Abstract
To take full advantage of locked nucleic acid (LNA) based molecular beacons (LNA-MBs) for a variety of applications including analysis of complex samples and intracellular monitoring, we have systematically synthesized a series of DNA/LNA chimeric MBs and studied the effect of DNA/LNA ratio in MBs on their thermodynamics, hybridization kinetics, protein binding affinity and enzymatic resistance. It was found that the LNA bases in a MB stem sequence had a significant effect on the stability of the hair-pin structure. The hybridization rates of LNA-MBs were significantly improved by lowering the DNA/LNA ratio in the probe, and most significantly, by having a shared-stem design for the LNA-MB to prevent sticky-end pairing. It was found that only MB sequences with DNA/LNA alternating bases or all LNA bases were able to resist nonspecific protein binding and DNase I digestion. Additional results showed that a sequence consisting of a DNA stretch less than three bases between LNA bases was able to block RNase H function. This study suggested that a shared-stem MB with a 4 base-pair stem and alternating DNA/LNA bases is desirable for intracellular applications as it ensures reasonable hybridization rates, reduces protein binding and resists nuclease degradation for both target and probes. These findings have implications on the design of LNA molecular probes for intracellular monitoring application, disease diagnosis and basic biological studies.
INTRODUCTION
Genetic information analyses have placed a strong demand for advanced biomolecular recognition probes with high sensitivity and excellent specificity. One such promising probe is the molecular beacon (MB) (1), a short hair-pin oligonucleotide probe (Figure 1A) that binds to a specific oligonucleotide sequence and produces fluorescence signal. With an inherent signal generation mechanism, MBs are able to detect nucleic acid targets without separation or the addition of extra reagents. This property makes MBs especially useful for real-time detection of DNA and RNA, which is of great significance to the study of gene expression in real-time and at the single cell level (2). However, when used for intracellular analyses, MBs tend to generate dramatic false-positive signals due to nuclease degradation, protein binding (3,4) and thermodynamic fluctuations (5). For example, it has been reported that unmodified phosphodiester oligonucleotides may possess a half-life as short as 15–20 min in living cells (6). In addition to nuclease degradation, DNA-MBs are subject to a myriad of nucleic acid binding proteins in cells. The interaction of these nucleic acid binding proteins with MBs can disrupt the stem-loop structure and cause nonspecific fluorescence signal (7–9).
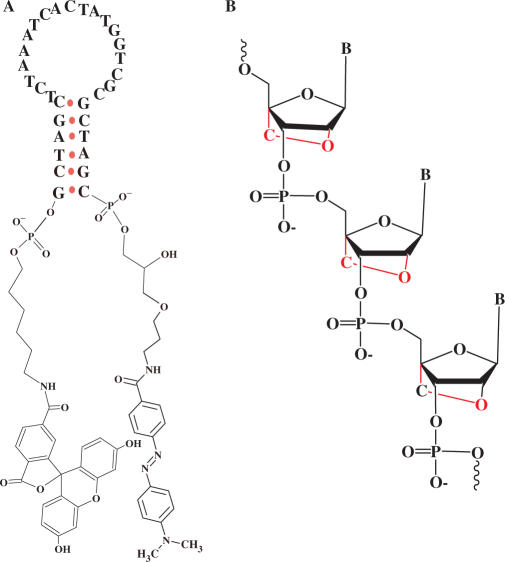
Figure 1.
Structure of a MB (A) and a LNA sequence (B). A LNA nucleotide contains a ribose ring that has a 2′-O, 4′-C methylene bridge.
To overcome the bioinstability problem, MBs have been synthesized with nuclease-resistant backbones such as phosphorothioate and 2′-O-methyl RNA bases (10–12). More drastically, various groups have considered neutral peptide nucleic acids (PNAs) as a scaffold for MBs (13–16).
Backbone modifications have both advantages and disadvantages. Those that retain the repeating charge continue to behave like natural nucleic acids in their hybridizing properties, balancing certain disadvantages, such as the toxicity occasionally associated with phosphorothioate-containing oligonucleotides (17). To the extent that MBs incorporating such modifications resemble natural nucleic acids, however, they still may be opened by intracellular DNA- and RNA-binding proteins, many of which are believed to recognize a repeating backbone negative charge as one pharmacophore. This can lead to a signal in the absence of analyte. For example, MBs with 2′-O-methyl RNA bases possess a better nuclease resistance, higher affinity, increased specificity, faster hybridization kinetics and a superior ability to bind to structured targets compared to their DNA counterparts. However, 2′-O-methyl modified MBs open up nonspecifically in cells, possibly due to protein binding (11,12).
Lacking repeating charges, PNAs are not degraded by nucleases and their hybridization products with RNA are thermally more stable compared with DNA–RNA and RNA–RNA duplexes. The neutral charged PNAs in a MB probe are not likely to be recognized by endogenous RNA- or DNA-binding proteins. Xi et al. (16) reported that using PNA-MBs instead of DNA-MBs for traditional fluorescent in situ hybridization probes would benefit cell detection under a wide range of conditions. The neutral backbone, however, creates other undesirable properties. For instance, PNAs have a well documented propensity to self-aggregate (18) and fold in a way that interferes with duplex formation (19). Furthermore, PNAs change their physical properties substantially (and unpredictably) with small changes in sequence (20,21), although adding charged appendages helps (22). Since the environmental conditions inside of a living cell can not be optimized for the solubility of PNA, intracellular applications of PNA are limited.
Considering these facts, we reasoned that a scaffold that differs as much as possible in the geometric and steric properties from ribose, but retains the repeating charge, might be the most likely to retain the desirable solubility and rule-based molecular recognition features of natural DNA, while avoiding binding to intracellular DNA- and RNA-binding proteins.
Locked nucleic acids (LNAs) (23–26) offer one possible implementation of this reasoning. LNA is a conformationally restricted nucleic acid analogue, in which the ribose ring is locked into a rigid C3′-endo (or Northern-type) conformation by a simple 2′-O, 4′-C methylene bridge (26,27) (Figure 1B). LNA has many attractive properties (26,27), such as high binding affinity, excellent base mismatch discrimination capability, and decreased susceptibility to nuclease digestion. Duplexes involving LNA (hybridized to either DNA or RNA) display a large increase in melting temperatures ranging from +3.0°C to +9.6°C per LNA modification, compared to corresponding unmodified reference duplexes. Furthermore, LNA oligonucleotides can be synthesized using conventional phosphoramidite chemistry, allowing automated synthesis of both fully modified LNA and chimeric oligonucleotides such as DNA/LNA and LNA/RNA. Other advantages of LNA include its close structural resemblance to native nucleic acids, which leads to very good solubility in physiological conditions and easy handling. In addition, owing to its charged phosphate backbone, LNA is nontoxic and can be delivered into cells using standard protocols that employ cationic lipid (28). All these properties are highly advantageous for a molecular tool for diagnostic applications. Several reports have revealed LNA as a most promising molecule for the development of oligonucleotide-based therapeutics.
