Abstract
The HOP2 and MND1 genes are indispensable for meiotic recombination. The products of these genes associate to form a stable heterodimeric complex that binds DNA and stimulates the recombinase activity of Rad51 and Dmc1. Here we conduct molecular studies to delineate the action mechanism of the Hop2–Mnd1 complex. We present evidence to implicate Hop2 as the major DNA-binding subunit and Mnd1 as the prominent Rad51 interaction entity. Hop2–Mnd1 stabilizes the Rad51–single-stranded DNA (ssDNA) nucleoprotein filament, the catalytic intermediate in recombination reactions. We also show that Hop2–Mnd1 enhances the ability of the Rad51–ssDNA nucleoprotein filament to capture duplex DNA, an obligatory step in the formation of the synaptic complex critical for DNA joint formation. Thus, our results unveil a bipartite mechanism of Hop2–Mnd1 in homologous DNA pairing: stabilization of the Rad51 presynaptic filament and duplex DNA capture to enhance synaptic complex formation.
Keywords: DNA repair, homologous recombination, Rad51 recombinase, synaptic complex
Studies in yeast and other organisms have implicated homologous recombination (HR) in the repair of DNA double-strand breaks (DSBs), other types of chromosome damage, and injured DNA replication forks (Pâques and Haber 1999; Symington 2002; Michel et al. 2004; McEachern and Haber 2006). HR also has a role in telomere maintenance (McEachern and Haber 2006). Importantly, dysfunction in HR or its mediators and regulators can lead to cancer-prone human diseases, including Bloom’s syndrome, Fanconi anemia, and breast, ovarian, and other cancers (Jasin 2002; Moynahan 2002; Hickson 2003; Surralles et al. 2004; Kennedy and D’Andrea 2005). In meiosis, programmed DSBs commit chromosome homologues to undergo recombination, forming crossovers that are essential for proper chromosome disjunction at the first meiotic division (Bishop and Zickler 2004; Neale and Keeney 2006).
HR is best understood in the context of DSB repair. Herein, 3′ single-stranded DNA (ssDNA) tails derived from the nucleolytic processing of the DSB are engaged by the HR machinery, leading to the targeting and invasion of a homologous chromatid to form a DNA joint called the D-loop. Subsequent steps include DNA synthesis, resolution of DNA intermediates, and ligation (Symington 2002; Sung and Klein 2006). In eukaryotes, the homologous DNA pairing reaction responsible for D-loop formation is catalyzed by one of two recombinase enzymes, Rad51 and Dmc1. Whereas Rad51 is needed for recombination in both mitotic and meiotic cells, the expression of Dmc1 is restricted to meiosis. These two recombinases yield inter-homolog crossovers necessary for bridging the homologous chromosomes to ensure their disjunction in meiosis I (Bishop and Zickler 2004; Neale and Keeney 2006).
Both Rad51 and Dmc1 are structurally related to the Escherichia coli recombinase RecA. Like RecA, Rad51 and Dmc1 polymerize on ssDNA in an ATP-dependent manner to form a filamentous structure, referred to as the presynaptic filament. The presynaptic filament engages the duplex DNA partner to yield a ternary complex consisting of the recombinase protein filament, ssDNA, and duplex DNA. DNA homology search leads to the formation of the synaptic complex in which the recombining DNA molecules are aligned in homologous registry, and DNA strand invasion occurs upon the location of a free DNA end in either of the DNA molecules (Radding 1982; Sung and Klein 2006).
Genetic studies in Saccharomyces cerevisiae and mice have found an important role of the HOP2 and MND1 genes in meiotic recombination. In S. cerevisiae, hop2 mutants arrest in the meiotic prophase and chromosomes are more frequently synapsed with a nonhomologous counterpart (Leu et al. 1998). Similarly, mnd1 mutants arrest before the first meiotic division and in general behave like dmc1 mutants (Gerton and DeRisi 2002; Chen et al. 2004; Zierhut et al. 2004; Henry et al. 2006). Further analysis revealed an epistatic relationship among HOP2, MND1, and DMC1, leading to the hypothesis that the Hop2 and Mnd1 proteins function with Dmc1 to ensure the timely formation of a DNA intermediate critical for the completion of meiotic recombination (Gerton and DeRisi 2002; Tsubouchi and Roeder 2002, 2003; Chen et al. 2004; Henry et al. 2006). The mouse hop2 mutant displays a variety of meiotic chromosomal abnormalities, such as meiotic arrest and a failure in chromosome synapsis (Petukhova et al. 2003). While the expression of HOP2 and MND1 genes are restricted to meiosis in S. cerevisiae, these genes are also expressed in somatic tissues in plants, mice, and humans (Ko et al. 2002; Zierhut et al. 2004; Domenichini et al. 2006; Henry et al. 2006). This latter observation hints at the possibility that in higher eukaryotes, aside from a functional interaction with Dmc1, the MND1- and HOP2-encoded products likely possess a mitotic function, possibly in influencing the recombinase activity of Rad51.
The Hop2 and Mnd1 proteins from S. cerevisiae, mice, and human are associated in a stable complex (Chen et al. 2004; Petukhova et al. 2005; Enomoto et al. 2006; Pezza et al. 2006). Consistent with the genetic evidence implicating a role of the Hop2–Mnd1 complex in HR, purified S. cerevisiae Hop2–Mnd1 complex enhances the ability of S. cerevisiae Dmc1 to make DNA joints (Chen et al. 2004), and the mouse or human Hop2–Mnd1 complex physically interacts with human Dmc1 (hDmc1) and Rad51 (hRad51) and promotes DNA strand invasion by both recombinases (Petukhova et al. 2005; Enomoto et al. 2006; Pezza et al. 2006; Ploquin et al. 2007).
Here we strive to define the action mechanism of the Hop2–Mnd1 complex by examining its interactions with hRad51 and DNA substrates. Our results indicate that Hop2–Mnd1 exerts two distinctive effects: It stabilizes the hRad51 presynaptic filament and also promotes the formation of the ternary complex consisting of the recombinase filament and the recombining DNA molecules. In an accompanying paper by Pezza et al. (2007), Hop2–Mnd1 is shown to influence the activity of the meiosis-specific recombinase hDmc1 in a similar manner. To our knowledge, Hop2–Mnd1 is the only essential meiotic HR factor that acts in such a bipartite fashion.
Results
Hop2, Mnd1, and the Hop2–Mnd1 complex
Hop2 and Mnd1 are associated in a tight complex (Chen et al. 2004; Petukhova et al. 2005; Enomoto et al. 2006; Pezza et al. 2006), which was purified (Fig. 1A) from extracts of bacteria tailored to coexpress N-terminally (His)6-tagged Hop2 and untagged Mnd1 using a newly developed procedure. N-terminally (His)6-tagged Hop2 and Mnd1 were individually expressed and purified as described (Pezza et al. 2006). Several independent preparations of the Hop2–Mnd1 complex and individually purified Hop2 and Mnd1 proteins gave the same results in all the biochemical experiments described below.
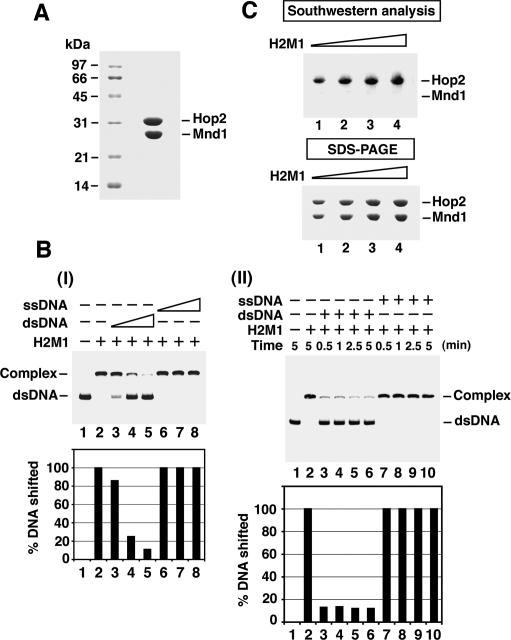
Figure 1.
DNA-binding properties of Hop2–Mnd1. (A) Purified Hop2–Mnd1 complex (10 μg) was subjected to SDS-PAGE with Coomassie blue staining. (B) In panel I, the turnover of Hop2–Mnd1 (H2M1) from dsDNA was examined by challenging a nucleoprotein complex of Hop2–Mnd1 and radiolabeled 80-mer dsDNA for 10 min with unlabeled 80-mer ssDNA or dsDNA. The concentrations of the competitor DNAs were 4.8, 14.4, and 24 μM nucleotides or base pairs. The results were plotted. In panel II, the nucleoprotein complex of Hop2–Mnd1 and radiolabeled 80-mer dsDNA was challenged for the indicated times with unlabeled 80-mer ssDNA or dsDNA (24 μM nucleotides or base pairs). The results were plotted. (C) Varying amounts (4, 6, 8, and 10 μg; lanes 1–4) of Hop2–Mnd1 were analyzed by Southwestern analysis or SDS-PAGE.
