Abstract
Although DNA repair is faster in the transcribed strand of active genes, little is known about the possible contribution of mRNP biogenesis and export in transcription-coupled repair (TCR). Interestingly, mutants of THO, a transcription complex involved in maintenance of genome integrity, mRNP biogenesis and export, were recently found to be deficient in nucleotide excision repair. In this study we show by molecular DNA repair analysis, that Sub2-Yra1 and Thp1-Sac3, two main mRNA export complexes, are required for efficient TCR in yeast. Careful analysis revealed that THO mutants are also specifically affected in TCR. Ribozyme-mediated mRNA self-cleavage between two hot spots for UV damage showed that efficient TCR does not depend on the nascent mRNA, neither in wild-type nor in mutant cells. Along with severe UV damage-dependent loss in processivity, RNAPII was found binding to chromatin upon UV irradiation in THO mutants, suggesting that RNAPII remains stalled at DNA lesions. Furthermore, Def1, a factor responsible for the degradation of stalled RNAPII, appears essential for the viability of THO mutants subjected to DNA damage. Our results indicate that RNAPII is not proficient for TCR in mRNP biogenesis and export mutants, opening new perspectives on our knowledge of TCR in eukaryotic cells.
INTRODUCTION
Nucleotide excision repair (NER) is an evolutionarily conserved DNA repair pathway that deals with severely distorting DNA lesions including intrastrand crosslinks such as UV-induced pyrimidine dimers [reviewed in (1,2)]. Within NER two damage-sensing pathways are recognized: one for the entire genome, global genome repair (GGR), and one for the transcribed strand of active genes, transcription-coupled repair (TCR). In yeast, GGR requires Rad7, a protein carrying leucine-rich repeats, and Rad16, a member of the SWI2/SNF2 subfamily of putative helicases (3). These proteins presumably act in a complex that might be required in chromatin remodeling to facilitate damage recognition by Rad4/Rad23 [reviewed in (1,4)]. As ongoing transcription is required for TCR, damage recognition is likely done by the elongating RNA polymerase (RNAP) itself. RNAP arrests at injuries in the template strand triggering, likely via additional specific factors, the recruitment of the DNA repair machinery [reviewed in (5–7)]. Interestingly, TCR appears to be functional once a low and basal rate of transcription is achieved, beyond which there is no simple correlation between transcription and repair rates (8).
In Escherichia coli, the stalled RNAP leads to the recruitment of the transcription-repair coupling factor (TCRF) Mfd, allowing for the release of RNAP and further recruitment of the repair factors (9,10). In eukaryotes the precise mechanism of TCR remains poorly understood. Mutations in proteins required for NER lead to severe disorders known as Xeroderma pigmentosum and Cockayne's syndrome [for review see (11)]. One of these proteins, Cockayne syndrome B protein (CSB), and its yeast ortholog Rad26, share conserved functions (12,13) and represent putative eukaryotic TCRF candidates. CSB and Rad26 belong to the SWI2/SNF2 helicase superfamily. Although CSB has been shown to have DNA-dependent ATPase activity, an ATPase-deficient mutant partially restores CSB activity in vivo (14). The putative function of CSB as a TCRF has been substantiated by in vitro reconstitution of the TCR initiation steps, in which an elongating RNAPII arrested at a DNA lesion was shown to mediate an ATP-dependent incision of the damaged DNA only in the presence of CSB (15). XPG, one of the structure-specific DNA endonuclease responsible for the removal of an oligonucleotide containing the DNA lesion in NER, is another protein involved in TCR. Recent results imply a coordinated recognition of stalled RNAPII by XPG and CSB in TCR initiation in mammalian cells and suggest that TFIIH-dependent remodeling of stalled RNAPII without release may be sufficient to allow repair (16). In yeast, the Rpb9 subunit of RNAPII has also been shown to contribute to TCR (17,18). Alternatively, and analogous to the mRNA-dependent loading of termination factors in E. coli (19,20), it is also conceivable that the nascent mRNA, or proteins bound to it, may be required to load repair enzymes at stalled polymerases.
On the other hand, RNAPII is subject to ubiquitylation and proteasome-mediated degradation in response to UV-generated DNA damage (21–24). It has been proposed that degradation of damage-stalled RNAPII complexes might be an alternative to TCR (25). Indeed, recent studies have shown that arrested RNAPII elongation complexes are the preferred substrate for ubiquitylation, which is dependent on the C-terminal repeat domain (CTD) of RNAPII and on the Def1 protein in yeast (26,27).
In eukaryotic cells, export of nuclear mRNA to the cytoplasm requires correct RNA-processing and the association of a number of RNA-binding proteins to form export-competent ribonucleoprotein particles (mRNP) [for review see (28–30)]. Although there is growing evidence for transcription-coupled mRNA export, the physical nature of this coupling is not known. A connection between mRNP biogenesis and transcription is provided by THO, a conserved four-protein complex composed of stoichiometric amounts of Tho2, Hpr1, Mft1 and Thp2 (31) that is recruited to active chromatin in vivo (32,33). Null mutation of any component of THO leads to similar phenotypes of transcription impairment and mRNA export defects, as well as to a strong transcription-associated hyper-recombination phenotype (31,32,34–36). Together with the mRNA export proteins Sub2/UAP56 and Yra1/Aly, THO forms a larger complex termed TREX in yeast and humans (32). However, even though deletion of any of the THO genes leads to complete depletion of the complex (37), THO remains stable in vivo in sub2 mutants (36), indicating that it forms a core complex independently of Sub2. Strikingly, mutants of the Mex67-Mtr2 mRNA export factor—a heterodimer that mediates the interaction of the mRNP with the nuclear pore complex (NPC)—show THO-like gene expression and recombination phenotypes (36). The idea of THO being functionally involved in mRNP biogenesis and export is further strengthened by the observation that mutants of the Thp1–Sac3 complex, which has been shown to function in mRNA export by docking the mRNP to specific nucleoporins at the nuclear pore entry (38,39) confer the same transcription, mRNA export, and hyper-recombination phenotypes as do THO/TREX mutations (40,41).
Previously, we showed that null mutations of the HPR1 and THO2 genes confer defects in NER (42). With this precedent and considering that RNA-binding proteins are concomitantly assembled on the nascent mRNA to generate a stable and export-competent mRNP [reviewed in (30,43,44)], we studied whether TCR might be connected to mRNP biogenesis and export. We found that THO, Sub2-Yra1, Mex67-Mtr2 and Thp1-Sac3 are required for efficient TCR in yeast, thus linking mRNP biogenesis and export to TCR. Using a construct in which a self-cleaving Hammerhead ribozyme was cloned between two hot spots for UV damage, we demonstrate that TCR does not depend on the nascent mRNA, neither in wild-type nor in THO and Thp1-Sac3 deficient strains. Chromatin immunoprecipitation (ChIP) analyses revealed that, beside a severe UV damage-dependent loss in processivity, RNAPII is found to be bound to chromatin upon UV irradiation in THO mutants. Interestingly, Def1, a factor responsible for the removal of stalled RNAPII from a DNA template, is essential for the viability of THO mutants subjected to DNA damage. Our results support a model in which mRNP biogenesis and export is required for efficient TCR by preventing the occurrence of defective RNAPII complexes, which may remain stalled at a DNA lesion.
MATERIALS AND METHODS
Strains and plasmids
We used strains W303-1A, rad1Δ::LEU2 (W839-5D, R. Rothstein), rad7Δ::URA3 (MGSC97, 3), rad26Δ::HIS3 (MGSC102, 13), hpr1Δ::HIS3 (U674-1C), tho2Δ::KAN (RK2-6D, 45), hpr1-101 (WH101-1A, 37), sub2Δ::HIS3 pCM185-sub2-206 (TRP1, DLY33), sub2Δ::HIS3 (Ura- segregant from DLY23, 46), mex67-5 (WMC1-1A, 36), thp1Δ::KAN (WFBE046, F. Fabre), which have been reported previously, and isogenic derivatives obtained by genetic crosses. The def1Δ::HYGR strains were obtained by replacement of the DEF1 gene in rad7Δ::URA3 hpr1Δ::HIS3 diploids and subsequent tetrad dissection. For ChIP analyses, the GAL1 promoter fused to the 5′-most 300 bp of the YLR454w open-reading frame was integrated at the YLR454w locus (32). Plasmids pHG001-Rib+ and pHG001-ribm were obtained by insertion of a fragment containing 39 bp yeast T-tracts (DED1 promoter), 52 bp Hammerhead ribozyme sequence (47), the Eco47III-BssHII LacZ fragment, and 39 bp yeast T-tracts (DED1 promoter) into the Eco47III-BssHII sites of plasmid pRWE005 (48).
UV survival curves
Yeast cells were grown in YPD-rich medium to an OD600 of 0.6. Plating, UV irradiation, and quantification were performed as described (42). All survival curves shown represent the average of at least three independent experiments.
