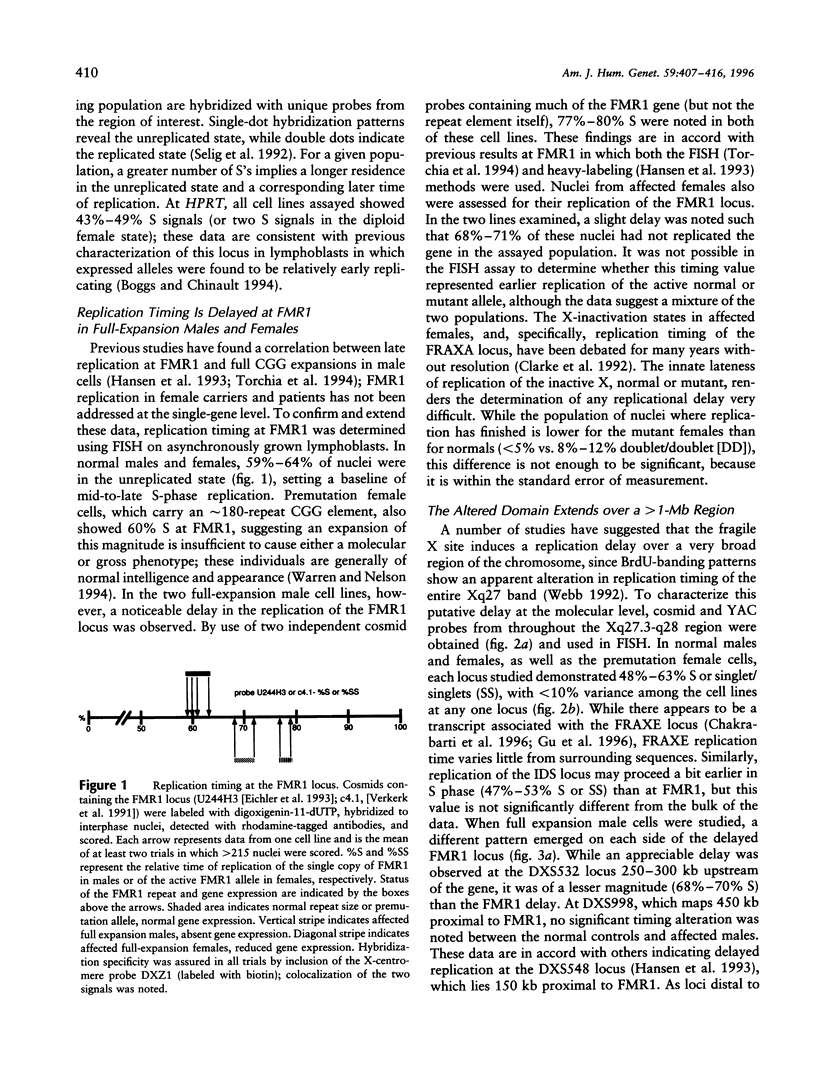
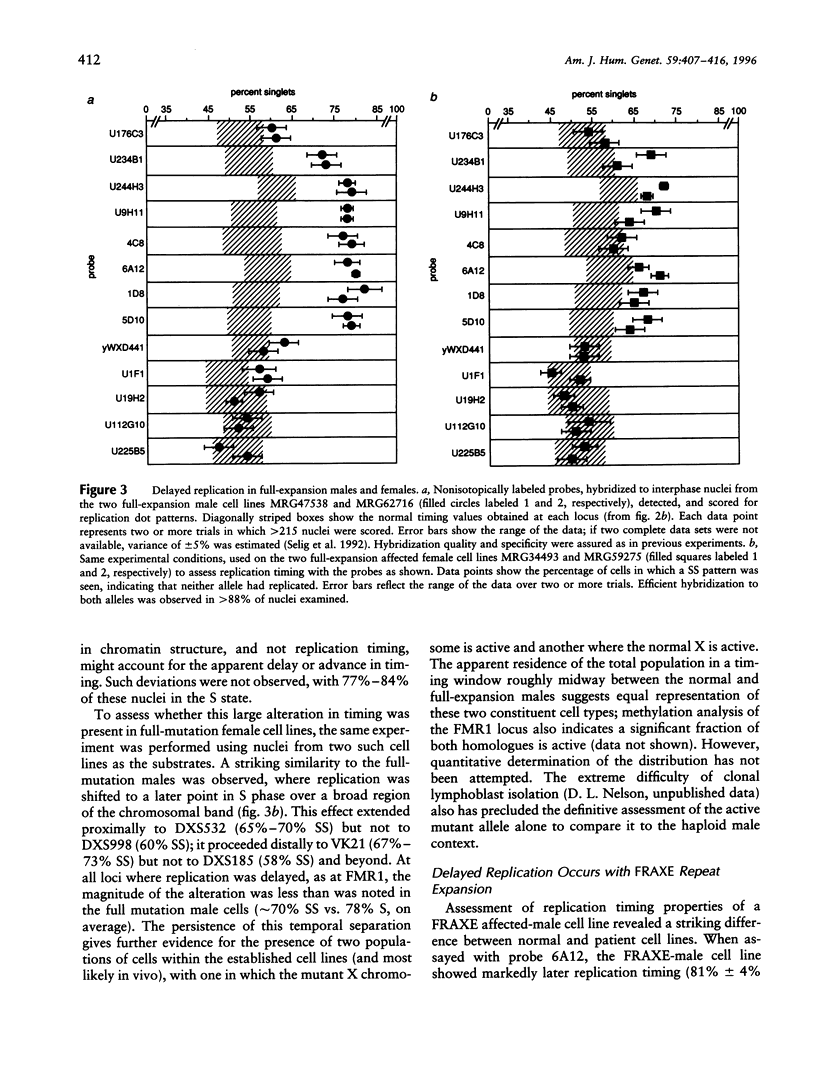
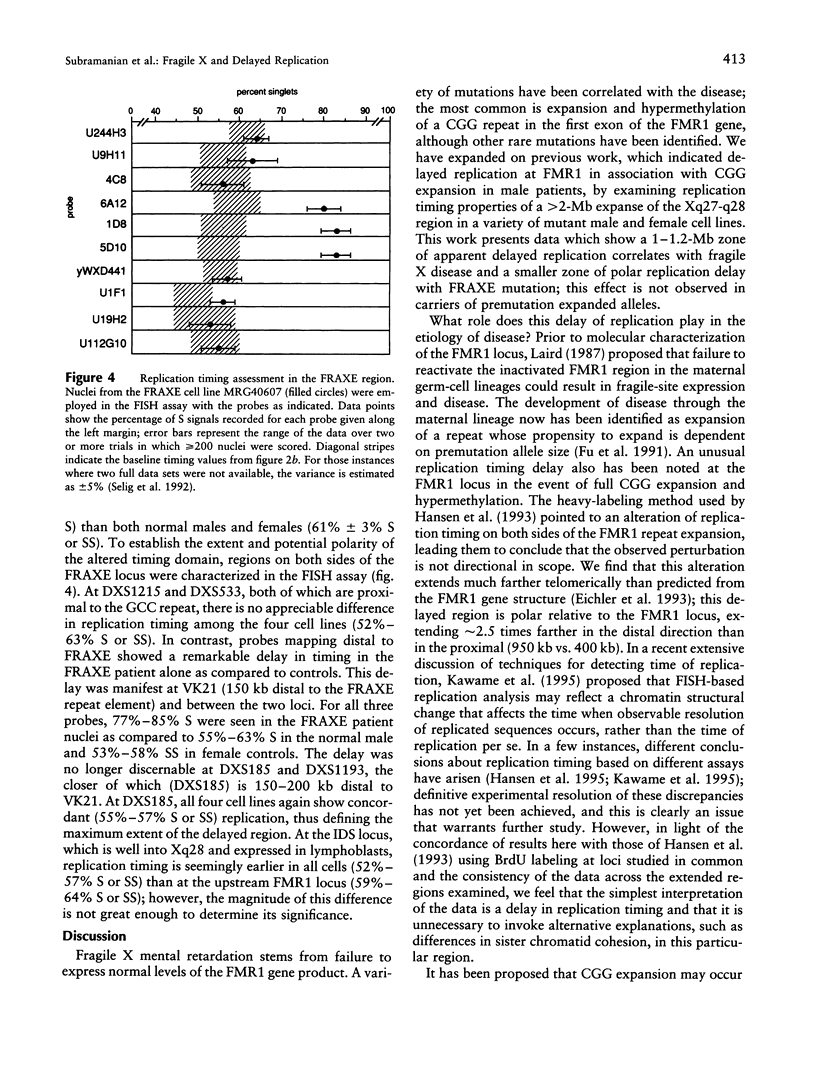
Trinucleotide repeat expansions have been implicated in the causation of a number of neurodegenerative disorders. In the case of fragile X syndrome, full expansion of the FMR1 repeat element (CGG)n has also been correlated with replication timing delay of the locus and proximal flanking sequences in male lymphoblasts. To define more extensively this altered region of DNA replication, as well as to extend these studies to female cells containing premutant and mutant alleles, study of the replication timing properties of a >2-Mb zone in the FRAXA region (Xq27.3-q28) was undertaken by using a FISH technique. In this assay, relative times of replication of specific loci are inferred from the ratios of singlet and doublet hybridization signals in interphase nuclei. In all individuals with a full expansion of the trinucleotide repeat, a large (1-1.2-Mb) region of delayed timing was observed; the apparent timing of the earlier-replicating allele in female cells in this region was intermediate between normal and affected alleles in males, which is in accordance with expectations of a mixed population of cells resulting from random X inactivation. In addition, expansion of the nearby FRAXE locus also was found to correlate with replication timing delay, although the extent of the altered region was somewhat less. Trinucleotide repeat expansion thus may be acting in the Xq27.3-q28 region to alter long-range chromatin structure that could influence transcription of gene sequences within the affected domain.
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Boggs B. A., Chinault A. C. Analysis of replication timing properties of human X-chromosomal loci by fluorescence in situ hybridization. Proc Natl Acad Sci U S A. 1994 Jun 21;91(13):6083–6087. doi: 10.1073/pnas.91.13.6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti L., Knight S. J., Flannery A. V., Davies K. E. A candidate gene for mild mental handicap at the FRAXE fragile site. Hum Mol Genet. 1996 Feb;5(2):275–282. doi: 10.1093/hmg/5.2.275. [DOI] [PubMed] [Google Scholar]