We have communicated the preparation of a fully modified LNA-MB and the investigation of its properties (29). In vitro experiments showed that the LNA-MB not only exhibits excellent thermal stability and single base discrimination capability, but also resists nuclease digestion and binding of single stranded DNA binding proteins. All of these properties are ideal for intracellular applications and studies. Unfortunately, the hybridization rate of this LNA-MB was relatively slow.
In this study, we evaluated different LNA-MB designs on their thermodynamics, binding kinetics and enzymatic resistance. Specifically, we have designed and synthesized MBs with different DNA/LNA ratios and stem lengths. Systematic studies on the thermostability and hybridization kinetics of the MBs were performed. The binding of these LNA-MBs to single stranded DNA binding protein (SSB) was investigated. In addition, the effects of incorporating LNA bases in a MB on the activity of DNase and RNase were explored. The effect of shortening stem length on the hybridization rate of LNA-MBs was studied. The design guidelines of LNA-MBs for intracellular applications were recommended.
MATERIALS AND METHODS
Chemicals and reagents
MBs prepared are listed in Tables 1 and 2. DNA and LNA synthesis reagents were purchased from Glen Research (Sterling,VA,USA). Deoxynuclease I, Ribonuclease H and SSB were purchased from Fisher.
Table 1.
DNA/LNA MBs with 6 bp in the stem
 |
Red letters represent LNA bases. Underlined letters are bases for MB stem. All MBs are labeled with DABCYL at 3′ ends and FAM at 5′ ends.
Table 2.
MBs with alternating DNA/LNA bases and different stem lengths
 |
Red letters represent LNA bases. Underlined letters are bases for MB stem. All MBs are labeled with DABCYL at 3′ ends and FAM at 5′ ends.
Instruments
An ABI3400 DNA/RNA synthesizer (Applied Biosystems, Foster City, CA, USA) was used for target DNA synthesis and MB probe preparation. Probe purification was performed with a ProStar HPLC (Varian, Walnut Creek, CA, USA), where a C18 column (Econosil, 5u, 250 × 4.6 mm) from Alltech (Deerfield, IL, USA) was used. UV-Vis measurements were performed with a Cary Bio-300 UV spectrometer (Varian, Walnut Creek, CA, USA) for probe quantitation. Fluorescence measurements were performed on a Fluorolog-Tau-3 spectrofluorometer (Jobin Yvon, Inc., Edison, NJ, USA).
Molecular beacon synthesis
MBs possessing locked nucleic acid bases were synthesized using LNA phorsphoramidites. The controlled-pore glass columns used for these syntheses introduced a DABCYL (4-(4-(dimethylamino) phenylazo) benzoic acid) molecule at the 3′ ends of the oligonucleotides. FAM (6-carboxyfluorescein) phorsphoramidite was used to couple FAM to the 5′ ends of the sequences. The complete MB sequences were then deprotected in concentrated ammonia overnight at 65°C and purified by high-pressure liquid chromatography. The collection from the first HPLC separation was then vacuum dried, incubated in 200 µl 80% acetic acid for 15 min, incubated with 200 µl ethanol and vacuum dried before the second round of HPLC.
Hybridization study
Unless otherwise indicated, hybridization experiments were conducted with 100 nM MBs, 500 nM complimentary target sequences in a total volume of 200 µl. All experiments were conducted in 20 mM Tris-HCl (pH 7.5) buffer containing 5 mM MgCl2 and 50 mM NaCl.
DNase I sensitivity
To test the nuclease sensitivity of MBs, the fluorescence of a 200 μl solution containing 20 mM Tris-HCl (pH 7.5), 5 mM MgCl2, 50 mM NaCl and 100 nM MBs was measured as a function of time at room temperature. One unit of ribonuclease-free DNase I was added, and the subsequent change in fluorescence was recorded.
RNase H sensitivity
To test the susceptibility of MB-RNA hybrids to ribonuclease H digestion, 100 nM of MBs were incubated with the same concentration of RNA target in the aforementioned buffer. The fluorescence intensity of the solution was monitored. After the hybridization reached equilibrium, 12 units of ribonuclease H were added, and the subsequent change in fluorescence was recorded as a function of time.
Thermal stability studies
The thermal stabilities of the MB samples were determined by a BioRad RT-PCR thermal cycler. The fluorescence intensity of 100 μl MBs (100 nM) in 20 mM Tris-HCl (pH 7.5) buffer containing 5 mM MgCl2, 50 mM NaCl was monitored as a function of temperature. The temperature was brought to 10°C and increased at 1°C increments to 95°C, with each step lasting for 2 min and fluorescence monitored during each step. The melting temperatures (Tm values) were obtained as the maxima of the first derivative of the melting curves.
Protein binding studies
Gel electrophoresis was performed to study the interaction between SSB and MBs. In 20 mM Tris-HCl buffer (pH 7.5, 5 mM MgCl2, 50 mM NaCl), 5 μM MB was incubated with the same concentration of SSB. After 1 h, the solution was analyzed in a 3% agarose gel in TBE buffer (100 V) for 50 min. The gel was then stained with Gelcode blue stain solution (Pierce) for 1 h and washed with deionized water for 30 min. The image of the resulting gel was obtained by scanning on a regular scanner.
RESULTS
Design and synthesis of LNA-MBs
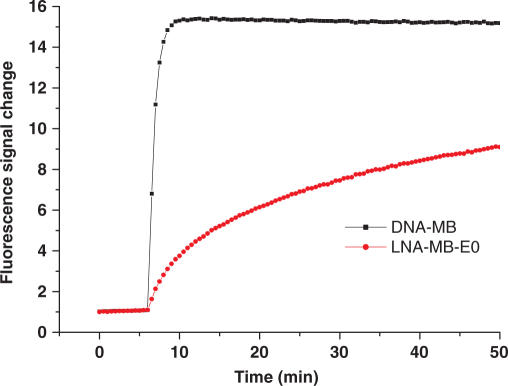
Compared to DNA-MBs, MBs fully modified with LNA respond slowly to complementary target DNA or RNA sequences. Figure 2 shows the hybridization of a DNA-MB and a fully modified LNA-MB (LNA-MB-E0) with the same loop target DNA. The fluorescence signal of DNA-MB reached equilibrium within minutes. In contrast, the fluorescence intensity of the fully modified LNA-MB-E0 increased slowly over time, indicating its slow hybridization kinetics. The reaction did not reach equilibrium even after 20 h under the same conditions. The slow hybridization kinetics would compromise temporal resolution when obtaining the dynamic information of RNA in living cells. In order to take advantage of LNA-MBs for intracellular analysis, it is necessary to expedite LNA-MB hybridization kinetics.
Figure 2.
