Abstract
Long-term facilitation of the connections between the sensory and motor neurons of the gill-withdrawal reflex in Aplysia requires five repeated pulses of serotonin (5-HT). The repeated pulses of 5-HT initiate a cascade of gene activation that leads ultimately to the growth of new synaptic connections. Several genes in this process have been identified, including the transcriptional regulators apCREB-1, apCREB-2, apC/EBP, and the cell adhesion molecule apCAM, which is thought to be involved in the formation of new synaptic connections. Here we report that the transcriptional regulators apCREB-2 and apC/EBP, as well as a peptide derived from the cytoplasmic domain of apCAM, are phosphorylated in vitro by Aplysia mitogen-activated protein kinase (apMAPK). We have cloned the cDNA encoding apMAPK and show that apMAPK activity is increased in sensory neurons treated with repeated pulses of 5-HT and by the cAMP pathway. These results suggest that apMAPK may participate with cAMP-dependent protein kinase during long-term facilitation in sensory cells by modifying some of the key elements involved in the consolidation of short- to long-lasting changes in synaptic strength.
In both humans and animals, memory has a short-term phase lasting minutes and a long-term phase lasting days or longer. In Aplysia, Drosophila, and mice, it has proven possible to explore this transition on the molecular level for different types of learning (1–6). In each case the short-term form has been found to involve covalent modifications of preexisting proteins. By contrast, the long-term form involves gene expression, protein synthesis, and the growth of new synapses. Although different types of learning recruit different kinases for short-term memory, they commonly recruit the activity of cAMP, the cAMP-dependent protein kinase (PKA), and the transcription regulator CREB for long-term memory.
The transition from short- to long-term memory has been studied most completely at the molecular level in the context of a simple form of nonassociative learning, sensitization of the gill-withdrawal reflex in Aplysia. In this reflex, one stimulus (electric shock) to the tail or one pulse of serotonin (5-hydroxytryptamine; 5-HT), a facilitatory transmitter released by such stimuli, produces short-term facilitation of the connections between sensory and motor neurons of this reflex. By contrast, five stimuli or five pulses of 5-HT produce a long-term facilitation accompanied by the growth of new connections (7, 8). This long-term facilitation requires cAMP, PKA, and CREB-1. In each of the three major synaptic pathways of the mammalian hippocampus there is a similar requirement for cAMP and PKA during late long-term potentiation (LTP) (9–12).
Although PKA is an essential component of the signal transduction pathway for consolidating certain types of long-term memory, it seems unlikely that PKA is the only kinase involved in the long-term process. For example, long-term facilitation requires not only the activation of CREB-1 but also the removal of the repressive action of CREB-2. However, CREB-2 lacks consensus sites for PKA phosphorylation (13). This led us to wonder whether other second messenger systems might be involved in long-term facilitation. One clue was provided by CREB-2, which has two conserved consensus sites for phosphorylation by mitogen-activated protein kinase (MAPK) (14). Similarly, C/EBP, a transcription factor downstream from CREB that is thought to be critical for growth, contains a MAPK phosphorylation site (15), and its mammalian homolog is a substrate for MAPK in fibroblasts (16). Furthermore, the transmembrane isoform of apCAM, the Aplysia NCAM-related cell adhesion molecule, lacks PKA sites but has two MAPK phosphorylation sites in its cytoplasmic tail (17), and as we have shown, mutations in the MAPK sites in the cytoplasmic tail of apCAM abolish its internalization in response to 5-HT treatment (18).
These several findings raise the possibility that MAPK, which is typically associated with growth and terminal differentiation, might participate, with PKA, in long-term facilitation. Unlike in many cells in which activation of PKA inhibits the MAPK pathway (19, 20), in the pheochromocytoma cell line PC-12, nerve growth factor initiates differentiation accompanied by sustained activation and nuclear translocation of MAPK (21). By contrast, epidermal growth factor (EGF) induces a transient, cytoplasmically restricted, activation of MAPK and triggers cell proliferation. However, if EGF is paired with elevations in intracellular cAMP, it leads to sustained activation and nuclear translocation of MAPK and results in neuronal differentiation (21).
We therefore asked: Can 5-HT activate MAPK in Aplysia? If so, does it activate MAPK by means of cAMP? Once MAPK is activated, what are its potential targets? We have now addressed these three questions in the sensory neurons of the gill-withdrawal reflex. We first report the cloning of an Aplysia MAPK (apMAPK). We then show that apMAPK is activated in isolated sensory cells in culture by repeated application of 5-HT and by forskolin. In addition, we find that apMAPK may act on targets in two different cellular compartments: at the surface membrane and in the nucleus. In vitro, apMAPK phosphorylates peptides derived from the cytoplasmic domain of apCAM and it also phosphorylates full-length apCREB-2 and apC/EBP. We have previously shown that apMAPK is required for long-term facilitation (22) and furthermore that the internalization of apCAM, an early feature of the growth process, is abolished in mutant forms that lack MAPK phosphorylation sites (17, 18, 23). Together, our findings suggest that long-term memory may require the coordinated activity of MAPK and PKA, and that in the sensory neurons, MAPK acts both in the nucleus and at the membrane.
MATERIALS AND METHODS
Antibodies, Peptides, and Purified Materials.
