SUMMARY
Reactive oxygen species, generated either by cellular respiration or upon exposure to environmental agents such as ionizing radiation (IR), attack DNA to form a variety of oxidized base and sugar modifications. Accumulation of oxidative DNA damage has been associated with age-related disease as well as the aging process. Single-strand breaks harboring oxidative 3' obstructive termini, e.g. 3' phosphates and 3' phosphoglycolates, must be removed prior to DNA repair synthesis or ligation. In addition, 3' tyrosyl-linked protein damage, resulting from therapeutic agents such as camptothecin (CPT), must be processed to initiate repair. Several nucleases participate in DNA repair and the excision of 3' obstructive ends. As the protein defective in the segmental progeroid Werner syndrome (WRN) possesses 3' to 5' exonuclease activity, and Werner syndrome cells are hypersensitive to IR and CPT, we examined for WRN exonuclease activity on 3' blocking lesions. Moreover, we compared side-by-side the activity of four prominent human 3' to 5' exonucleases (WRN, APE1, TREX1, and p53) on substrates containing 3' phosphates, phosphoglycolates, and tyrosyl residues. Our studies reveal that while WRN degrades 3' hydroxyl containing substrates in a nonprocessive manner, it does not excise 3' phosphate, phosphoglycolate, or tyrosyl groups. In addition, we found that APE1 was most active at excising 3' blocking termini in comparison to the disease-related exonucleases TREX1, WRN, and p53 under identical physiological reaction conditions, and that TREX1 was the most powerful 3' to 5' exonuclease on undamaged oligonucleotide substrates.
Keywords: exonuclease, WRN, Werner syndrome, 3' damage repair
INTRODUCTION
DNA damage arises via many routes, including spontaneous decomposition, reactions with endogenous or exogenous agents, and as intermediates during normal cellular processes (Lindahl 1993). If unrepaired, such damage can promote genetic mutations or gross chromosomal rearrangements, which in turn can lead to cell death, human disease, and/or aging. To avert the cytotoxic or mutagenic effects of DNA damage, cells are equipped with DNA repair systems (Krokan et al. 2004;Sancar et al. 2004). Nucleases operate as central components of nearly all DNA repair processes, executing or initiating removal of the DNA damage to promote cell survival and ensure genetic integrity (Marti and Fleck 2004).
Reactive oxygen species are generated during normal cellular metabolism, or are induced upon exposure to certain environmental hazards (Lindahl 1993). These reactive chemicals, if not scavenged, can attack or modify any cellular constituent, including lipids, proteins, and nucleic acids. Accumulation of such damage has been suggested to be a major underlying mechanism of the aging process and several age-related diseases (Ames and Shigenaga 1992). Prominent forms of oxidative DNA damage include single-strand breaks harboring obstructive 3' termini such as phosphates and phosphoglycolates (Dizdaroglu et al. 2002). Moreover, these DNA strand breaks are major products of exposure to ionizing radiation (IR) (Ward 1988). Other 3' terminal damages include protein-DNA intermediates formed during DNA relaxation. Specifically, type IB topoisomerases, such as human topoisomerase I (topoI), cleave DNA to adjust supercoiled states, and in the process, form a transient covalent intermediate with the 3' terminus of the nucleic acid (Wang 2002). Certain oxidative damages, such as abasic sites, or therapeutic agents, such as camptothecin (CPT), can trap these protein-DNA intermediates, giving rise to stable 3'-tyrosyl-linked protein damage (Pommier et al. 2003;Connelly and Leach 2004). In each of the cases above, the 3' blocking end requires removal prior to DNA synthesis or ligation, and for the initiation of repair (Wilson, III et al. 2003).
In humans, several key 3' to 5' exonucleases have been identified and characterized to varying degrees. These include apurinic/apyrimidinic endonuclease 1 (APE1), three prime repair exonuclease (TREX1), the damage response tumor suppressor protein p53, and Werner syndrome protein (WRN). APE1 is the major AP endonuclease, but also contributes to the repair of 3'-phosphates, -phosphoglycolates, -tyrosyl groups, and -mismatched nucleotides (Demple and Harrison 1994;Winters et al. 1994;Suh et al. 1997;Chou and Cheng 2002;Hadi et al. 2002; Wilson, III 2003). TREX1, originally designated DNase III, was isolated as the major nuclear, DNA-specific 3' to 5' exonuclease (Hoss et al. 1999;Mazur and Perrino 1999;Mazur and Perrino 2001b). This protein displays sequence similarity to known DNA editing enzymes, and has been shown to exhibit a slight preference for 3' mismatched nucleotides, which can arise by misincorporation via a proofreading-deficient DNA polymerase (Mazur and Perrino 1999;Mazur and Perrino 2001b). Previous work reported that 3' phosphate and phosphoglycolate termini are resistant to TREX1 degradation (Inamdar et al. 2002b). Notably, recent work indicates that inactivating mutations in TREX1 give rise to the human neurological disorder Aicardi-Goutières syndrome (Crow et al. 2006). p53 is a tumor suppressor gene that has been found to be mutated in >50% of all human tumors (Oren 2003). Prior work demonstrated that p53 possesses an intrinsic Mg2+-dependent 3' to 5' exonuclease activity (Mummenbrauer et al. 1996). Subsequent studies have shown that p53 has preferential activity on 3' mismatched nucleotides, suggesting a proofreading role for this enzyme (Huang1998;Skalski et al. 2000;Bakhanashvili 2001). Its activity on 3' blocking termini has not been examined. WRN is a multifunctional enzyme defective in Werner syndrome (WS), a disease characterized by the premature appearance of age-associated phenotypes including cataracts, diabetes, osteoporosis, and some cancers. WRN operates as a 3' to 5' exonuclease, as well as a 3' to 5' helicase. It has been suggested that during certain situations, such as telomere maintenance, that these two functions may operate cooperatively (Opresko et al. 2001). Prior studies have suggested that WRN has the ability to excise 3' phosphate damages and has some preference for 3' mismatched nucleotides (Kamath-Loeb et al. 1998). The amino-terminal exonuclease domain of WRN shows similarity to the proofreading domain of E. coli DNA polymerase I (Mushegian et al. 1997).