DNA-binding characteristics of Hop2–Mnd1
We used 32P-labeled oligonucleotide-based single-stranded and double-stranded substrates in a mobility shift assay to examine the DNA-binding properties of the purified Hop2–Mnd1 complex. As reported previously (Chen et al. 2004; Petukhova et al. 2005; Enomoto et al. 2006; Pezza et al. 2006), Hop2–Mnd1 binds both ssDNA and double-stranded DNA (dsDNA) (Supplementary Fig. S1A, panels I,II) but has a strong preference for the ds species (Supplementary Fig. S1A, panels III,IV). We asked whether Hop2–Mnd1 turns over from the bound dsDNA by challenging a nucleoprotein complex of Hop2–Mnd1 and radiolabeled dsDNA with either unlabeled ssDNA or dsDNA. The preformed nucleoprotein complex could be easily chased away by the unlabeled dsDNA but not by ssDNA within 10 min (Fig. 1B, panel I), indicating that Hop2–Mnd1 turns over from the bound dsDNA. A time-course analysis revealed that Hop2–Mnd1 dissociates from dsDNA within 30 sec (Fig. 1B, panel II). We also used a magnetic bead-based assay to ask whether Hop2–Mnd1 turns over from bound dsDNA and to examine the rapidity of the process. Herein, Hop2–Mnd1 complex was loaded onto a biotinylated dsDNA substrate coupled to streptavidin magnetic beads, followed by a challenge with nonbiotinylated dsDNA. We found again that Hop2–Mnd1 turns over from the bound dsDNA within 30 sec (data not shown).
The DNA-binding ability of Hop2–Mnd1 is derived mainly from Hop2
A Southwestern blot assay was performed to query which of the subunits of the Hop2–Mnd1 complex binds DNA. In this assay, purified Hop2–Mnd1 was fractionated in a SDS–polyacrylamide gel, followed by electroblotting of the separated Hop2 and Mnd1 onto a sheet of nitrocellulose and protein renaturation in situ. The membrane was incubated with 32P-labeled dsDNA, washed, and then analyzed by phosphorimaging analysis to reveal DNA binding. As shown in Figure 1C, binding of the DNA by Hop2, but not by Mnd1, was seen. However, it remained possible that DNA binding by Mnd1 might not have been recoverable by renaturation in situ. Therefore, we also examined individually purified Hop2 and Mnd1 proteins (Pezza et al. 2006) for their DNA-binding properties. Consistent with results from the Southwestern assay, while Hop2 readily bound the radiolabeled dsDNA in the mobility shift assay, a much higher amount of Mnd1 was required to see shifting of the substrate (Supplementary Fig. S1B). Half-maximal binding of the DNA was seen at 300 nM of the Hop2–Mnd1 complex and 300 nM Hop2; however, a much smaller fraction (<15%) of the DNA was bound at 300 nM Mnd1. Thus, the apparent affinity of Hop2 for dsDNA is similar to that of the Hop2–Mnd1 complex. The already modest DNA-binding activity of Mnd1 appears to be high-salt sensitive, such that 100 mM KCl was enough to diminish DNA binding of Mnd1 by fourfold, as compared with <10% drop in the case of Hop2 (Supplementary Fig. S1C, panel III). The DNA-binding activity of the Hop2–Mnd1 complex is not at all affected by 100 mM KCl. Using the Southwestern analysis, we have verified that the ssDNA-binding activity of Hop2–Mnd1 is attributable to Hop2 (Supplementary Fig. S1D).
Taken together, the results indicate that the DNA-binding capability of the Hop2–Mnd1 complex is derived mainly from Hop2, with Mnd1 making a minor contribution.
Mnd1 is the primary interaction partner of hRad51 in Hop2–Mnd1
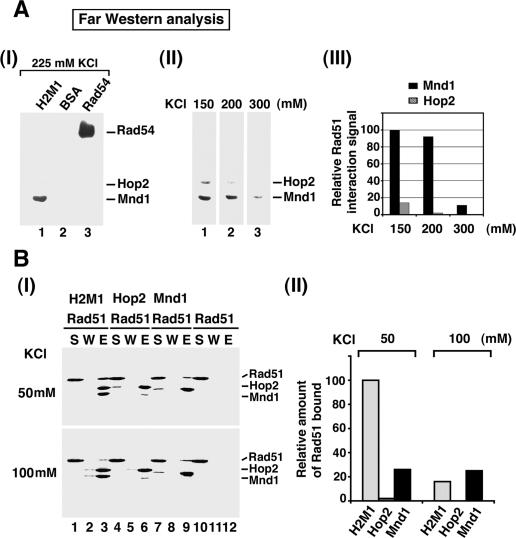
We used affinity pull-down through the (His)6 tag on Hop2 to verify that Hop2–Mnd1 binds hRad51 but not RecA (see below; Supplementary Fig. S2A). Next, we employed the Far Western assay to ask which subunit of this complex interacts with hRad51. For this, a nitrocellulose membrane containing Hop2 and Mnd1 that had previously been resolved by SDS-PAGE but renatured in situ was incubated with hRad51 to allow for complex formation, followed by probing with anti-Rad51 antibodies in a standard Western blot assay to reveal hRad51 association. We varied the reaction stringency by including 150, 200, 225, or 300 mM KCl during the incubation of the nitrocellulose membrane with hRad51. Figure 2A shows that Mnd1 bound hRad51 more avidly than Hop2 at 150 and 200 mM of KCl (panels II,III), but at 225 and 300 mM KCl, only Mnd1 was capable of binding hRad51 (panels I–III). As expected, (1) hRad51 did not interact with bovine serum albumin (BSA) but bound hRad54 (Golub et al. 1997; Ristic et al. 2001; Sigurdsson et al. 2002), and (2) the reactivity of Hop2, Mnd1, and hRad54 with the antibodies was contingent on preincubation of the nitrocellulose membrane with hRad51 (data not shown). However, it remained possible that weak interaction of Hop2 with hRad51 might have resulted from an inability to fully renature Hop2 protein in situ. Therefore, aside from the Far Western analysis, we employed affinity pull-down with individually purified Hop2 and Mnd1 to examine which of these proteins has a higher affinity for hRad51, with the Hop2–Mnd1 complex being included for comparison. The experiments were done either at 50 or 100 mM KCl. As shown in Figure 2B, at the lower stringency, Mnd1 reproducibly bound more hRad51 than did Hop2, with an even higher level of hRad51 being pulled down through the Hop2–Mnd1 complex. Under increased stringency (100 mM KCl), while the ability of Mnd1 to bind hRad51 was not significantly diminished, complex formation of Hop2–Mnd1 with hRad51 was reduced to the level seen with Mnd1 alone, and little, if any, binding of hRad51 to Hop2 could be detected.
Figure 2.
Examination of protein–protein interactions. (A) For the Far Western analysis, (His)6-Hop2–Mnd1 (H2M1), BSA, and hRad54 were resolved by SDS-PAGE, transferred onto nitrocellulose, and renatured in situ. (Panel I) The nitrocellulose membrane was incubated with hRad51 followed by probing with anti-Rad51 antibodies; 225 mM KCl was included in all of the buffers. In panel II, strips of nitrocellulose membrane containing resolved Hop2–Mnd1 were subjected to the Far Western analysis in the presence of the indicated concentration of KCl. The results were plotted in panel III. (B) For affinity pull-down, purified (His)6-Hop2, (His)6-Mnd1, and (His)6-Hop2–Mnd1 were incubated with hRad51 in the presence of either 50 or 100 mM KCl, and the reaction mixtures were mixed with nickel NTA agarose beads to capture protein complexes through the (His)6 tag. (Panel I) The supernatant (S), wash (W), and SDS eluate (E) from the pull-down reactions were analyzed by SDS-PAGE with Coomassie blue staining. The results were plotted in panel II, with the amount of hRad51 bound to Hop2–Mnd1 at the lowest ionic strength being regarded as 100% binding.
In aggregate, the results from the Far Western and affinity pull-down analyses indicate that the ability of Hop2–Mnd1 to interact with hRad51 is derived from both subunits of the complex, with Mnd1 playing a much more prominent role.
Stabilization of the hRad51 presynaptic filament by Hop2–Mnd1
The Hop2–Mnd1 complex stimulates the ability of hRad51 to make DNA joints (Petukhova et al. 2005; Enomoto et al. 2006; Pezza et al. 2006). The documented stimulatory activity of Hop2–Mnd1 is faithfully reproduced using a system that monitors the pairing and exchange of strands between oligonucleotide-based homologous ssDNA and dsDNA substrates. Over the range of Hop2–Mnd1 to hRad51 ratios of 1:2 and 1:5, a marked stimulation of product formation was seen (data not shown). We obtained similar results using a D-loop assay that monitors the assimilation of an oligonucleotide into supercoiled homologous plasmid DNA (data not shown). In contrast, RecA-mediated homologous pairing was not stimulated by Hop2–Mnd1 (Supplementary Fig. S2B,C).