Polyclonal anti-peptide (BSA-conjugated) antibodies were raised in rabbits. The 7884 antibody was raised against mammalian MAPK subdomain-XI peptide (C)RRITVEEALAHPYLEQYYDPTDE (24). The D8 antibody was raised against the peptide (CG)ELIFQETLQIQDKNLEHS from the C terminus of apMAPK. Peptides were coupled to a sulfo-link column (Pierce) to affinity-purify anti-peptide antibodies. The peptide derived from apCAM was (RRR)PAPAPTTEIKITPETPEKP(A). ERK2 was purified and activated as described (25). MKP1 (a gift from H. Sun, Cold Spring Harbor Laboratories) was 95% pure with an activity of 500 nmol/min per ml against MAPK. Glutathione S-transferase (GST) fusion proteins were prepared as described (26).
Isolation and Treatment of Pleural-Pedal Ganglia.
Both pleural-pedal ganglia from each animal were rapidly isolated in sea water containing high Mg2+ and low Ca2+ concentrations (7) at 12–14°C. Ganglia from one side served as controls. Paired ganglia were washed twice with pH = 7.6–7.7 artificial sea water (IO) (Instant Ocean; Coralife Marine Products) and then placed in L15/IO medium (1:1) pH = 7.6 with or without 5-HT or forskolin for the indicated period at 18°C. L15 medium (Sigma) was supplemented as described (8). The 5-HT (Sigma), forskolin, and 1,9-dideoxyforskolin (Calbiochem) were freshly made.
Aplysia Sensory Cell Cultures and Treatments.
Sensory cells (about 50) were isolated from pleural ganglia of adult animals (80–100 g) and cultured (7). Three- to 4-day-old cultures were used for experiments. Cells were cultured in L15/IO (1:1) without hemolymph, pH 7.6, with two or three changes of solution for 2 hr before the experiments. Control cultures were incubated with L15/IO medium only. Cells were scraped with 50% strength standard protein sample buffer.
Preparation of Cell Extracts.
Aplysia pleural-pedal ganglia were washed twice with cold H buffer (50 mM β-glycerophosphate, pH 7.3/1 mM EDTA/1.5 mM EGTA/0.2 mM sodium vanadate) and then processed in a glass homogenizer in lysis buffer [H buffer with 1 mM DTT, 1 mM benzamidine, aprotinin and leupeptin (10 μg/ml each), and pepstatin A (2 μg/ml)]. After centrifugation at 100,000 × g for 45 min, the supernatant was used for biochemical fractionation. In the experiments shown in Figs. 3A, 4, and 5, a brief sonication step was introduced, the second centrifugation step was eliminated, and Triton X-100 (0.5%) was used.
Figure 3.
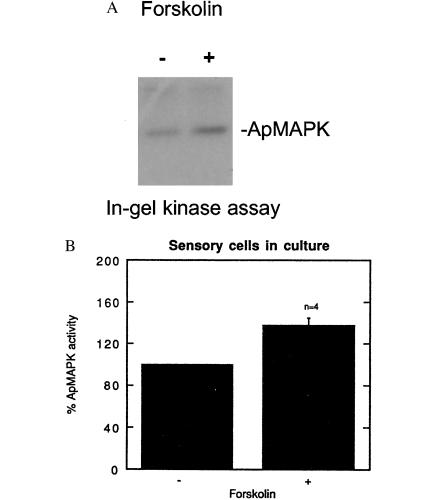
Forskolin increases MAPK activity in pleural-pedal ganglia and in isolated cells in culture. (A) Paired pleural-pedal ganglia were treated with either 150 mM forskolin (+) or its inactive analog 1,9-dideoxyforskolin (−), for 3 hr, and a kinase assay was performed in MBP-containing gels by using fractionated apMAPK (fraction F7). (B) A similar kinase assay was performed by using sensory cells in culture that were treated with forskolin (100 μM) for 10 min (n = 2), 30 min (n = 1), or 1 hr (n = 1). Control cultures were treated with 100 μM 1,9-dideoxyforskolin. Shown is the mean percentage ±SE.
Figure 4.
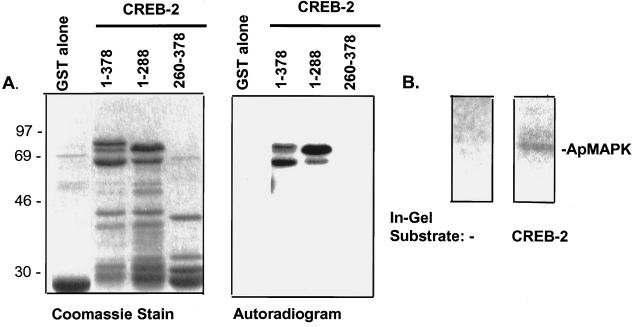
Phosphorylation of CREB-2 by MAPK. (A) Purified activated rat ERK2 was incubated with various CREB-2 fusion proteins, including GST alone, full-length GST-CREB-2-(1–378), and two mutants encoding the N-terminal sequence (1–288) or the C-terminal sequence (260–378) of CREB-2. The relative amounts of protein used are shown by Coomassie stain in Left. Purified active rat ERK2 was allowed to phosphorylate for 30 min. Subsequently, substrates were separated by gel electrophoresis; relative phosphorylation efficiencies are depicted in Right (2.5-hr exposure). Quantitation revealed that CREB-2 (1–378) and its low molecular weight derivatives contained 0.015 pmol of phosphate per pmol of substrate, and that CREB-2-(1–288) and its derivative contained 0.016 pmol/pmol. (B) In-gel kinase assays using no substrate, or CREB-2 as a substrate and Aplysia CNS-derived fraction F7 as the source of kinase activities. The 43-kDa kinase activity (apMAPK) was capable of phosphorylating CREB-2.
Figure 5.