UV irradiation and repair
Irradiation and repair was carried out as described (49) with minor modifications. Yeast cells were grown in 400 ml YPD-rich medium, or SG supplemented with the appropriate amino acids in the case of cells harboring the plasmid pHG001, to an OD600 of 0.8, harvested, and resuspended in SD or SG to an OD600 of 1.2. A 200 ml aliquot was irradiated with 230 J/m2 UV light using germicidal lamps (Philips T UV 15 W). The medium was supplemented to YPD-rich or with the appropriate amino acids and the cells incubated at 30°C in the dark for recovery. Fifty milliliter aliquots were taken at the indicated repair times, chilled on ice and DNA purified using the described CTAB protocol (48). All steps from UV irradiation to DNA extraction were carried out in red light (Philips T LD 18 W RED).
Gene- and strand-specific DNA repair assays
CPDs were mapped by indirect end-labeling and quantified as described (49). DNA was cut with appropriate restriction enzymes and aliquots were cut at CPDs with T4-endonuclease V (T4endoV, Epicentre) or mock treated. The DNA was electrophoresed in 1.3% alkaline agarose gels, blotted to Nylon membranes and hybridized with radioactively labeled strand-specific DNA probes. Strand-specific probes were generated by primer extension. Primers to generate DNA templates and probes that hybridize to the TS- and NTS-strand, respectively, were: Rpb2-A: 5′-TCTTGGAATAATAACTTCGCGGC-3′; Rpb2-B: 5′-GGTGGATGACAAGATACATGCC-3′; pHG001-A: 5′-ATTTTTGACACCAGACCAACTG-3′; pHG001-B: 5′-TCTGCCATTGTCAGACATGTAT-3′.
Membranes were analyzed and quantified with a PhosphorImager (Fuji FLA3000). The CPD content was calculated using the Poisson distribution, -ln(RFa/RFb), where RFa and RFb represent the signal intensities of the intact restriction fragment of the T4endoV- and mock-treated DNA, respectively. Region-specific damage was calculated as the signal of that region in the T4endoV-treated DNA divided by the signal of the whole lane. The corresponding signal of the mock-treated DNA was subtracted as background. The average of the initial damage generated with 230 J/m2 was 0.3 CPD/kb. To allow direct comparison between different strains, repair curves were calculated as the fraction of CPDs removed versus repair time. The initial damage was set to 0% repair.
Northern analyses
RNA was extracted and northern analyses performed according to standard procedures. For RNA synthesis recovery analyses, filters were hybridized with a 324-bp long RPB2 fragment obtained by PCR using primers RPB2 A, RPB2 B. Northern blots were quantified using a Fuji FLA 3000 and normalized to the rRNA levels of each samples. For pHG001 ribozyme cleavage analyses, filters were hybridized with a 314-bp long LacZ fragment obtained by PCR using primers HG001-A and HG001-B.
ChIP analyses
Cells were grown and irradiated as described above. Forty milliliter aliquots were taken at the indicated repair times and cross-linked with a 1% formaldehyde solution for 15 min at RT. Glycine was added to a final concentration of 125 mM, and the cell pellets frozen in liquid nitrogen and kept at −80°C. ChIP assays were performed as described (50). Monoclonal 8WG16 antibody (COVANCE) and protein A-Sepharose were used to immunoprecipitate RNAPII. The GFX purification system (GE Healthcare) was used for the last purification step. All samples were treated with 200 ng photolyase (TREVIGEN) for 30 min under photoreactivating light (Sylvania F15T8 BLB) prior to real-time quantitative PCR analysis. We used 20–30-bp oligonucleotides for the PCR amplification of two fragments of YLR454c (3–43 and 7621–7674) and the 9716–9863 intergenic region of chromosome V, which was used as non-transcribed control. Real-time quantitative PCR was performed using SYBR green dye in the 7500 Real Time PCR system (Applied Biosystems). Standard curves for all three pairs of primers were performed for each PCR analysis, all PCR reactions being performed in triplicate. The enrichment of each PCR amplification of interest was calculated as the ratio between the region-specific signal and the intergenic signal of the precipitated fractions normalized with respect to the corresponding ratios of the input fractions. At least three independent experiments were performed for each condition. Primer sequences are available upon request.
RESULTS
mRNA export-deficient cells are sensitive to UV in the absence of global genome repair
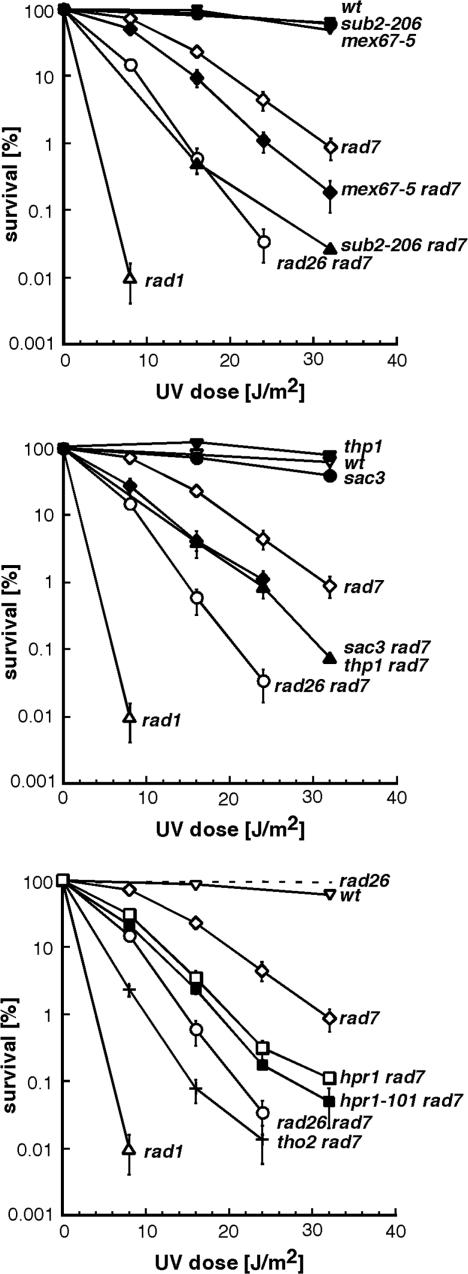
To analyze whether defects in mRNP biogenesis and export result in impaired TCR, we studied mutants defective in both GGR and mRNA export. Isogenic sub2-206, mex67-5, rad7Δ, sub2-206 rad7Δ, mex67-5 rad7Δ mutants were generated and survival after UV irradiation was determined (Figure 1, upper panel). Isogenic repair-proficient W303-1A, repair-deficient rad1Δ and TCR-deficient rad26Δ rad7Δ strains were used as controls. The sub2-206 and mex65-7 single mutants show no increased UV sensitivity as compared to wild-type cells. However, upon UV irradiation viability of the sub2-206 rad7Δ and mex67-5 rad7Δ double mutants dropped below the levels of the rad7Δ single mutant. Survival of the sub2-206 rad7Δ strain was similar to survival of the rad26Δ rad7Δ strain, whereas mex67-5 rad7Δ was less affected. Next, we analyzed mutants of the Thp1-Sac3 complex, which acts downstream of Mex67-Mtr2 on the mRNP biogenesis and export route. Isogenic thp1Δ, sac3Δ, thp1Δ rad7Δ, and sac3Δ rad7Δ mutants were generated and UV survival was determined (Figure 1, middle panel). The thp1Δ and sac3Δ single mutants show no increased UV sensitivity as compared with wild-type cells. However, viability of the thp1Δ rad7Δ and sac3Δ rad7Δ double mutants was below the level of the rad7Δ single mutant upon UV irradiation.
Figure 1.
mRNA export deficient cells are sensitive to UV in the absence of global genome repair. UV sensitivity curves of sub2-206, mex67-5, thp1Δ, sac3Δ, hpr1-101, and rad7Δ single and double mutants (plain symbols). Isogenic wild-type, NER-deficient rad1Δ, and TCR- and GGR-deficient and rad26Δ rad7Δ served as controls (empty symbols). Double mutants carrying the hpr1Δ and tho2Δ mutations in combination with the rad7Δ mutations were used as marker of phenotype strength. Data for the rad26Δ control strain were taken from (42) (dash line). Average values from three independent experiments are plotted for each genotype.
Since THO mutants have been shown to be sensitive to UV irradiation in the absence of GGR (42), we performed UV survival curves of hpr1Δ rad7Δ and tho2Δ rad7Δ strains for comparison of phenotype strength (Figure 1, lower panel). As expected, hpr1Δ rad7Δ and tho2Δ rad7Δ survival were reduced below the levels of the rad7Δ single mutant upon UV irradiation. UV sensitivity of sub2-206 rad7Δ cells was stronger than hpr1Δ rad7Δ cells, and weaker than tho2Δ rad7Δ cells, whereas thp1Δ rad7Δ and hpr1Δ rad7Δ showed similar UV sensitivity, consistent with the individual phenotype of the single mutant in other assays (36,40,45,51). We have recently described the hpr1-101 mutant allele, which exhibits severe transcription defects but weak hyper-recombination (37). We tested the hpr1-101 allele for UV sensitivity in the absence of GGR to check whether the UV sensitivity of THO mutants was rather linked to their transcription deficiencies or to the formation of recombinogenic structures. Survival of hpr1-101 rad7Δ and hpr1Δ rad7Δ cells were similar (Figure 1, lower panel), indicating that the observed UV sensitivity was linked to the transcription defects of THO mutants.