In the work here, we have expanded the characterization of WRN exonuclease activity, examining specifically for degradation of substrates containing either a 3' phosphate, phosphoglycolate, or tyrosyl residue. In addition, we have more broadly defined the comparative substrate specificities of four major human 3' to 5' exonucleases (i.e. WRN, APE1, TREX1, and p53) and determined (side-by-side) their relative efficiencies under common, physiologically-relevant, reaction conditions. Our studies reveal that WRN is a non-processive exonuclease which degrades 3' hydroxyl containing substrates, but not 3' phosphate, phosphoglycolate, or tyrosyl-containing DNAs. In addition, under identical reaction platforms, we found that APE1 removed all 3' blocking termini, while TREX1, WRN, and p53 displayed limited or no exonuclease activity on these lesions. As discussed herein, our findings have novel implications for the biological functions of these prominent human exonucleases.
MATERIALS AND METHODS
Proteins and Oligonucleotides
Recombinant APE1 (Erzberger et al. 1998), WRN (Orren et al. 1999), and p53 (Sun et al. 2003) were purified as previously described. A human TREX1 protein fragment (spanning amino acids 1-242) was used in these studies (Supplemental Fig. 1). Prior analysis of purified full-length TREX1 by SDS-PAGE and by mass spectrometry (Mazur and Perrino 2001b) consistently revealed a major TREX1 fragment of 26,162 daltons, corresponding to the N-terminal 242 amino acids of the protein. Alignments of the TREX1 sequence with TREX2 indicated that the C-terminal residue of the 26,162 dalton fragment was positioned precisely at the C-terminal end of TREX2 (Mazur and Perrino 2001a;Perrino et al. 2005), indicating that a stable catalytic fragment of TREX1 was produced. To create this stable fragment, the complete TREX1 coding sequence (Mazur and Perrino 2001a) was first cloned into the pMYC vector (Perrino et al. 2004) to generate the expression construct pMYCT1 that encodes the fusion protein MBP-Intein(CBD)-TREX1. Next, a TAA stop codon was introduced into the TREX1 intein-based construct at position 243 via site-directed mutagenesis to create the TREX1P243Stop protein. Overexpression and purification of TREX1P243Stop was performed as described for TREX2 (Perrino, Krol, Harvey, Zheng, Horita, Hollis, Meyers, Isaacs, and Xu 2004). The 3' excision activities of the human TREX1P243Stop are similar to those described for the mouse Trex1 (Mazur and Perrino 2001b).
All oligonucleotides (Table 1) were purchased from Midland Certified Reagent Co. (Midland, TX), except 21-pg, which was obtained from Trevigen (Gaithersburg, MD). Substrate oligonucleotides were 5' end-labeled with [γ-32P]ATP and 3'-phosphatase-minus T4 polynucleotide kinase (PNK, Fermentas), and annealed to the respective unlabeled complementary strand.
TABLE 1.
Oligonucleotides used in this study
WRN exonuclease assays
For examining the processivity of WRN exonuclease activity, reactions were performed in a final volume of 10 μl containing Exo buffer (40 mM Tris, pH 8, 5 mM DTT, and 0.1 mg/ml BSA). WRN (100 fmol) was preincubated with the labeled substrate (10 fmol) on ice. MgCl2 (4 mM) and then unlabeled DNA trap (1000 fmol, Flap 10/34G) were added. The reactions were incubated at 37°C for the indicated times. An equal volume of formamide stop dye (80% formamide, 0.5X TBE, 0.1% bromophenol blue, and 0.1% xylene cyanol) was added to terminate the reactions. Following heat denaturation at 95°C for 5 min, products were run on a 20% denaturing polyacrylamide gel, and visualized using a PhosphorImager.
Exonuclease reactions containing WRN or APE1, and various recessed substrates containing 3' hydroxyl or 3' blocking lesions as indicated, were performed in Exo buffer containing 4 mM MgCl2 in a final volume of 10 μl. Samples were incubated for 15 min at 37°C and terminated by the addition of an equal volume of formamide stop dye. Reaction products were visualized as described above.
Comparative exonuclease assays
Exonuclease reactions (10 μl) containing 250 fmol of APE1, TREX1, WRN, or p53 were incubated in Exo-P buffer (25 mM MOPS, pH 7.2, 100 mM KCl, 1 mM MgCl2, 1 mM DTT, and 50 μg/ml BSA) in the presence of various recessed substrates (250 fmol) containing 3' hydroxyl or 3' blocking lesions as indicated. Samples were incubated for 15 min at 37°C and terminated by the addition of an equal volume of formamide stop dye. Reaction products were visualized as described above. Quantitation was performed using ImageQuant TL v2003 software (Amersham Biosciences Corp, Piscataway, NJ) and expressed relative to APE1 exonuclease activity for each substrate.
EXPERIMENTAL RESULTS
WRN is a non-processive exonuclease
To further characterize WRN 3' to 5' exonuclease activity, we examined the processivity of WRN degradation on recessed oligonucleotide substrates using a “trapping assay” similar to that previously published (Wilson, III 2003). Briefly, a radiolabeled 3' recessed substrate was incubated with WRN on ice. Subsequently, MgCl2 and a 100-fold excess (relative to labeled DNA) of unlabeled fork substrate (“trap”) were added, and WRN was assayed for exonuclease degradation. As shown in Figure 1, the presence of the trap inhibited progression of WRN exonuclease activity. Similar results were obtained with other 3' recessed substrates containing different primer/template lengths and sequence contexts (data not shown). Collectively, the data suggests that WRN is a non-processive enzyme, which apparently dissociates from DNA prior to significant nuclease degradation.
Fig. 1.