Hybridization of DNA-MB and LNA-MB-E0 to 10-fold excess of loop cDNA (GCG ACC ATA GTG ATT TAG A). Both MBs shared the same sequence FAM-CCTAGCTCTAAATCACTATGGTCGCGCTA GG-DABCYL. DNA-MB was synthesized with DNA bases, while the LNA-MB-E0 was fully modified with LNA bases. The hybridization experiments were performed at room temperature in 20 mM Tris-HCl (pH 7.5) buffer containing 5 mM MgCl2 and 50 mM NaCl.
It is believed that two factors could lead to the slow hybridization kinetics: slow stem dehybridization rate and tendency of hybridized LNAMBs to form sticky-end pairs. First of all, hybridization of MBs with target sequences competes with the dehybridization of the MB stem. The strong binding affinity of the LNA bases in the stem is unfavorable for stem dehybridization. It has been reported that replacing DNA with LNA bases in one of the oligonucleotide sequences in an octamer duplex had no evident effect on the association rate (30), but significantly decreased the dissociation rate of the octamer sequence from its complementary sequence. The dissociation rate dropped as much as 2-fold for a single base replacement and 5-fold for two base replacements. More than 30-fold decrease in dissociation rate was observed after replacing three DNA/DNA base pairs with DNA/LNA base pairs (30). Therefore, inserting several LNA bases in a MB stem greatly enhances the energy barrier of opening the LNA-MB stem and significantly slows down its hybridization kinetics.
Secondly, because of the excellent binding affinity of LNA, hybridized MB sequences are more likely to form sticky-end pairs (31). DNA sticky-end pairing (SEP) plays an important role in cellular processes and has been well used in various biotechnological applications. For MBs, SEP is defined by the intermolecular interaction between the stems of two or more MBs when loop target DNA is present. When hybridized to its loop target DNA, a MB opens up, exposing two complimentary stem sequences. The two complementary sticky ends from two MB/target hybrids can pair to form a short double helix, leading to the association of two hybrids at one end. These two MB hybrids can form a closed structure, ([MB]2), by pairing the other two sticky ends or polymerize into a multimolecular structure, [MB]n (n > 3), by pairing with more hybrids. With sticky-end pairing, the fluorophore and quencher are brought together again (but from different sequences), causing the quenching of the fluorescence. Any parameters that stabilize the short double helix of the sticky-end pairs will lead to a more severe loss of the fluorescence intensity. For example, the higher the [Mg2+] in the solution, the more signal is lost (31). The direct result of SEP is the quenching of the fluorophore in opened MBs. Therefore, when the opening of MB via target hybridization is immediately followed by SEP, only slow signal intensity increase is observed.
Considering the two factors discussed earlier, we believe that the hybridization kinetics of LNA-MBs could be improved by preventing SEP, or by reducing the LNA percentage in MB sequences, especially in the stem, to lower the reaction energy barrier and speed up the opening rate of MB hair-pin structure. A series of DNA/LNA chimeric MB with a 6-mer stem were prepared as listed in Table 1. DNA-MB is a probe constructed solely from DNA bases. The LNA-MB probes were prepared by gradually replacing every base, every other base, every second base, every third base, every fourth base and every fifth base of the DNA-MB with LNA bases to produce LNA-MB-E0, LNA-MB-E1, LNA-MB-E2, LNA-MB-E3, LNA-MB-E4 and LNA-MB-E5, respectively, as shown in Table 1.
Thermodynamics of LNA-MBs
Stem melting temperatures (Tm) of LNA-MBs were measured to investigate the effect of LNA percentage on the stability of MB hair-pin structure. Table 1 lists the melting temperature of the MBs. It has been reported that introduction of LNA raises the Tm of the oligonucleotide and a complementary DNA or RNA by as much as 9.6°C per LNA modification (32). The incorporation of LNA bases in a MB sequence dramatically stabilized the stem duplex of the probe. When one DNA/DNA base pair was replaced by one LNA/LNA pair in the stem, as in LNA-MB-E5, the melting temperature of the MB stem increased as high as 20°C. The stem Tm increased about 27°C when the number of LNA/LNA base pair increased to two (as in LNA-MB-E2, LNA-MB-E4).
The melting temperature change was found weakly dependent on the nature of base pair change. For example, Tm of LNA-MB-E4 was about 1°C higher than that of LNA-MB-E2. The difference between the stems of these two sequences was that the former had two G: C LNA pairs, while the latter contained one G: C and one A: T LNA pairs.
Insertion of three LNA: DNA pairs in the MB stem had similar effects on the stability of MB stem as two LNA: LNA pairs did, as indicated by the Tm of LNA-MB-E4 and LNA-MB-E3. No fluorescence signal change was observed for LNA-MB-E0, suggesting an extremely high Tm of the probe.
Shared-stem cDNA blocks SEP
The hybridization of the 6-mer-stem MBs to their target DNA sequence that is complementary to the MBs’ loop sequence was investigated. A very slow increase of fluorescence signal was observed, when target DNA was introduced into the LNA-MB-E0 solution. Lowering the percentage of LNA in the probe sequences significantly improved the initial hybridization rate. For LNA-MB-E2, E3 and E4, interesting fluorescence traces were observed. The fluorescence of the solution initially had a sharp increase and then suddenly began to decay. The blue trace in Figure 3 shows a typical response of LNA-MB-E3 to the addition of target DNA. Similar trace was observed for DNA-MBs at higher [Mg2+] in previous studies and was attributed to SEP of MBs in the presence of target (31). Incorporation of LNA bases in MBs significantly enhanced the affinity of the stem, as indicated by the elevated melting temperature of the LNA-MBs. Consequently, intermolecular hybridization of LNA-MBs was evident for LNA-MB-E2, LNA-MB-E3 and LNA-MB-E4. The traces of LNA-MB-E1 and LNA-MB-E0, however, were somewhat different, with no clear drop in fluorescence after the initial signal increase. The signal of these two beacon solutions increased much slower because of more LNAs in the stem, resulting in a higher tendency to form sticky-end pairs at the initial hybridization state.
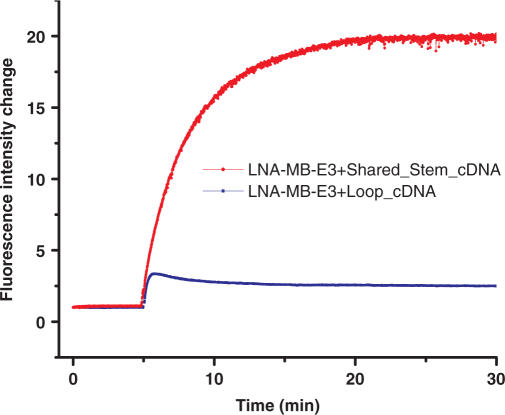
Figure 3.
Hybridization of 100 nM MB-LNA-E3 to 500 nM loop cDNA(GCG ACC ATA GTG ATT TAG A) (blue) and shared-stem cDNA(CCT AGC GCG ACC ATA GTG ATT TAG A) (red). The sequence of MB-LNA-E3 was FAM-CCTAGC TCTAAATCACTATGGTCGCGCTAGG-DABCYL, with red letters representing LNA bases. The hybridization experiments were performed at room temperature in 20 mM Tris-HCl (pH 7.5) buffer containing 5 mM MgCl2 and 50 mM NaCl.