Thus, because UV sensitivity in the absence of GGR is a phenotype mostly associated with TCR deficiencies, we decided to test whether Sub2-Yra1 and Thp1-Sac3 complexes are required for proficient TCR.
TCR is impaired in cells defective in Sub2-Yra1, Thp1-Sac3 and THO
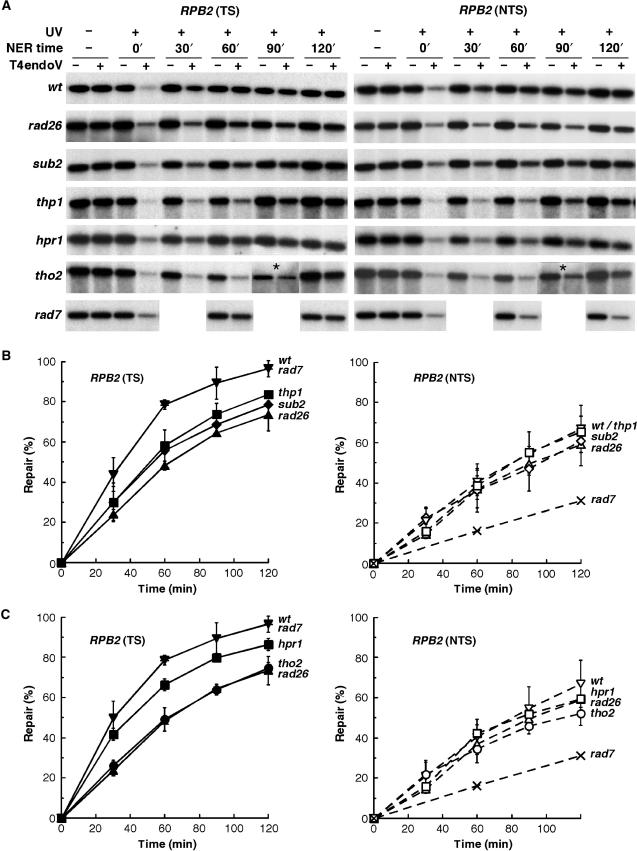
Next, we analyzed removal of UV photoproducts in sub2Δ and thp1Δ cells at the molecular level. Isogenic repair-proficient wild-type, TCR-deficient rad26Δ and GGR-deficient rad7Δ were used as controls. Repair after UV irradiation was determined in a 4.4-kb restriction fragment containing the constitutively expressed RPB2 gene by alkaline electrophoresis and indirect end-labeling (Figure 2). As previously reported (3), rad7Δ cells showed wild-type repair levels in the transcribed strand (TS) while repair of the non-transcribed strand (NTS) was strongly reduced. In rad26Δ cells, repair of the TS was significantly reduced while repair of the NTS almost reached wild-type levels (13). In sub2Δ and thp1Δ cells, repair of the TS was severely impaired while repair of the NTS did not exhibit significant repair defects (Figure 2B). Thus, our results indicate that sub2Δ and thp1Δ show defects in TCR, but not in GGR.
Figure 2.
Transcription coupled repair is impaired in mRNA export deficient cells. Southern blot analysis of representative experiments showing repair of a 4.4 kb (NsiI/PvuI) RPB2 fragment in W303 isogenic sub2Δ, thp1Δ, hpr1Δ, tho2Δ, rad26Δ, rad7Δ, and wt cells after UV irradiation (230 J/m2). The initial damage averaged 0.28 ± 0.05 CPD/Kb in the transcribed strand (TS, left) and 0.26 ± 0.04 CPD/Kb in the non-transcribed strand (NTS, right). The remaining intact restriction fragment after treatment of damaged DNA with T4endoV (+UV, +T4endoV) corresponds to the fraction of undamaged DNA. Non-irradiated DNA (-UV) and DNA not treated with T4endoV (-T4endoV) were used as controls. The 90 min time point of tho2 cells was taken from a different gel derived from the same experiment (*). As rad7 was merely used as a negative control, the analysis was restrained to the 60 and 120 min time points. (B and C) Graphical representation of the repair analysis as shown in A. Repair of TS (full symbols and lines) and NTS (empty symbols and dashed lines) are shown for each strain. The percentage of repair was determined from the signal intensities as described in ‘Materials and Methods’. Average values derived from at least two independent experiments are plotted.
In a previous report, molecular analysis of DNA repair in hpr1Δ and tho2Δ cells indicated general defects in NER (42). In these studies, the UV doses used had produced extensive DNA damage, in contrast to the conditions used in this study, in which one repair event is sufficient to restore the intact DNA (about 1 CPD per restriction fragment). The observations that mutants of the Sub2-Yra1 and Thp1-Sac3 complexes, which act downstream of THO in mRNP export, are specifically affected in TS repair lead us to examine CPD removal in tho2Δ and hpr1Δ cells, using our UV irradiation conditions (Figure 2A and C). Repair of the TS was significantly reduced in both strains. As observed by UV sensitivity assays in the absence of GGR (Figure 1), tho2Δ cells were more strongly affected in TS repair than hpr1Δ cells, reaching levels similar to rad26Δ cells. Likewise, repair of hpr1Δ and hpr1-101 cells were equally affected in DNA repair (data not shown). In the NTS, the repair levels of hpr1Δ, tho2Δ and rad26Δ cells were similar, in contrast to previous results obtained with extensive DNA damage (42), indicating that GGR is not significantly affected in hpr1Δ and tho2Δ cells in our conditions.
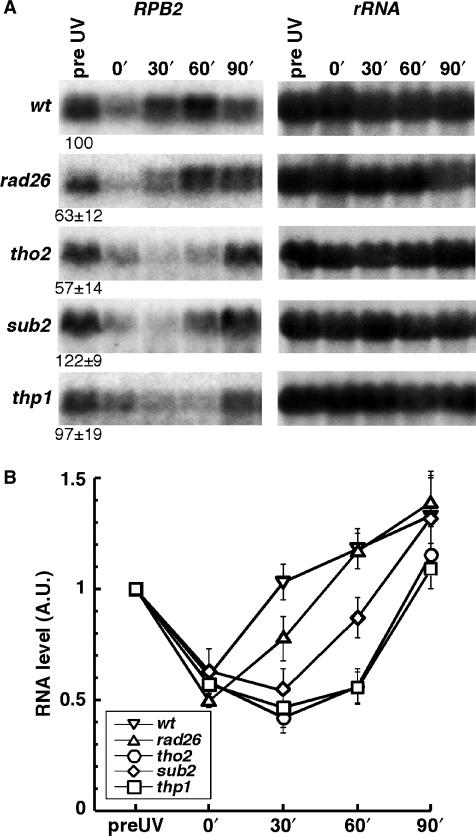
In yeast, RNA synthesis is inhibited shortly after UV irradiation, probably due to the presence of CPDs in the TS of active genes. The ability of wild-type and mutant yeast cells to recover RNAPII synthesis in individual genes has been shown to mirror their strand-specific repair capacity (52). To gain additional information on the connection between the recovery of mRNA levels and TCR, kinetics of the RPB2 transcript levels were determined by northern analysis in rad26Δ, tho2Δ, sub2Δ, thp1Δ, and wild-type cells (Figure 3). A direct correlation between the RPB2 expression levels prior to UV irradiation and repair rates was not apparent, in agreement with previous work showing the absence of simple correlation between transcription and repair rates (8).
Figure 3.
Delayed RNA synthesis recovery after UV irradiation in mRNA export deficient cells. (A) Northern blot analysis of RPB2 expression prior and after UV irradiation (230 J/m2) in the indicated strains. Transcript levels prior to UV irradiation (pre UV) expressed in percent of the wild-type level are shown below the gels. (B) Graphical representation of RPB2 synthesis kinetics after UV irradiation as shown in A. Northern blots were quantified using a Fuji FLA 3000 and normalized to the rRNA levels of each samples. The transcripts levels during DNA damage recovery were normalized to the pre UV values for each strain. Average values derived from at least two independent experiments are plotted.
However, upon UV irradiation, RPB2 transcript recovery appeared to be most efficient in rad26Δ cells, while transcript recovery was clearly affected in all other mutant strains. This result points to THO, Sub2-Yra1 and Thp1-Sac3 behaving differently from Rad26 in response to UV damage, since they appear to undergo severe transcription impairment upon UV irradiation, in addition to their TCR deficiencies.
Taken together, our results place THO, Sub2-Yra1, Mex67-Mtr2 and Thp1-Sac3 as new factors needed for efficient TCR. As these factors play a role at the interface between transcription and RNA export, it was important to determine whether the observed TCR defect was mediated by the RNAPII or by the nascent mRNA.