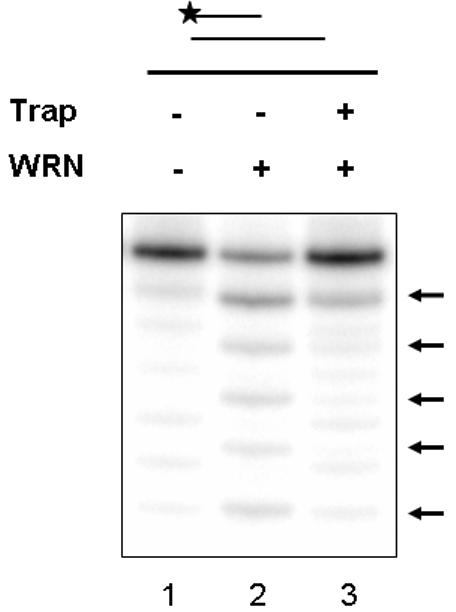
The WRN exonuclease is not processive.
A 32P-labeled 3' recessed substrate (21P/PG comp, 10 fmol) was incubated with WRN (100 fmol) in Exo buffer in the absence or presence of an unlabeled fork substrate (Flap10/34G, ie., “trap” 1000 fmol) for 10 min at 37°C. Following heat denaturation, samples were loaded on a 20% denaturing gel and visualized using a PhosphorImager. Arrows denote location of exonuclease degradation products.
WRN does not excise 3' phosphate, phosphoglycolate, or tyrosyl residues
Oxidative DNA damage, such as that induced by IR, can result in DNA strand breaks with 3' blocking groups, namely phosphates and phosphoglycolates (Henle and Linn 1997;Demple and DeMott 2002). As WS cells display elevated sensitivity to IR (Yannone et al. 2001;Saintigny et al. 2002), we aimed to further characterize WRN exonuclease activity on 3' blocking termini, including phosphates, using conditions previously established to be optimal for WRN degradation (Cooper et al. 2000;Opresko et al. 2001). To ensure that the 3' phosphate was not removed during the labeling reaction, a 3'-phosphatase-minus T4 PNK was utilized. At WRN concentrations in which we saw significant removal of nucleotides with 3' hydroxyl groups (Fig. 2A, lanes 4-5), we did not observe excision of the 3' phosphate ends (Fig. 2A, lanes 9-10). The small amount of apparent product observed in lanes containing WRN (Fig. 2A, lanes 9-10) likely reflects degradation of a small percentage of contaminating oligonucleotide that does not contain the 3' phosphate modification. Consistently, (i) this shorter product did not increase with increasing WRN concentration (suggestive of a limiting amount of degradable oligonucleotide), and (ii) we observed approximately 5% extension of the 3' phosphate-containing substrate by DNA polymerase β (pol β, Supplemental Fig. 2) indicative of the presence of 3' hydroxyl (primer) DNA. Furthermore, mass spectrometry analysis performed by the manufacturer indicated a similar percentage of non-phosphorylated species (Midland Certified Reagent Co., data not shown).
Fig. 2.
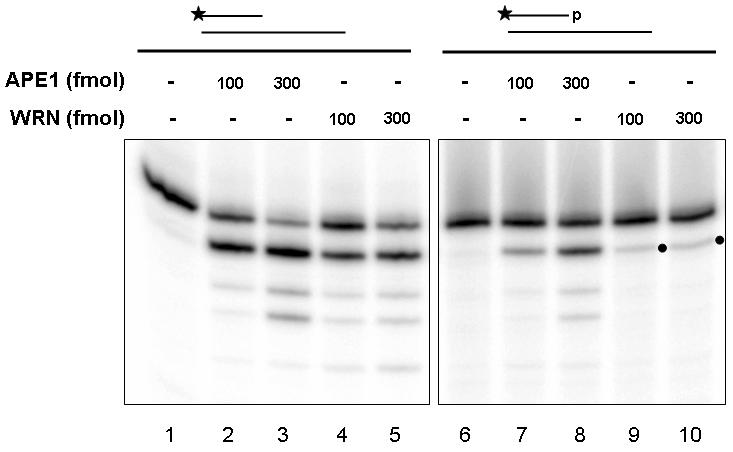
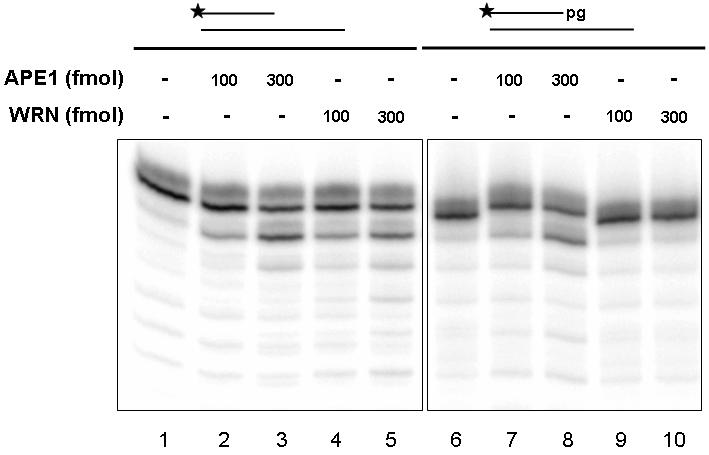
WRN does not remove 3' phosphates or 3' phosphoglycolates.
(A) A recessed DNA substrate (250 fmol) containing a 3' hydroxyl (15P/34G, lanes 1-5) or a 3' phosphate (15-p/34G, lanes 6-10) was incubated in the presence of WRN or APE1 in Exo buffer for 15 min at 37°C. Following heat denaturation at 90°C for 5 min, samples were loaded on a 20% denaturing gel and visualized using a PhosphorImager. The dot indicates position of a contaminant band. (B) A recessed DNA substrate (250 fmol) containing a 3' hydroxyl (21P/PG comp, lanes 1-5) or a 3' phosphoglycolate (21-pg/PG comp, lanes 6-10) was incubated in the presence of WRN or APE1 in Exo buffer for 15 min at 37°C. Reaction products were analyzed as described above.
