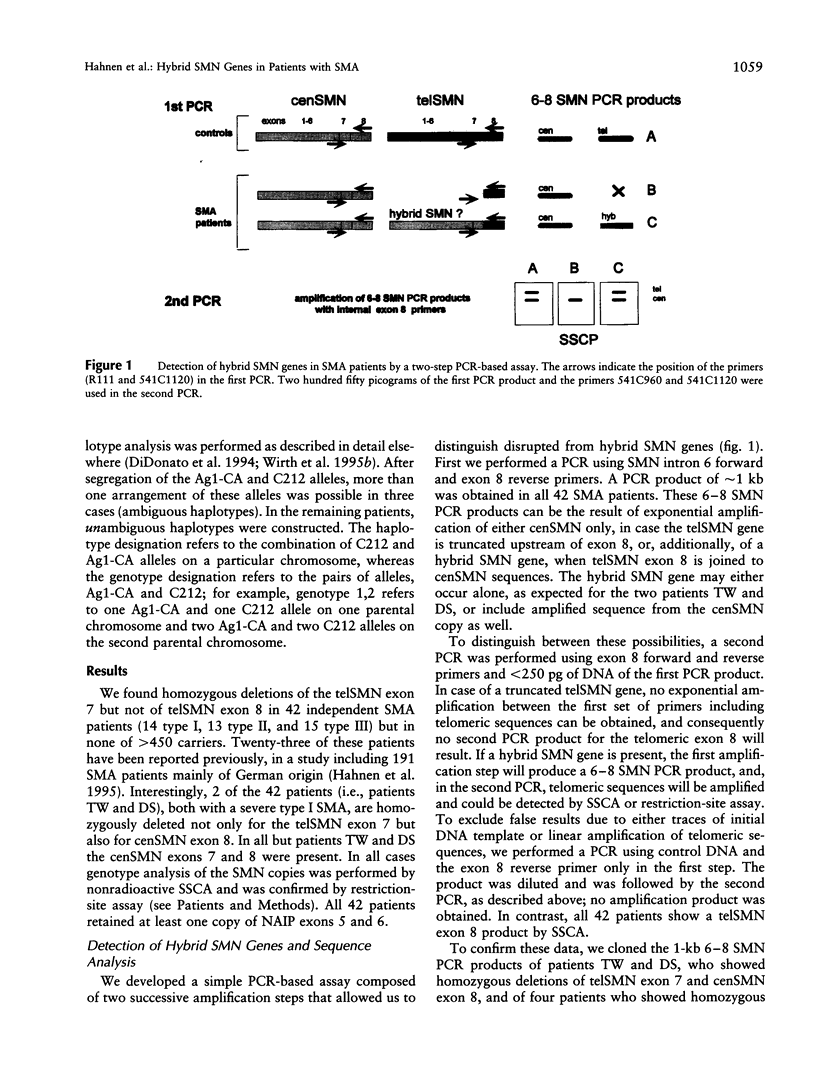
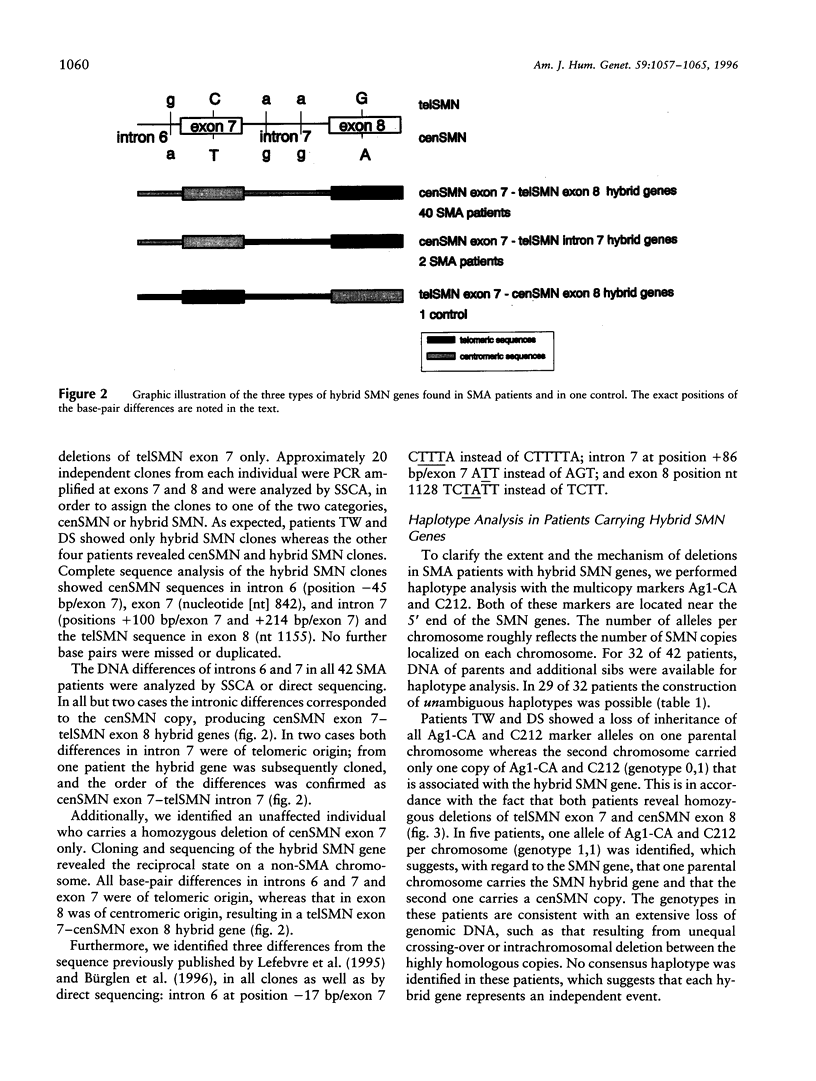
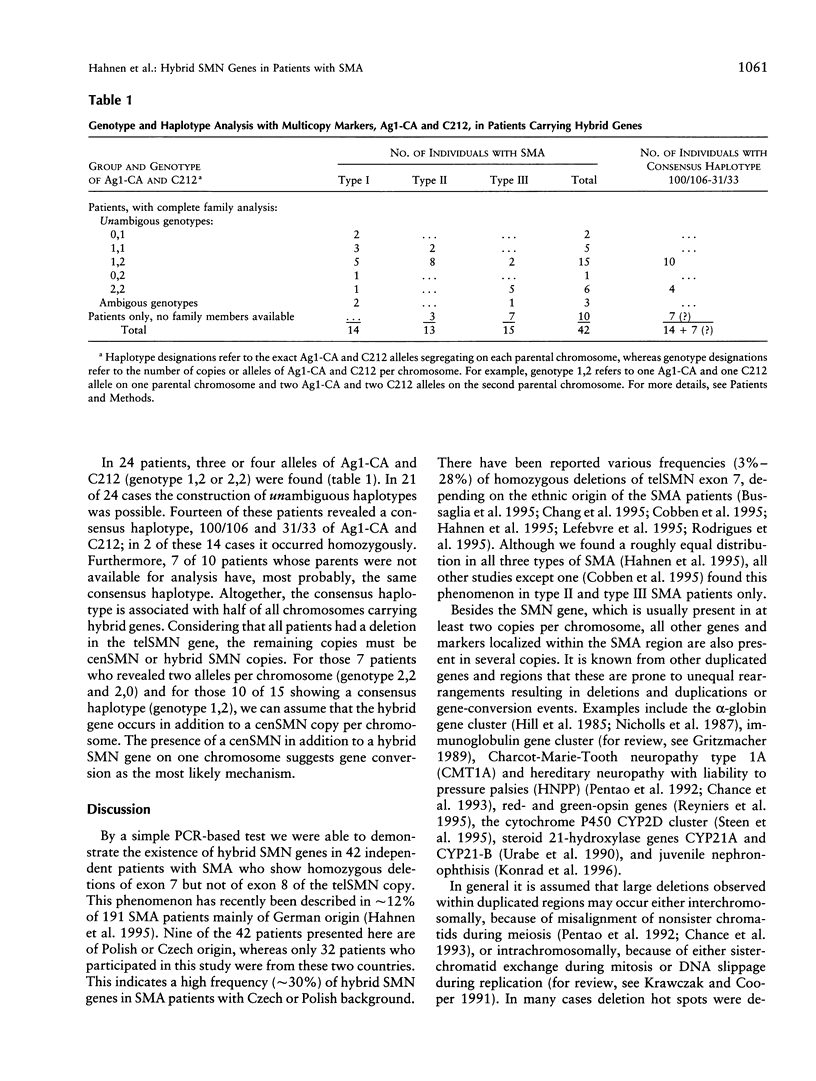
Abstract
Spinal muscular atrophy (SMA) is a frequent autosomal recessive neurodegenerative disorder leading to weakness and atrophy of voluntary muscles. The survival motor-neuron gene (SMN), a strong candidate for SMA, is present in two highly homologous copies (telSMN and cenSMN) within the SMA region. Only five nucleotide differences within the region between intron 6 and exon 8 distinguish these homologues. Independent of the severity of the disease, 90%-98% of all SMA patients carry homozygous deletions in telSMN, affecting either exon 7 or both exons 7 and 8. We present the molecular analysis of 42 SMA patients who carry homozygous deletions of telSMN exon 7 but not of exon 8. The question arises whether in these cases the telSMN is truncated upstream of exon 8 or whether hybrid SMN genes exist that are composed of centromeric and telomeric sequences. By a simple PCR-based assay we demonstrate that in each case the remaining telSMN exon 8 is part of a hybrid SMN gene. Sequencing of cloned hybrid SMN genes from seven patients, as well as direct sequencing and single-strand conformation analysis of all patients, revealed the same composition in all but two patients: the base-pair differences in introns 6 and 7 and exon 7 are of centromeric origin whereas exon 8 is of telomeric origin. Nonetheless, haplotype analysis with polymorphic multicopy markers, Ag1-CA and C212, localized at the 5' end of the SMN genes suggests different mechanisms of occurrence, unequal rearrangements, and gene conversion involving both copies of the SMN genes. In approximately half of all patients, we identified a consensus haplotype, suggesting a common origin. Interestingly, we identified a putative recombination hot spot represented by recombination-stimulating elements (TGGGG and TGAGGT) in exon 8 that is homologous to the human deletion-hot spot consensus sequence in the immunoglobulin switch region, the alpha-globin cluster, and the polymerase alpha arrest sites. This may explain why independent hybrid SMN genes show identical sequences.