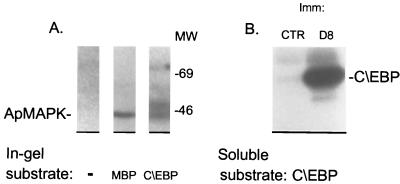
Aplysia C/EBP is a MAPK substrate. (A) To identify Aplysia kinase(s) that phosphorylate C/EBP, an in-gel kinase assay was performed with a partially purified kinase-containing fraction (fraction F7). The 43-kDa apMAPK activity was directed against C/EBP and MBP. In addition, a 50-kDa kinase activity was directed only against C/EBP. (B) Immune-complex kinase assays were performed with antibodies against apMAPK or control nonimmune serum. The immunoprecipitated material from extracts of isolated ganglia was incubated in each case with pure GST-C/EBP under conditions that favor MAPK activity, followed by gel electrophoresis. The autoradiogram demonstrates phosphorylation of C/EBP by immunoprecipitated apMAPK.
Fractionation of Cell Extracts.
To obtain partially purified MAPK, extracts were fractionated by using fast-flow Q resin (FFQ, Pharmacia). Resin (150 μl) was equilibrated with FFQ buffer (buffer H plus 1 mM DTT). Proteins in lysis buffer were diluted 1:1 with FFQ buffer, 300–600 μg of protein was loaded by twice applying 350 μl, and the resulting fractions were numbered F1 and F2. The column was then washed twice with 750 μl of FFQ buffer (fractions F3 and F4), and subsequently an increasing NaCl gradient in FFQ buffer was applied. F5 (500 μl of FFQ/40 mM NaCl), F6 (500 μl of FFQ/60 mM NaCl), F7 and F8 (500 μl of FFQ/150 mM NaCl each), F9 (250 μl of FFQ/150 mM NaCl), fractions 10 and 11 (250 μl of FFQ/165 mM NaCl each), F12 and F13 (500 μl of FFQ/230 mM NaCl each), F14 (500 μl of FFQ/280 mM NaCl), F15 (500 μl of FFQ/600 mM NaCl), and F16 (500 μl of FFQ/1 M NaCl) were collected on ice in the cold room.
Immunoprecipitation, SDS/PAGE, and Kinase Assays.
For immunoprecipitation, cell extracts were incubated with 0.5% SDS and heated at 90°C for 3 min. Subsequently, extracts were made to contain 0.1% SDS, 0.1% deoxycholate, and 0.06 μg/μl BSA. Proteins were resolved by using SDS/PAGE; gels were made of 10% acrylamide and 0.8% bisacrylamide at pH 8.8. Proteins were loaded in standard protein sample buffer. Soluble kinase assays were performed as previously described (27). Alternatively, for in vitro phosphorylation of GST and GST fusion proteins, “on beads” kinase assays were done using the same buffers (27). Purified recombinant ERK2 was activated using fractionated MEK (25) and employed in soluble kinase assays.
For the immunocomplex kinase assay, immunoprecipitated kinase was made with 8 μg of myelin basic protein (MBP) or 6–12 μg of GST fusion protein in Hepes (50 mM, pH 7.4) along with 7 μl of FFQ buffer, 6 μl of reaction mixture (28), and 0.5 μl of [γ-32P]ATP per reaction and incubated at 30°C for 20–30 min.
The “in-gel” kinase assay was performed essentially as previously described (29, 30). Proteins were renatured in the gel and kinases were allowed to phosphorylate in-gel, pre-embedded, substrate. The substrates were 0.8 mg/ml MBP (Sigma) or 0.25 mg/ml GST fusion proteins.
Densitometry and Statistical Analysis.
Densitometric analysis was done with a Bio-Rad densitometer model 670. Statistical significance was determined by Student’s t test, using the gb-stat software (Dynamic Microsystems).
RESULTS
The Aplysia Homolog of MAPK and the Cloning of Its cDNA.
MAPKs are highly conserved serine/threonine kinases (31). We therefore used an anti-rat MAPK antibody, the 7884 antibody (24), in a Western analysis of Aplysia pleural-pedal ganglia, and we detected one prominent 43-kDa band (Fig. 1A).
Figure 1.
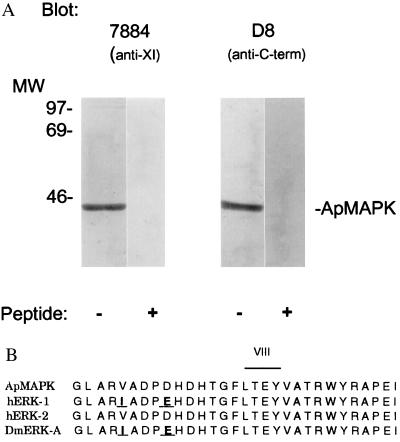
Immunoreactivity and cloning of apMAPK. (A) Antibody 7884 against rat MAPK (24) was affinity purified and used in immoblots of Aplysia central nervous system (CNS) extracts. The antibody specifically recognized a protein of 43 kDa. A λZAP II (Stratagene) expression library containing cDNAs from Aplysia total CNS was screened with the 7884 antibody. The sequence of an isolated cDNA clone was found to be 85% identical to human ERK2 and we called it apMAPK (GenBank accession no. U40484). (B) The sequence of the activation domain of apMAPK (domain VIII) is compared with the human and Drosophila homologs. The D8 antibody was made against the C-terminal 18 amino acids of the molecule, and it specifically recognized a 43-kDa protein in the Western blot analysis shown in A.