We next explored WRN exonuclease activity on 3' phosphates in the context of a 1 nt gap, and similar to what was observed with 3' recessed substrates, we did not detect degradation of 3' phosphate-containing oligonucleotides by WRN (data not shown). Based on the inability of WRN to remove 3' phosphates under our assay conditions, we compared directly WRN activity to that of APE1, an enzyme known to excise 3' phosphates (Chen et al. 1991). In addition to exonuclease activity on 3' hydroxyl-containing substrates (Fig. 2A, lanes 2-3), APE1, unlike WRN, degraded the 3' phosphate-containing strand of the recessed primer-template duplex in an enzyme concentration dependent manner (Fig. 2A, lanes 7-8). In these experiments, since the 3' hydroxyl and 3' phosphate oligonucleotides migrated similarly, we were unable to determine if APE1 excised the phosphate residue prior to further exonuclease degradation.
We next examined WRN exonuclease activity on substrates harboring 3' phosphoglycolates. As shown in Figure 2B, using a primer/template substrate (21P or 21-pg/PG comp) that differed in sequence context from the one employed above (15P or 15-p/34G) (see Table 1), WRN was similarly found to degrade recessed DNAs with 3' hydroxyl groups (lanes 4-5), but not 3' phosphoglycolates (lanes 9-10). Conversely, the APE1 exonuclease was active on both 21-mer substrates, where in the case of the phosphoglycolate-containing DNA, the enzyme appears to first remove the phosphoglycolate residue (slower migrating bands), prior to more extensive degradation (Fig. 2B, lanes 2-3 and 7-8). As seen previously, the variation in APE1 activity between Fig. 2A and 2B likely reflects different degradation efficiency depending on nucleotide sequence context (Hadi et al. 2002). In conclusion, WRN did not excise 3' phosphates or 3' phosphoglycolates under presumably optimal reaction conditions, while APE1 proved to be a more powerful 3' repair enzyme in comparison.
Another 3' obstructive terminus can be produced when topo I cleavage reactions are aborted. The covalent topo I-DNA intermediates formed during chromosome relaxation are referred to as “cleavable complexes”, and are typically transient (Wang 1996). However, these cleavable complexes can be stabilized by chemotherapeutic agents, such as CPT, which specifically inhibit the topo I religation step (Chen and Liu 1994), or by the presence of existing nearby DNA damage (Pourquier and Pommier 2001). Since WS cells are sensitive to CPT (Pichierri et al. 2000), we examined the ability of WRN to excise 3' tyrosyl groups, the covalent intermediate formed between DNA and the topo I active site. Using a 3' recessed substrate, we detected WRN exonuclease activity on the 3' hydroxyl containing version of the substrate (Fig. 3, lane 3), but not on the substrate harboring a 3' tyrosyl residue (Fig. 3, lane 6). At the same concentration of protein, APE1 substantially degraded both 3' hydroxyl and 3' tyrosyl containing substrates (Fig. 3, lanes 2 and 5, respectively), as reported previously (Wilson, III 2003).
Fig. 3.
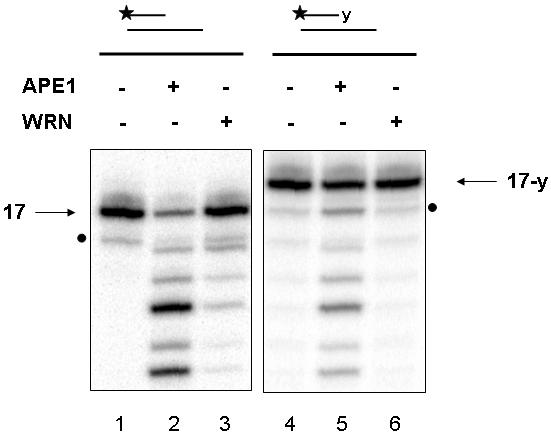
WRN does not excise 3' tyrosyl groups.
Reactions containing APE1 (500 fmol, lanes 2 and 5) or WRN (500 fmol, lanes 3 and 6) were incubated with a recessed DNA substrate (250 fmol) containing a 3' hydroxyl (17P/34G, lanes 1-3) or a 3' tyrosyl residue (17-y/34G, lanes 4-6) in Exo buffer for 15 min at 37°C. Following heat denaturation at 90°C for 5 min, samples were loaded on a 20% denaturing gel and visualized using a PhosphorImager. The 3' recessed primer is indicated. The dot indicates position of a contaminant band.
We next explored whether WRN exonuclease activity on 3' phosphates, phosphoglycolates, or tyrosyl groups could be stimulated by Ku. This heterodimer complex has been shown to stimulate WRN exonuclease activity on substrates it does not normally degrade alone, including several oxidatively modified DNA bases (Orren et al. 2001). However, while Ku stimulated WRN exonuclease activity on all recessed substrates containing 3' hydroxyl groups, we did not observe degradation of 3' phosphates, phosphoglycolates, or tyrosyls by WRN in the presence of Ku (data not shown).
Direct comparison of four human exonucleases
We wanted to directly compare the nuclease efficiencies of four prominent human exonucleases, i.e. WRN, APE1, TREX1, and p53, under the same experimental conditions using a “physiological” reaction buffer (i.e. neutral pH, a reducing environment, 100 mM K+, and 1 mM Mg2+). We employed here a TREX1 protein fragment (see Materials and Methods), as full-length protein proved to be unstable using many expression systems and production strategies. As shown in Figure 4 (lanes 1-5), TREX1 was the most active of the four exonucleases on a recessed substrate containing a 3' hydroxyl group (completely degrading the DNA substrate), even under reaction conditions which are not optimal for TREX1 exonuclease activity in vitro (Mazur and Perrino 2001b). APE1 also displayed robust exonuclease activity, whereas the exonuclease activity of WRN and p53 was essentially undetectable (Fig. 4, lanes 1-5). In contrast to the results observed with 3' hydroxyl groups, APE1 was the most active nuclease at degrading 3' phosphate-containing DNA substrates; TREX1 had some activity, although the percent degradation was roughly equivalent to the contaminating non-phosphate containing DNA (Fig. 4, lanes 6-10; see also Supplemental Fig. 2 for comparable 3' hydroxyl substrate). Neither WRN nor p53 digested duplexes harboring recessed 3' phosphates groups under these conditions (Fig. 4, lanes 6-10). A similar result was obtained with each of the exonucleases using other 3' hydroxyl-containing substrates (see for instance Supplemental Fig 3) and comparable DNAs containing either 3' phosphoglycolate (Fig. 4, lanes 11-15) or 3' tyrosyl blocking groups (Fig. 4, lanes 16-20). That is, under identical physiological reaction conditions, APE1 was considerably more active than TREX1, WRN, and p53 in digesting DNAs with 3' obstructive termini, whereas TREX1 was the most robust “natural” 3' to 5' exonuclease (see relative comparison summarized in Table 2).