Full text
PDF

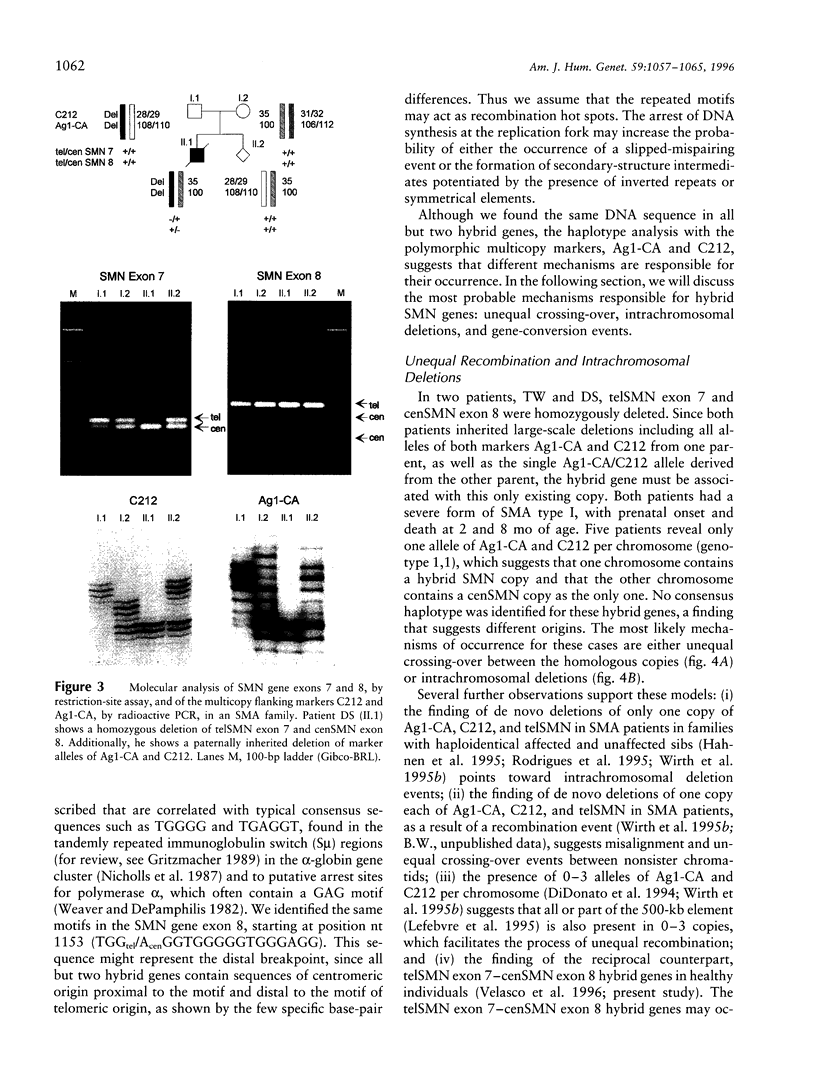



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brahe C., Velonà I., van der Steege G., Zappata S., van de Veen A. Y., Osinga J., Tops C. M., Fodde R., Khan P. M., Buys C. H. Mapping of two new markers within the smallest interval harboring the spinal muscular atrophy locus by family and radiation hybrid analysis. Hum Genet. 1994 May;93(5):494–501. doi: 10.1007/BF00202811. [DOI] [PubMed] [Google Scholar]
- Burghes A. H., Ingraham S. E., McLean M., Thompson T. G., McPherson J. D., Kote-Jarai Z., Carpten J. D., DiDonato C. J., Ikeda J. E., Surh L. A multicopy dinucleotide marker that maps close to the spinal muscular atrophy gene. Genomics. 1994 May 15;21(2):394–402. doi: 10.1006/geno.1994.1282. [DOI] [PubMed] [Google Scholar]
- Bussaglia E., Clermont O., Tizzano E., Lefebvre S., Bürglen L., Cruaud C., Urtizberea J. A., Colomer J., Munnich A., Baiget M. A frame-shift deletion in the survival motor neuron gene in Spanish spinal muscular atrophy patients. Nat Genet. 1995 Nov;11(3):335–337. doi: 10.1038/ng1195-335. [DOI] [PubMed] [Google Scholar]
- Bürglen L., Lefebvre S., Clermont O., Burlet P., Viollet L., Cruaud C., Munnich A., Melki J. Structure and organization of the human survival motor neurone (SMN) gene. Genomics. 1996 Mar 15;32(3):479–482. doi: 10.1006/geno.1996.0147. [DOI] [PubMed] [Google Scholar]
- Chance P. F., Alderson M. K., Leppig K. A., Lensch M. W., Matsunami N., Smith B., Swanson P. D., Odelberg S. J., Disteche C. M., Bird T. D. DNA deletion associated with hereditary neuropathy with liability to pressure palsies. Cell. 1993 Jan 15;72(1):143–151. doi: 10.1016/0092-8674(93)90058-x. [DOI] [PubMed] [Google Scholar]