Several strategies could be exploited to prevent the formation of sticky-end pairs (31). For example, lowering Mg2+ concentration is proven to be an effective way to prevent SEP for DNA-MBs. However, this approach is not practical for intracellular applications. Another approach is to use a target complementary to the loop plus one of the stem sequences. The hybridization of the so-called shared-stem targets to MBs results in a MB/target hybrid with only one sticky end, making SEP impossible. The red trace in Figure 3 shows the hybridization of a shared-stem target (CCT AGC GCG ACC ATA GTG ATT TAG A) to LNA-MB-E3. Faster hybridization kinetics and greater signal enhancement were observed.
The formation of sticky-end pairs was further confirmed by studying the thermal denaturation profiles of MBs in equilibrium with their targets. The fluorescence of the solutions of LNA-MB-E3 incubated with no target, loop target and shared-stem target were monitored at different temperatures.
The results (Figure 4A, blue trace) demonstrated that at lower temperatures the MB was in a closed state, the fluorophore and the quencher were held in close proximity to each other by the stem, and the MB did not fluoresce. However, at high temperature the helical order of the stem gave way to a random-coil configuration, separating the fluorophore from the quencher, restoring fluorescence (Figure 4B1) (33). This transition, defined as the melting temperature of the stem, occurred at 84°C.
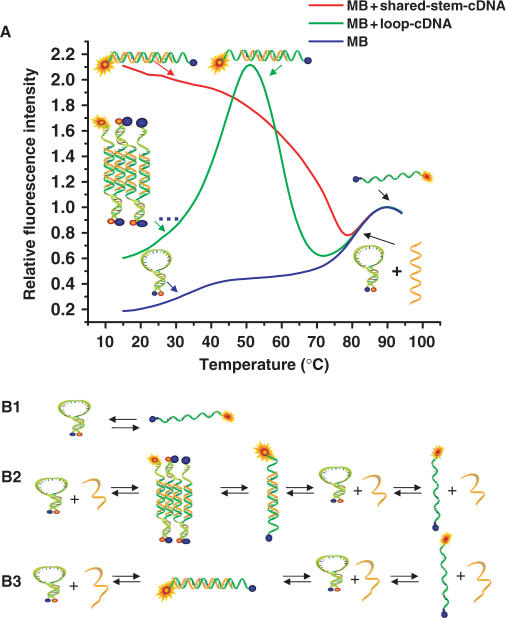
Figure 4.
Blocking LNA-MB sticky-end pairing with shared-stem targets. (A) Thermal denaturation profiles of solutions containing LNA-MB-E3 in the absence of target (blue), in the presence of 5-fold excess of loop complementary DNA target (green), and in the presence of 5-fold excess of shared-stem complementary DNA target (red). (B) Schematic representation of the phases for the MB solutions in the absence of target DNA (B1), in the presence of loop target DNA (B2) and in the presence of shared-stem target DNA(B3).
In the presence of 5-fold excess of single-stranded loop target, the MB fluoresced weakly (Figure 4A, green trace) at room temperature, but the fluorescence intensity increased significantly as the temperature was slowly raised. The signal peaked at around 50°C and then diminished significantly followed by an increase in fluorescence at high temperature. Figure 4B2 summarizes the phase transitions occurred. At low temperatures, MBs hybridized spontaneously to the target sequences that were complementary to the loop sequences. The formation of MB/target hybrids exposed the sticky ends of MBs, leading to the formation of [MB]n complex through the SEP process. As a result, the fluorescence intensity of the MB remained low even though the probe had hybridized with its target. Because the structure stability of the sticky-end pairs was weaker than that of the probe-target duplex and the hair-pin structure, the [MB]n complexes were disrupted and separated into individual MB/target duplexes as the temperature increased. Consequently, the fluorophores were unquenched and the fluorescence intensity increased accordingly. Continuing to raise the solution temperature destabilized probe-target duplexes, thus the MBs were released. The free MBs returned to their closed conformation and the fluorescence decreased. As the temperature was raised further, the closed MBs melted into random coils (Figure 4B2), restoring their fluorescence. This transition occurred at the same temperature (84°C) as in the MB solution free of targets.
When the MB was incubated with 5-fold excess of shared-stem target DNA that was complementary to the loop plus one of the stem sequences, different changes in fluorescence were observed (Figure 4A, red trace). The results showed that the MBs fluoresced brightly at low temperatures, but as the temperature is slowly raised fluorescence diminished significantly, followed by an increase in fluorescence at high temperatures. Figure 4B3 summarizes the phase transitions occurred. The difference between B2 and B3 is the formation of the [MB]n complexes in the case of using loop target sequences. Clearly, the SEP process decreased the overall signal change even when the target sequences were in excess and most MBs were in the open state. The use of shared-stem target sequences effectively blocked SEP formation, leading to a higher signal enhancement and significantly faster apparent hybridization kinetics. When incubated with shared-stem target DNA, the fluorescence signal of the MB was three times the intensity of the same MB with loop target DNA (Figure 4A).
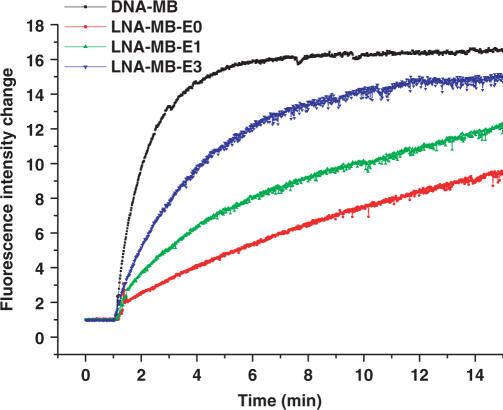
The hybridizations of all 6-mer-stem LNA-MBs with shared-stem targets were much faster and gave much higher fluorescence signal as compared to the hybridizations with loop target DNA. The hybridization results of these probes also indicated that the less the LNA bases in a beacon sequence, the faster the hybridization rate the beacon had. Figure 5 compares the response of the DNA-MB, LNA-MB-E0, LNA-MB-E1 and LNA-MB-E3 to the addition of target DNA. With less DNA bases being replaced by LNA bases, the hybridization rates of MBs were significantly increased.
Figure 5.
Hybridization of DNA-MB and LNA-MBs to shared-stem target sequences. All MBs had the same sequences. All bases in DNA-MB were DNA and LNA-MB-E0 was fully modified by LNA bases. Every other base in LNA-MB-E1 was a LNA base and every other third base in LNA-MB-E3 was a LNA base. The concentrations of the probes were 100 nM, and the concentration of the target sequence (CCT AGC GCG ACC ATA GTG ATT TAG A) was 500 nM. The hybridization experiments were performed at room temperature in 20 mM Tris-HCl (pH 7.5) buffer containing 5 mM MgCl2 and 50 mM NaCl.