Fig. 4.
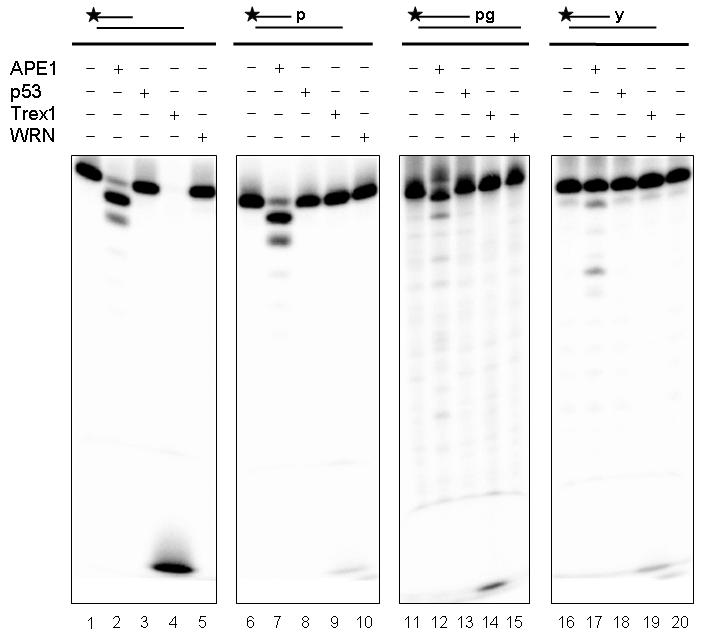
Comparative analysis of human exonucleases on 3' blocking termini.
Reactions containing 250 fmol of APE1, p53, TREX1, or WRN were incubated in Exo-P buffer with a recessed DNA substrate (250 fmol) containing a 3' hydroxyl (15P/34G, lanes 1-5), a 3' phosphate (15-p/34G, lanes 6-10), a 3' phosphoglycolate (21-pg/PG comp, lanes 11-15), or a 3' tyrosyl residue (17-y/34G, lanes 16-20) for 15 min at 37°C. Following heat denaturation at 90°C for 5 min, samples were loaded on a 18% denaturing gel and visualized using a PhosphorImager. See Supplemental Figure 3 for the assays with the comparable 3' hydroxyl primer/template substrates to the phosphoglycolate (21P/PG comp) and tyrosyl (17P/34G) containing DNAs.
TABLE 2.
Relative 3'-5' exonuclease activity under physiological buffer conditions
| 15/34 |
21/40 |
17/34 |
||||
|---|---|---|---|---|---|---|
| Protein | 3'-OH | 3'-p | 3'-OH | 3'-pg | 3'-OH | 3'-y |
| APE1 | 1 | 1 (0.93)a | 1 | 1 (0.89) | 1 | 1 (0.22) |
| p53 | UDb | UD | UD | UD | UD | UD |
| TREX1 | ≥1.1 | 0.02 | ≥1.3 | 0.14 | ≥1.6 | 0.21 |
| WRN | UD | UD | UD | UD | UD | UD |
Parentheses indicate APE1 activity relative to the 3'-OH containing substrate
UD, undetectable (i.e. <0.001). For each enzyme, digestion of the substrate was calculated using arbitrary PhosphorImager units. The amount of digestion by APE1 for each substrate was set to 1, and exonuclease activities of p53, TREX1, or WRN were expressed relative to APE1. The corresponding oligonucleotides are detailed in Table 1.
DISCUSSION AND CONCLUSIONS
Exonucleases are critical to the successful execution of nearly all DNA damage responses. In particular, these proteins recognize and excise specific forms of genetic damage, or minimally, initiate removal of a lesion-containing DNA strand. As damage excision is essential for the maintenance of genetic integrity, we examined further the 3' damage repair properties of the WRN 3' to 5' exonuclease, a protein defective in the segmental progeroid syndrome WS. Additionally, we compared simultaneously WRN exonuclease capacity on 3' damage-containing substrates to three other prominent human exonucleases, APE1, TREX1, and p53, under defined physiologically-relevant reaction parameters.
Under optimal reaction conditions for WRN (Exo buffer), we did not observe excision of 3' phosphates, phosphoglycolates, or tyrosyl residues, although we did observe significant degradation of substrates harboring 3' hydroxyl ends. These findings contrast the results of Kamath-Loeb et al., (Kamath-Loeb et al. 1998), who reported digestion of 3'-phosphate containing substrates by WRN alone. While the reason(s) for this discrepancy is not clear, one simple explanation could be the use of a phosphatase-free T4 PNK in the labeling reactions here. Regardless, our observations suggest that WRN does not contribute substantially to the repair of 3' blocking damages in vivo, and that the absence of this activity is not likely responsible for the mild IR sensitivity exhibited by WS cells. It is also noteworthy that overall WRN 3' to 5' exonuclease efficiency is reduced substantially in reaction conditions designed to mimic the in vivo environment (Exo-P buffer), suggesting that this activity may contribute in a limited manner to the pathophysiology of WS patients.
As with WRN, we did not observe detectable removal of any of the 3' blocking termini by p53. These results suggest that p53 does not play a significant role in the excision of 3' phosphates, phosphoglycolates, or tyrosyl groups. Furthermore, our data argue that p53 is a comparatively poor exonuclease (regardless of the 3' nucleotide composition), recognizing that p53 may preferentially degrade single-stranded DNA (Mummenbrauer et al. 1996;Huang 1998;Skalski et al. 2000;Bakhanashvili2001) or excise 3' mismatches (Huang 1998;Skalski et al. 2000;Bakhanashvili 2001). That withstanding, our results raise uncertainty about the biological significance of the 3' to 5' exonuclease function of p53 in cells, and support others' claims that p53 primarily operates as a transcriptional regulator (Hainaut1995;Schuler and Green 2005).