TCR depends on RNAPII rather than on the nascent mRNA
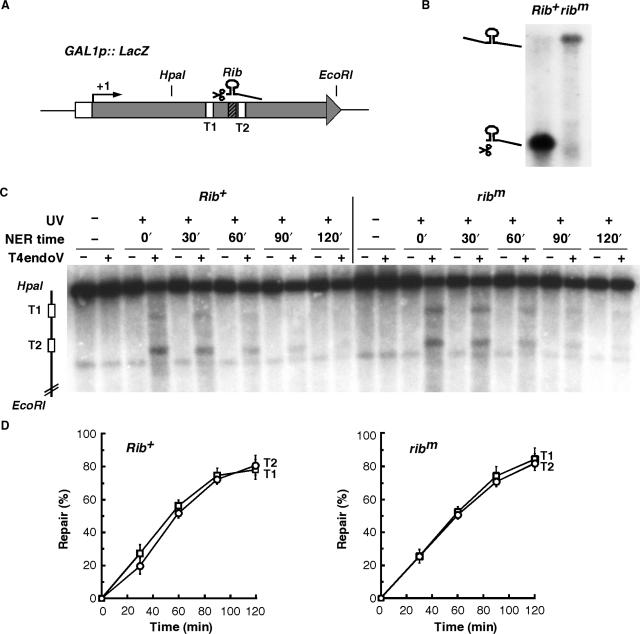
The molecular basis underlying the requirement of functional mRNP biogenesis and export factors for TCR might rely on the proper packaging of the nascent transcript or on their effect on transcription. In repair-proficient cells, the nascent mRNA could mediate the TCR reaction in response to the transcriptional stalling occurring at DNA lesions. To test this possibility, we designed a construct containing two 39-bp long T-tract sequences inserted at different sites within the GAL1 promoter-driven LacZ ORF. Between the two T-tract sequences, 52 bp encoding either an active self-cleaving Hammerhead ribozyme or an inactive mutated form (47) were inserted (Figure 4A). Northern analysis confirmed a complete disappearance of the full length mRNA in the construct carrying the active ribozyme (Figure 4B), indicating that the nascent mRNA was efficiently cleaved between the T-tracts.
Figure 4.
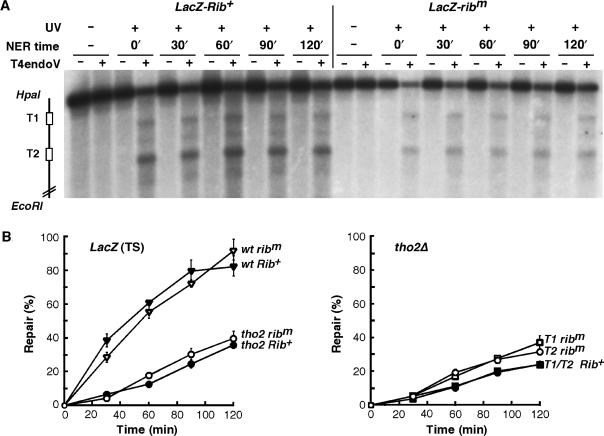
An intact nascent mRNA is not required for TCR in wild-type yeast cells. (A) Expression of a modified LacZ containing the self-cleaving Hammerhead ribozyme (Rib) between two 39-bp long T-tracts was placed under the control of the GAL1 promoter. The first (T1) and the second (T2) T-tracts as well as the Rib sequence are indicated. (B) Northern analysis of LacZ mRNA isolated from plasmids containing either an active (Rib+) or a mutated form of Rib (ribm). The apparent difference in signal intensity between the two constructs probably results from distinct transcript stability, unequal transfer efficiency of short and long RNA, as well as different hybridization efficiency due to the long transcript migrating very close to the abundant rRNA. (C) Southern blot analysis of representative experiments showing repair of a 2.2 kb (HpaI/EcoRI) LacZ fragment containing either the active Rib+ (LacZ-Rib+) or the mutated ribm (LacZ-ribm) in W303 cells after UV irradiation (230 J/m2). Description is as in Figure 2A, except that genomic DNA was probed for the TS of LacZ. The initial damage averaged 0.3 ± 0.005 CPD/Kb. In the T-tracts, the initial damage corresponded to 0.54% (T1, Rib+), 1.08% (T2, Rib+), 0.79% (T1, ribm) and 1.03% (T2, ribm) of the respective total lane signal. (D) Graphical representation of the T-tracts repair analysis as shown in C. The percentage of repair was determined from the signal intensities as described in Materials and Methods. Average values derived from two independent experiments are plotted.
We first assessed whether TCR efficiency depends on the integrity of the nascent mRNA in yeast wild-type cells by comparing repair rates in the T-tracts situated upstream (T1) and downstream (T2) of the ribozyme (Figure 4C and D). The initial damage was higher in T2 than in T1, likely reflecting local differences in chromatin structure. Importantly, our results show no significant difference between repair of T1 and T2, neither in the active (Rib+) nor in the mutated (ribm) ribozyme constructs, indicating that an intact and 5′-capped nascent mRNA is not required for efficient TCR in wild-type cells.
Nevertheless, in contrast to wild-type cells, the occurrence of sub-optimal mRNP might impede the process of TCR in mutants of THO/TREX and Thp1-Sac3. To test this possibility, we used the ribozyme system to assess whether wild-type TCR can be restored in T2 after ribozyme self-cleavage of the nascent mRNA in tho2Δ and thp1Δ cells (Figure 5). Repair in the full length LacZ fragment was clearly below repair levels achieved in wild-type cells, confirming the TCR deficiency of tho2Δ cells. Comparison of repair efficiencies in T1 and T2 did not show any significant difference, neither in the active (Rib+) nor in the mutated (ribm) ribozyme constructs. Similar results were obtained in thp1Δ cells (data not shown). Thus, our results indicate that cleavage of the nascent mRNA does not restore TCR in THO and Thp1 mutants. Consequently, we assume that the key player in the organization of TCR is the RNAPII itself, or some associated factors, rather than the nascent mRNA.
Figure 5.
Cleavage of the nascent mRNA does not restore TCR in cells defective in THO. (A) Southern blot analysis of strand-specific repair of a 2.2-kb LacZ fragment containing either the active (Rib+) or mutated Rib (ribm) between two T-tracts in tho2Δ cells after UV irradiation (230 J/m2). The initial damage averaged 0.5 ± 0.07 CPD/Kb. In the T-tracts, the initial damage corresponded to 1.05% (T1, Rib+), 2.51% (T1, Rib+), 1.55% (T1, ribm) and 2.2% (T2, ribm) of the respective total lane signal. Description is as in Figure 4C. (B) Graphical representation of the repair analysis as shown in A and in Figure 4D. Repair of the intact restriction fragment in wild-type and tho2 cells (left panel) and of the T-tracts in tho2 cells (right panel) are plotted. The percentage of repair was determined from the signal intensities as described in ‘Materials and Methods’. Average values derived from two independent experiments are plotted.
Def1 is required for removal of stalled RNAPII complexes in THO mutants
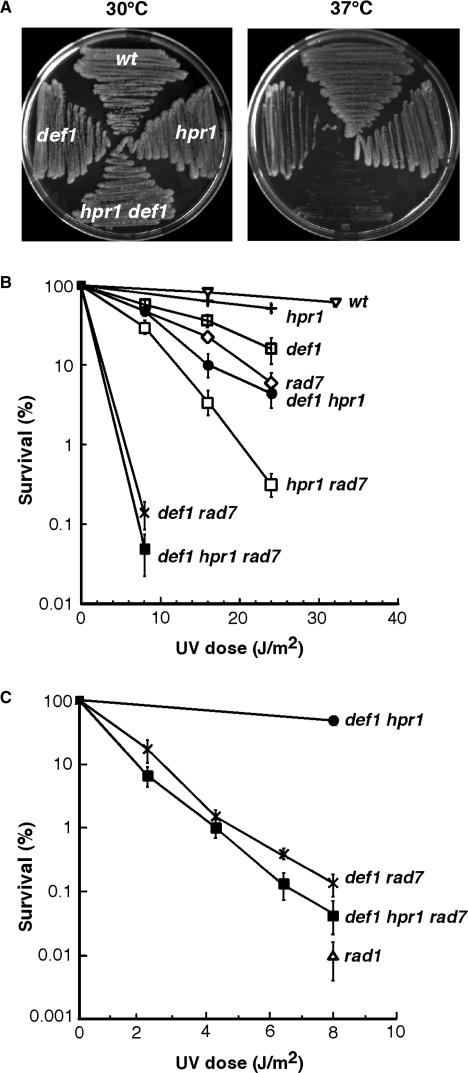
Since we can rule out an active role of the nascent mRNA in TCR, it is conceivable that an intact RNAPII complex is sufficient to mediate proficient TCR. Recently, a protein called Def1 was shown to trigger ubiquitylation and degradation of RNAPII in response to UV damage as an alternative pathway to DNA repair (26,27,53). To study the possible requirement for Def1 in the removal of trapped RNAPII presumably present in THO mutants, hpr1Δ def1Δ double mutants were generated and analyzed. hpr1Δ def1Δ double mutants were viable, but very slow growing. Since all THO/TREX mutants grow poorly at 37°C, we first investigated whether deletion of DEF1 might increase the temperature sensitivity (ts) phenotype of hpr1Δ cells (Figure 6A). An additive effect that made cells inviable at 37°C was observed in the double mutant as compared to the single mutant. Next, we performed UV survival curves in isogenic def1Δ, hpr1Δ and def1Δ hpr1Δ mutants (Figure 6B). Viability of the def1Δ hpr1Δ double mutants was reduced below the levels of the def1Δ and hpr1Δ single mutants upon UV irradiation, indicating a synergistic effect of the two mutations on UV sensitivity. In the absence of GGR, def1Δ has been shown to be highly sensitive to UV irradiation (53). Nevertheless, the UV sensitivity of def1Δ hpr1Δ rad7Δ cells was increased as compared to def1Δ rad7Δ cells (Figure 6B and C). These genetic interactions between DEF1 and HPR1 indicate that Def1 is important for the viability of THO mutants subjected to stress and DNA damage.