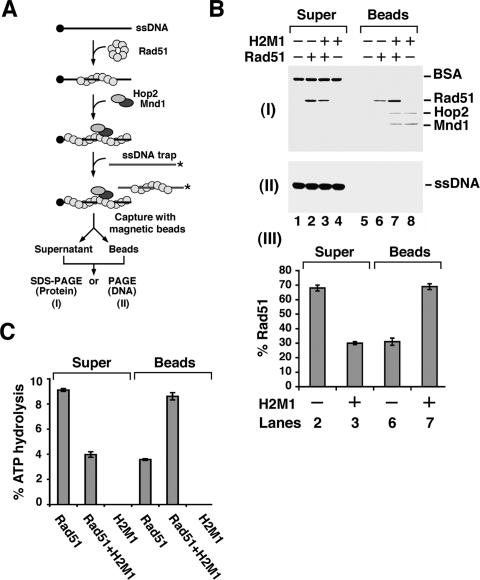
Assembly of the presynaptic filament requires only ATP binding (Sung and Stratton 1996), and ATP hydrolysis prompts the dissociation of the presynaptic filament, resulting in attenuation of the recombinase activity (Bugreev and Mazin 2004; Ristic et al. 2005; Chi et al. 2006). We asked whether Hop2–Mnd1 would stabilize the presynaptic filament when ATP is used as the nucleotide cofactor. To do this, the presynaptic filament was assembled on oligo dT immobilized on magnetic beads through a biotin–streptavidin linkage (Fig. 3A) and then challenged with 10 M excess of 32P-labeled oligo dT, done either in the presence or absence of Hop2–Mnd1. The beads and associated proteins were captured with a magnet, followed by treatment with SDS to elute the bound proteins and SDS-PAGE analysis. As shown in Figure 3B, while most of the input hRad51 was found on the ssDNA trap without Hop2–Mnd1 (Fig. 3B, lanes 2,6), the majority of it remained associated with the magnetic bead-bound oligo dT upon the inclusion of a substoichiometric quantity of Hop2–Mnd1 (Fig. 3B, lanes 3,7). Experiments that employed an extended range of Hop2–Mnd1 concentration revealed a highly significant effect with a 1:3 molar ratio of Hop2–Mnd1 to hRad51 (Supplementary Fig. S3A). We have verified that neither Hop2 nor Mnd1 alone is capable of stabilizing the hRad51 presynaptic filament (data not shown). Importantly, this attribute of Hop2–Mnd1 appears to be highly specific for hRad51, as Hop2–Mnd1 exerts little or no stabilizing effect on either the RecA or yeast Rad51 presynaptic filament under the same reaction conditions (Supplementary Fig. S4). Since hRad51 has a DNA-dependent ATPase activity, but Hop2–Mnd1 neither hydrolyzes ATP nor stimulates the hRad51 ATPase activity (Enomoto et al. 2006; data not shown), we could independently verify the level of hRad51 on the magnetic bead-bound oligo dT by examining ATPase activity. Results from the ATPase assay also showed a stabilizing effect of Hop2–Mnd1 on the hRad51 presynaptic filament (Fig. 3C). In addition to the magnetic bead-based assay, we used an exonuclease protection assay to independently verify the stabilizing effect of Hop2–Mnd1 on the hRad51 presynaptic filament. In this assay, hRad51 is incubated with 5′-radiolabeled ssDNA with or without Hop2–Mnd1 and then treated with E. coli Exonuclease I. The results are again indicative of stabilization of the hRad51 presynaptic filament by a substoichiometric quantity of Hop2–Mnd1 (Supplementary Fig. S3B).
Figure 3.
Stabilization of the hRad51 presynaptic filament by Hop2–Mnd1. (A) Assay to examine the effect of Hop2–Mnd1 on hRad51 presynaptic filament stability. See the main text and Materials and Methods for a detailed description. (B) The presynaptic filament was challenged with the DNA trap with or without Hop2–Mnd1 being present. The appropriate controls were included. (Panel I) The supernatant (Super) fractions containing proteins trapped on the radiolabeled ssDNA and the bead (Beads) fractions containing proteins associated with the biotinylated ssDNA were analyzed by SDS-PAGE with Coomassie blue staining. (Panel II) These fractions were also subjected to electrophoresis in a polyacrylamide gel followed by PhosphorImaging analysis to visualize the radiolabeled DNA trap. (Panel III) The results from lanes 2, 3, 6, and 7 from panel I were plotted. (C) Supernatant and bead fractions from B were examined for their hRad51 content by ATPase analysis.
A distinct function of Hop2–Mnd1 in hRad51 stimulation
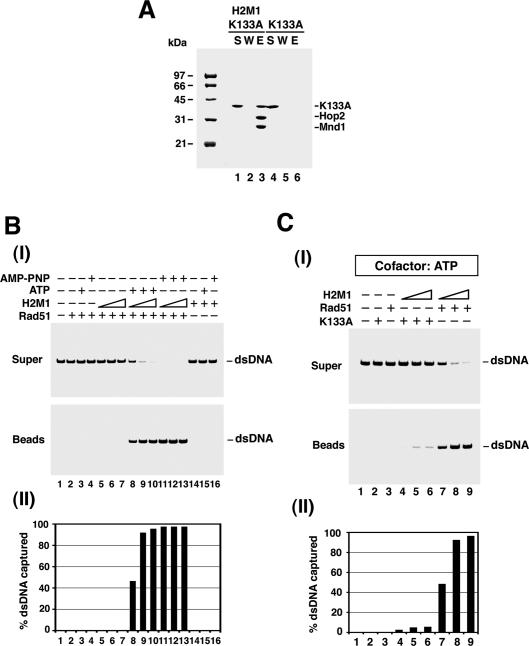
The results above revealed that Hop2–Mnd1 enhances the stability of the presynaptic filament. We were interested in determining whether Hop2–Mnd1 functions in a later step of hRad51-mediated homologous DNA pairing as well. To answer this query, we sought to eliminate presynaptic filament stability as a complicating factor and then assess the effect of Hop2–Mnd1 on homologous pairing efficiency. Since the dissociation of hRad51 from the presynaptic filament is intimately linked to ATP hydrolysis (Bugreev and Mazin 2004; Ristic et al. 2005; Chi et al. 2006), we could make a highly stable presynaptic filament by using the hRad51 K133R variant that binds but does not hydrolyze ATP (Morrison et al. 1999; Chi et al. 2006).
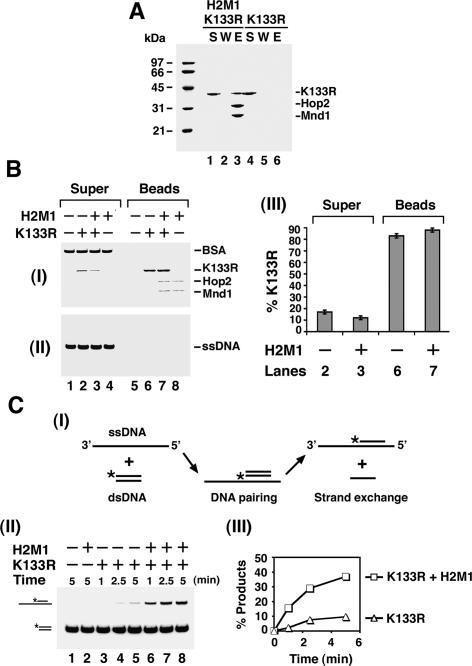
Figure 4A shows that hRad51 K133R binds the Hop2–Mnd1 complex as well as hRad51. Using our assay system (see Fig. 4B), we found that (1) there was very little (<20%) turnover of hRad51 K133R from the magnetic bead-bound oligo dT substrate, and (2) Hop2–Mnd1 slightly reduced the already low level of turnover of the hRad51 K133R presynaptic filament. Importantly, Hop2–Mnd1 stimulated the homologous DNA pairing activity of hRad51 K133R just as efficiently as it did that of hRad51 (Fig. 4C; data not shown). The results thus indicated that Hop2–Mnd1, in addition to enhancing the stability of the hRad51 presynaptic filament, must exert a positive influence in another step of the homologous DNA pairing reaction. This conclusion is further corroborated by the observations that (1) when AMP-PNP, a nonhydrolyzable ATP analog, was used as nucleotide cofactor, hRad51 makes a highly stable presynaptic filament, but the ability to perform homologous DNA pairing is still enhanced by Hop2–Mnd1 to the same degree as when ATP was used; and (2) in the presence of calcium ion, which suppresses the turnover of the hRad51 presynaptic filament (Bugreev and Mazin 2004), Hop2–Mnd1 also enhances the homologous pairing efficiency of hRad51 to the same extent (data not shown).
Figure 4.
Physical and functional interactions of Hop2–Mnd1 with the hRad51 K133R protein. (A) Affinity pull-down showing interaction of hRad51 K133R (K133R) with (His)6-Hop2–Mnd1 (H2M1), conducted in the presence of 50 mM KCl as described for Figure 2B. (B, panels I,II) The effect of Hop2–Mnd1 on the stability of the hRad51 K133R presynaptic filament was examined as described for hRad51 in Figure 3B. (Panel III) The results from lanes 2, 3, 6, and 7 of panel I were plotted. (C) The schematic of the homologous DNA pairing assay is shown in panel I. The hRad51 K133R presynaptic filament assembled on the 150-mer ssDNA was incubated with Hop2–Mnd1 or not, followed by the incorporation of homologous 60-mer dsDNA. The reaction mixtures were incubated for varying lengths of time and resolved by electrophoresis in a polyacrylamide gel. (Panel II) The radiolabeled DNA species were visualized by PhosphorImaging analysis. (Panel III) The results were plotted.