We next used the antibody to screen an expression library derived from Aplysia CNS. One of two clones encoded a 351-amino acid polypeptide with 85% homology to the sequence of human ERK2, and we have named this protein apMAPK. By comparing the catalytic domains of different kinases, we found that all the general subdomains of kinases are present (32). Of particular interest is the 100% identity of a region of apMAPK with subdomain VIII of human ERK2 (Fig. 1B). Phosphorylation at this domain of both the threonine and tyrosine in the T-E-Y motif is required for maximal kinase activity (28, 33, 34). This dual phosphorylation, which is directly catalyzed by the dual-specificity kinase MAPK kinase (MAPKK or MEK), can be triggered by many distinct stimuli (27, 35, 36). A lower degree of conservation is found at the 18 amino acids at the C terminus of apMAPK (only 38% identity with ERK2), and a peptide containing this sequence was used to generate a polyclonal anti-apMAPK antibody. This antibody, D8, like antibody 7884, specifically recognized a single band of 43-kDa molecular mass, apMAPK (Fig. 1A).
Five Pulses of 5-HT Activate MAPK in Isolated Sensory Neurons.
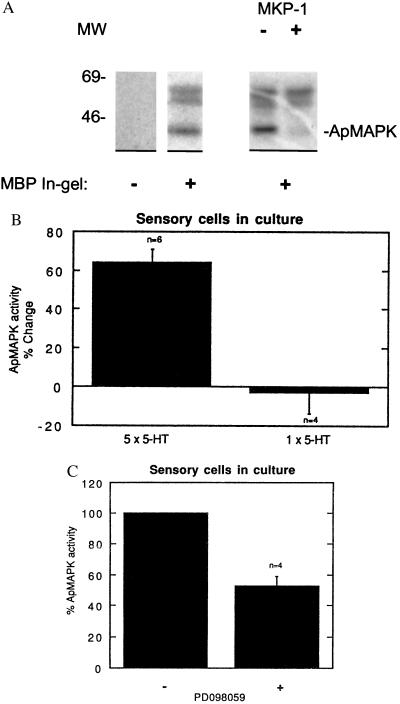
The MAPK pathway can be activated by agonists that act through heterotrimeric G-protein-coupled receptors (34, 37, 38). Because many 5-HT receptors are coupled to G proteins, we initially sought to characterize possible effects of 5-HT on MAPK activity in sensory cells in culture. To do this, we carried out in-gel kinase assays using extracts of sensory cells in culture and MBP, a good MAPK substrate in vitro. We detected three major non-autophosphorylating kinase activities against MBP, migrating as proteins of 43, 58, and 63 kDa molecular mass (Fig. 2A). The 43-kDa kinase activity represents authentic apMAPK activity, as, unlike the 58- and 63-kDa kinase activities, it was sensitive to pretreatment with the specific MAPK phosphatase MKP-1 (39, 40).
Figure 2.
Repeated application of 5-HT onto sensory cells in culture activates apMAPK. Protein extracts from approximately 50 cells in culture were subjected to a kinase assay in MBP-containing gels (A–C, except where otherwise indicated). (A) An extract of Aplysia sensory cells in buffer A and 0.3 mg/ml BSA was treated with 1 μl of the pure MAPK phosphatase 1 (MKP-1) prior to the kinase assay in MBP-containing gels. (B) Five pulses of 10 μM 5-HT were given to sensory cells for 5 min, each pulse at 20-min intervals, while control cultures were given regular medium at the same intervals. An in-gel kinase assay was subsequently performed. The kinase activities at 58 and 63 kDa were used as references to correct and normalize for the apMAPK activities. Normalized MAPK activity as detected by the in-gel kinase assay was 64% higher in cultures treated with five pulses of 5-HT compared with control (mock treated) cultures. Also, similar experiments were performed with one pulse of 5-HT that lasted 5 min. The mean apMAPK activity after one pulse compared with control cultures was 2.7 ± 10.8%; P < 0.01 vs. five pulses of 5-HT. (C) Sensory cells in culture were treated with 30 μM PD098059 for 30 min or with vehicle (0.1% dimethyl sulfoxide) and apMAPK activity was determined by kinase analysis in MBP-containing gels. Shown is mean percentage ±SE.
In sensory-motor cocultures one application of 5-HT produces short-term facilitation, whereas five spaced applications result in long-term facilitation. As shown in Fig. 2B, five repeated applications of 5-HT significantly increased apMAPK activity. In six independent experiments five pulses of 5-HT increased apMAPK activity by 64% ± 6.7%. By contrast, one pulse of 5-HT produced no activation: the mean apMAPK activity after one pulse compared with control cultures was −2.7% ± 10.8%, n = 4; (Fig. 2B, P < 0.01 one pulse vs. five pulses of 5-HT). Taken together, the data in Fig. 2 indicate that apMAPK is active in the basal state and can be further activated by repeated application of 5-HT in cultured sensory cells.
To inhibit MAPK in vivo we used PD098059, an inhibitor of MEK and the downstream MAPK cascade, which inhibits MEK in vitro and in vivo (41, 42) without affecting PKA or protein kinase C (42, 43). As shown in an in-gel kinase assay in Fig. 2C, we observed a decrease of 47.3% ± 6.4% in MAPK activity after 30-min incubation with PD098059 (n = 4, P < 0.01). This provides further evidence that the 43-kDa kinase activity represents authentic MAPK activity, and it also indicates that, in the basal state, MAPK is constitutively activated by MEK in sensory neurons.
Forskolin Stimulates apMAPK Activity in Sensory Neurons.