- Chang J. G., Jong Y. J., Huang J. M., Wang W. S., Yang T. Y., Chang C. P., Chen Y. J., Lin S. P. Molecular basis of spinal muscular atrophy in Chinese. Am J Hum Genet. 1995 Dec;57(6):1503–1505. [PMC free article] [PubMed] [Google Scholar]
- Cobben J. M., van der Steege G., Grootscholten P., de Visser M., Scheffer H., Buys C. H. Deletions of the survival motor neuron gene in unaffected siblings of patients with spinal muscular atrophy. Am J Hum Genet. 1995 Oct;57(4):805–808. [PMC free article] [PubMed] [Google Scholar]
- DiDonato C. J., Morgan K., Carpten J. D., Fuerst P., Ingraham S. E., Prescott G., McPherson J. D., Wirth B., Zerres K., Hurko O. Association between Ag1-CA alleles and severity of autosomal recessive proximal spinal muscular atrophy. Am J Hum Genet. 1994 Dec;55(6):1218–1229. [PMC free article] [PubMed] [Google Scholar]
- Gritzmacher C. A. Molecular aspects of heavy-chain class switching. Crit Rev Immunol. 1989;9(3):173–200. [PubMed] [Google Scholar]
- Hahnen E. T., Wirth B. Frequent DNA variant in exon 2a of the survival motor neuron gene (SMN): a further possibility for distinguishing the two copies of the gene. Hum Genet. 1996 Jul;98(1):122–123. doi: 10.1007/s004390050172. [DOI] [PubMed] [Google Scholar]
- Hahnen E., Forkert R., Marke C., Rudnik-Schöneborn S., Schönling J., Zerres K., Wirth B. Molecular analysis of candidate genes on chromosome 5q13 in autosomal recessive spinal muscular atrophy: evidence of homozygous deletions of the SMN gene in unaffected individuals. Hum Mol Genet. 1995 Oct;4(10):1927–1933. doi: 10.1093/hmg/4.10.1927. [DOI] [PubMed] [Google Scholar]
- Hill A. V., Nicholls R. D., Thein S. L., Higgs D. R. Recombination within the human embryonic xi-globin locus: a common xi-xi chromosome produced by gene conversion of the psi xi gene. Cell. 1985 Oct;42(3):809–819. doi: 10.1016/0092-8674(85)90277-6. [DOI] [PubMed] [Google Scholar]