Consistent with the overall findings of others (Perrino et al. 1994;Inamdar et al. 2002b), TREX1 was found within to, at best, marginally degrade oligonucleotides harboring 3' phosphate or 3' phosphoglycolate groups. Similarly, TREX1 exhibited poor degradation activity on 3' tyrosyl-containing substrates. Thus, while TREX1 displays a robust 3' to 5' exonuclease activity on several 3' hydroxyl-containing substrates, the enzyme appears to play a minor role in the repair of 3' blocking lesions in vivo. In support, Trex1−/− MEFs do not show increased sensitivity to IR (Morita et al. 2004). We note that TREX1 was recently identified as a component of the SET complex (Chowdhury et al. 2006), which is important in the granzyme A activated caspase-independent cell death pathway, and has been causally linked to the human disease Aicardi-Goutières syndrome, characterized by severe neurological brain disease and an aberrant immune response (Crow et al. 2006). Significantly, abrogation of TREX1 exonuclease activity appears to be responsible for disease manifestation.
In contrast to the weak or non-detectable exonuclease activity of WRN, p53, and TREX1 on 3' blocking termini, we found that under identical physiologically-relevant reaction conditions, APE1 displayed substantial degradation activity on each of the three lesion-containing substrates examined. Our findings are consistent with the reports of others showing that APE1 is able to remove 3' phosphates (Demple et al. 1991;Seki et al. 1992;Winters et al. 1994), 3' phosphoglycolates (Suh et al. 1997), and 3' tyrosyl groups (Wilson, III 2003), importantly, under a wide variety of assay conditions. Thus, although the in vitro 3' to 5' exonuclease activity of APE1 is approximately 300-fold lower than its AP endonuclease activity, the data in total argue that APE1 is a likely candidate to remove 3' phosphates, phosphoglycolates, and tyrosyl residues during relevant DNA repair processes, particularly in comparison to the other human exonucleases examined here. Nonetheless, substantial biochemical and cellular evidence indicates that PNK is the predominant 3'-DNA phosphatase in vivo (Karimi-Busheri et al. 1998; Wiederhold et al. 2004).
In contrast to 3' phosphates, APE1 was identified as possessing the major activity for the removal of 3' phosphoglycolates in human cell extracts when positioned within a single-strand break (Parsons et al. 2004). Specifically, the major 3' phosphoglycolate activity co-purified with APE1 and immunodepletion of APE1 resulted in complete loss of 3' phosphoglycolate removal (Parsons et al. 2004). While APE1 has been shown to remove phosphoglycolates from blunt and recessed 3' ends (Suh et al. 1997), removal of phosphoglycolates from 3' protruding ends was refractory to both APE1 and PNK. However, tyrosyl-DNA phosphodiesterase (TDP1) is capable of removing such lesions (Inamdar et al. 2002a). Furthermore, extracts from patients with spinocerebellar ataxia with axonal neuropathy (SCAN1), containing a homozygous mutation in the active site of TDP1, were not capable of processing protruding 3' phosphoglycolates (Zhou et al. 2005). Thus, APE1 is likely the major repair enzyme for single-strand breaks harboring 3' phosphoglycolates, whereas TDP1, in cooperation with the 3' phosphatase activity of PNK, functions to remove phosphoglycolates from DNA double-strand break ends. TDP1 is also likely the major activity for the removal of 3' tyrosyl residues in vivo (Yang et al. 1996;Interthal et al. 2005;El-Khamisy et al. 2005), although APE1 may excise these lesions under certain circumstances as evidenced by its significant activity on 3' tyrosyls in vitro and its central role in single-strand break repair in vivo.
In closing, the importance of removing 3' blocking lesions during DNA repair events, i.e. for subsequent DNA synthesis and religation, is evident by the multiple mechanisms employed by cells to maintain genome integrity. In particular, there appears to be primary enzymes responsible for the removal of each specific 3' blocking termini, yet in their absence, a back-up activity can often compensate. In addition, microenvironments within a cell or certain DNA configurations may favor the involvement of one enzyme over another. We have demonstrated that the disease-associated human exonucleases TREX1, WRN, and p53 exhibit little to no activity on 3' damage-containing DNA substrates, whereas APE1 is a comparatively robust 3' repair enzyme. The studies herein (summarized in Table 2 and discussed above) shed novel insights into the relative contributions of four primary 3' to 5' human exonucleases in the repair of 3' phosphate, phosphoglycolate, and tyrosyl residues, as well as the degradation of undamaged DNA.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Drs. Mohammad Hedayati and Meltem Muftuoglu (NIA) for critical reading of the manuscript. This research was supported by the Intramural Research Program of the NIH, NIA, and by the NIH grant GM069962 (FWP).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ames BN, Shigenaga MK. Oxidants are a major contributor to aging. Ann N Y Acad Sci. 1992;663:85–96. doi: 10.1111/j.1749-6632.1992.tb38652.x. [DOI] [PubMed] [Google Scholar]