To determine the nature of the relationship between PKA and MAPK in Aplysia CNS, we tested the effect of forskolin, an adenylyl cyclase activator, on apMAPK activity in pleural-pedal ganglia. Extracts from ganglia were fractionated on an anion-exchange minicolumn, and we found that forskolin stimulated kinase activity against MBP by a factor of 2 as compared with ganglia treated with dideoxyforskolin (Fig. 3A). We next exposed cultured sensory cells to 100 μM forskolin, which resulted in an average increase of 38% ± 4% (n = 4) in MAPK activity against MBP in an in-gel kinase assay as compared with control cells (Fig. 3B; P < 0.01, Student’s t test). Thus, PKA appears to activate MAPK in sensory cells.
apMAPK Can Phosphorylate Several Key Substrates for Long-Term Facilitation.
We next asked: Does apMAPK phosphorylate any of the proteins that participate in the consolidation of long-term facilitation? The transcriptional switch from short- to long-term facilitation at the transcriptional level requires CREB-1, CREB-2, and C/EBP (13, 15, 44), and a stabilization phase involving the growth of new synapses, during which internalization of apCAM is thought to play a critical role (17, 23).
CREB-2 has two consensus sites for phosphorylation by MAPK (at amino acids 150–153 and 234–237). Using purified rat MAPK, we observed that full-length apCREB-2 as well as its N-terminal segment (amino acids 1–288), which contains the two consensus sites for phosphorylation by MAPK, were efficiently phosphorylated (Fig. 4A). No phosphorylation of a C-terminal fragment of apCREB-2 (amino acids 260–378) was detected, consistent with the lack of MAPK phosphorylation consensus sites. We then performed a kinase assay in gels containing Aplysia CREB-2, as shown in Fig. 4B. An apMAPK-containing fraction from isolated Aplysia ganglia was allowed to phosphorylate apCREB-2, and the radioactive band at 43 kDa corresponds to apMAPK activity (Fig. 4B). The results indicate that Aplysia CREB-2 is a substrate of apMAPK in vitro and that the phosphorylation occurs N-terminal to the DNA-binding domain.
Aplysia C/EBP (apC/EBP) is a leucine zipper transcription factor that is critical for the growth of new synaptic connections (15). We first established that in an in-gel kinase assay, apC/EBP was phosphorylated by apMAPK (lower band in Fig. 5A). An additional, as yet unidentified, kinase activity against apC/EBP was also detected in this assay, corresponding to the higher molecular weight band in Fig. 5A. We next showed that immunoprecipitated apMAPK was capable of phosphorylating apC/EBP in an “immune-complex” kinase assay (Fig. 5B).
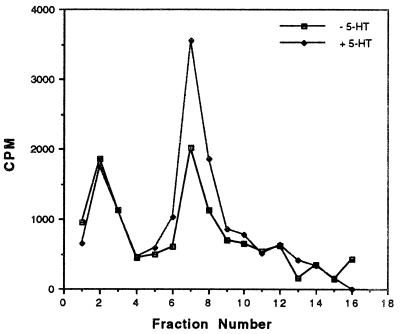
Finally, because the cytoplasmic domain of apCAM contains two consensus sites for phosphorylation by MAPK, we used a peptide containing one of these sites to probe for apMAPK activity before and after treatment of ganglia with 20 μM 5-HT for 30 min. Cell extracts were fractionated as described in Fig. 3 and eluted kinase activity was assayed using the apCAM peptide as substrate (Fig. 6). The first peak of activity eluted in fraction F2 did not contain apMAPK activity (in-gel kinase assay, data not shown) and was not modulated by 5-HT. However, the second peak of activity directed against apCAM eluted in fractions 7–10 contained more than 95% of apMAPK antigenicity (data not shown). This peak of activity was modulated by 5-HT, with an average increase of 73% ± 4.2% in three independent experiments. Thus, apCAM appears to be a good in vitro substrate for apMAPK. As reported by Bailey et al. (18), mutation of the consensus sites of phosphorylation by MAPK blocks internalization of apCAM. Together, these results suggest that apCAM phosphorylation is critical for internalization.
Figure 6.
Effect of 5-HT on the phosphorylation of an apCAM-derived peptide by fractionated MAPK. Kinase activity against apCAM peptides was determined in control extracts or extracts from 5-HT-treated ganglia. Paired pleural-pedal ganglia were incubated in the absence (− 5-HT) or presence of 20 μM 5-HT for 30 min and cell extracts were subjected to fractionation on anion-exchange FFQ minicolumns. Samples from the individual fractions were allowed to phosphorylate apCAM-derived peptide. This experiment was repeated (n = 3) and quantitation (mean percentage change in fractions F7 and fraction F8, combined) revealed that 5-HT caused an increase in kinase activity against apCAM by 73% ± 4% (SE) (P < 0.01).
DISCUSSION
We have cloned an Aplysia MAPK (apMAPK) and find it to be 85% identical to the human MAPK isoform ERK2. The activity of apMAPK is modulated by both 5-HT and cAMP in cultured sensory neurons of the gill-withdrawal reflex. In sensory cells, apMAPK activity is increased by five spaced applications of 5-HT, which produce long-term facilitation. An inhibitor of MEK, the upstream activator of MAPK, not only inhibits MAPK in vivo (Fig. 2C) but also blocks long-term but not short-term facilitation (22). This suggests that apMAPK may have a selective role in long-term facilitation. Consistent with this view, we find that a number of target proteins important for long-term facilitation, including apCAM, apCREB-2, and apC/EBP, are phosphorylated in vitro by apMAPK (Figs. 4–6).