- Clarke A., Bradley D., Gillespie K., Rees D., Holland A., Thomas N. S. Fragile X mental retardation and the iduronate sulphatase locus: testing Laird's model of fra(X) inheritance. 1992 Apr 15-May 1Am J Med Genet. 43(1-2):299–306. doi: 10.1002/ajmg.1320430146. [DOI] [PubMed] [Google Scholar]
- De Boulle K., Verkerk A. J., Reyniers E., Vits L., Hendrickx J., Van Roy B., Van den Bos F., de Graaff E., Oostra B. A., Willems P. J. A point mutation in the FMR-1 gene associated with fragile X mental retardation. Nat Genet. 1993 Jan;3(1):31–35. doi: 10.1038/ng0193-31. [DOI] [PubMed] [Google Scholar]
- Devys D., Lutz Y., Rouyer N., Bellocq J. P., Mandel J. L. The FMR-1 protein is cytoplasmic, most abundant in neurons and appears normal in carriers of a fragile X premutation. Nat Genet. 1993 Aug;4(4):335–340. doi: 10.1038/ng0893-335. [DOI] [PubMed] [Google Scholar]
- Dhar V., Skoultchi A. I., Schildkraut C. L. Activation and repression of a beta-globin gene in cell hybrids is accompanied by a shift in its temporal replication. Mol Cell Biol. 1989 Aug;9(8):3524–3532. doi: 10.1128/mcb.9.8.3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler E. E., Holden J. J., Popovich B. W., Reiss A. L., Snow K., Thibodeau S. N., Richards C. S., Ward P. A., Nelson D. L. Length of uninterrupted CGG repeats determines instability in the FMR1 gene. Nat Genet. 1994 Sep;8(1):88–94. doi: 10.1038/ng0994-88. [DOI] [PubMed] [Google Scholar]
- Eichler E. E., Richards S., Gibbs R. A., Nelson D. L. Fine structure of the human FMR1 gene. Hum Mol Genet. 1993 Aug;2(8):1147–1153. doi: 10.1093/hmg/2.8.1147. [DOI] [PubMed] [Google Scholar]
- Feng Y., Zhang F., Lokey L. K., Chastain J. L., Lakkis L., Eberhart D., Warren S. T. Translational suppression by trinucleotide repeat expansion at FMR1. Science. 1995 May 5;268(5211):731–734. doi: 10.1126/science.7732383. [DOI] [PubMed] [Google Scholar]
- Flynn G. A., Hirst M. C., Knight S. J., Macpherson J. N., Barber J. C., Flannery A. V., Davies K. E., Buckle V. J. Identification of the FRAXE fragile site in two families ascertained for X linked mental retardation. J Med Genet. 1993 Feb;30(2):97–100. doi: 10.1136/jmg.30.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fryns J. P., Van den Berghe H. Inactivation pattern of the fragile X in heterozygous carriers. Am J Med Genet. 1988 May-Jun;30(1-2):401–406. doi: 10.1002/ajmg.1320300140. [DOI] [PubMed] [Google Scholar]
