Abstract
Six polymorphisms across the NF1 gene have been adapted for genotyping through application of PCR-based assays. Three exon-based polymorphisms--at positions 702, 2034, and 10647 in the NF1 cDNA--were genotyped by mutagenically separated PCR (MS-PCR). A fourth polymorphism, DV1.9, is an L1 insertion element in intron 30, and the other two polymorphisms, GXAlu and EVI-20, are short tandem repeats in intron 27b. All the polymorphisms were evaluated in a cohort of 110 CEPH individuals who previously had been analyzed by use of eight RFLPs at the NF1 locus. Pairwise linkage-disequilibrium analyses with the six PCR-based polymorphisms and their flanking markers demonstrated disequilibrium between all tested loci. Genotypes of the four diallelic polymorphisms (702, 2034, 10647, and DV1.9) were also evaluated in cohorts from the CEPH, African, and Japanese populations. The CEPH and Japanese cohorts showed similar heterozygosities and linkage-disequilibrium coefficients. The African cohort showed a higher degree of heterozygosity and lower linkage-disequilibrium values, compared with the CEPH and Japanese cohorts.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andersen L. B., Tarlé S. A., Marchuk D. A., Legius E., Collins F. S. A compound nucleotide repeat in the neurofibromatosis (NF1) gene. Hum Mol Genet. 1993 Jul;2(7):1083–1083. doi: 10.1093/hmg/2.7.1083-a. [DOI] [PubMed] [Google Scholar]
- Bell G. I., Karam J. H., Rutter W. J. Polymorphic DNA region adjacent to the 5' end of the human insulin gene. Proc Natl Acad Sci U S A. 1981 Sep;78(9):5759–5763. doi: 10.1073/pnas.78.9.5759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleyl S., Ainsworth P., Nelson L., Viskochil D., Ward K. An ancient Ta subclass L1 insertion results in an intragenic polymorphism in an intron of the NF1 gene. Hum Mol Genet. 1994 Mar;3(3):517–518. doi: 10.1093/hmg/3.3.517. [DOI] [PubMed] [Google Scholar]
- Bowcock A. M., Ruiz-Linares A., Tomfohrde J., Minch E., Kidd J. R., Cavalli-Sforza L. L. High resolution of human evolutionary trees with polymorphic microsatellites. Nature. 1994 Mar 31;368(6470):455–457. doi: 10.1038/368455a0. [DOI] [PubMed] [Google Scholar]
- Dams E., Van de Kelft E. J., Martin J. J., Verlooy J., Willems P. J. Instability of microsatellites in human gliomas. Cancer Res. 1995 Apr 1;55(7):1547–1549. [PubMed] [Google Scholar]
- Deka R., Jin L., Shriver M. D., Yu L. M., DeCroo S., Hundrieser J., Bunker C. H., Ferrell R. E., Chakraborty R. Population genetics of dinucleotide (dC-dA)n.(dG-dT)n polymorphisms in world populations. Am J Hum Genet. 1995 Feb;56(2):461–474. [PMC free article] [PubMed] [Google Scholar]
- Elbein S. C. Linkage disequilibrium among RFLPs at the insulin-receptor locus despite intervening Alu repeat sequences. Am J Hum Genet. 1992 Nov;51(5):1103–1110. [PMC free article] [PubMed] [Google Scholar]
- Heim R. A., Kam-Morgan L. N., Binnie C. G., Corns D. D., Cayouette M. C., Farber R. A., Aylsworth A. S., Silverman L. M., Luce M. C. Distribution of 13 truncating mutations in the neurofibromatosis 1 gene. Hum Mol Genet. 1995 Jun;4(6):975–981. doi: 10.1093/hmg/4.6.975. [DOI] [PubMed] [Google Scholar]