The finding that elevations of cAMP produce long-term facilitation (45) led us to examine whether elevations of cAMP also activated MAPK. We find that exposure of cultured Aplysia sensory cells to forskolin increases MAPK activity (Fig. 3). How might the PKA and MAPK pathways converge in Aplysia sensory neurons? In the simplest case, the MAPK pathway would be downstream from the PKA pathway. In many cells, PKA negatively regulates the MAPK pathway by phosphorylating Raf-1 (19, 20). However, in B-Raf-containing cells, PKA activates the MAPK pathway by signaling through Rap1, a member of the Ras family of small G proteins (46). PKA may thus activate MAPK by phosphorylating the Aplysia homolog of Rap1, thereby activating B-Raf, MEK, and MAPK.
In addition to acting in series, MAPK might also act in parallel with PKA during long-term facilitation. The growth factors brain-derived growth factor (BDNF) and transforming growth factor β (TGF-β) have been found to produce long-term facilitation of Aplysia sensory-motor synapses (47, 48), raising the possibility that 5-HT treatment triggers release of endogenous growth factors, and that these growth factors act back on the sensory neurons through growth factor receptor kinases to activate MAPK.
As described by Martin et al. (22), MAPK is normally present in the cytoplasm, but translocates to the nucleus after repeated exposure to 5-HT. We thus tested critical nuclear and cytoplasmic molecules for their ability to be phosphorylated by apMAPK in vitro. The CREB-1 transcription factor has been shown to be necessary in Drosophila (49) and both necessary and sufficient in Aplysia (D. Bartsch and A. Casadio, personal communication) for the conversion of short- to long-lasting forms of memory. Because the MAPK pathway is critical for activating CREB-1 in many cell types (50, 51), it may contribute to long-lasting synaptic plasticity by modulating this activator of the long-term process. Similarly, the immediate early gene product C/EBP, which is required for the consolidation of short- to long-term facilitation in Aplysia (15), is phosphorylated by apMAPK in vitro (Fig. 5). Phosphorylation of apC/EBP by MAPK greatly enhances its ability to bind DNA (52). Thus, MAPK may contribute to long-term facilitation by promoting the ability of C/EBP to convert the short-term to the long-term process.
In contrast to the activators apC/EBP and CREB-1, the transcription factor CREB-2 appears to act as a repressor of long-term facilitation (13). While CREB-2 lacks PKA phosphorylation sites, it can be phosphorylated in vitro by apMAPK (Fig. 4). Thus, MAPK may also contribute to long-term facilitation by phosphorylating CREB-2 and thereby relieve its ability to inhibit the long-term process.
Finally, MAPK acts at the membrane to phosphorylate the cell adhesion molecule apCAM, which appears to act as a restraint on growth. Internalization of apCAM during long-term facilitation leads to defasciculation, which may be required for new synaptic growth (17, 23). As shown in Fig. 6, an apCAM peptide is targeted by a MAPK preparation from 5-HT-treated cells. As recently demonstrated by Bailey et al. (18), site-directed mutagenesis of the MAPK phosphorylation sites present in the cytoplasmic tail of apCAM prevents its internalization in response to 5-HT. Exposure to the MEK inhibitor PD098059 has a similar effect. Taken together, these data strongly suggest that apCAM phosphorylation by apMAPK in vivo triggers its internalization and thereby relieves a restraint on growth.
Interestingly, MAPK activity has been found to play a critical role in long-term potentiation in CA1 hippocampal cells (53). We have shown by immunocytochemistry that application of forskolin or SP-cAMPs activates MAPK in mouse hippocampus (22), and we have also found that forskolin increases MAPK activity in the hippocampus (data not shown). In many cell types, the MAPK pathway acting alone or in conjunction with PKA activates CREB-mediated transcription (50, 51), which is known to be a central component of synaptic plasticity in Aplysia, Drosophila, and mice. In fact, the MAPK pathway is commonly activated by growth factors and neurotransmitters (54–58). Taken together, the coordinated actions of PKA and MAPK may serve as a general signaling mechanism for long-term plasticity.
Acknowledgments
We thank Hiuxiang Zhu for Aplysia sensory cell cultures, Min Zhou for experiments on MAPK in rat hippocampus, Mark Barad for critical reading of the manuscript, and Harriet Ayers and Irma Trumpet for typing the manuscript. We thank Hong Sun for purified MKP1 and Alan Saltiel and Parke-Davis for PD098059.
ABBREVIATIONS
- PKA
cAMP-dependent protein kinase
- 5-HT
serotonin (5-hydroxytryptamine)
- MAPK
mitogen-activated protein kinase
- apMAPK
Aplysia MAPK
- apCAM
Aplysia cell adhesion molecule
- GST
glutathione S-transferase
- MBP
myelin basic protein
- CNS
central nervous system
- MEK
MAPK kinase
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. U40484).