Hop2–Mnd1 promotes duplex DNA capture by the presynaptic filament
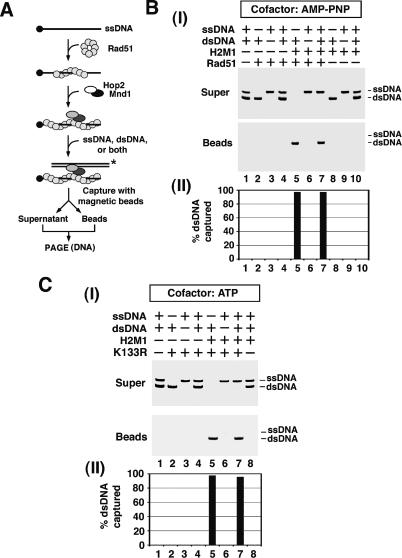
In the homologous DNA pairing reaction, the presynaptic filament must first engage the duplex DNA molecule for DNA homology search and DNA joint formation to occur (Bianco et al. 1998; Sung and Klein 2006). We devised an assay system to examine the ability of the presynaptic filament to engage duplex DNA. Herein, either hRad51 with AMP-PNP as the nucleotide cofactor or hRad51 K133R with ATP as the nucleotide cofactor was incubated with magnetic beads that harbored a ssDNA molecule in order to assemble a stable presynaptic filament, followed by mixing of the magnetic bead-immobilized ensemble with radiolabeled ssDNA or duplex DNA. The bead-bound hRad51 or hRad51 K133R and associated DNA were eluted with SDS and subjected to electrophoresis in a nondenaturing polyacrylamide gel, followed by phosphorimaging analysis to quantify the amount of radiolabeled DNA captured by the presynaptic filament (see Fig. 5A for schematic). As shown in Figure 5, B and C, the presynaptic filament assembled with hRad51 or hRad51 K133R has little ability to engage either ssDNA or dsDNA. Similar results were obtained when the presynaptic filament was assembled with hRad51 and ATP as cofactor (data not shown).
Figure 5.
Promotion of DNA capture by Hop2–Mnd1. (A) Assay for examining DNA capture. See the main text and Materials and Methods for a detailed description. (B) The hRad51 presynaptic filament was incubated with Hop2–Mnd1 (H2M1) and then with either radiolabeled 80-mer ssDNA, dsDNA, or both DNAs, as indicated. The appropriate controls were included. (Panel I) The supernatant (Super) fractions containing free radiolabeled DNA and the bead (Beads) fractions containing bound radiolabeled DNA were subjected to electrophoresis in a polyacrylamide gel and PhosphorImaging analysis. (Panel II) The results were plotted. (C, panel I) The same analysis as in B was conducted but with hRad51 K133R replacing hRad51. (Panel II) The results were plotted.
Since Hop2–Mnd1 physically interacts with hRad51 and binds dsDNA, we asked whether its inclusion would enhance DNA capture by the presynaptic filament. Importantly, amounts of Hop2–Mnd1 that exerted a strong stimulation of the homologous DNA pairing efficiency (Fig. 4C) likewise enhanced the capture of duplex DNA. For instance, while <1% of the dsDNA associated with the hRad51 or hRad51 K133R presynaptic filament, inclusion of Hop2–Mnd1 led to 99% of the duplex molecule being captured (Fig. 5B,C). As expected, duplex DNA capture requires both hRad51 or hRad51 K133R and Hop2–Mnd1. Experiments with an extended range of Hop2–Mnd1 concentrations revealed a substoichiometric requirement of Hop2–Mnd1 relative to hRad51 (Supplementary Fig. S5). Notably, Hop2–Mnd1 did not enhance the ability of the hRad51 or hRad51 K133R presynaptic filament to capture ssDNA (Fig. 5B,C). We obtained very similar results when the presynaptic filament was assembled with hRad51 and ATP as the nucleotide cofactor (see Fig. 6B). As anticipated, we found that neither Hop2 nor Mnd1 alone enhances duplex capture by hRad51 or hRad51 K133R (data not shown). Importantly, the Hop2–Mnd1 complex does not enhance duplex DNA capture by the RecA or RecA K72R presynaptic filament (Supplementary Fig. S6).
Figure 6.
A functional hRad51 presynaptic filament is needed for duplex capture. (A) Purified hRad51 K133A (K133A) was assayed in the presence of 50 mM KCl for complex formation with Hop2–Mnd1, as in Figure 2B. (B, panel I) To examine the nucleotide requirement in duplex capture, hRad51 was incubated with magnetic bead-bound biotinylated 83-mer oligo dT in the presence of ATP or AMP-PNP, or without any nucleotide, as indicated. Following the incorporation of Hop2–Mnd1 (H2M1; lanes 5,8,11, 0.9 μM; lanes 6,9,12, 1.3 μM; lanes 7,10,13, 1.8 μM), radiolabeled 80-mer dsDNA was added to complete the reaction. The remainder of the experiment followed the procedure described in Figure 5B. (Panel II) The results were plotted. (C, panel I) hRad51 K133A was subjected to the same analysis as in B in conjunction with Hop2–Mnd1 (lanes 4–6,7–9, 0.9, 1.3, and 1.8 μM, respectively). hRad51 was included as a positive control in this experiment. (Panel II) The results were plotted.
DNA homology is not a prerequisite for duplex capture
The duplex molecule in the previous experiments was heterologous to the ssDNA used for assembling the presynaptic filament, indicating that DNA homology is not a prerequisite for duplex capture to occur. However, it was of interest to ask whether the presence of homology would facilitate duplex capture. We prepared two duplex substrates, with one bearing no significant homology with the ssDNA and the other harboring a central region of homology but with heterology on either end to prevent plectonemic DNA joint formation (Supplementary Fig. S7A). The capture of the duplex substrates was examined over a range of Hop2–Mnd1 concentrations to see if DNA homology would have an impact. The results showed the same level of capture in both cases, thus demonstrating that the reaction efficiency is not affected by DNA homology (Supplementary Fig. S7B).
Duplex capture is not seen with several other HR factors
Physical and functional interactions between hRad51 and several HR factors—including hRad52, hRad54, and Rad54B—have been reported (McIlwraith et al. 2000; Sigurdsson et al. 2002; Tanaka et al. 2002; Mazina and Mazin 2004; Wesoly et al. 2006). None of these HR factors are capable of promoting duplex capture under the same reaction conditions used for examining Hop2–Mnd1 (Supplementary Fig. S8).
A functional hRad51 presynaptic filament is needed for duplex capture
hRad51 binds DNA in the absence of a nucleotide (Benson et al. 1994; De Zutter and Knight 1999; Tombline et al. 2002; Chi et al. 2006; Graneli et al. 2006), but ATP is required for the recombinase to form a functional presynaptic filament capable of homologous DNA pairing (Sung 1994; Baumann et al. 1996; Sung and Stratton 1996; Gupta et al. 1997; Sigurdsson et al. 2001; Chi et al. 2006). To investigate whether duplex DNA capture requires a functional Rad51 presynaptic filament, we either omitted ATP or used the hRad51 K133A protein, which does not bind ATP (Sung and Stratton 1996; Chi et al. 2006) but retains the ability to interact with Hop2–Mnd1 (Fig. 6A). As shown in Figure 6, B and C, duplex capture was greatly diminished in both cases. Thus, aside from Hop2–Mnd1, a functional presynaptic filament represents a prerequisite for duplex DNA capture.
Discussion
Several recent studies have provided evidence that Hop2–Mnd1 enhances the recombinase activity of hRad51 and hDmc1 (Chen et al. 2004; Petukhova et al. 2005; Enomoto et al. 2006; Henry et al. 2006; Pezza et al. 2006). However, the mechanism that underlies this Hop2–Mnd1 action has not yet been determined. We have presented evidence that Hop2–Mnd1 exerts a stimulatory effect on hRad51 at two distinct stages of the HR reaction: in stabilizing the recombinase presynaptic filament and also in facilitating the capture of duplex DNA substrate by the presynaptic filament. Of the HR factors—hRad52, hRad54, Rad54B, and Hop2–Mnd1—that were examined in our current study, Hop2–Mnd1 is uniquely capable of enhancing dsDNA capture. Pezza et al. (2007) provide evidence in an accompanying paper that Hop2–Mnd1 influences the activity of hDmc1 in a similar bipartite manner. Together, the results from the two studies shed light on the mechanistic role of Hop2–Mnd1 in mammalian meiosis.
Biochemical attributes of Hop2 and Mnd1 are germane for their HR function
Hop2–Mnd1 binds dsDNA preferentially over ssDNA (Chen et al. 2004; Enomoto et al. 2006; Pezza et al. 2006) and physically interacts with hRad51 (Petukhova et al. 2005; Pezza et al. 2006). Complex formation between hRad51 and Hop2–Mnd1 is specific, as the related E. coli RecA shows little or no affinity for the complex. We have shown here that Hop2–Mnd1 turns over readily from dsDNA, a property that is likely germane for the HR role of Hop2–Mnd1 (see below). Published work has found that both Hop2 and Mnd1 bind DNA (Pezza et al. 2006). However, in our Southwestern analysis, only Hop2 shows an appreciable ability to bind DNA. The premise that Hop2 makes a more prominent contribution to the overall DNA-binding activity of the Hop2–Mnd1 complex is corroborated by the observations that (1) in the electrophoretic mobility shift assay, Hop2 binds DNA more avidly than does Mnd1; (2) the DNA-binding isotherm obtained with Hop2 parallels that of the Hop2–Mnd1 complex; and (3) the Hop2 DNA-binding function is much more salt-resistant than is that of Mnd1. On the other hand, even though both Hop2 and Mnd1 physically interact with hRad51, the latter appears to be much more capable of doing so. Specifically, in the affinity pull-down and the Far Western assays, significantly more hRad51 is bound by Mnd1 than by Hop2 at a moderate reaction stringency, but only Mnd1 seems to interact with hRad51 when the reaction stringency is elevated. In fact, as much hRad51 was bound by Mnd1 as by the Hop2–Mnd1 complex under the more stringent condition. Future studies involving specific mutant variants of Hop2 and Mnd1 will ascertain our expectation that the former protein provides the DNA-binding function while the latter harbors the hRad51 interaction ability of the Hop2–Mnd1 complex.