Figure 6.
Synergistic increase of temperature and UV sensitivity phenotypes in def1 hpr1 double mutants. (A) Growth of isogenic W303 yeast strains carrying single and double combinations of the hpr1 and def1 mutations at 30°C (left panel) and at 37°C (right panel). (B and C) UV sensitivity curves of isogenic hpr1Δ, def1Δ, and rad7Δ single, double and triple mutants. Isogenic wild-type and NER-deficient rad1Δ strain were used as controls. Note that the UV dose used in panel C was lower than in panel B. Description is as in Figure 1.
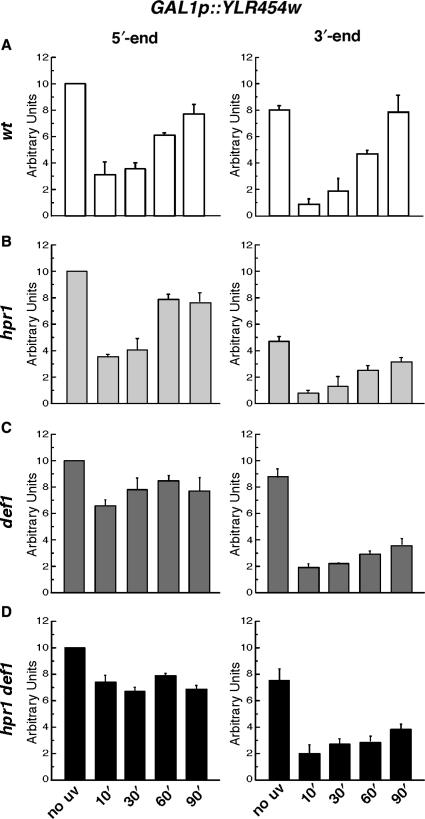
Given the exacerbated ts and UV sensitivity phenotypes of hpr1Δ def1Δ double mutant (Figure 6) and the reduced transcription processivity of THO mutants (54), we determined the kinetics of RNAPII distribution after UV damage in hpr1Δ, def1Δ, def1Δ hpr1Δ and wild-type cells. For this purpose, the levels of RNAPII at distal positions within the large (8 kb) YLR454w gene driven by the GAL1 promoter were analyzed by ChIPs with antibody 8WG16 directed against RNAPII (Figure 7). In wild-type cells, we observed a drop in the overall amount of RNAPII after UV irradiation that was accompanied by a significant loss of processivity along the transcribed unit (Figure 7A). Recovery of both the amount of RNAPII and processivity occurred at similar rates and were nearly complete 90 min after UV irradiation. In hpr1Δ cells, a drop in the overall amount of RNAPII was observed after induction of UV lesions (Figure 7B). Both the drop in RNAPII density and its recovery were comparable to wild-type cells. However, the loss of processivity observed in hpr1Δ cells was much stronger than in wild-type cells and its recovery very slow, 50% of the polymerases being lost between the 5′- and the 3′-end of the gene 90 min after UV irradiation. Therefore, the amount of RNAPII loaded on a transcribed unit is not the limiting factor for TCR in THO mutants. Importantly, the recovery of RNAPII association toward the end of the YLR454w coding region (3′-end) with increasing repair time is consistent with the repair rates observed previously (Figure 2).
Figure 7.
RNAPII kinetics after UV irradiation in wt, hpr1, def1, and hpr1 def1 strains. Kinetics of RNAPII occupancy at the 3′- and the 5′- ends of the GAL1p-driven YLR454w gene in (A) wt, (B) hpr1Δ, (C) def1Δ, and (D) hpr1Δ def1Δ strains after UV irradiation (230 J/m2). Soluble chromatin was prepared and immunoprecipitated with antibody 8WG16 against RNAPII. Real-time PCR using specific primers was performed to test the relative enrichment for either the 3′- or the 5′-regions of the gene as compared to the non-irradiated (pre-UV) samples. Average values and standard deviations derived from three independent experiments are shown for each genotype.
While the amount of RNAPII loaded on the YLR454w gene was strongly decreased after UV irradiation in wild-type and hpr1Δ cells, only a weak drop in the amount of RNAPII was observed in def1Δ cells (Figure 7C), probably reflecting the defects in RNAPII degradation of this mutant strain. Recovery of RNAPII on the 3′-end of the gene was slow as compared to wild-type, with kinetics similar to those of hpr1Δ, indicating a DNA damage-dependent processivity defect. In hpr1Δ def1Δ cells (Figure 7D), a weak drop in the amount of RNAPII loaded on the gene was observed after UV irradiation. The amount of RNAPII localized on the 5′-end of the gene was similar to the levels found in def1Δ cells, suggesting that the hpr1Δ mutant did not significantly alter the accumulation of RNAPII observed in the def1Δ mutant. Noteworthy, the amount of RNAPII associated with the 5′-end of the gene was repeatedly higher at 60 min than at 90 min after UV irradiation in both strains. This observation was also made in hpr1Δ strains but never in wild-type cells. Thus, in mutant strains in which TCR and/or RNAPII degradation is impeded, RNAPII tends to accumulate on the template with a peak 60 min after UV irradiation, this effect being stronger in def1Δ mutants which fail to ubiquitylate and degrade the stalled RNAPII. In hpr1Δ def1Δ cells, recovery of RNAPII at the 3′-end of the gene after UV irradiation was slow, reflecting a DNA damage-dependent loss of processivity.
Taken together, our data indicate that RNAPII is indeed associated with transcribed genes after UV irradiation in THO mutants, even if repair of the lesions and transcription recovery are impeded. However, no accumulation of RNAPII was detected at the 5′ of the gene in THO mutants, in contrast to def1Δ and hpr1Δ def1Δ cells, in which RNAPII accumulated at the 5′-end after UV irradiation as a result of the lack of RNAPII degradation (Figure 7). These results imply that TCR-deficient RNAPII complexes remain stalled at DNA lesions in THO mutant and have to be removed from the template in order to provide access for DNA repair machineries, a process that depends on Def1.
DISCUSSION
TCR and mRNP biogenesis are coupled
The nuclear envelope marks a fundamental difference between eukaryotic and prokaryotic cells. The obligatory passage of eukaryotic mRNA through the nuclear envelope to reach protein translation machineries is tightly controlled by mRNA export systems. TCR has to cope with mRNA export in eukaryotes, possibly explaining why it cannot take advantage of one transcription-repair coupling factor as found in prokaryotic cells. This study unveils that the repair of active genes indeed depends on the correct interplay between transcription and mRNA export. We found that removal of proteins all along the mRNP export route, including the transcription-associated THO and Sub2–Yra1 complexes, the Mex67-Mtr2 export receptor as well as the Thp1-Sac3 nuclear pore-associated complex, confer TCR-deficient phenotypes. Thus, our results establish a general dependency of TCR on proficient mRNP biogenesis and export.
The inability of human Cockayne's syndrome cells to recover RNA synthesis after DNA damage was recently found not to be due solely to the failure of these mutants to remove lesions in the transcribed strand of active genes, but to defects in the re-initiation of the transcriptional program after UV irradiation (55). Although TCR of the constitutively expressed RPB2 gene is strongly affected in cells depleted of Rad26—the yeast CSB homologue—we found that, in rad26 mutants, mRNA recovery is much faster as compared to THO, Sub2-Yra1 and Thp1-Sac3 mutants (Figure 3). The mRNA export-dependent TCR-deficient phenotype could reflect a substantial lag in transcription initiation. This is not the case, as ChIP analysis indicated that, in contrast to CSB, RNAPII loading after UV irradiation was not affected in THO mutants (Figure 7). However, the amount of polymerases reaching the 3′-end of the YLR454w gene was substantially lower in hpr1. Thus, the RNAPII processivity defects of hpr1 appear to be strengthened upon UV-irradiation, presumably leading to severe delays in mRNA recovery. Even though it is a priori conceivable that poor transcription rate and/or low RNAPII processivity alone might lead to defective TCR, several lines of evidence argue against this possibility. First, no simple correlation could be found between repair and transcription rates neither in different mutants (Figure 3) nor at different loci (8). Second, transcription-elongation mutants like spt4, in which transcription elongation is impaired and RNAPII processivity significantly reduced (54,56), do not show defects in TCR (57). On the contrary, spt4 suppresses the TCR defects of rad26, indicating that reduced RNAPII processivity can even act positively on TCR.