- Hoffmeyer S., Assum G. An RsaI polymorphism in the transcribed region of the neurofibromatosis (NF1)-gene. Hum Genet. 1994 Apr;93(4):481–482. doi: 10.1007/BF00201684. [DOI] [PubMed] [Google Scholar]
- Hoffmeyer S., Assum G., Griesser J., Kaufmann D., Nürnberg P., Krone W. On unequal allelic expression of the neurofibromin gene in neurofibromatosis type 1. Hum Mol Genet. 1995 Aug;4(8):1267–1272. doi: 10.1093/hmg/4.8.1267. [DOI] [PubMed] [Google Scholar]
- Hofman K. J., Boehm C. D. Familial neurofibromatosis type 1: clinical experience with DNA testing. J Pediatr. 1992 Mar;120(3):394–398. doi: 10.1016/s0022-3476(05)80903-5. [DOI] [PubMed] [Google Scholar]
- Honchel R., Halling K. C., Thibodeau S. N. Genomic instability in neoplasia. Semin Cell Biol. 1995 Feb;6(1):45–52. doi: 10.1016/1043-4682(95)90014-4. [DOI] [PubMed] [Google Scholar]
- Jorde L. B., Bamshad M. J., Watkins W. S., Zenger R., Fraley A. E., Krakowiak P. A., Carpenter K. D., Soodyall H., Jenkins T., Rogers A. R. Origins and affinities of modern humans: a comparison of mitochondrial and nuclear genetic data. Am J Hum Genet. 1995 Sep;57(3):523–538. doi: 10.1002/ajmg.1320570340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorde L. B., Watkins W. S., Viskochil D., O'Connell P., Ward K. Linkage disequilibrium in the neurofibromatosis 1 (NF1) region: implications for gene mapping. Am J Hum Genet. 1993 Nov;53(5):1038–1050. [PMC free article] [PubMed] [Google Scholar]
- Leppig K. A., Viskochil D., Neil S., Rubenstein A., Johnson V. P., Zhu X. L., Brothman A. R., Stephens K. The detection of contiguous gene deletions at the neurofibromatosis 1 locus with fluorescence in situ hybridization. Cytogenet Cell Genet. 1996;72(1):95–98. doi: 10.1159/000134171. [DOI] [PubMed] [Google Scholar]
- Li Y., O'Connell P., Breidenbach H. H., Cawthon R., Stevens J., Xu G., Neil S., Robertson M., White R., Viskochil D. Genomic organization of the neurofibromatosis 1 gene (NF1). Genomics. 1995 Jan 1;25(1):9–18. doi: 10.1016/0888-7543(95)80104-t. [DOI] [PubMed] [Google Scholar]
- Lázaro C., Gaona A., Estivill X. Two CA/GT repeat polymorphisms in intron 27 of the human neurofibromatosis (NF1) gene. Hum Genet. 1994 Mar;93(3):351–352. doi: 10.1007/BF00212039. [DOI] [PubMed] [Google Scholar]
- Lázaro C., Gaona A., Ravella A., Volpini V., Casals T., Fuentes J. J., Estivill X. Novel alleles, hemizygosity and deletions at an Alu-repeat within the neurofibromatosis type 1 (NF1) gene. Hum Mol Genet. 1993 Jun;2(6):725–730. doi: 10.1093/hmg/2.6.725. [DOI] [PubMed] [Google Scholar]
- Lázaro C., Gaona A., Xu G., Weiss R., Estivill X. A highly informative CA/GT repeat polymorphism in intron 38 of the human neurofibromatosis type 1 (NF1) gene. Hum Genet. 1993 Oct;92(4):429–430. doi: 10.1007/BF01247353. [DOI] [PubMed] [Google Scholar]
- Marchuk D. A., Saulino A. M., Tavakkol R., Swaroop M., Wallace M. R., Andersen L. B., Mitchell A. L., Gutmann D. H., Boguski M., Collins F. S. cDNA cloning of the type 1 neurofibromatosis gene: complete sequence of the NF1 gene product. Genomics. 1991 Dec;11(4):931–940. doi: 10.1016/0888-7543(91)90017-9. [DOI] [PubMed] [Google Scholar]