Stabilization of the hRad51 presynaptic filament by Hop2–Mnd1
Upon ATP hydrolysis, hRad51 dissociates from ssDNA, resulting in destabilization of the presynaptic filament. Accordingly, the uncoupling of ATP hydrolysis from ATP binding, such as the use of the nonhydrolyzable ATP analog AMP-PNP or the hRad51 K133R protein, results in an enhanced level of homologous DNA pairing (Bianco et al. 1998; Cox 2003; Bugreev and Mazin 2004; Ristic et al. 2005; Chi et al. 2006). We have presented biochemical evidence that Hop2–Mnd1 stabilizes the hRad51 filament when ATP is used as the nucleotide cofactor. This effect of Hop2–Mnd1 is specific, as it is not seen with E. coli RecA. Previous work has shown that calcium stabilizes the hRad51 presynaptic filament by attenuating ATP hydrolysis (Bugreev and Mazin 2004). In contrast, the presynaptic filament stabilizing function of Hop2–Mnd1 does not appear to involve the attenuation of ATP hydrolysis within the microenvironment of the hRad51 presynaptic filament, because the ATPase activity of hRad51 is not affected by Hop2–Mnd1 (Enomoto et al. 2006; data not shown). We speculate that through its ability to bind DNA and to interact with hRad51, Hop2–Mnd1 retards or prevents the exit of hRad51 from ssDNA. We note that S. cerevisiae Rad54 can stabilize the presynaptic filament assembled with the ScRad51 protein (Mazin et al. 2003). It remains to be determined whether ScRad54 accomplishes this feat by attenuating the ability of the ScRad51 presynaptic filament to hydrolyze ATP or through another means. Regardless, it remains a formal possibility that Rad54 and Hop2–Mnd1 both contribute toward stabilization of the hRad51 presynaptic filament during HR.
Hop2–Mnd1 promotes assembly of a synaptic complex
The DNA-binding properties of Hop2–Mnd1 suggested to us that aside from its presynaptic filament stabilizing role, it may also serve another function that involves an interaction with duplex DNA. In our attempt to uncover the putative second function in Hop2–Mnd1, we (1) substituted ATP with AMP-PNP when wild-type hRad51 was used (Ristic et al. 2005; Chi et al. 2006), (2) replaced hRad51 with the hRad51 K133R mutant (Morrison et al. 1999; Chi et al. 2006), and (3) included calcium ion in the reaction (Bugreev and Mazin 2004), so as to eliminate filament stabilization as a complicating factor in our analysis. Even under these conditions, Hop2–Mnd1 still greatly enhances homologous DNA pairing, thus revealing a second role of Hop2–Mnd1. By biochemical means, Hop2–Mnd1 is shown to promote the capture of duplex DNA to facilitate the assembly of a synaptic complex. The duplex capture reaction is independent of DNA homology and does not occur when ATP is omitted or when hRad51 is substituted with the hRad51 K133A mutant (Chi et al. 2006). These results indicate that synaptic complex assembly requires cooperation between the presynaptic filament and Hop2–Mnd1. Under the same reaction conditions, hRad52, hRad54, and Rad54B are inactive in the duplex capture reaction. However, it is interesting to note that Mazina and Mazin (2004) have seen an ability of hRad54 to recruit duplex DNA to the hRad51 presynaptic filament under a different set of reaction conditions (Mazina and Mazin 2004). Thus, it remains possible that Rad54 synergizes with Hop2–Mnd1 and the hRad51 presynaptic filament to provide the optimal duplex capture capability during HR processes.
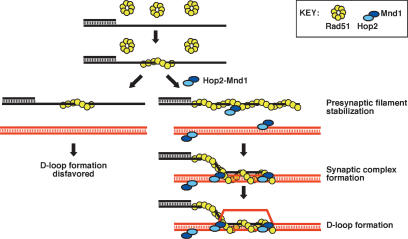
A model for Hop2–Mnd1 action
Our working model concerning the bipartite action of Hop2–Mnd1 is shown in Figure 7. First, Hop2–Mnd1 engages the hRad51 presynaptic filament, resulting in its stabilization. Second, Hop2–Mnd1 helps assemble a synaptic complex via duplex DNA recruitment. These attributes rely on the specific affinities of Hop2–Mnd1 for hRad51 and dsDNA. Our results have also shown that Hop2–Mnd1 turns over readily from dsDNA and that promotion of duplex capture by Hop2–Mnd1 does not require DNA homology. These properties of Hop2–Mnd1 likely permit the efficient sampling of potential partner chromosomes for DNA homology during the HR reaction.
Figure 7.
Model depicting the bipartite action of Hop2–Mnd1 on hRad51. In the absence of Hop2–Mnd1, hRad51 forms a presynaptic filament that is prone to dissociation, a situation disadvantageous for D-loop formation. In the presence of Hop2–Mnd1, the hRad51 presynaptic filament is stabilized. Furthermore, Hop2–Mnd1 promotes synaptic complex formation via duplex DNA recruitment.
Materials and methods
DNA substrates
All the oligonucleotides were obtained from Oligos Etc., Inc., and their sequences are listed in Supplementary Table S1. The oligonucleotides were purified as described (Chi et al. 2006). The procedures for radiolabeling DNA and linking biotinylated DNA to streptavidin magnetic beads are described in the Supplemental Material.
Recombination proteins
The Hop2–Mnd1 complex was purified from E. coli cells tailored to coexpress the two subunits as described in the Supplemental Material. (His)6-tagged Hop2 and Mnd1 proteins were expressed in E. coli and purified as described (Pezza et al. 2006). hRad51 and the hRad51 K133A and hRad51 K133R mutants were expressed in E. coli and purified as described (Sigurdsson et al. 2001; Chi et al. 2006). ScRad51 was expressed in yeast and purified as described (Sung and Stratton 1996). RecA and RecA K72R were a kind gift from Michael Cox (University of Wisconsin, Madison, WI). Rad54B and Flag-tagged hRad54 were expressed in insect cells and purified as described (Sigurdsson et al. 2002; Wesoly et al. 2006). (His)6-tagged hRad52 was expressed in E. coli and purified as described (Motycka et al. 2004).
Turnover of Hop2–Mnd1 from dsDNA
32P-labeled 80-mer dsDNA (2.4 μM base pairs [bp]) was incubated with Hop2–Mnd1 (0.54 μM) in buffer A (35 mM Tris-HCl at pH 7.5, 1 mM DTT, 100 ng/μL BSA, 50 mM KCl, 1 mM MgCl2, 2 mM ATP) for 5 min at 37°C, followed by a challenge with unlabeled 80-mer ssDNA or dsDNA at 37°C for the times indicated in Figure 1B (panel II). The reaction mixtures were run in 10% nondenaturing polyacrylamide gels in TAE buffer (40 mM Tris-acetate at pH 7.5, 0.5 mM EDTA) at 4°C. The gels were dried onto DE81 paper (Whatman) and analyzed in an FX PhosphorImager (Bio-Rad).
Southwestern analysis
Hop2–Mnd1 was mixed with 10 μL of SDS-PAGE buffer, warmed for 1 min at 37°C, and electrophoresed in an 11% SDS-PAGE gel at 4°C. The resolved proteins were electrotransferred onto nitrocellulose (Whatman, Inc.) at 4°C that was soaked in buffer B (25 mM Tris-HCl at pH 7.5, 2 mg/mL BSA, 1 mM DTT) for 20 h at 4°C. The membrane was rinsed twice with buffer C (25 mM Tris-HCl at pH 7.5, 200 μg/mL BSA, 50 mM KCl, 4 mM MgCl2, 1 mM DTT) and incubated in 10 mL of buffer C containing 32P-labeled 80-mer duplex DNA (200 nM bp) for 1 h at 25°C. After being washed four times with 10 mL of buffer C, the membrane was subjected to PhosphorImaging analysis.
Far Western analysis
Hop2–Mnd1 (6 μg), BSA (3 μg), and hRad54 (6 μg) were subjected to 10% SDS-PAGE and then electrotransferred to nitrocellulose, as described for the Southwestern analysis. All the subsequent steps were carried out at 25°C. The membrane was soaked for 2 h in buffer D (10 mM KH2PO4 at pH 7.4, containing 15 mg/mL BSA, 2 mM 2-mercaptoethanol, 0.05% [vol/vol] Tween 20) containing 150 mM KCl before being incubated with 5 μg/mL hRad51 and the indicated concentration of KCl in buffer D for 2 h. The membrane was washed twice with 10 mL of buffer D, incubated with anti-Rad51 antibodies (200 ng/mL) (Sung 1994) for 2 h in buffer D, washed twice with buffer D, and then incubated with anti-rabbit IgG conjugated to horseradish peroxidase (10 ng/mL; Pierce) for 1 h in buffer D. The membrane was developed with the SuperSignal Substrate Kit (Pierce).