There is growing evidence for transcription-coupled mRNA export [for review see (28–30)], though the physical nature of this coupling is not known. The possible functional and physical connection between a subset of transcribed genes and the NPC (58–60) suggest that deficient mRNA export could negatively affect RNAPII transcription. Our results, providing evidence for a functional link between TCR and mRNA export, support the existence of such a feedback mechanism, which would alter transcription so that it is no longer proficient for TCR. A major player in the coupling of transcription and mRNA processing is the CTD of the largest subunit of RNAPII, which acts as a loading platform for pre-mRNA processing factors [reviewed in (28,61,62)]. Several lines of evidence indicate that CTD phosphorylation occurs upon UV irradiation and might be involved in the signaling of stalled RNAPII for TCR (24,63,64). These features place the CTD tail as a potential target for feedback modifications of RNAPII in response to mRNP export defects.
TCR depends on the RNAPII machinery rather than on the nascent mRNA
Given the association between mRNA export and efficient TCR, the nascent mRNA could act as a mediator for TCR. However, we discard the possibility that the nascent mRNA actively supports the TCR process by monitoring the repair of UV-lesions encountered by a RNAPII-complex containing either the full-length nascent mRNA or about 70 bp left over after ribozyme-mediated cleavage (Figure 4). It is worth noticing that the available nascent mRNA might be even shorter than 70 bp considering that 15 to 20 bp are covered by the RNAPII holoenzyme (65,66). Furthermore, our results indicate that RNA 5′-capping is dispensable for TCR. Thus, our results support the idea of the RNAPII machinery being the main mediator for an active DNA damage response.
Different evidence led to the proposal that the hyper-recombination phenotype of THO mutants is mediated by the formation of R-loops (RNA:DNA hybrids) behind the elongating RNAPII, as well as impaired replication fork progression (48,67). Here, we show that ribozyme-mediated cleavage of the nascent mRNA does not suppress the TCR deficiency of tho2 (Figure 5), suggesting that the hyper-recombination and TCR phenotypes might be mediated by different intermediates. This conclusion is further supported by the observation that the non-hyper-recombinant hpr1-101 allele shares the transcription, mRNA export and TCR phenotypes of hpr1 cells (Figure 1) (37).
Def1 is required for removal of defective RNAPII complexes in THO mutants
Recently, RNAPII processivity has been shown to be significantly reduced in THO mutants (54). Our ChIP analysis of UV-irradiated cells revealed that RNAPII processivity was further impaired while RNAPII loading remained unaffected in hpr1 cells (Figure 7). A conclusion of these findings is that the TCR defect of hpr1 cells is a consequence of damage-trapped RNAPII. Since transcription is a one-track copying process, a trapped RNAPII can only be resolved by repairing the damage so that transcription can resume or by physically removing the stalled RNAPII [reviewed in (25)]. Thus, it is understandable that RNAPII degradation activities, such as Def1, might be important in backgrounds in which aberrant stalled RNAPII are formed. Indeed, we observed that the hpr1 def1 mutant exhibited profound growth defects and was highly temperature- as well as UV-sensitive (Figure 6). In contrast to wild-type and hpr1 cells, RNAPII accumulated at the 5′-end of YLR454w upon UV-irradiation in def1 mutant strains (Figure 7). Hence, Def1-mediated RNAPII degradation appears to be important for the removal of stalled RNAPII after UV damage, in agreement with previous works showing that UV-dependent RNAPII ubiquitylation depends on Def1 (26,27,53). Worthy of note, we find that the amount of RNAPII tends to decrease 90 min after UV irradiation in backgrounds in which TCR or RNAPII degradation is compromised. This decrease might reflect the physiological turnover of stalled RNAPII in yeast cells. Such an interpretation implies that endogenous dissociation of stalled RNAPII would be much faster in vivo than in vitro, where human RNAPII stalled at a CPD remains stable for days (68). Alternatively, the reduction in the amount of RNAPII at 90 min could be due to a down-regulation of the promoter activity or to the action of alternative RNAPII degradation activities.
Given that RNAPII distribution obtained at the 5′-end of YLR454w was similar in def1 and hpr1 def1 mutants, it is conceivable that RNAPII removal by Def1 serves as an essential backup mechanism to allow repair of active genes in hpr1 mutants. Our results suggest that the low processivity and mRNA export defects of hpr1 mutants lead to stalled RNAPII-complexes in adverse transcription conditions, which are subjected to Def1 mediated degradation if transcription elongation cannot be rescued. Further investigation of the hpr1 def1 mutants will help us to understand the mechanisms of the different phenotypes of THO mutants, from hyper-recombination to DNA polymerase stalling, and how they might depend on each other.
CONCLUDING REMARKS
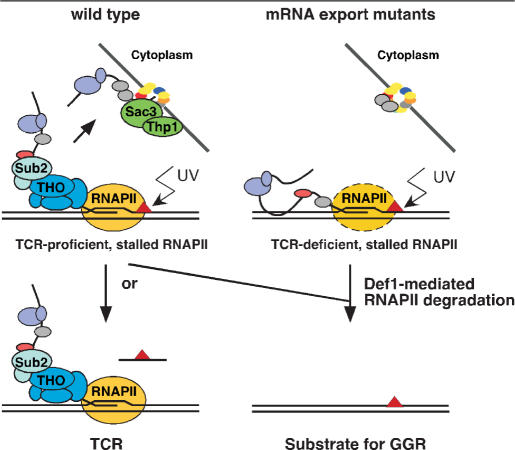
Our results present a model by which TCR-deficient RNAPII complexes remain stalled at DNA lesions in THO, Sub2-Yra1, Mex67-Mtr2 and Thp1-Sac3 mutants, thereby preventing repair (Figure 8). On the mRNA export route, THO works in immediate proximity to the ternary complex formed by the polymerase holoenzyme, template DNA and nascent RNA, whereas Sub2-Yra1, Mex67-Mtr2 and Thp1-Sac3 work downstream in the mRNP biogenesis and export route. Here, we show that cleavage of the nascent mRNA, which is the only known physical link between all these complexes, is not sufficient to rescue TCR. Thus, yet to be discovered bridging factors, whether or not functionally connected to the RNAPII, could mediate the TCR phenotype of mRNA export mutants. Alteration of these factors might result in an incomplete or modified transcription apparatus, which in turn would be incompetent to promote TCR, possibly as a consequence of its impaired transcription processivity (Figure 8). A future new challenge will be to unravel the configuration of the RNAPII machinery in mRNP biogenesis and export mutants.
Figure 8.
Model to explain the response to UV damage within a transcribed gene both in wild-type and in mRNA export mutants. In wild-type, mRNP biogenesis and export occur co-transcriptionally. RNAPII encountering a UV damage gets stalled, triggering the TCR reaction by yet unknown mechanisms. Alternatively, the stalled RNAPII might get ubiquitylated and degraded via Def1. In mRNA export mutants, co-transcriptional mRNP biogenesis fails and mRNA are not exported. A feedback effect acts on RNAPII, impeding it to trigger TCR in the case of encountering a UV lesion, possibly as a consequence of impaired transcription processivity. The stalled TCR-deficient RNAPII has to be removed by Def1-mediated degradation to allow accessibility of the damage to repair enzymes.
ACKNOWLEDGEMENTS
Research was funded by grants from the Spanish Ministry of Science and Education (SAF2003-00204 and BFU2006-05260) and from the Junta de Andalucia (CVI102). HG was granted by the Swiss National Science Foundation (PBEZA-100700 and PA00A-105027) and the Novartis Foundation.
We thank D. Fitzgerald and S. González-Barrera for critical reading of the manuscript and D. Haun for style supervision. Funding to pay the Open Access publication charges for this article was provided by Spanish Ministry of Science and Education grant BFU2006-05260.
Conflict of interest statement. None declared.