- Fu Y. H., Kuhl D. P., Pizzuti A., Pieretti M., Sutcliffe J. S., Richards S., Verkerk A. J., Holden J. J., Fenwick R. G., Jr, Warren S. T. Variation of the CGG repeat at the fragile X site results in genetic instability: resolution of the Sherman paradox. Cell. 1991 Dec 20;67(6):1047–1058. doi: 10.1016/0092-8674(91)90283-5. [DOI] [PubMed] [Google Scholar]
- Gedeon A. K., Baker E., Robinson H., Partington M. W., Gross B., Manca A., Korn B., Poustka A., Yu S., Sutherland G. R. Fragile X syndrome without CCG amplification has an FMR1 deletion. Nat Genet. 1992 Aug;1(5):341–344. doi: 10.1038/ng0892-341. [DOI] [PubMed] [Google Scholar]
- Gedeon A. K., Meinänen M., Adès L. C., Käriäinen H., Gécz J., Baker E., Sutherland G. R., Mulley J. C. Overlapping submicroscopic deletions in Xq28 in two unrelated boys with developmental disorders: identification of a gene near FRAXE. Am J Hum Genet. 1995 Apr;56(4):907–914. [PMC free article] [PubMed] [Google Scholar]
- Gu Y., Lugenbeel K. A., Vockley J. G., Grody W. W., Nelson D. L. A de novo deletion in FMR1 in a patient with developmental delay. Hum Mol Genet. 1994 Sep;3(9):1705–1706. doi: 10.1093/hmg/3.9.1705. [DOI] [PubMed] [Google Scholar]
- Gu Y., Shen Y., Gibbs R. A., Nelson D. L. Identification of FMR2, a novel gene associated with the FRAXE CCG repeat and CpG island. Nat Genet. 1996 May;13(1):109–113. doi: 10.1038/ng0596-109. [DOI] [PubMed] [Google Scholar]
- Hamel B. C., Smits A. P., de Graaff E., Smeets D. F., Schoute F., Eussen B. H., Knight S. J., Davies K. E., Assman-Hulsmans C. F., Oostra B. A. Segregation of FRAXE in a large family: clinical, psychometric, cytogenetic, and molecular data. Am J Hum Genet. 1994 Nov;55(5):923–931. [PMC free article] [PubMed] [Google Scholar]
- Hansen R. S., Canfield T. K., Gartler S. M. Reverse replication timing for the XIST gene in human fibroblasts. Hum Mol Genet. 1995 May;4(5):813–820. doi: 10.1093/hmg/4.5.813. [DOI] [PubMed] [Google Scholar]
- Hansen R. S., Canfield T. K., Lamb M. M., Gartler S. M., Laird C. D. Association of fragile X syndrome with delayed replication of the FMR1 gene. Cell. 1993 Jul 2;73(7):1403–1409. doi: 10.1016/0092-8674(93)90365-w. [DOI] [PubMed] [Google Scholar]