Minimal binding of single stranded binding protein with LNA-MBs
Normal MBs are subject to nonspecific binding of proteins, such as the ubiquitous SSB. Such a binding causes normal DNA-MBs to open and give false-positive signal. In addition, protein binding lowers the accessibility of MBs for hybridization to their target sequences. A nucleic acid probe immune to protein binding will perform its analysis with higher accuracy and better targeting efficiency in vivo, where such proteins are highly abundant.
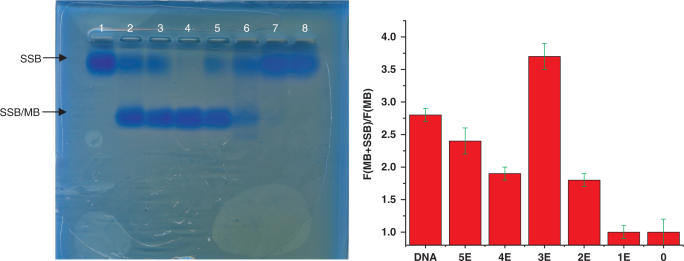
The hybridization results of 6-mer-stem MBs showed that the hybridization rate of a MB was tunable by adjusting the density of LNA bases in the probe sequence. We have reported that MBs with fully modified LNA bases had no response to the addition of excess SSB. With the decrease of DNA/LNA ratio in the probe, it became a concern that the probe might be subject to DNA binding proteins. To find out how the ratio of DNA/LNA in a probe sequence would affect the binding affinity of the probe to DNA binding proteins, fluorescence measurements and gel electrophoresis experiments were carried out to study the binding of SSB to LNA-MBs. Agarose gel (3%) was used to visualize the SSB/MB complex. Due to the high molecular weight of SSB (74 kD), SSB migrates slowly in the gel. The binding of the MB to SSB would significantly expedite the migration rate of the protein because of the multiple negative charges of nucleic acids. As shown in Figure 6, only one band was observed for the sample containing only SSB. The SSB barely migrated in the gel under the experimental conditions. When the DNA-MB was added to the SSB solution, an extra protein band with a faster migration rate appeared, while the intensity of the first band decreased. This clearly indicated the binding of SSB to DNA-MB. The SSB/MB complex band was observed for LNA-MB-E5, LNA-MB-E4, LNA-MB-E3 and LNA-MB-E2. Under the same experimental condition, no visible SSB/MB band was seen for LNA-MB-E1 and LNA-MB-E0. The binding of SSB to MBs was further confirmed by using fluorescence measurements. The disruption of MB secondary structure due to SSB binding displaces the fluorophore from the proximity of the quencher molecule, resulting in fluorescence increase. All the MB sequences except LNA-MB-E1 and LNA-MB-E0 had higher fluorescence intensity when excess SSB was added. For example, an approximate 4-fold increase in fluorescence signal was observed for LNA-MB-E3. Unfortunately, the signal enhancement was also affected by the purity and the stem stability of MBs; thus, the binding affinity could not be directly evaluated with the fluorescence intensity change. Further measurements of the signal changes at different concentrations of an individual MB need to be performed in order to measure the Kd of the corresponding probe to SSB. Nonetheless, the fluorescence measurements were consistent with the gel imaging results. Results from both assays suggested no observable binding of SSB to MBs with all LNA or alternating DNA/LNA bases.
Figure 6.
Interactions between MBs and SSBs. (Left) Gel electrophoresis (3% agarose gel) of SSB solutions containing no MB (lane 1), MB-DNA (lane 2), LNA-MB-E5 (lane 3), LNA-MB-E4 (lane 4), LNA-MB-E3 (lane 5), LNA-MB-E2 (lane 6), LNA-MB-E1 (lane 7) and LNA-MB-E0 (lane 8). (Right) Signal enhancement of MBs to the addition of the same concentration of SSB. [MB] = [SSB] = 5 μM.
Resistance to nuclease digestion
Ideal probes for targets in living cells should be stable inside the cell, should not induce the destruction or perturbation of their target, and should signal only at the presence of their target (34). MBs constructed from natural deoxyribonucleotides are not suitable for this purpose, because cellular nucleases can degrade them and cellular ribonuclease H can digest the target mRNA in the region where the probe is bound.
The sensitivity of the 6-mer-stem LNA-MBs to nuclease digestion were tested using DNase I. As shown in Figure 7, immediate increase in fluorescence signal was observed, when DNase I was added to a solution containing DNA-MB. This result excludes the viability of using MBs constructed from natural deoxyribonucleotides for intracellular applications. With the introduction of LNA in a MB sequence, the digestion process slowed down, as seen in the case of LNA-MB-E5, LNA-MB-E4, LNA-MB-E3 and LNA-MB-E2. In contrast, MBs with DNA/LNA alternating bases (LNA-MB-E1) in the sequence or with all LNA (MB-LNA-E0) gave no observable signal change, indicating their resistance to DNase I after more than 2 h incubation with the nuclease. To test the susceptibility of LNA-MBs to nucleases found in cells, the stability of LNA-MB-E1 and LNA-MB-E0 were further tested inside living cells containing no target sequences. After injection into cells, both MBs remained close during the observation period (more than 1 h), as indicated by no observable fluorescence signal change over time.
Figure 7.
Response of MBs to DNase I. To 100 nM of MB in 20 mM Tris-HCl (pH 7.5, 5 mM MgCl2, 50 mM NaCl), one unit of ribonuclease-free DNase I was added and the subsequent change in fluorescence was recorded at room temperature.
The results imply the necessity of designing MBs with DNA/LNA alternating bases or with all LNA bases, in order for successful intracellular applications.
Effect of LNA-MB on ribonuclease H activity
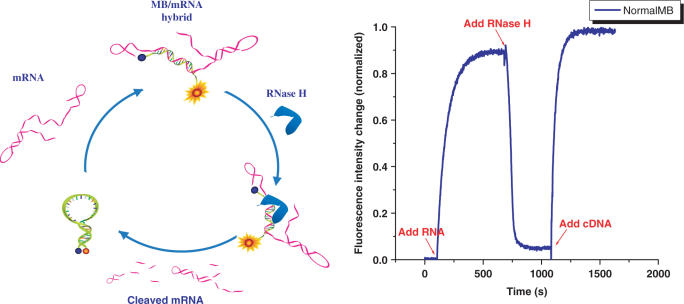
RNase H (Ribonuclease H) is an endoribonuclease that specifically hydrolyzes the phosphodiester bonds of RNA when it is hybridized to DNA. This enzyme does not digest single or double-stranded RNA. RNase H is found in both the nucleus and the cytoplasm of all cells and its normal function is to remove RNA primers from Okazaki fragments during DNA replication. Antisense RNase H activation has proved not only to be a powerful weapon in assessing gene function but is emerging as the method of choice for antisense therapeutics as well. While desirable in antisense technology, RNase H activation is not favored in RNA monitoring. The action of RNase H on MB: RNA duplexes will lead to destruction of the target RNA and loss of signal due to reformation of MB hair-pin structure (Figure 8, left).