REFERENCES
- 1.Prakash S, Prakash L. Nucleotide excision repair in yeast. Mutat. Res. 2000;451:13–24. doi: 10.1016/s0027-5107(00)00037-3. [DOI] [PubMed] [Google Scholar]
- 2.Hanawalt PC. Controlling the efficiency of excision repair. Mutat. Res. 2001;485:3–13. doi: 10.1016/s0921-8777(00)00071-9. [DOI] [PubMed] [Google Scholar]
- 3.Verhage R, Zeeman AM, de Groot N, Gleig F, Bang DD, van de Putte P, Brouwer J. The RAD7 and RAD16 genes, which are essential for pyrimidine dimer removal from the silent mating type loci, are also required for repair of the nontranscribed strand of an active gene in Saccharomyces cerevisiae. Mol. Cell. Biol. 1994;14:6135–6142. doi: 10.1128/mcb.14.9.6135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Laat WL, Jaspers NG, Hoeijmakers JH. Molecular mechanism of nucleotide excision repair. Genes Dev. 1999;13:768–785. doi: 10.1101/gad.13.7.768. [DOI] [PubMed] [Google Scholar]
- 5.Svejstrup JQ. Mechanisms of transcription-coupled DNA repair. Nat. Rev. Mol. Cell. Biol. 2002;3:21–29. doi: 10.1038/nrm703. [DOI] [PubMed] [Google Scholar]
- 6.Mellon I. Transcription-coupled repair: a complex affair. Mutat. Res. 2005;577:155–161. doi: 10.1016/j.mrfmmm.2005.03.016. [DOI] [PubMed] [Google Scholar]
- 7.Tornaletti S. Transcription arrest at DNA damage sites. Mutat. Res. 2005;577:131–145. doi: 10.1016/j.mrfmmm.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 8.Bedoyan J, Gupta R, Thoma F, Smerdon MJ. Transcription, nucleosome stability, and DNA repair in a yeast minichromosome. J. Biol. Chem. 1992;267:5996–6005. [PubMed] [Google Scholar]
- 9.Selby CP, Sancar A. Transcription preferentially inhibits nucleotide excision repair of the template DNA strand in vitro. J. Biol. Chem. 1990;265:21330–21336. [PubMed] [Google Scholar]
- 10.Selby CP, Sancar A. Molecular mechanism of transcription-repair coupling. Science. 1993;260:53–58. doi: 10.1126/science.8465200. [DOI] [PubMed] [Google Scholar]
- 11.de Boer J, Hoeijmakers JH. Nucleotide excision repair and human syndromes. Carcinogenesis. 2000;21:453–460. doi: 10.1093/carcin/21.3.453. [DOI] [PubMed] [Google Scholar]
- 12.Venema J, Mullenders LH, Natarajan AT, van Zeeland AA, Mayne LV. The genetic defect in Cockayne syndrome is associated with a defect in repair of UV-induced DNA damage in transcriptionally active DNA. Proc. Natl Acad. Sci. USA. 1990;87:4707–4711. doi: 10.1073/pnas.87.12.4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Gool AJ, Verhage R, Swagemakers SM, van de Putte P, Brouwer J, Troelstra C, Bootsma D, Hoeijmakers JH. RAD26, the functional S. cerevisiae homolog of the Cockayne syndrome B gene ERCC6. EMBO J. 1994;13:5361–5369. doi: 10.1002/j.1460-2075.1994.tb06871.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Citterio E, Rademakers S, van der Horst GT, van Gool AJ, Hoeijmakers JH, Vermeulen W. Biochemical and biological characterization of wild-type and ATPase-deficient Cockayne syndrome B repair protein. J. Biol. Chem. 1998;273:11844–11851. doi: 10.1074/jbc.273.19.11844. [DOI] [PubMed] [Google Scholar]
- 15.Laine JP, Egly JM. Initiation of DNA repair mediated by a stalled RNA polymerase IIO. EMBO J. 2006;25:387–397. doi: 10.1038/sj.emboj.7600933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sarker AH, Tsutakawa SE, Kostek S, Ng C, Shin DS, Peris M, Campeau E, Tainer JA, Nogales E, et al. Recognition of RNA polymerase II and transcription bubbles by XPG, CSB, and TFIIH: insights for transcription-coupled repair and Cockayne Syndrome. Mol. Cell. 2005;20:187–198. doi: 10.1016/j.molcel.2005.09.022. [DOI] [PubMed] [Google Scholar]
- 17.Li S, Smerdon MJ. Rpb4 and Rpb9 mediate subpathways of transcription-coupled DNA repair in Saccharomyces cerevisiae. EMBO J. 2002;21:5921–5929. doi: 10.1093/emboj/cdf589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li S, Chen X, Ruggiero C, Ding B, Smerdon MJ. Modulation of Rad26 and Rpb9 mediated DNA repair by different promoter elements. J. Biol. Chem. 2006;281:36643–36651. doi: 10.1074/jbc.M604885200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.von Hippel PH. An integrated model of the transcription complex in elongation, termination, and editing. Science. 1998;281:660–665. doi: 10.1126/science.281.5377.660. [DOI] [PubMed] [Google Scholar]
- 20.Watnick RS, Gottesman ME. Binding of transcription termination protein nun to nascent RNA and template DNA. Science. 1999;286:2337–2339. doi: 10.1126/science.286.5448.2337. [DOI] [PubMed] [Google Scholar]
- 21.Bregman DB, Halaban R, van Gool AJ, Henning KA, Friedberg EC, Warren SL. UV-induced ubiquitination of RNA polymerase II: a novel modification deficient in Cockayne syndrome cells. Proc. Natl Acad. Sci. USA. 1996;93:11586–11590. doi: 10.1073/pnas.93.21.11586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ratner JN, Balasubramanian B, Corden J, Warren SL, Bregman DB. Ultraviolet radiation-induced ubiquitination and proteasomal degradation of the large subunit of RNA polymerase II. Implications for transcription-coupled DNA repair. J. Biol. Chem. 1998;273:5184–5189. doi: 10.1074/jbc.273.9.5184. [DOI] [PubMed] [Google Scholar]
- 23.Beaudenon SL, Huacani MR, Wang G, McDonnell DP, Huibregtse JM. Rsp5 ubiquitin-protein ligase mediates DNA damage-induced degradation of the large subunit of RNA polymerase II in Saccharomyces cerevisiae. Mol. Cell. Biol. 1999;19:6972–6979. doi: 10.1128/mcb.19.10.6972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luo Z, Zheng J, Lu Y, Bregman DB. Ultraviolet radiation alters the phosphorylation of RNA polymerase II large subunit and accelerates its proteasome-dependent degradation. Mutat. Res. 2001;486:259–274. doi: 10.1016/s0921-8777(01)00097-0. [DOI] [PubMed] [Google Scholar]
- 25.Svejstrup JQ. Rescue of arrested RNA polymerase II complexes. J. Cell Sci. 2003;116:447–451. doi: 10.1242/jcs.00271. [DOI] [PubMed] [Google Scholar]
- 26.Reid J, Svejstrup JQ. DNA damage-induced Def1-RNA polymerase II interaction and Def1 requirement for polymerase ubiquitylation in vitro. J. Biol. Chem. 2004;279:29875–29878. doi: 10.1074/jbc.C400185200. [DOI] [PubMed] [Google Scholar]
- 27.Somesh BP, Reid J, Liu WF, Sogaard TM, Erdjument-Bromage H, Tempst P, Svejstrup JQ. Multiple mechanisms confining RNA polymerase II ubiquitylation to polymerases undergoing transcriptional arrest. Cell. 2005;121:913–923. doi: 10.1016/j.cell.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 28.Proudfoot NJ, Furger A, Dye MJ. Integrating mRNA processing with transcription. Cell. 2002;108:501–512. doi: 10.1016/s0092-8674(02)00617-7. [DOI] [PubMed] [Google Scholar]
- 29.Reed R. Coupling transcription, splicing and mRNA export. Curr. Opin. Cell Biol. 2003;15:326–331. doi: 10.1016/s0955-0674(03)00048-6. [DOI] [PubMed] [Google Scholar]
- 30.Aguilera A. Cotranscriptional mRNP assembly: from the DNA to the nuclear pore. Curr. Opin. Cell Biol. 2005;17:242–250. doi: 10.1016/j.ceb.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 31.Chavez S, Beilharz T, Rondon AG, Erdjument-Bromage H, Tempst P, Svejstrup JQ, Lithgow T, Aguilera A. A protein complex containing Tho2, Hpr1, Mft1 and a novel protein, Thp2, connects transcription elongation with mitotic recombination in Saccharomyces cerevisiae. EMBO J. 2000;19:5824–5834. doi: 10.1093/emboj/19.21.5824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Strasser K, Masuda S, Mason P, Pfannstiel J, Oppizzi M, Rodriguez-Navarro S, Rondon AG, Aguilera A, Struhl K, et al. TREX is a conserved complex coupling transcription with messenger RNA export. Nature. 2002;417:304–308. doi: 10.1038/nature746. [DOI] [PubMed] [Google Scholar]
- 33.Zenklusen D, Vinciguerra P, Wyss JC, Stutz F. Stable mRNP formation and export require cotranscriptional recruitment of the mRNA export factors Yra1p and Sub2p by Hpr1p. Mol. Cell. Biol. 2002;22:8241–8253. doi: 10.1128/MCB.22.23.8241-8253.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rondon AG, Jimeno S, Garcia-Rubio M, Aguilera A. Molecular evidence that the eukaryotic THO/TREX complex is required for efficient transcription elongation. J. Biol. Chem. 2003;278:39037–39043. doi: 10.1074/jbc.M305718200. [DOI] [PubMed] [Google Scholar]