Affinity pull-downs
hRad51 (4.8 μg) was incubated with either 8 μg of (His)6-Hop2–Mnd1, 4 μg of (His)6-Hop2, or 4 μg of (His)6-Mnd1 in 30 μL of buffer E (25 mM Tris-HCl at pH 7.5, 10% glycerol, 0.5 mM EDTA, 0.01% Igepal CA-630 [Sigma], 1 mM 2-mercaptoethanol, 5 mM imidazole) with either 50 or 100 mM KCl for 30 min at 4°C. After being mixed with 7 μL of Ni2+-NTA agarose beads (Qiagen) for 30 min at 4°C to capture the (His)6-tagged proteins and associated hRad51, the beads were washed three times with 30 μL of buffer E containing 10 mM imidazole with either 50 or 100 mM KCl and then treated with 20 μL of 2% SDS to elute the proteins. The supernatant, last wash, and SDS eluate, 10 μL each, were analyzed by SDS-PAGE. In the experiments shown in Figures 4A and 6A and Supplementary Figure S2A, 4.8 μg of hRad51 K133R, hRad51 K133A, or RecA was incubated with 8 μg of (His)6-Hop2–Mnd1 in 30 μL of buffer E with 50 mM KCl and subjected to the same analysis.
Homologous DNA pairing assay
hRad51 K133R or RecA, 1.5 μM each, was incubated with the 150-mer Oligo 3 (4.5 μM nucleotides) with Hop2–Mnd1 (0.3 μM) in 10.5 μL of buffer A for 5 min. The reaction mixture was completed by adding spermidine (4 mM) and the homologous 32P-labeled 60-mer duplex (4.5 μM bp) to 12.5 μL final volume. At the indicated times (shown in Fig. 4C, panel II), a 5-μL aliquot was removed and mixed with an equal volume of 1% SDS containing proteinase K (1 mg/mL) and incubated for 5 min at 37°C. The samples were resolved in 10% polyacrylamide gels run in TAE buffer at 4°C, and the DNA species were quantified by PhosphorImaging analysis.
Recombinase turnover from ssDNA
To assemble the presynaptic filament, 4 μL of magnetic beads containing biotinylated 83-mer oligo dT (80 ng, corresponding to 12 μM nucleotides final concentration) were mixed with 2.7 μM hRad51, hRad51 K133R, RecA, or RecA K72R in 19 μL of buffer A containing 0.1 mM ATP for 5 min at 37°C. Following the addition of Hop2–Mnd1 (0.9 μM) in 0.4 μL and a 5-min incubation, the reaction was completed by adding 32P-labeled 83-mer oligo dT ssDNA (100 μM nucleotides) in 0.6 μL as a protein trap. The reaction mixtures (20 μL final volume) were mixed gently for 10 min at 37°C, followed by capture of the beads with the Magnetic Particle Separator; the supernatant was set aside for later analysis. After the beads were washed twice with 20 μL of buffer A containing 0.01% Igepal, proteins were eluted with 20 μL of 2% SDS. The SDS eluates and supernatants were analyzed by SDS-PAGE to determine their protein content and also by polyacrylamide gel electrophoresis in TAE buffer and PhosphorImaging analysis to follow the radiolabeled DNA. To examine hRad51 turnover by the ATPase assay (Fig. 3C), the reaction was set up as described above, except that the ssDNA trap used was unlabeled 83-mer oligo dT and the sample without treated SDS. The ATPase reaction mixture contained a 10-μL portion of the supernatant or bead fraction (suspended in 20 μL of buffer A) and 0.1 mM [γ-32P]ATP and was incubated for 1 h at 37°C. The level of ATP hydrolysis was determined by thin-layer chromatography as described (Petukhova et al. 1998).
Assay to measure duplex DNA capture
To assemble the presynaptic filament, 4 μL of magnetic beads containing 5′-biotinylated 83-mer dT ssDNA (80 ng, corresponding to the final concentration of 12 μM nucleotides) were incubated with 2.7 μM hRad51, hRad51 K133R, hRad51 K133A, RecA, or RecA K72R in 18 μL of buffer A containing either 2 mM ATP, AMP-PNP, ATP-γ-S, or dATP, or no nucleotide for 5 min at 37°C. The beads were captured with the Magnetic Particle Separator, washed once with buffer A, and then resuspended in the same buffer. Following the incorporation of Hop2–Mnd1 (0.9 μM) and a 5-min incubation at 30°C, the beads were again captured, washed, and resuspended. The reaction was completed by adding either radiolabeled 80-mer ssDNA (4 μM nucleotides), radiolabeled dsDNA (4 μM bp), or the combination of both DNA species. The reaction mixtures (20 μL final volume) were incubated for 10 min at 30°C with gentle mixing every 30 sec. The beads were captured, and the supernatant was set aside for analysis later. After the beads were washed twice with 20 μL of buffer A containing 0.01% Igepal, the bound proteins and radiolabeled DNA were eluted with 20 μL of 2% SDS. The SDS eluates were resolved in a 10% polyacrylamide gel in TAE buffer, and the DNA species were visualized and quantified by PhosphorImaging analysis. The supernatant fractions were first treated with SDS and proteinase K (0.5% and 0.5 mg/mL, respectively) for 5 min at 37°C before being subjected to the same analytical procedure.
Acknowledgments
We are very grateful to Roberto Pezza and Daniel Camerini-Otero for providing the Hop2–Mnd1 expression plasmid and other material, for communicating unpublished results, for experimental advice, and for a critical reading of our manuscript. We thank Michael Cox (University of Wisconsin, Madison, WI) for kindly providing RecA and RecA K72R, and members of the Sung laboratory for reading the manuscript and making suggestions. This work was supported by US National Institutes of Health research grants RO1CA110415 and RO1ES07061 and also by Susan G. Komen Breast Cancer Foundation Research Grant BCTR503471 and Post-doctoral Fellowship PDF0503471.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1563007
References
- Baumann P., Benson F.E., West S.C., Benson F.E., West S.C., West S.C. Human Rad51 protein promotes ATP-dependent homologous pairing and strand transfer reactions in vitro. Cell. 1996;87:757–766. doi: 10.1016/s0092-8674(00)81394-x. [DOI] [PubMed] [Google Scholar]
- Benson F.E., Stasiak A., West S.C., Stasiak A., West S.C., West S.C. Purification and characterization of the human Rad51 protein, an analogue of E. coli RecA. EMBO J. 1994;13:5764–5771. doi: 10.1002/j.1460-2075.1994.tb06914.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco P.R., Tracy R.B., Kowalczykowski S.C., Tracy R.B., Kowalczykowski S.C., Kowalczykowski S.C. DNA strand exchange proteins: A biochemical and physical comparison. Front. Biosci. 1998;3:D570–D603. doi: 10.2741/a304. [DOI] [PubMed] [Google Scholar]
- Bishop D.K., Zickler D., Zickler D. Early decision; meiotic crossover interference prior to stable strand exchange and synapsis. Cell. 2004;117:9–15. doi: 10.1016/s0092-8674(04)00297-1. [DOI] [PubMed] [Google Scholar]
- Bugreev D.V., Mazin A.V., Mazin A.V. Ca2+ activates human homologous recombination protein Rad51 by modulating its ATPase activity. Proc. Natl. Acad. Sci. 2004;101:9988–9993. doi: 10.1073/pnas.0402105101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.K., Leng C.H., Olivares H., Lee M.H., Chang Y.C., Kung W.M., Ti S.C., Lo Y.H., Wang A.H., Chang C.S., Leng C.H., Olivares H., Lee M.H., Chang Y.C., Kung W.M., Ti S.C., Lo Y.H., Wang A.H., Chang C.S., Olivares H., Lee M.H., Chang Y.C., Kung W.M., Ti S.C., Lo Y.H., Wang A.H., Chang C.S., Lee M.H., Chang Y.C., Kung W.M., Ti S.C., Lo Y.H., Wang A.H., Chang C.S., Chang Y.C., Kung W.M., Ti S.C., Lo Y.H., Wang A.H., Chang C.S., Kung W.M., Ti S.C., Lo Y.H., Wang A.H., Chang C.S., Ti S.C., Lo Y.H., Wang A.H., Chang C.S., Lo Y.H., Wang A.H., Chang C.S., Wang A.H., Chang C.S., Chang C.S., et al. Heterodimeric complexes of Hop2 and Mnd1 function with Dmc1 to promote meiotic homolog juxtaposition and strand assimilation. Proc. Natl. Acad. Sci. 2004;101:10572–10577. doi: 10.1073/pnas.0404195101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi P., Van Komen S., Sehorn M.G., Sigurdsson S., Sung P., Van Komen S., Sehorn M.G., Sigurdsson S., Sung P., Sehorn M.G., Sigurdsson S., Sung P., Sigurdsson S., Sung P., Sung P. Roles of ATP binding and ATP hydrolysis in human Rad51 recombinase function. DNA Repair (Amst.) 2006;5:381–391. doi: 10.1016/j.dnarep.2005.11.005. [DOI] [PubMed] [Google Scholar]