- Hirst M., Grewal P., Flannery A., Slatter R., Maher E., Barton D., Fryns J. P., Davies K. Two new cases of FMR1 deletion associated with mental impairment. Am J Hum Genet. 1995 Jan;56(1):67–74. [PMC free article] [PubMed] [Google Scholar]
- Holmquist G. P. Role of replication time in the control of tissue-specific gene expression. Am J Hum Genet. 1987 Feb;40(2):151–173. [PMC free article] [PubMed] [Google Scholar]
- Kawame H., Gartler S. M., Hansen R. S. Allele-specific replication timing in imprinted domains: absence of asynchrony at several loci. Hum Mol Genet. 1995 Dec;4(12):2287–2293. doi: 10.1093/hmg/4.12.2287. [DOI] [PubMed] [Google Scholar]
- Knight S. J., Voelckel M. A., Hirst M. C., Flannery A. V., Moncla A., Davies K. E. Triplet repeat expansion at the FRAXE locus and X-linked mild mental handicap. Am J Hum Genet. 1994 Jul;55(1):81–86. [PMC free article] [PubMed] [Google Scholar]
- Kunst C. B., Warren S. T. Cryptic and polar variation of the fragile X repeat could result in predisposing normal alleles. Cell. 1994 Jun 17;77(6):853–861. doi: 10.1016/0092-8674(94)90134-1. [DOI] [PubMed] [Google Scholar]
- Laird C. D. Proposed mechanism of inheritance and expression of the human fragile-X syndrome of mental retardation. Genetics. 1987 Nov;117(3):587–599. doi: 10.1093/genetics/117.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichter P., Boyle A. L., Cremer T., Ward D. C. Analysis of genes and chromosomes by nonisotopic in situ hybridization. Genet Anal Tech Appl. 1991 Feb;8(1):24–35. doi: 10.1016/1050-3862(91)90005-c. [DOI] [PubMed] [Google Scholar]
- Lugenbeel K. A., Peier A. M., Carson N. L., Chudley A. E., Nelson D. L. Intragenic loss of function mutations demonstrate the primary role of FMR1 in fragile X syndrome. Nat Genet. 1995 Aug;10(4):483–485. doi: 10.1038/ng0895-483. [DOI] [PubMed] [Google Scholar]
- Meijer H., de Graaff E., Merckx D. M., Jongbloed R. J., de Die-Smulders C. E., Engelen J. J., Fryns J. P., Curfs P. M., Oostra B. A. A deletion of 1.6 kb proximal to the CGG repeat of the FMR1 gene causes the clinical phenotype of the fragile X syndrome. Hum Mol Genet. 1994 Apr;3(4):615–620. doi: 10.1093/hmg/3.4.615. [DOI] [PubMed] [Google Scholar]