- Bakhanashvili M. Exonucleolytic proofreading by p53 protein. European Journal of Biochemistry. 2001;268:2047–2054. doi: 10.1046/j.1432-1327.2001.02075.x. [DOI] [PubMed] [Google Scholar]
- Chen AY, Liu LF. DNA topoisomerases: essential enzymes and lethal targets. Annu.Rev.Pharmacol.Toxicol. 1994;34:191–218. doi: 10.1146/annurev.pa.34.040194.001203. [DOI] [PubMed] [Google Scholar]
- Chen DS, Herman T, Demple B. Two distinct human DNA diesterases that hydrolyze 3′-blocking deoxyribose fragments from oxidized DNA. Nucleic Acids Res. 1991;19:5907–5914. doi: 10.1093/nar/19.21.5907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou KM, Cheng YC. An exonucleolytic activity of human apurinic/apyrimidinic endonuclease on 3′ mispaired DNA. Nature. 2002;415:655–659. doi: 10.1038/415655a. [DOI] [PubMed] [Google Scholar]
- Chowdhury D, Beresford PJ, Zhu P, Zhang D, Sung JS, Demple B, Perrino FW, Lieberman J. The exonuclease TREX1 is in the SET complex and acts in concert with NM23-H1 to degrade DNA during granzyme A-mediated cell death. Mol.Cell. 2006;23:133–142. doi: 10.1016/j.molcel.2006.06.005. [DOI] [PubMed] [Google Scholar]
- Connelly JC, Leach DR. Repair of DNA covalently linked to protein. Mol.Cell. 2004;13:307–316. doi: 10.1016/s1097-2765(04)00056-5. [DOI] [PubMed] [Google Scholar]
- Cooper MP, Machwe A, Orren DK, Brosh RM, Ramsden D, Bohr VA. Ku complex interacts with and stimulates the Werner protein. Genes Dev. 2000;14:907–912. [PMC free article] [PubMed] [Google Scholar]
- Crow YJ, Hayward BE, Parmar R, Robins P, Leitch A, Ali M, Black DN, van BH, Brunner HG, Hamel BC, Corry PC, Cowan FM, Frints SG, Klepper J, Livingston JH, Lynch SA, Massey RF, Meritet JF, Michaud JL, Ponsot G, Voit T, Lebon P, Bonthron DT, Jackson AP, Barnes DE, Lindahl T. Mutations in the gene encoding the 3′-5′ DNA exonuclease TREX1 cause Aicardi-Goutieres syndrome at the AGS1 locus. Nat.Genet. 2006 doi: 10.1038/ng1845. [DOI] [PubMed] [Google Scholar]
- Demple B, DeMott MS. Dynamics and diversions in base excision DNA repair of oxidized abasic lesions. Oncogene. 2002;21:8926–8934. doi: 10.1038/sj.onc.1206178. [DOI] [PubMed] [Google Scholar]
- Demple B, Harrison L. Repair of oxidative damage to DNA: Enzymology and biology. Annu Rev Biochem. 1994;63:915–948. doi: 10.1146/annurev.bi.63.070194.004411. [DOI] [PubMed] [Google Scholar]
- Demple B, Herman T, Chen DS. Cloning and expression of APE, the cDNA encoding the major human apurinic endonuclease: definition of a family of DNA repair enzymes. Proc.Natl.Acad.Sci.U.S.A. 1991;88:11450–11454. doi: 10.1073/pnas.88.24.11450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dizdaroglu M, Jaruga P, Birincioglu M, Rodriguez H. Free radical-induced damage to DNA: mechanisms and measurement. Free Radic.Biol.Med. 2002;32:1102–1115. doi: 10.1016/s0891-5849(02)00826-2. [DOI] [PubMed] [Google Scholar]
- El-Khamisy SF, Saifi GM, Weinfeld M, Johansson F, Helleday T, Lupski JR, Caldecott KW. Defective DNA single-strand break repair in spinocerebellar ataxia with axonal neuropathy-1. Nature. 2005;434:108–113. doi: 10.1038/nature03314. [DOI] [PubMed] [Google Scholar]
- Erzberger JP, Barsky D, Scharer OD, Colvin ME, Wilson DM., III Elements in abasic site recognition by the major human and Escherichia coli apurinic/apyrimidinic endonucleases. Nucleic Acids Res. 1998;26:2771–2778. doi: 10.1093/nar/26.11.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadi MZ, Ginalski K, Nguyen LH, Wilson DM., III Determinants in nuclease specificity of Ape1 and Ape2, human homologues of Escherichia coli exonuclease III. J.Mol.Biol. 2002;316:853–866. doi: 10.1006/jmbi.2001.5382. [DOI] [PubMed] [Google Scholar]
- Hainaut P. The tumor suppressor protein p53: a receptor to genotoxic stress that controls cell growth and survival. Curr.Opin.Oncol. 1995;7:76–82. [PubMed] [Google Scholar]
- Henle ES, Linn S. Formation, prevention, and repair of DNA damage by Iron/hydrogen peroxide. J.Biol.Chem. 1997;272:19095–19098. doi: 10.1074/jbc.272.31.19095. [DOI] [PubMed] [Google Scholar]
- Hoss M, Robins P, Naven TJ, Pappin DJ, Sgouros J, Lindahl T. A human DNA editing enzyme homologous to the Escherichia coli DnaQ/MutD protein. EMBO J. 1999;18:3868–3875. doi: 10.1093/emboj/18.13.3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P. Excision of mismatched nucleotides from DNA: a potential mechanism for enhancing DNA replication fidelity by the wild-type p53 protein. Oncogene. 1998;17:261–270. doi: 10.1038/sj.onc.1201946. [DOI] [PubMed] [Google Scholar]
- Inamdar KV, Pouliot JJ, Zhou T, Lees-Miller SP, Rasouli-Nia A, Povirk LF. Conversion of phosphoglycolate to phosphate termini on 3′ overhangs of DNA double strand breaks by the human tyrosyl-DNA phosphodiesterase hTdp1. J.Biol.Chem. 2002a;277:27162–27168. doi: 10.1074/jbc.M204688200. [DOI] [PubMed] [Google Scholar]
- Inamdar KV, Yu Y, Povirk LF. Resistance of 3′-phosphoglycolate DNA ends to digestion by mammalian DNase III. Radiat.Res. 2002b;157:306–311. doi: 10.1667/0033-7587(2002)157[0306:ropdet]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Interthal H, Chen HJ, Kehl-Fie TE, Zotzmann J, Leppard JB, Champoux JJ. SCAN1 mutant Tdp1 accumulates the enzyme--DNA intermediate and causes camptothecin hypersensitivity. EMBO J. 2005;24:2224–2233. doi: 10.1038/sj.emboj.7600694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath-Loeb AS, Shen JC, Loeb LA, Fry M. Werner Syndrome Protein. Ii. characterization of the integral 3′ --> 5′ dna exonuclease. J.Biol.Chem. 1998;273:34145–34150. doi: 10.1074/jbc.273.51.34145. [DOI] [PubMed] [Google Scholar]