References
- 1.Davis H P, Squire L R. Psychol Bull. 1984;96:518–559. [PubMed] [Google Scholar]
- 2.Goelet P, Castellucci V F, Schacher S, Kandel E R. Nature (London) 1986;322:419–422. doi: 10.1038/322419a0. [DOI] [PubMed] [Google Scholar]
- 3.Dezazzo J, Tully T. Trends Neurosci. 1995;18:212–218. doi: 10.1016/0166-2236(95)93905-d. [DOI] [PubMed] [Google Scholar]
- 4.Alberini C M, Ghirardi M, Huang Y-Y, Nguyen P, Kandel E R. Ann NY Acad Sci. 1995;758:261–286. doi: 10.1111/j.1749-6632.1995.tb24833.x. [DOI] [PubMed] [Google Scholar]
- 5.Carew T J. Neuron. 1996;16:5–8. doi: 10.1016/s0896-6273(00)80016-1. [DOI] [PubMed] [Google Scholar]
- 6.Bailey C H, Bartsch D, Kandel E R. Proc Natl Acad Sci USA. 1996;93:13445–13452. doi: 10.1073/pnas.93.24.13445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Montarolo P G, Goelet P, Castellucci V F, Morgan J, Kandel E R, Schacher S. Science. 1986;234:1249–1254. doi: 10.1126/science.3775383. [DOI] [PubMed] [Google Scholar]
- 8.Glanzman D L, Kandel E R, Schacher S. Science. 1990;249:799–802. doi: 10.1126/science.2389145. [DOI] [PubMed] [Google Scholar]
- 9.Frey U, Huang Y-Y, Kandel E R. Science. 1993;260:1661–1664. doi: 10.1126/science.8389057. [DOI] [PubMed] [Google Scholar]
- 10.Huang Y-Y, Kandel E R. Learning Memory. 1994;1:74–82. [PubMed] [Google Scholar]
- 11.Huang Y-Y, Li X-C, Kandel E R. Cell. 1994;79:69–79. doi: 10.1016/0092-8674(94)90401-4. [DOI] [PubMed] [Google Scholar]
- 12.Weisskopf M G, Castillo P E, Zalutsky R A, Nicoll R A. Science. 1994;265:1878–1882. doi: 10.1126/science.7916482. [DOI] [PubMed] [Google Scholar]
- 13.Bartsch D, Ghirardi M, Skehel P A, Karl K A, Herder S P, Chen M, Bailey C H, Kandel E R. Cell. 1995;83:979–992. doi: 10.1016/0092-8674(95)90213-9. [DOI] [PubMed] [Google Scholar]
- 14.Gonzalez F A, Raden D L, Davis R J. J Biol Chem. 1991;266:22159–22163. [PubMed] [Google Scholar]
- 15.Alberini C M, Ghirardi M, Metz R, Kandel E R. Cell. 1994;76:1099–1114. doi: 10.1016/0092-8674(94)90386-7. [DOI] [PubMed] [Google Scholar]
- 16.Nakajima T, Kinoshita S, Sasagawa T, Sasaki K, Naruto M, Kishimoto T, Akira S. Proc Natl Acad Sci USA. 1993;90:2207–2211. doi: 10.1073/pnas.90.6.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mayford M, Barzilai A, Keller F, Schacher S, Kandel E R. Science. 1992;256:638–644. doi: 10.1126/science.1585176. [DOI] [PubMed] [Google Scholar]
- 18.Bailey C H, Kaang B-K, Chen M, Martin K C, Lim C-S, Casadio A, Kandel E R. Neuron. 1997;18:913–924. doi: 10.1016/s0896-6273(00)80331-1. [DOI] [PubMed] [Google Scholar]
- 19.Cook S J, McCormick F. Science. 1993;262:1069–1072. doi: 10.1126/science.7694367. [DOI] [PubMed] [Google Scholar]
- 20.Wu J, Dent P, Jelinek T, Wolfman A, Weber M J, Sturgill T W. Science. 1993;262:1065–1068. doi: 10.1126/science.7694366. [DOI] [PubMed] [Google Scholar]
- 21.Marshall C J. Cell. 1995;80:179–185. doi: 10.1016/0092-8674(95)90401-8. [DOI] [PubMed] [Google Scholar]
- 22.Martin K C, Michael D, Rose J C, Barad M, Casadio A, Zhu H, Kandel E R. Neuron. 1997;18:899–912. doi: 10.1016/s0896-6273(00)80330-x. [DOI] [PubMed] [Google Scholar]
- 23.Bailey C H, Chen M, Keller F, Kandel E R. Science. 1992;256:645–649. doi: 10.1126/science.1585177. [DOI] [PubMed] [Google Scholar]
- 24.Gause K C, Homma M K, Licciardi K A, Seger R, Ahn N G, Peterson M J, Krebs E G, Meier K E. J Biol Chem. 1993;268:16124–16129. [PubMed] [Google Scholar]
- 25.Seger R, Seger D, Reszka A A, Munar E S, Eldar-Finkelman H, Dobrowolska G, Jensen A M, Campbell J S, Fischer E H, Krebs E G. J Biol Chem. 1994;269:25699–25709. [PubMed] [Google Scholar]
- 26.Frangioni J V, Neel B G. Anal Biochem. 1993;210:179–187. doi: 10.1006/abio.1993.1170. [DOI] [PubMed] [Google Scholar]
- 27.Ahn N G, Seger R, Bratlien R L, Diltz C D, Tonks N K, Krebs E G. J Biol Chem. 1991;266:4220–4227. [PubMed] [Google Scholar]
- 28.Seger R, Ahn N G, Boulton T G, Yancopoulos G D, Panayotatos N, Radziejewska E, Ericsson L, Bratlien R L, Cobb M H, Krebs E G. Proc Natl Acad Sci USA. 1991;88:6142–6146. doi: 10.1073/pnas.88.14.6142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kameshita I, Fujisawa H. Anal Biochem. 1989;183:139–143. doi: 10.1016/0003-2697(89)90181-4. [DOI] [PubMed] [Google Scholar]