- Cox M.M. The bacterial RecA protein as a motor protein. Annu. Rev. Microbiol. 2003;57:551–577. doi: 10.1146/annurev.micro.57.030502.090953. [DOI] [PubMed] [Google Scholar]
- De Zutter J.K., Knight K.L., Knight K.L. The hRad51 and RecA proteins show significant differences in cooperative binding to single-stranded DNA. J. Mol. Biol. 1999;293:769–780. doi: 10.1006/jmbi.1999.3200. [DOI] [PubMed] [Google Scholar]
- Domenichini S., Raynaud C., Ni D.A., Henry Y., Bergounioux C., Raynaud C., Ni D.A., Henry Y., Bergounioux C., Ni D.A., Henry Y., Bergounioux C., Henry Y., Bergounioux C., Bergounioux C. Atmnd1-δ1 is sensitive to γ-irradiation and defective in meiotic DNA repair. DNA Repair (Amst.) 2006;5:455–464. doi: 10.1016/j.dnarep.2005.12.007. [DOI] [PubMed] [Google Scholar]
- Enomoto R., Kinebuchi T., Sato M., Yagi H., Kurumizaka H., Yokoyama S., Kinebuchi T., Sato M., Yagi H., Kurumizaka H., Yokoyama S., Sato M., Yagi H., Kurumizaka H., Yokoyama S., Yagi H., Kurumizaka H., Yokoyama S., Kurumizaka H., Yokoyama S., Yokoyama S. Stimulation of DNA strand exchange by the human TBPIP/Hop2–Mnd1 complex. J. Biol. Chem. 2006;281:5575–5581. doi: 10.1074/jbc.M506506200. [DOI] [PubMed] [Google Scholar]
- Gerton J.L., DeRisi J.L., DeRisi J.L. Mnd1p: An evolutionarily conserved protein required for meiotic recombination. Proc. Natl. Acad. Sci. 2002;99:6895–6900. doi: 10.1073/pnas.102167899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golub E.I., Kovalenko O.V., Gupta R.C., Ward D.C., Radding C.M., Kovalenko O.V., Gupta R.C., Ward D.C., Radding C.M., Gupta R.C., Ward D.C., Radding C.M., Ward D.C., Radding C.M., Radding C.M. Interaction of human recombination proteins Rad51 and Rad54. Nucleic Acids Res. 1997;25:4106–4110. doi: 10.1093/nar/25.20.4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graneli A., Yeykal C.C., Robertson R.B., Greene E.C., Yeykal C.C., Robertson R.B., Greene E.C., Robertson R.B., Greene E.C., Greene E.C. Long-distance lateral diffusion of human Rad51 on double-stranded DNA. Proc. Natl. Acad. Sci. 2006;103:1221–1226. doi: 10.1073/pnas.0508366103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R.C., Bazemore L.R., Golub E.I., Radding C.M., Bazemore L.R., Golub E.I., Radding C.M., Golub E.I., Radding C.M., Radding C.M. Activities of human recombination protein Rad51. Proc. Natl. Acad. Sci. 1997;94:463–468. doi: 10.1073/pnas.94.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry J.M., Camahort R., Rice D.A., Florens L., Swanson S.K., Washburn M.P., Gerton J.L., Camahort R., Rice D.A., Florens L., Swanson S.K., Washburn M.P., Gerton J.L., Rice D.A., Florens L., Swanson S.K., Washburn M.P., Gerton J.L., Florens L., Swanson S.K., Washburn M.P., Gerton J.L., Swanson S.K., Washburn M.P., Gerton J.L., Washburn M.P., Gerton J.L., Gerton J.L. Mnd1/Hop2 facilitates Dmc1-dependent interhomolog crossover formation in meiosis of budding yeast. Mol. Cell. Biol. 2006;26:2913–2923. doi: 10.1128/MCB.26.8.2913-2923.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickson I.D. RecQ helicases: Caretakers of the genome. Nat. Rev. Cancer. 2003;3:169–178. doi: 10.1038/nrc1012. [DOI] [PubMed] [Google Scholar]
- Jasin M. Homologous repair of DNA damage and tumorigenesis: The BRCA connection. Oncogene. 2002;21:8981–8993. doi: 10.1038/sj.onc.1206176. [DOI] [PubMed] [Google Scholar]
- Kennedy R.D., D’Andrea A.D., D’Andrea A.D. The Fanconi anemia/BRCA pathway: New faces in the crowd. Genes & Dev. 2005;19:2925–2940. doi: 10.1101/gad.1370505. [DOI] [PubMed] [Google Scholar]
- Ko L., Cardona G.R., Henrion-Caude A., Chin W.W., Cardona G.R., Henrion-Caude A., Chin W.W., Henrion-Caude A., Chin W.W., Chin W.W. Identification and characterization of a tissue-specific coactivator, GT198, that interacts with the DNA-binding domains of nuclear receptors. Mol. Cell. Biol. 2002;22:357–369. doi: 10.1128/MCB.22.1.357-369.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leu J.Y., Chua P.R., Roeder G.S., Chua P.R., Roeder G.S., Roeder G.S. The meiosis-specific Hop2 protein of S. cerevisiae ensures synapsis between homologous chromosomes. Cell. 1998;94:375–386. doi: 10.1016/s0092-8674(00)81480-4. [DOI] [PubMed] [Google Scholar]
- Mazin A.V., Alexeev A.A., Kowalczykowski S.C., Alexeev A.A., Kowalczykowski S.C., Kowalczykowski S.C. A novel function of Rad54 protein. Stabilization of the Rad51 nucleoprotein filament. J. Biol. Chem. 2003;278:14029–14036. doi: 10.1074/jbc.M212779200. [DOI] [PubMed] [Google Scholar]
- Mazina O.M., Mazin A.V., Mazin A.V. Human Rad54 protein stimulates DNA strand exchange activity of hRad51 protein in the presence of Ca2+ J. Biol. Chem. 2004;279:52042–52051. doi: 10.1074/jbc.M410244200. [DOI] [PubMed] [Google Scholar]
- McEachern M.J., Haber J.E., Haber J.E. Break-induced replication and recombinational telomere elongation in yeast. Annu. Rev. Biochem. 2006;75:111–135. doi: 10.1146/annurev.biochem.74.082803.133234. [DOI] [PubMed] [Google Scholar]
- McIlwraith M.J., Van Dyck E., Masson J.Y., Stasiak A.Z., Stasiak A., West S.C., Van Dyck E., Masson J.Y., Stasiak A.Z., Stasiak A., West S.C., Masson J.Y., Stasiak A.Z., Stasiak A., West S.C., Stasiak A.Z., Stasiak A., West S.C., Stasiak A., West S.C., West S.C. Reconstitution of the strand invasion step of double-strand break repair using human Rad51 Rad52 and RPA proteins. J. Mol. Biol. 2000;304:151–164. doi: 10.1006/jmbi.2000.4180. [DOI] [PubMed] [Google Scholar]
- Michel B., Grompone G., Flores M.J., Bidnenko V., Grompone G., Flores M.J., Bidnenko V., Flores M.J., Bidnenko V., Bidnenko V. Multiple pathways process stalled replication forks. Proc. Natl. Acad. Sci. 2004;101:12783–12788. doi: 10.1073/pnas.0401586101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison C., Shinohara A., Sonoda E., Yamaguchi-Iwai Y., Takata M., Weichselbaum R.R., Takeda S., Shinohara A., Sonoda E., Yamaguchi-Iwai Y., Takata M., Weichselbaum R.R., Takeda S., Sonoda E., Yamaguchi-Iwai Y., Takata M., Weichselbaum R.R., Takeda S., Yamaguchi-Iwai Y., Takata M., Weichselbaum R.R., Takeda S., Takata M., Weichselbaum R.R., Takeda S., Weichselbaum R.R., Takeda S., Takeda S. The essential functions of human Rad51 are independent of ATP hydrolysis. Mol. Cell. Biol. 1999;19:6891–6897. doi: 10.1128/mcb.19.10.6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motycka T.A., Bessho T., Post S.M., Sung P., Tomkinson A.E., Bessho T., Post S.M., Sung P., Tomkinson A.E., Post S.M., Sung P., Tomkinson A.E., Sung P., Tomkinson A.E., Tomkinson A.E. Physical and functional interaction between the XPF/ERCC1 endonuclease and hRad52. J. Biol. Chem. 2004;279:13634–13639. doi: 10.1074/jbc.M313779200. [DOI] [PubMed] [Google Scholar]
- Moynahan M.E. The cancer connection: BRCA1 and BRCA2 tumor suppression in mice and humans. Oncogene. 2002;21:8994–9007. doi: 10.1038/sj.onc.1206177. [DOI] [PubMed] [Google Scholar]
- Neale M.J., Keeney S., Keeney S. Clarifying the mechanics of DNA strand exchange in meiotic recombination. Nature. 2006;442:153–158. doi: 10.1038/nature04885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pâques F., Haber J.E., Haber J.E. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petukhova G., Stratton S., Sung P., Stratton S., Sung P., Sung P. Catalysis of homologous DNA pairing by yeast Rad51 and Rad54 proteins. Nature. 1998;393:91–94. doi: 10.1038/30037. [DOI] [PubMed] [Google Scholar]