- 35.Schneiter R, Guerra CE, Lampl M, Gogg G, Kohlwein SD, Klein HL. The Saccharomyces cerevisiae hyperrecombination mutant hpr1Delta is synthetically lethal with two conditional alleles of the acetyl coenzyme A carboxylase gene and causes a defect in nuclear export of polyadenylated RNA. Mol. Cell. Biol. 1999;19:3415–3422. doi: 10.1128/mcb.19.5.3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jimeno S, Rondon AG, Luna R, Aguilera A. The yeast THO complex and mRNA export factors link RNA metabolism with transcription and genome instability. EMBO J. 2002;21:3526–3535. doi: 10.1093/emboj/cdf335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huertas P, Garcia-Rubio ML, Wellinger RE, Luna R, Aguilera A. An hpr1 point mutation that impairs transcription and mRNP biogenesis without increasing recombination. Mol. Cell. Biol. 2006;26:7451–7465. doi: 10.1128/MCB.00684-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fischer T, Strasser K, Racz A, Rodriguez-Navarro S, Oppizzi M, Ihrig P, Lechner J, Hurt E. The mRNA export machinery requires the novel Sac3p-Thp1p complex to dock at the nucleoplasmic entrance of the nuclear pores. EMBO J. 2002;21:5843–5852. doi: 10.1093/emboj/cdf590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lei EP, Stern CA, Fahrenkrog B, Krebber H, Moy TI, Aebi U, Silver PA. Sac3 is an mRNA export factor that localizes to cytoplasmic fibrils of nuclear pore complex. Mol. Biol. Cell. 2003;14:836–847. doi: 10.1091/mbc.E02-08-0520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gallardo M, Aguilera A. A new hyperrecombination mutation identifies a novel yeast gene, THP1, connecting transcription elongation with mitotic recombination. Genetics. 2001;157:79–89. doi: 10.1093/genetics/157.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gallardo M, Luna R, Erdjument-Bromage H, Tempst P, Aguilera A. Nab2p and the Thp1p-Sac3p complex functionally interact at the interface between transcription and mRNA metabolism. J. Biol. Chem. 2003;278:24225–24232. doi: 10.1074/jbc.M302900200. [DOI] [PubMed] [Google Scholar]
- 42.Gonzalez-Barrera S, Prado F, Verhage R, Brouwer J, Aguilera A. Defective nucleotide excision repair in yeast hpr1 and tho2 mutants. Nucleic Acids Res. 2002;30:2193–2201. doi: 10.1093/nar/30.10.2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reed R, Cheng H. TREX, SR proteins and export of mRNA. Curr. Opin. Cell Biol. 2005;17:269–273. doi: 10.1016/j.ceb.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 44.Ares M, Jr, Proudfoot NJ. The spanish connection: transcription and mRNA processing get even closer. Cell. 2005;120:163–166. doi: 10.1016/j.cell.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 45.Piruat JI, Aguilera A. A novel yeast gene, THO2, is involved in RNA pol II transcription and provides new evidence for transcriptional elongation-associated recombination. EMBO J. 1998;17:4859–4872. doi: 10.1093/emboj/17.16.4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jensen TH, Boulay J, Rosbash M, Libri D. The DECD box putative ATPase Sub2p is an early mRNA export factor. Curr. Biol. 2001;11:1711–1715. doi: 10.1016/s0960-9822(01)00529-2. [DOI] [PubMed] [Google Scholar]
- 47.Samarsky DA, Ferbeyre G, Bertrand E, Singer RH, Cedergren R, Fournier MJ. A small nucleolar RNA:ribozyme hybrid cleaves a nucleolar RNA target in vivo with near-perfect efficiency. Proc. Natl Acad. Sci. USA. 1999;96:6609–6614. doi: 10.1073/pnas.96.12.6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wellinger RE, Prado F, Aguilera A. Replication fork progression is impaired by transcription in hyperrecombinant yeast cells lacking a functional THO complex. Mol. Cell. Biol. 2006;26:3327–3334. doi: 10.1128/MCB.26.8.3327-3334.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Suter B, Livingstone-Zatchej M, Thoma F. Mapping cyclobutane-pyrimidine dimers in DNA and using DNA-repair by photolyase for chromatin analysis in yeast. Methods Enzymol. 1999;304:447–461. doi: 10.1016/s0076-6879(99)04027-6. [DOI] [PubMed] [Google Scholar]
- 50.Hecht A, Grunstein M. Mapping DNA interaction sites of chromosomal proteins using immunoprecipitation and polymerase chain reaction. Methods Enzymol. 1999;304:399–414. doi: 10.1016/s0076-6879(99)04024-0. [DOI] [PubMed] [Google Scholar]
- 51.Libri D, Graziani N, Saguez C, Boulay J. Multiple roles for the yeast SUB2/yUAP56 gene in splicing. Genes Dev. 2001;15:36–41. doi: 10.1101/gad.852101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reagan MS, Friedberg EC. Recovery of RNA polymerase II synthesis following DNA damage in mutants of Saccharomyces cerevisiae defective in nucleotide excision repair. Nucleic Acids Res. 1997;25:4257–4263. doi: 10.1093/nar/25.21.4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Woudstra EC, Gilbert C, Fellows J, Jansen L, Brouwer J, Erdjument-Bromage H, Tempst P, Svejstrup JQ. A Rad26-Def1 complex coordinates repair and RNA pol II proteolysis in response to DNA damage. Nature. 2002;415:929–933. doi: 10.1038/415929a. [DOI] [PubMed] [Google Scholar]
- 54.Mason PB, Struhl K. Distinction and relationship between elongation rate and processivity of RNA polymerase II in vivo. Mol. Cell. 2005;17:831–840. doi: 10.1016/j.molcel.2005.02.017. [DOI] [PubMed] [Google Scholar]
- 55.Proietti-De-Santis L, Drane P, Egly JM. Cockayne syndrome B protein regulates the transcriptional program after UV irradiation. EMBO J. 2006;25:1915–1923. doi: 10.1038/sj.emboj.7601071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rondon AG, Garcia-Rubio M, Gonzalez-Barrera S, Aguilera A. Molecular evidence for a positive role of Spt4 in transcription elongation. EMBO J. 2003;22:612–620. doi: 10.1093/emboj/cdg047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jansen LE, den Dulk H, Brouns RM, de Ruijter M, Brandsma JA, Brouwer J. Spt4 modulates Rad26 requirement in transcription-coupled nucleotide excision repair. EMBO J. 2000;19:6498–6507. doi: 10.1093/emboj/19.23.6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Casolari JM, Brown CR, Komili S, West J, Hieronymus H, Silver PA. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell. 2004;117:427–439. doi: 10.1016/s0092-8674(04)00448-9. [DOI] [PubMed] [Google Scholar]
- 59.Rodriguez-Navarro S, Fischer T, Luo MJ, Antunez O, Brettschneider S, Lechner J, Perez-Ortin JE, Reed R, Hurt E. Sus1, a functional component of the SAGA histone acetylase complex and the nuclear pore-associated mRNA export machinery. Cell. 2004;116:75–86. doi: 10.1016/s0092-8674(03)01025-0. [DOI] [PubMed] [Google Scholar]
- 60.Dieppois G, Iglesias N, Stutz F. Cotranscriptional recruitment to the mRNA export receptor Mex67p contributes to nuclear pore anchoring of activated genes. Mol. Cell. Biol. 2006;26:7858–7870. doi: 10.1128/MCB.00870-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Meinhart A, Kamenski T, Hoeppner S, Baumli S, Cramer P. A structural perspective of CTD function. Genes Dev. 2005;19:1401–1415. doi: 10.1101/gad.1318105. [DOI] [PubMed] [Google Scholar]
- 62.Buratowski S. Connections between mRNA 3′ end processing and transcription termination. Curr. Opin. Cell Biol. 2005;17:257–261. doi: 10.1016/j.ceb.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 63.Tijsterman M, Tasseron-de Jong JG, Verhage RA, Brouwer J. Defective Kin28, a subunit of yeast TFIIH, impairs transcription-coupled but not global genome nucleotide excision repair. Mutat. Res. 1998;409:181–188. doi: 10.1016/s0921-8777(98)00060-3. [DOI] [PubMed] [Google Scholar]
- 64.McKay BC, Chen F, Clarke ST, Wiggin HE, Harley LM, Ljungman M. UV light-induced degradation of RNA polymerase II is dependent on the Cockayne's syndrome A and B proteins but not p53 or MLH1. Mutat. Res. 2001;485:93–105. doi: 10.1016/s0921-8777(00)00064-1. [DOI] [PubMed] [Google Scholar]
- 65.Kettenberger H, Armache KJ, Cramer P. Complete RNA polymerase II elongation complex structure and its interactions with NTP and TFIIS. Mol. Cell. 2004;16:955–965. doi: 10.1016/j.molcel.2004.11.040. [DOI] [PubMed] [Google Scholar]
- 66.Westover KD, Bushnell DA, Kornberg RD. Structural basis of transcription: nucleotide selection by rotation in the RNA polymerase II active center. Cell. 2004;119:481–489. doi: 10.1016/j.cell.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 67.Huertas P, Aguilera A. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol. Cell. 2003;12:711–721. doi: 10.1016/j.molcel.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 68.Selby CP, Drapkin R, Reinberg D, Sancar A. RNA polymerase II stalled at a thymine dimer: footprint and effect on excision repair. Nucleic Acids Res. 1997;25:787–793. doi: 10.1093/nar/25.4.787. [DOI] [PMC free article] [PubMed] [Google Scholar]