Figure 8.
Degradation of mRNA by RNase H. (Left)The hybridization of DNA-MB to its target mRNA forms MB/mRNA duplex, initiating RNase H action. The enzymatic cleavage breaks RNA in the duplex into pieces, releasing MB to restore to hair-pin structure and participate in next cycle of hybridization and cleavage until all mRNA sequences are cleaved. (Right)The response of 100 nM DNA-MB to sequential addition of 100 nM RNA targets, 12 units of RNase H and 100 nM of DNA target.
Use of MBs does allow real-time monitoring of RNase H activity. Figure 8 right shows the response of a DNA-MB to the sequential addition of target RNA, RNase H and target DNA, respectively. The probe lit up with the introduction of RNA in the solution. After reaching the hybridization equilibrium, RNase H was added. An immediate decrease of the fluorescence signal from the probe was observed, which indicated the degradation of RNA on the MB: RNA duplex through enzyme cleavage and the reformation of MB hair-pin structure. After enzyme digestion, the DNA-MB remained intact, as evidenced by its response to the addition of target DNA. Thus, the MB will act as a catalyst inside the cell to destroy all target RNA sequences through RNase H degradation.
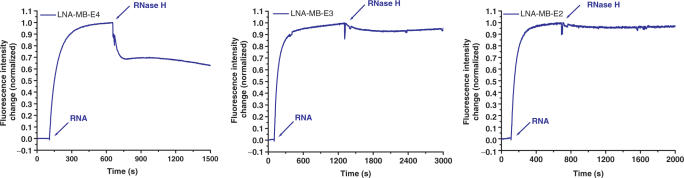
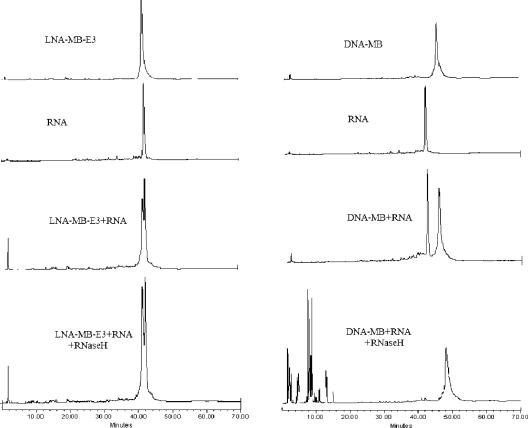
The series of DNA/LNA chimeric MBs enabled the systematic study of the effect of LNA on the RNase H activity using fluorescence measurements. Insertion of LNA bases in DNA MB sequence slowed down the degradation of RNA in the probe: target duplex as compared to the DNA-MB. As seen in Figure 9, as the DNA gap between LNA bases in a MB was shortened, the activity of RNase was inhibited more. For example, about 30% of RNA target bound to LNA-MB-E4 was immediately destroyed after the introduction of RNase H (Figure 9). No significance signal decrease was observed, when the enzyme was added to the duplexes of RNA with LAN-MB-E3, LNA-MB-E2, LNA-MB-E1 and LNA-MB-E0. Ion exchange HPLC analysis of a solution containing LNA-MB-E3, RNA, and RNase H further confirmed the protection of RNA after overnight incubation of LNA-MB-E3 with RNA. There were no small pieces of RNA cleavage product observed in HPLC, as compared to the appearance of different lengths of RNA pieces for the mixture of DNA-MB, RNA and RNAse H (Figure 10). Our results suggested that the number of DNA bases in a stretch for an LNA-MB should be less than three, in order to prevent significant cleavage of the target RNA.
Figure 9.
Effect of LNA composition in a MB on the activity of RNase H. To 100 nM LNA-MB in 20 mM Tris-HCl (5 mM MgCl2, 50 mM NaCl), RNA target was added to reach a final concentration of 100 nM. After the hybridization reached its steady-state, 12 units of RNase H was added.
Figure 10.
Monitor RNase H cleavage of RNA in LNA-MB-E3/RNA duplex (left) and in DNA-MB/RNA duplex (right) using Ion Exchange HPLC. The concentrations of MBs and RNA were 1 µM. All samples were incubated overnight at room temperature before analyzed by ion-exchange HPLC (Dionex DNAPac™ PA-100 column (4 × 250 mm), 30%–70%, 45 min gradient 1 M NaCl/20 mM NaOH, pH 12).
Stem length effect on hybridization kinetics
The results from the 6-mer-stem LNA-MBs with different DNA/LNA compositions suggest that a MB with DNA/LNA alternating bases is desirable for intracellular mRNA imaging applications, which require minimum protein binding and enzymatic degradation. In addition, a shared-stem MB is necessary to prevent SEP and ensures a faster hybridization rate and higher target-induced signal change. Lowering DNA/LNA ratio in a MB does improve the hybridization rate of the probe. Unfortunately, this improvement is limited because of the protein binding and enzyme degradation concerns. For example, although LNA-MB-E5 had a hybridization profile similar to that of DNA-MB, it was a target to SSB; was rapidly digested by DNase I; and had little inhibition to RNase H.
Changing DNA/LNA ratio can affect the stem stability of a MB sequence, the length of the MB stem can likewise be tailored to such a degree that it is stable enough to maintain a hair-pin structure, while presenting a low energy barrier in order for a fast hybridization kinetics. We have reported that shortening the stem length can significantly improve the LNA-MB hybridization rate. In the current study, DNA/LNA alternating MBs with 5-mer, 4-mer and 3-mer stems, designated as LNA-MB-E1-5S, LNA-MB-E1-4S, LNA-MB-E1-3S as listed in Table 2, respectively, were prepared to investigate the optimal stem length of DNA/LNA MB.
As expected, the melting temperature of MB stem decreased with the decrease of probe stem length. With a 5-mer stem, LNA-MB-E1-5S had a Tm of 68.6°C, which was at least 28°C lower than that of a 6-mer stem LNA-MB-E1. The 4-mer-stem probe LNA-MB-E1-4S had a melting temperature of about 52.3°C. Further decrease of stem length to three base pairs did not render any stable hair-pin structure, as indicated by the negative response of MB-LNA-E1-3S to its complementary DNA and heating.
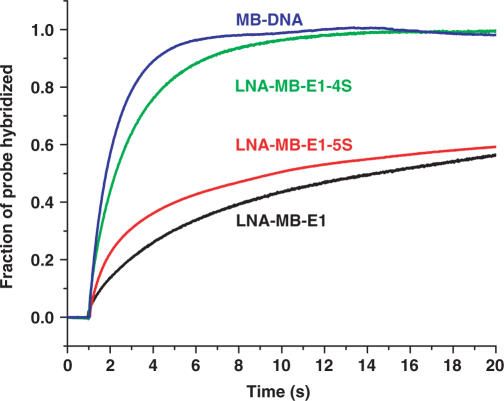
As a result of decreasing stem stability, MBs with shorter stems hybridize to their target DNA much faster. For example, in the presence of 5-fold excess of shared-stem target DNA, it took about 4 h for LNA-MB -E1 to reach its hybridization equilibrium. Under the same condition, the hybribrization of LNA-MB-E1-5S reached equilibrium after about 1 h, while the 4-mer-stem probe LNA-MB-E1-4S completely opened up in less than 15 min. As shown in Figure 11, the hybridization rate of LNA-MB-E1-4S was comparable to a regular DNA-MB. More than 80% of the 4-mer LNA-MB hybridized to the target DNA within 3 min, which makes it an excellent probe for introceullar imaging applications.