- Ottini L., Esposito D. L., Richetta A., Carlesimo M., Palmirotta R., Verí M. C., Battista P., Frati L., Caramia F. G., Calvieri S. Alterations of microsatellites in neurofibromas of von Recklinghausen's disease. Cancer Res. 1995 Dec 1;55(23):5677–5680. [PubMed] [Google Scholar]
- Purandare S. M., Huntsman Breidenbach H., Li Y., Zhu X. L., Sawada S., Neil S. M., Brothman A., White R., Cawthon R., Viskochil D. Identification of neurofibromatosis 1 (NF1) homologous loci by direct sequencing, fluorescence in situ hybridization, and PCR amplification of somatic cell hybrids. Genomics. 1995 Dec 10;30(3):476–485. doi: 10.1006/geno.1995.1268. [DOI] [PubMed] [Google Scholar]
- Pykett M. J., Murphy M., Harnish P. R., George D. L. Identification of a microsatellite instability phenotype in meningiomas. Cancer Res. 1994 Dec 15;54(24):6340–6343. [PubMed] [Google Scholar]
- Rodenhiser D. I., Ainsworth P. J., Coulter-Mackie M. B., Singh S. M., Jung J. H. A genetic study of neurofibromatosis type 1 (NF1) in south-western Ontario. II. A PCR based approach to molecular and prenatal diagnosis using linkage. J Med Genet. 1993 May;30(5):363–368. doi: 10.1136/jmg.30.5.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
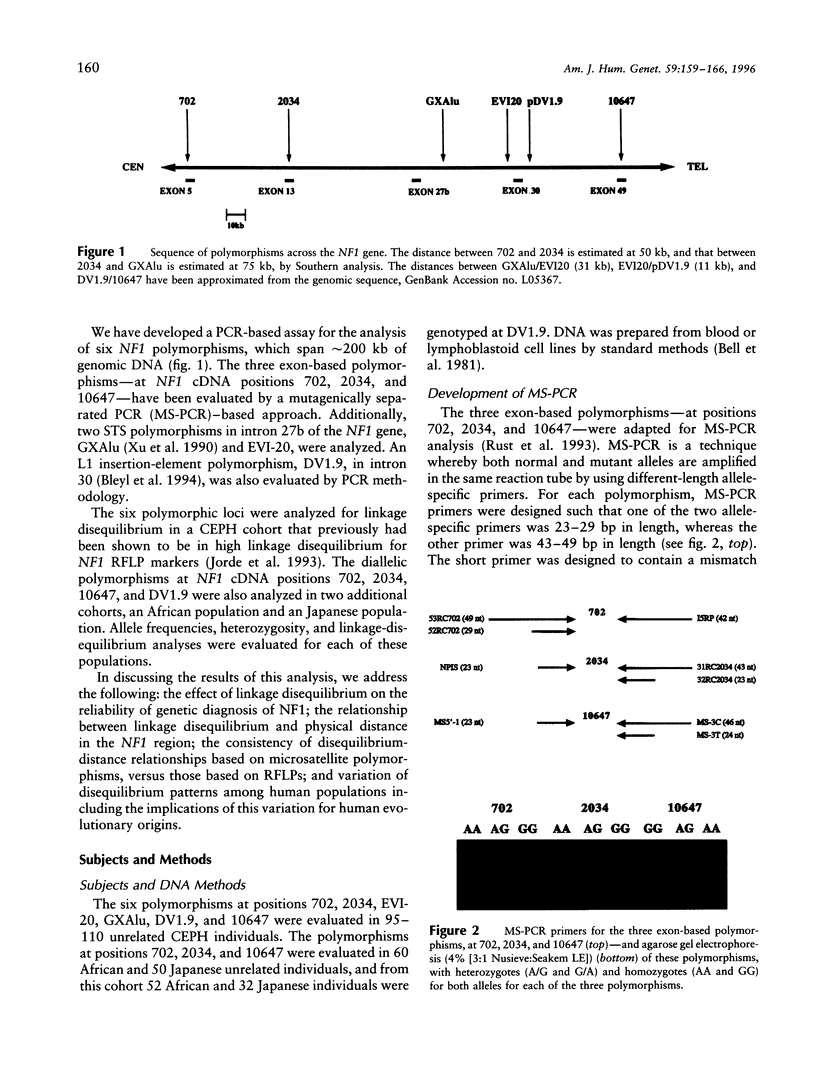
- Rust S., Funke H., Assmann G. Mutagenically separated PCR (MS-PCR): a highly specific one step procedure for easy mutation detection. Nucleic Acids Res. 1993 Aug 11;21(16):3623–3629. doi: 10.1093/nar/21.16.3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Régnier V., Danglot G., Nguyen V. C., Bernheim A. A Tsp509I variant in exon 13 of the neurofibromatosis type 1 (NF1) gene allows the identification of both alleles at the mRNA level. Hum Genet. 1995 Jul;96(1):131–132. doi: 10.1007/BF00214202. [DOI] [PubMed] [Google Scholar]
- Tishkoff S. A., Dietzsch E., Speed W., Pakstis A. J., Kidd J. R., Cheung K., Bonné-Tamir B., Santachiara-Benerecetti A. S., Moral P., Krings M. Global patterns of linkage disequilibrium at the CD4 locus and modern human origins. Science. 1996 Mar 8;271(5254):1380–1387. doi: 10.1126/science.271.5254.1380. [DOI] [PubMed] [Google Scholar]
- Ward K., O'Connell P., Carey J. C., Leppert M., Jolley S., Plaetke R., Ogden B., White R. Diagnosis of neurofibromatosis I by using tightly linked, flanking DNA markers. Am J Hum Genet. 1990 May;46(5):943–949. [PMC free article] [PubMed] [Google Scholar]
- Weir B. S. Inferences about linkage disequilibrium. Biometrics. 1979 Mar;35(1):235–254. [PubMed] [Google Scholar]
- Xu G. F., Nelson L., O'Connell P., White R. An Alu polymorphism intragenic to the neurofibromatosis type 1 gene (NF1). Nucleic Acids Res. 1991 Jul 11;19(13):3764–3764. doi: 10.1093/nar/19.13.3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G. F., O'Connell P., Viskochil D., Cawthon R., Robertson M., Culver M., Dunn D., Stevens J., Gesteland R., White R. The neurofibromatosis type 1 gene encodes a protein related to GAP. Cell. 1990 Aug 10;62(3):599–608. doi: 10.1016/0092-8674(90)90024-9. [DOI] [PubMed] [Google Scholar]
- Xu W., Liu L., Ponder M., Ponder B. A. A TaqI polymorphism in the human NF1 gene. Nucleic Acids Res. 1991 Aug 25;19(16):4570–4570. doi: 10.1093/nar/19.16.4570. [DOI] [PMC free article] [PubMed] [Google Scholar]