- 30.Gotoh Y, Nishida E, Yamashita T, Hoshi M, Kawakami M, Sakai H. Eur J Biochem. 1970;193:661–669. doi: 10.1111/j.1432-1033.1990.tb19384.x. [DOI] [PubMed] [Google Scholar]
- 31.Errede B, Levin D E. Curr Opin Cell Biol. 1993;5:254–260. doi: 10.1016/0955-0674(93)90112-4. [DOI] [PubMed] [Google Scholar]
- 32.Hanks S K, Quinn A M, Hunter T. Science. 1988;241:42–52. doi: 10.1126/science.3291115. [DOI] [PubMed] [Google Scholar]
- 33.Boulton T G, Cobb M H. Cell Reg. 1991;2:357–371. doi: 10.1091/mbc.2.5.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Payne D M, Rossomando A J, Martino P, Erickson A K, Her J-H, Shabanowitz J, Hunt D F, Weber M J, Sturgill T W. EMBO J. 1991;10:885–892. doi: 10.1002/j.1460-2075.1991.tb08021.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Crews C M, Alessandrini A, Erikson R L. Science. 1992;258:477–480. doi: 10.1126/science.1411546. [DOI] [PubMed] [Google Scholar]
- 36.Seger R, Seger D, Lozeman F J, Ahn N G, Graves L M, Campbell J S, Ericsson L, Harrylock M, Jensen A M, Krebs E G. J Biol Chem. 1992;267:25628–25631. [PubMed] [Google Scholar]
- 37.Anderson N G, Kilgour E, Sturgill T W. J Biol Chem. 1991;266:10131–10135. [PubMed] [Google Scholar]
- 38.Crews C M, Alessandrini A, Erikson R L. Cell Growth Diff. 1992;3:135–142. [PubMed] [Google Scholar]
- 39.Sun H, Charles C H, Lau L F, Tonks N K. Cell. 1993;75:487–493. doi: 10.1016/0092-8674(93)90383-2. [DOI] [PubMed] [Google Scholar]
- 40.Qian Z, Gilbert M, Kandel E R. Learning Memory. 1994;1:180–188. [PubMed] [Google Scholar]
- 41.Pang L, Sawada T, Decker S J, Saltiel A R. J Biol Chem. 1995;270:13585–13588. doi: 10.1074/jbc.270.23.13585. [DOI] [PubMed] [Google Scholar]
- 42.Dudley D T, Pang L, Decker S J, Bridges A J, Saltiel A R. Proc Natl Acad Sci USA. 1995;92:7686–7689. doi: 10.1073/pnas.92.17.7686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alessi D R, Cuenda A, Cohen P, Dudley D T, Saltiel A R. J Biol Chem. 1995;270:27489–27494. doi: 10.1074/jbc.270.46.27489. [DOI] [PubMed] [Google Scholar]
- 44.Dash P K, Hochner B, Kandel E R. Nature (London) 1990;345:718–721. doi: 10.1038/345718a0. [DOI] [PubMed] [Google Scholar]
- 45.Schacher S, Castellucci V F, Kandel E R. Science. 1988;240:1667–1669. doi: 10.1126/science.2454509. [DOI] [PubMed] [Google Scholar]
- 46.Vossler M R, Yao H, York R D, Pan M-G, Rim C S, Stork P J S. Cell. 1997;88:73–82. doi: 10.1016/s0092-8674(00)80184-1. [DOI] [PubMed] [Google Scholar]
- 47.McKay S E, Carew T J. Soc Neurosci Abstr. 1996;22:695. [Google Scholar]
- 48.Zhang F, Endo S, Cleary L J, Eskin A, Byrne J H. Science. 1997;275:1318–1320. doi: 10.1126/science.275.5304.1318. [DOI] [PubMed] [Google Scholar]
- 49.Yin J C P, Del Vecchio M, Zhou H, Tully T. Cell. 1995;81:107–115. doi: 10.1016/0092-8674(95)90375-5. [DOI] [PubMed] [Google Scholar]
- 50.Nakajima T, Fukamizu A, Takahashi J, Gage F H, Fisher T, Blenis J, Montminy M R. Cell. 1996;86:465–474. doi: 10.1016/s0092-8674(00)80119-1. [DOI] [PubMed] [Google Scholar]
- 51.Xing J, Ginty D D, Greenberg M E. Science. 1996;273:959–963. doi: 10.1126/science.273.5277.959. [DOI] [PubMed] [Google Scholar]
- 52.Yamamoto N, Hegde A N, Chain D G, Schwartz J H. Soc Neurosci Abstr. 1996;22:1445. [Google Scholar]
- 53.English J D, Sweatt J D. J Biol Chem. 1997;272:19103–19106. doi: 10.1074/jbc.272.31.19103. [DOI] [PubMed] [Google Scholar]
- 54.Bading H, Greenberg M E. Science. 1991;253:912–914. doi: 10.1126/science.1715095. [DOI] [PubMed] [Google Scholar]
- 55.Frödin M, Feraldi P, Van Obberghen E. J Biol Chem. 1994;269:6207–6214. [PubMed] [Google Scholar]
- 56.Murphy T H, Blatter L A, Bhat R V, Fiore R S, Wier W G, Baraban J M. J Neurosci. 1994;14:1320–1331. doi: 10.1523/JNEUROSCI.14-03-01320.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rosen L B, Ginty D D, Weber M J, Greenberg M E. Neuron. 1994;12:1207–1221. doi: 10.1016/0896-6273(94)90438-3. [DOI] [PubMed] [Google Scholar]
- 58.Zhong Y. Nature (London) 1995;375:588–592. doi: 10.1038/375588a0. [DOI] [PubMed] [Google Scholar]