Figure 11.
Hybridization of LNA molecular beacons with alternating DNA/LNA bases and different stem lengths to their shared-stem cDNA. The hybridization of MB-DNA was also included as a reference for comparison purpose. Both MB-DNA and LNA-MB-E1 had a 6-mer stem, while LNA-MB-E1-5S had a 5-mer stem and the LNA-MB-E1-4S had a 4-mer stem. The hybridization experiments were performed at room temperature in 20 mM Tris-HCl (pH 7.5) buffer containing 5 mM MgCl2 and 50 mM NaCl. [MB] = 100 nM, [cDNA] = 500 nM.
CONCLUSIONS
Developing stable, sensitive and selective molecular probes is of great significance for the deciphering of life processes inside cells. LNA has showed its interesting properties, such as higher affinity, greater selectivity and better stability. To take full advantage of LNA for intracellular applications and gain a better understanding of the LNA effect on the behavior of MBs, we have synthesized and investigated a series of DNA/LNA chimeric MBs. These series of MBs allow us to systematically study the effect of the DNA/LNA ratio in molecular beacons on their thermal stability, hybridization kinetics, protein binding affinity and nuclease resistance.
The number of LNA bases in a MB stem sequence has a significant effect on the stability of the hair-pin structure. The MB stem melting temperature was elevated by as high as 20°C by simply replacing one DNA pair to an LNA/LNA pair. A MB with 6-mer LNA stem had a melting temperature higher than 95°C. The melting temperature of MBs was found to decrease with lower number of LNA bases in the stem. Due to the high affinity of LNA, LNA-MBs tend to form multiple-probe complexes through a SEP process, when hybridized to loop target DNA, resulting in a very low signal response and slow apparent hybridization kinetics. SEP can be prevented by using shared-stem target DNA that is complementary not only to the loop section of the probe, but also to one of the stem sequences, as evidenced by the higher signal enhancements and faster hybridization rates. The hybridization rate of LNA-MBs could also be significantly improved by lowering the DNA/LNA ratio in the probe. The binding of LNA-MBs to SSB was also studied. Under the experiment condition, it was found that only MB sequences with DNA/LNA alternating bases and all LNA bases were able to resist SSB binding. Further detailed studies need to be conducted on how the LNA modification changes the affinity of SSB binding to nucleic acids. The prepared LNA-MBs with different numbers of DNA bases between two LNA bases enable a systematic study on the effect of LNA modification on the susceptibility of the probe to nuclease and the activity of RNase H function. It was found that the only MB sequences with DNA/LNA alternating bases, or fully modified with LNA were not subject to DNase I digestion and stable inside living cells. Our results showed that a sequence consisting of a DNA stretch less than three bases between two LNA bases were able to block RNase H function. Hybridization results from DNA/LNA-alternating MBs with different stem lengths indicated that satisfactory hybridization kinetics could be achieved from an alternating DNA/LNA MB with a 4-mer stem.
The results from this study suggest a guideline for designing MBs for intracellular applications. A shared-stem MB with a 4-mer stem and alternating DNA/LNA bases ensures reasonable hybridization rates, reduced protein binding, and resistance to nuclease degradation for both target and probes. These findings will also have implications on the design of other LNA molecular probes for intracellular diagnosis, therapeutic and basic biological studies.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
This work was supported by NIH NIGMS Grants and NIH Center of Excellence in Genomic Science. C.J.Y. received support as an ACS Division of Analytical Chemistry Fellow in 2005, sponsored by Merck. L.W. received support as an ACS Division of Analytical Chemistry Fellow in 2006, sponsored by GlaxoSmithKline. The authors would like to thank Dr Steven Benner and Dr Zunyi Yang from the Foundation for Applied Molecular Evolution for their assistance in Ion Exchange HPLC analysis and stimulating discussions. Funding to pay the Open Access Publication charge was provided by the NIH.
Conflict of interest. None declared.
REFERENCES
- 1.Tyagi S, Kramer FR. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 2.Tan WH, Wang KM, Drake TJ. Molecular beacons. Curr. Opin. Chem. Biol. 2004;8:547–553. doi: 10.1016/j.cbpa.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 3.Li JWJ, Fang XH, Schuster SM, Tan WH. Molecular beacons: a novel approach to detect protein - DNA interactions. Angewandte Chemie-International Edition. 2000;39:1049–1052. doi: 10.1002/(sici)1521-3773(20000317)39:6<1049::aid-anie1049>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 4.Tan WH, Fang XH, Li J, Liu XJ. Molecular beacons: a novel DNA probe for nucleic acid and protein studies. Chem. Eur. J. 2000;6:1107–1111. doi: 10.1002/(sici)1521-3765(20000403)6:7<1107::aid-chem1107>3.3.co;2-0. [DOI] [PubMed] [Google Scholar]
- 5.Santangelo PJ, Nix B, Tsourkas A, Bao G. Dual FRET molecular beacons for mRNA detection in living cells. Nucleic Acids Res. 2004;32:e57. doi: 10.1093/nar/gnh062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Uchiyama H, Hirano K, KashiwasakeJibu M, Taira K. Detection of undegraded oligonucleotides in vivo by fluorescence resonance energy transfer. J. Biol. Chem. 1996;271:380–384. doi: 10.1074/jbc.271.1.380. [DOI] [PubMed] [Google Scholar]
- 7.Fang XH, Li JJ, Tan WH. Using molecular beacons to probe molecular interactions between lactate dehydrogenase and single-stranded DNA. Anal. Chem. 2000;72:3280–3285. doi: 10.1021/ac991434j. [DOI] [PubMed] [Google Scholar]
- 8.Yang CYJ, Medley CD, Tan WH. Monitoring nucleic acids using molecular beacons. Curr. Pharm. Biotech. 2005;6:445–452. doi: 10.2174/138920105775159322. [DOI] [PubMed] [Google Scholar]