- Kleyn P. W., Wang C. H., Lien L. L., Vitale E., Pan J., Ross B. M., Grunn A., Palmer D. A., Warburton D., Brzustowicz L. M. Construction of a yeast artificial chromosome contig spanning the spinal muscular atrophy disease gene region. Proc Natl Acad Sci U S A. 1993 Jul 15;90(14):6801–6805. doi: 10.1073/pnas.90.14.6801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konrad M., Saunier S., Heidet L., Silbermann F., Benessy F., Calado J., Le Paslier D., Broyer M., Gubler M. C., Antignac C. Large homozygous deletions of the 2q13 region are a major cause of juvenile nephronophthisis. Hum Mol Genet. 1996 Mar;5(3):367–371. doi: 10.1093/hmg/5.3.367. [DOI] [PubMed] [Google Scholar]
- Krawczak M., Cooper D. N. Gene deletions causing human genetic disease: mechanisms of mutagenesis and the role of the local DNA sequence environment. Hum Genet. 1991 Mar;86(5):425–441. doi: 10.1007/BF00194629. [DOI] [PubMed] [Google Scholar]
- Lefebvre S., Bürglen L., Reboullet S., Clermont O., Burlet P., Viollet L., Benichou B., Cruaud C., Millasseau P., Zeviani M. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995 Jan 13;80(1):155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- Melki J., Lefebvre S., Burglen L., Burlet P., Clermont O., Millasseau P., Reboullet S., Bénichou B., Zeviani M., Le Paslier D. De novo and inherited deletions of the 5q13 region in spinal muscular atrophies. Science. 1994 Jun 3;264(5164):1474–1477. doi: 10.1126/science.7910982. [DOI] [PubMed] [Google Scholar]
- Miller S. A., Dykes D. D., Polesky H. F. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988 Feb 11;16(3):1215–1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls R. D., Fischel-Ghodsian N., Higgs D. R. Recombination at the human alpha-globin gene cluster: sequence features and topological constraints. Cell. 1987 May 8;49(3):369–378. doi: 10.1016/0092-8674(87)90289-3. [DOI] [PubMed] [Google Scholar]
- Pentao L., Wise C. A., Chinault A. C., Patel P. I., Lupski J. R. Charcot-Marie-Tooth type 1A duplication appears to arise from recombination at repeat sequences flanking the 1.5 Mb monomer unit. Nat Genet. 1992 Dec;2(4):292–300. doi: 10.1038/ng1292-292. [DOI] [PubMed] [Google Scholar]