- Petukhova G.V., Romanienko P.J., Camerini-Otero R.D., Romanienko P.J., Camerini-Otero R.D., Camerini-Otero R.D. The Hop2 protein has a direct role in promoting interhomolog interactions during mouse meiosis. Dev. Cell. 2003;5:927–936. doi: 10.1016/s1534-5807(03)00369-1. [DOI] [PubMed] [Google Scholar]
- Petukhova G.V., Pezza R.J., Vanevski F., Ploquin M., Masson J.Y., Camerini-Otero R.D., Pezza R.J., Vanevski F., Ploquin M., Masson J.Y., Camerini-Otero R.D., Vanevski F., Ploquin M., Masson J.Y., Camerini-Otero R.D., Ploquin M., Masson J.Y., Camerini-Otero R.D., Masson J.Y., Camerini-Otero R.D., Camerini-Otero R.D. The Hop2 and Mnd1 proteins act in concert with Rad51 and Dmc1 in meiotic recombination. Nat. Struct. Mol. Biol. 2005;12:449–453. doi: 10.1038/nsmb923. [DOI] [PubMed] [Google Scholar]
- Pezza R.J., Petukhova G.V., Ghirlando R., Camerini-Otero R.D., Petukhova G.V., Ghirlando R., Camerini-Otero R.D., Ghirlando R., Camerini-Otero R.D., Camerini-Otero R.D. Molecular activities of meiosis-specific proteins Hop2, Mnd1, and the Hop2–Mnd1 complex. J. Biol. Chem. 2006;281:18426–18434. doi: 10.1074/jbc.M601073200. [DOI] [PubMed] [Google Scholar]
- Pezza R.J., Voloshin O.N., Vanevski F., Camerini-Otero R.D., Voloshin O.N., Vanevski F., Camerini-Otero R.D., Vanevski F., Camerini-Otero R.D., Camerini-Otero R.D. Hop2/Mnd1 acts on two critical steps in Dmc1-promoted homologous pairing. Genes & Dev. 2007 doi: 10.1101/gad.1562907. (this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploquin M., Petukhova G.V., Morneau D., Déry U., Bransi A., Stasiak A., Camerini-Otero R.D., Masson J.-Y., Petukhova G.V., Morneau D., Déry U., Bransi A., Stasiak A., Camerini-Otero R.D., Masson J.-Y., Morneau D., Déry U., Bransi A., Stasiak A., Camerini-Otero R.D., Masson J.-Y., Déry U., Bransi A., Stasiak A., Camerini-Otero R.D., Masson J.-Y., Bransi A., Stasiak A., Camerini-Otero R.D., Masson J.-Y., Stasiak A., Camerini-Otero R.D., Masson J.-Y., Camerini-Otero R.D., Masson J.-Y., Masson J.-Y. Stimulation of fission yeast and mouse Hop2–Mnd1 of the Dmc1 and Rad51 recombinases. Nucleic Acids Res. 2007;35:2719–2733. doi: 10.1093/nar/gkm174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radding C.M. Homologous pairing and strand exchange in genetic recombination. Annu. Rev. Genet. 1982;16:405–437. doi: 10.1146/annurev.ge.16.120182.002201. [DOI] [PubMed] [Google Scholar]
- Ristic D., Wyman C., Paulusma C., Kanaar R., Wyman C., Paulusma C., Kanaar R., Paulusma C., Kanaar R., Kanaar R. The architecture of the human Rad54–DNA complex provides evidence for protein translocation along DNA. Proc. Natl. Acad. Sci. 2001;98:8454–8460. doi: 10.1073/pnas.151056798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ristic D., Modesti M., van der Heijden T., van Noort J., Dekker C., Kanaar R., Wyman C., Modesti M., van der Heijden T., van Noort J., Dekker C., Kanaar R., Wyman C., van der Heijden T., van Noort J., Dekker C., Kanaar R., Wyman C., van Noort J., Dekker C., Kanaar R., Wyman C., Dekker C., Kanaar R., Wyman C., Kanaar R., Wyman C., Wyman C. Human Rad51 filaments on double- and single-stranded DNA: Correlating regular and irregular forms with recombination function. Nucleic Acids Res. 2005;33:3292–3302. doi: 10.1093/nar/gki640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigurdsson S., Trujillo K., Song B., Stratton S., Sung P., Trujillo K., Song B., Stratton S., Sung P., Song B., Stratton S., Sung P., Stratton S., Sung P., Sung P. Basis for avid homologous DNA strand exchange by human Rad51 and RPA. J. Biol. Chem. 2001;276:8798–8806. doi: 10.1074/jbc.M010011200. [DOI] [PubMed] [Google Scholar]
- Sigurdsson S., Van Komen S., Petukhova G., Sung P., Van Komen S., Petukhova G., Sung P., Petukhova G., Sung P., Sung P. Homologous DNA pairing by human recombination factors Rad51 and Rad54. J. Biol. Chem. 2002;277:42790–42794. doi: 10.1074/jbc.M208004200. [DOI] [PubMed] [Google Scholar]
- Sung P. Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science. 1994;265:1241–1243. doi: 10.1126/science.8066464. [DOI] [PubMed] [Google Scholar]
- Sung P., Klein H., Klein H. Mechanism of homologous recombination: Mediators and helicases take on regulatory functions. Nat. Rev. Mol. Cell Biol. 2006;7:739–750. doi: 10.1038/nrm2008. [DOI] [PubMed] [Google Scholar]
- Sung P., Stratton S.A., Stratton S.A. Yeast Rad51 recombinase mediates polar DNA strand exchange in the absence of ATP hydrolysis. J. Biol. Chem. 1996;271:27983–27986. doi: 10.1074/jbc.271.45.27983. [DOI] [PubMed] [Google Scholar]
- Surralles J., Jackson S.P., Jasin M., Kastan M.B., West S.C., Joenje H., Jackson S.P., Jasin M., Kastan M.B., West S.C., Joenje H., Jasin M., Kastan M.B., West S.C., Joenje H., Kastan M.B., West S.C., Joenje H., West S.C., Joenje H., Joenje H. Molecular cross-talk among chromosome fragility syndromes. Genes & Dev. 2004;18:1359–1370. doi: 10.1101/gad.1216304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington L.S. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 2002;66:630–670. doi: 10.1128/MMBR.66.4.630-670.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K., Kagawa W., Kinebuchi T., Kurumizaka H., Miyagawa K., Kagawa W., Kinebuchi T., Kurumizaka H., Miyagawa K., Kinebuchi T., Kurumizaka H., Miyagawa K., Kurumizaka H., Miyagawa K., Miyagawa K. Human Rad54B is a double-stranded DNA-dependent ATPase and has biochemical properties different from its structural homolog in yeast, Tid1/Rdh54. Nucleic Acids Res. 2002;30:1346–1353. doi: 10.1093/nar/30.6.1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tombline G., Heinen C.D., Shim K.S., Fishel R., Heinen C.D., Shim K.S., Fishel R., Shim K.S., Fishel R., Fishel R. Biochemical characterization of the human RAD51 protein. III. Modulation of DNA binding by adenosine nucleotides. J. Biol. Chem. 2002;277:14434–14442. doi: 10.1074/jbc.M109917200. [DOI] [PubMed] [Google Scholar]
- Tsubouchi H., Roeder G.S., Roeder G.S. The Mnd1 protein forms a complex with hop2 to promote homologous chromosome pairing and meiotic double-strand break repair. Mol. Cell. Biol. 2002;22:3078–3088. doi: 10.1128/MCB.22.9.3078-3088.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubouchi H., Roeder G.S., Roeder G.S. The importance of genetic recombination for fidelity of chromosome pairing in meiosis. Dev. Cell. 2003;5:915–925. doi: 10.1016/s1534-5807(03)00357-5. [DOI] [PubMed] [Google Scholar]
- Wesoly J., Agarwal S., Sigurdsson S., Bussen W., Van Komen S., Qin J., van Steeg H., van Benthem J., Wassenaar E., Baarends W.M., Agarwal S., Sigurdsson S., Bussen W., Van Komen S., Qin J., van Steeg H., van Benthem J., Wassenaar E., Baarends W.M., Sigurdsson S., Bussen W., Van Komen S., Qin J., van Steeg H., van Benthem J., Wassenaar E., Baarends W.M., Bussen W., Van Komen S., Qin J., van Steeg H., van Benthem J., Wassenaar E., Baarends W.M., Van Komen S., Qin J., van Steeg H., van Benthem J., Wassenaar E., Baarends W.M., Qin J., van Steeg H., van Benthem J., Wassenaar E., Baarends W.M., van Steeg H., van Benthem J., Wassenaar E., Baarends W.M., van Benthem J., Wassenaar E., Baarends W.M., Wassenaar E., Baarends W.M., Baarends W.M., et al. Differential contributions of mammalian Rad54 paralogs to recombination, DNA damage repair, and meiosis. Mol. Cell. Biol. 2006;26:976–989. doi: 10.1128/MCB.26.3.976-989.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zierhut C., Berlinger M., Rupp C., Shinohara A., Klein F., Berlinger M., Rupp C., Shinohara A., Klein F., Rupp C., Shinohara A., Klein F., Shinohara A., Klein F., Klein F. Mnd1 is required for meiotic interhomolog repair. Curr. Biol. 2004;14:752–762. doi: 10.1016/j.cub.2004.04.030. [DOI] [PubMed] [Google Scholar]