- Nelson D. L., Ledbetter S. A., Corbo L., Victoria M. F., Ramírez-Solis R., Webster T. D., Ledbetter D. H., Caskey C. T. Alu polymerase chain reaction: a method for rapid isolation of human-specific sequences from complex DNA sources. Proc Natl Acad Sci U S A. 1989 Sep;86(17):6686–6690. doi: 10.1073/pnas.86.17.6686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussbaum R. L., Airhart S. D., Ledbetter D. H. Recombination and amplification of pyrimidine-rich sequences may be responsible for initiation and progression of the Xq27 fragile site: an hypothesis. Am J Med Genet. 1986 Jan-Feb;23(1-2):715–721. doi: 10.1002/ajmg.1320230162. [DOI] [PubMed] [Google Scholar]
- Ohashi H., Kuwano A., Tsukahara M., Arinami T., Kajii T. Replication patterns of the fragile X in heterozygous carriers: analysis by a BrdUrd antibody method. Am J Hum Genet. 1990 Dec;47(6):988–993. [PMC free article] [PubMed] [Google Scholar]
- Quan F., Zonana J., Gunter K., Peterson K. L., Magenis R. E., Popovich B. W. An atypical case of fragile X syndrome caused by a deletion that includes the FMR1 gene. Am J Hum Genet. 1995 May;56(5):1042–1051. [PMC free article] [PubMed] [Google Scholar]
- Reyniers E., Vits L., De Boulle K., Van Roy B., Van Velzen D., de Graaff E., Verkerk A. J., Jorens H. Z., Darby J. K., Oostra B. The full mutation in the FMR-1 gene of male fragile X patients is absent in their sperm. Nat Genet. 1993 Jun;4(2):143–146. doi: 10.1038/ng0693-143. [DOI] [PubMed] [Google Scholar]
- Riggs A. D. DNA methylation and late replication probably aid cell memory, and type I DNA reeling could aid chromosome folding and enhancer function. Philos Trans R Soc Lond B Biol Sci. 1990 Jan 30;326(1235):285–297. doi: 10.1098/rstb.1990.0012. [DOI] [PubMed] [Google Scholar]
- Rosenberg C., Vianna-Morgante A. M., Otto P. A., Navajas L. Effect of X inactivation on fragile X frequency and mental retardation. Am J Med Genet. 1991 Feb-Mar;38(2-3):421–424. doi: 10.1002/ajmg.1320380255. [DOI] [PubMed] [Google Scholar]
- Selig S., Okumura K., Ward D. C., Cedar H. Delineation of DNA replication time zones by fluorescence in situ hybridization. EMBO J. 1992 Mar;11(3):1217–1225. doi: 10.1002/j.1460-2075.1992.tb05162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe J. S., Nelson D. L., Zhang F., Pieretti M., Caskey C. T., Saxe D., Warren S. T. DNA methylation represses FMR-1 transcription in fragile X syndrome. Hum Mol Genet. 1992 Sep;1(6):397–400. doi: 10.1093/hmg/1.6.397. [DOI] [PubMed] [Google Scholar]