- Reyniers E., Van Thienen M. N., Meire F., De Boulle K., Devries K., Kestelijn P., Willems P. J. Gene conversion between red and defective green opsin gene in blue cone monochromacy. Genomics. 1995 Sep 20;29(2):323–328. doi: 10.1006/geno.1995.9998. [DOI] [PubMed] [Google Scholar]
- Rodrigues N. R., Owen N., Talbot K., Ignatius J., Dubowitz V., Davies K. E. Deletions in the survival motor neuron gene on 5q13 in autosomal recessive spinal muscular atrophy. Hum Mol Genet. 1995 Apr;4(4):631–634. doi: 10.1093/hmg/4.4.631. [DOI] [PubMed] [Google Scholar]
- Rodrigues N. R., Owen N., Talbot K., Patel S., Muntoni F., Ignatius J., Dubowitz V., Davies K. E. Gene deletions in spinal muscular atrophy. J Med Genet. 1996 Feb;33(2):93–96. doi: 10.1136/jmg.33.2.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy N., Mahadevan M. S., McLean M., Shutler G., Yaraghi Z., Farahani R., Baird S., Besner-Johnston A., Lefebvre C., Kang X. The gene for neuronal apoptosis inhibitory protein is partially deleted in individuals with spinal muscular atrophy. Cell. 1995 Jan 13;80(1):167–178. doi: 10.1016/0092-8674(95)90461-1. [DOI] [PubMed] [Google Scholar]
- Steen V. M., Molven A., Aarskog N. K., Gulbrandsen A. K. Homologous unequal cross-over involving a 2.8 kb direct repeat as a mechanism for the generation of allelic variants of human cytochrome P450 CYP2D6 gene. Hum Mol Genet. 1995 Dec;4(12):2251–2257. doi: 10.1093/hmg/4.12.2251. [DOI] [PubMed] [Google Scholar]
- Theodosiou A. M., Morrison K. E., Nesbit A. M., Daniels R. J., Campbell L., Francis M. J., Christodoulou Z., Davies K. E. Complex repetitive arrangements of gene sequence in the candidate region of the spinal muscular atrophy gene in 5q13. Am J Hum Genet. 1994 Dec;55(6):1209–1217. [PMC free article] [PubMed] [Google Scholar]