- 9.Yang CYJ, Martinez K, Lin H, Tan WH. Hybrid molecular probe for nucleic acid analysis in biological samples. J. Am. Chem. Soc. 2006;128:9986–9987. doi: 10.1021/ja0618346. [DOI] [PubMed] [Google Scholar]
- 10.Mhlanga MM, Vargas DY, Fung CW, Kramer FR, Tyagi S. tRNA-linked molecular beacons for imaging mRNAs in the cytoplasm of living cells. Nucleic Acids Res. 2005;33:1902–1912. doi: 10.1093/nar/gki302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Molenaar C, Marras SA, Slats JCM, Truffert JC, Lemaitre M, Raap AK, Dirks RW, Tanke HJ. Linear 2′ O-Methyl RNA probes for the visualization of RNA in living cells. Nucleic Acids Res. 2001;29 doi: 10.1093/nar/29.17.e89. art-e89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsourkas A, Behlke MA, Bao G. Hybridization of 2′-O-methyl and 2′-deoxy molecular beacons to RNA and DNA targets. Nucleic Acids Res. 2002;30:5168–5174. [PMC free article] [PubMed] [Google Scholar]
- 13.Kuhn H, Demidov VV, Gildea BD, Fiandaca MJ, Coull JC, Frank-Kamenetskii MD. PNA beacons for duplex DNA. Antisense Nucleic A. 2001;11:265–270. doi: 10.1089/108729001317022269. [DOI] [PubMed] [Google Scholar]
- 14.Petersen K, Vogel U, Rockenbauer E, Nielsen KV, Kolvraa S, Bolund L, Nexo B. Short PNA molecular beacons for real-time PCR allelic discrimination of single nucleotide polymorphisms. Mol. Cell. Probes. 2004;18:117–122. doi: 10.1016/j.mcp.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 15.Seitz O. Solid-phase synthesis of doubly labeled peptide nucleic acids as probes for the real-time detection of hybridization. Angewandte Chemie-International Edition. 2000;39:3249–3253. doi: 10.1002/1521-3773(20000915)39:18<3249::aid-anie3249>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 16.Xi CW, Balberg M, Boppart SA, Raskin L. Use of DNA and peptide nucleic acid molecular beacons for detection and quantification of rRNA in solution and in whole cells. Appl. Environ. Microbiol. 2003;69:5673–5678. doi: 10.1128/AEM.69.9.5673-5678.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Braasch DA, Corey DR. Locked nucleic acid (LNA): fine-tuning the recognition of DNA and RNA. Chem. Biol. 2001;8:1–7. doi: 10.1016/s1074-5521(00)00058-2. [DOI] [PubMed] [Google Scholar]
- 18.Egholm M, Nielsen PE, Buchardt O, Berg RH. Recognition of guanine and adenine in Dna by cytosine and thymine containing peptide nucleic-acids (Pna) J. Am. Chem. Soc. 1992;114:9677–9678. [Google Scholar]
- 19.Dueholm KL, Egholm M, Behrens C, Christensen L, Hansen HF, Vulpius T, Petersen KH, Berg RH, Nielsen PE, et al. Synthesis of peptide nucleic-acid monomers containing the 4 natural nucleobases - thymine, cytosine, adenine, and guanine and their oligomerization. J. Org. Chem. 1994;59:5767–5773. [Google Scholar]
- 20.Bergmann F, Bannwarth W, Tam S. Solid-phase synthesis of directly linked pna-dna-hybrids. Tetrahedron Lett. 1995;36:6823–6826. [Google Scholar]
- 21.Gildea BD, Casey S, MacNeill J, Perry-O’Keefe H, Sorensen D, Coull JM. PNA solubility enhancers. Tetrahedron Lett. 1998;39:7255–7258. [Google Scholar]
- 22.Gangamani BP, Kumar VA, Ganesh KN. Spermine conjugated peptide nucleic acids (spPNA): UV and fluorescence studies of PNA-DNA hybrids with improved stability. Biochem. Biophys. Res. Commun. 1997;240:778–782. doi: 10.1006/bbrc.1997.7745. [DOI] [PubMed] [Google Scholar]
- 23.Koshkin AA, Rajwanshi VK, Wengel J. Novel convenient syntheses of LNA [2.2.1]bicyclo nucleosides. Tetrahedron Lett. 1998;39:4381–4384. [Google Scholar]
- 24.Koshkin AA, Nielsen P, Meldgaard M, Rajwanshi VK, Singh SK, Wengel J. LNA (locked nucleic acid): an RNA mimic forming exceedingly stable LNA: LNA duplexes. J. Am. Chem. Soc. 1998;120:13252–13253. [Google Scholar]
- 25.Wengel J. Synthesis of 3′-C- and 4′-C-branched oligodeoxynucleotides and the development of locked nucleic acid (LNA) Acc. Chem. Res. 1999;32:301–310. [Google Scholar]
- 26.Vester B, Wengel J. LNA (Locked nucleic acid): high-affinity targeting of complementary RNA and DNA. Biochemistry (Mosc) 2004;43:13233–13241. doi: 10.1021/bi0485732. [DOI] [PubMed] [Google Scholar]
- 27.Orum H, Wengel J. Locked nucleic acids: a promising molecular family for gene-function analysis and antisense drug development. Curr. Opin. Mol. Ther. 2001;3:239–243. [PubMed] [Google Scholar]
- 28.Hertoghs KML, Ellis JH, Catchpole IR. Use of locked nucleic acid oligonucleotides to add functionality to plasmid DNA. Nucleic Acids Res. 2003;31:5817–5830. doi: 10.1093/nar/gkg801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang L, Yang CYJ, Medley CD, Benner SA, Tan WH. Locked nucleic acid molecular beacons. J. Am. Chem. Soc. 2005;127:15664–15665. doi: 10.1021/ja052498g. [DOI] [PubMed] [Google Scholar]
- 30.Christensen U, Jacobsen N, Rajwanshi VK, Wengel J, Koch T. Stopped-flow kinetics of lacked nucleic acid (LNA)-oligonucleotide duplex formation: studies of LNA-DNA and DNA-DNA interactions. Biochem. J. 2001;354:481–484. doi: 10.1042/0264-6021:3540481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li JWJ, Tan WH. A real-time assay for DNA sticky-end pairing using molecular beacons. Anal. Biochem. 2003;312:251–254. doi: 10.1016/s0003-2697(02)00375-5. [DOI] [PubMed] [Google Scholar]
- 32.Bondensgaard K, Petersen M, Singh SK, Rajwanshi VK, Kumar R, Wengel J, Jacobsen JP. Structural studies of LNA: RNA duplexes by NMR: conformations and implications for RNase H activity. Chem. Eur. J. 2000;6:2687–2695. doi: 10.1002/1521-3765(20000804)6:15<2687::aid-chem2687>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 33.Bonnet G, Tyagi S, Libchaber A, Kramer FR. Thermodynamic basis of the enhanced specificity of structured DNA probes. Proc. Natl Acad. Sci. USA. 1999;96:6171–6176. doi: 10.1073/pnas.96.11.6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bratu DP, Cha BJ, Mhlanga MM, Kramer FR, Tyagi S. Visualizing the distribution and transport of mRNAs in living cells. Proc. Natl Acad. Sci. USA. 2003;100:13308–13313. doi: 10.1073/pnas.2233244100. [DOI] [PMC free article] [PubMed] [Google Scholar]