- Sutherland G. R., Baker E. Characterisation of a new rare fragile site easily confused with the fragile X. Hum Mol Genet. 1992 May;1(2):111–113. doi: 10.1093/hmg/1.2.111. [DOI] [PubMed] [Google Scholar]
- Tarleton J., Richie R., Schwartz C., Rao K., Aylsworth A. S., Lachiewicz A. An extensive de novo deletion removing FMR1 in a patient with mental retardation and the fragile X syndrome phenotype. Hum Mol Genet. 1993 Nov;2(11):1973–1974. doi: 10.1093/hmg/2.11.1973. [DOI] [PubMed] [Google Scholar]
- Torchia B. S., Call L. M., Migeon B. R. DNA replication analysis of FMR1, XIST, and factor 8C loci by FISH shows nontranscribed X-linked genes replicate late. Am J Hum Genet. 1994 Jul;55(1):96–104. [PMC free article] [PubMed] [Google Scholar]
- Verkerk A. J., Pieretti M., Sutcliffe J. S., Fu Y. H., Kuhl D. P., Pizzuti A., Reiner O., Richards S., Victoria M. F., Zhang F. P. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991 May 31;65(5):905–914. doi: 10.1016/0092-8674(91)90397-h. [DOI] [PubMed] [Google Scholar]
- Warren S. T., Nelson D. L. Advances in molecular analysis of fragile X syndrome. JAMA. 1994 Feb 16;271(7):536–542. [PubMed] [Google Scholar]
- Webb T. Delayed replication of Xq27 in individuals with the fragile X syndrome. Am J Med Genet. 1992 Aug 1;43(6):1057–1062. doi: 10.1002/ajmg.1320430633. [DOI] [PubMed] [Google Scholar]
- Wilhelm D., Froster-Iskenius U., Paul J., Schwinger E. Fra(X) frequency on the active X-chromosome and phenotype in heterozygous carriers of the fra(X) form of mental retardation. Am J Med Genet. 1988 May-Jun;30(1-2):407–415. doi: 10.1002/ajmg.1320300141. [DOI] [PubMed] [Google Scholar]
- Wöhrle D., Hennig I., Vogel W., Steinbach P. Mitotic stability of fragile X mutations in differentiated cells indicates early post-conceptional trinucleotide repeat expansion. Nat Genet. 1993 Jun;4(2):140–142. doi: 10.1038/ng0693-140. [DOI] [PubMed] [Google Scholar]
- Yu W. D., Wenger S. L., Steele M. W. X chromosome imprinting in fragile X syndrome. Hum Genet. 1990 Oct;85(6):590–594. doi: 10.1007/BF00193580. [DOI] [PubMed] [Google Scholar]