- Urabe K., Kimura A., Harada F., Iwanaga T., Sasazuki T. Gene conversion in steroid 21-hydroxylase genes. Am J Hum Genet. 1990 Jun;46(6):1178–1186. [PMC free article] [PubMed] [Google Scholar]
- Velasco E., Valero C., Valero A., Moreno F., Hernández-Chico C. Molecular analysis of the SMN and NAIP genes in Spanish spinal muscular atrophy (SMA) families and correlation between number of copies of cBCD541 and SMA phenotype. Hum Mol Genet. 1996 Feb;5(2):257–263. doi: 10.1093/hmg/5.2.257. [DOI] [PubMed] [Google Scholar]
- Wang C. H., Xu J., Carter T. A., Ross B. M., Dominski M. K., Bellcross C. A., Penchaszadeh G. K., Munsat T. L., Gilliam T. C. Characterization of survival motor neuron (SMNT) gene deletions in asymptomatic carriers of spinal muscular atrophy. Hum Mol Genet. 1996 Mar;5(3):359–365. doi: 10.1093/hmg/5.3.359. [DOI] [PubMed] [Google Scholar]
- Weaver D. T., DePamphilis M. L. Specific sequences in native DNA that arrest synthesis by DNA polymerase alpha. J Biol Chem. 1982 Feb 25;257(4):2075–2086. [PubMed] [Google Scholar]
- Wirth B., Hahnen E., Morgan K., DiDonato C. J., Dadze A., Rudnik-Schöneborn S., Simard L. R., Zerres K., Burghes A. H. Allelic association and deletions in autosomal recessive proximal spinal muscular atrophy: association of marker genotype with disease severity and candidate cDNAs. Hum Mol Genet. 1995 Aug;4(8):1273–1284. doi: 10.1093/hmg/4.8.1273. [DOI] [PubMed] [Google Scholar]
- Wirth B., el-Agwany A., Baasner A., Burghes A., Koch A., Dadze A., Piechaczeck-Wappenschmidt B., Rudnik-Schöneborn S., Zerres K., Schönling J. Mapping of the spinal muscular atrophy (SMA) gene to a 750-kb interval flanked by two new microsatellites. Eur J Hum Genet. 1995;3(1):56–60. doi: 10.1159/000472274. [DOI] [PubMed] [Google Scholar]
- van der Steege G., Grootscholten P. M., van der Vlies P., Draaijers T. G., Osinga J., Cobben J. M., Scheffer H., Buys C. H. PCR-based DNA test to confirm clinical diagnosis of autosomal recessive spinal muscular atrophy. Lancet. 1995 Apr 15;345(8955):985–986. [PubMed] [Google Scholar]