- Karimi-Busheri F, Lee J, Tomkinson AE, Weinfeld M. Repair of DNA strand gaps and nicks containing 3′-phosphate and 5′-hydroxyl termini by purified mammalian enzymes. Nucleic Acids Res. 1998;26:4395–4400. doi: 10.1093/nar/26.19.4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krokan HE, Kavli B, Slupphaug G. Novel aspects of macromolecular repair and relationship to human disease. J.Mol.Med. 2004;82:280–297. doi: 10.1007/s00109-004-0528-1. [DOI] [PubMed] [Google Scholar]
- Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- Marti TM, Fleck O. DNA repair nucleases. Cell Mol.Life Sci. 2004;61:336–354. doi: 10.1007/s00018-003-3223-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazur DJ, Perrino FW. Structure and expression of the TREX1 and TREX2 3′ --> 5′ exonuclease genes. J.Biol.Chem. 2001a;276:14718–14727. doi: 10.1074/jbc.M010051200. [DOI] [PubMed] [Google Scholar]
- Mazur DJ, Perrino FW. Excision of 3′ termini by the Trex1 and TREX2 3′-->5′ exonucleases. Characterization of the recombinant proteins. J.Biol.Chem. 2001b;276:17022–17029. doi: 10.1074/jbc.M100623200. [DOI] [PubMed] [Google Scholar]
- Mazur DJ, Perrino FW. Identification and expression of the TREX1 and TREX2 cDNA sequences encoding mammalian 3′-->5′ exonucleases. J.Biol.Chem. 1999;274:19655–19660. doi: 10.1074/jbc.274.28.19655. [DOI] [PubMed] [Google Scholar]
- Morita M, Stamp G, Robins P, Dulic A, Rosewell I, Hrivnak G, Daly G, Lindahl T, Barnes DE. Gene-targeted mice lacking the Trex1 (DNase III) 3′-->5′ DNA exonuclease develop inflammatory myocarditis. Mol.Cell Biol. 2004;24:6719–6727. doi: 10.1128/MCB.24.15.6719-6727.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mummenbrauer T, Janus F, Muller B, Wiesmuller L, Deppert W, Grosse F. p53 Protein exhibits 3′-to-5′ exonuclease activity. Cell. 1996;85:1089–1099. doi: 10.1016/s0092-8674(00)81309-4. [DOI] [PubMed] [Google Scholar]
- Mushegian AR, Bassett DE, Jr., Boguski MS, Bork P, Koonin EV. Positionally cloned human disease genes: patterns of evolutionary conservation and functional motifs. Proc.Natl.Acad.Sci.U.S.A. 1997;94:5831–5836. doi: 10.1073/pnas.94.11.5831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opresko PL, Laine JP, Brosh RM, Jr., Seidman MM, Bohr VA. Coordinate Action of the Helicase and 3′ to 5′ Exonuclease of Werner Syndrome Protein. J.Biol.Chem. 2001;276:44677–44687. doi: 10.1074/jbc.M107548200. [DOI] [PubMed] [Google Scholar]
- Oren M. Decision making by p53: life, death and cancer. Cell Death.Differ. 2003;10:431–442. doi: 10.1038/sj.cdd.4401183. [DOI] [PubMed] [Google Scholar]
- Orren DK, Brosh RM, Jr, Nehlin JO, Machwe A, Gray MD, Bohr VA. Enzymatic and DNA binding properties of purified WRN protein: high affinity binding to single-stranded DNA but not to DNA damage induced by 4NQO. Nucleic Acids Res. 1999;27:3557–3566. doi: 10.1093/nar/27.17.3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orren DK, Machwe A, Karmakar P, Piotrowski J, Cooper MP, Bohr VA. A functional interaction of Ku with Werner exonuclease facilitates digestion of damaged DNA. Nucleic Acids Res. 2001;29:1926–1934. doi: 10.1093/nar/29.9.1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons JL, Dianova II, Dianov GL. APE1 is the major 3′-phosphoglycolate activity in human cell extracts. Nucleic Acids Res. 2004;32:3531–3536. doi: 10.1093/nar/gkh676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrino FW, Harvey S, McMillin S, Hollis T. The human TREX2 3′ -> 5′-exonuclease structure suggests a mechanism for efficient nonprocessive DNA catalysis. J.Biol.Chem. 2005;280:15212–15218. doi: 10.1074/jbc.M500108200. [DOI] [PubMed] [Google Scholar]
- Perrino FW, Krol A, Harvey S, Zheng SL, Horita DA, Hollis T, Meyers DA, Isaacs WB, Xu J. Sequence variants in the 3′-->5′ deoxyribonuclease TREX2: identification in a genetic screen and effects on catalysis by the recombinant proteins. Adv.Enzyme Regul. 2004;44:37–49. doi: 10.1016/j.advenzreg.2003.11.010. [DOI] [PubMed] [Google Scholar]
- Perrino FW, Miller H, Ealey KA. Identification of a 3′-->5′-exonuclease that removes cytosine arabinoside monophosphate from 3′ termini of DNA. J.Biol.Chem. 1994;269:16357–16363. [PubMed] [Google Scholar]
- Pichierri P, Franchitto A, Mosesso P, Palitti F. Werner's syndrome cell lines are hypersensitive to camptothecin-induced chromosomal damage. Mutat.Res. 2000;456:45–57. doi: 10.1016/s0027-5107(00)00109-3. [DOI] [PubMed] [Google Scholar]
- Pommier Y, Redon C, Rao VA, Seiler JA, Sordet O, Takemura H, Antony S, Meng L, Liao Z, Kohlhagen G, Zhang H, Kohn KW. Repair of and checkpoint response to topoisomerase I-mediated DNA damage. Mutat.Res. 2003;532:173–203. doi: 10.1016/j.mrfmmm.2003.08.016. [DOI] [PubMed] [Google Scholar]
- Pourquier P, Pommier Y. Topoisomerase I-mediated DNA damage. Adv.Cancer Res. 2001;80:189–216. doi: 10.1016/s0065-230x(01)80016-6. [DOI] [PubMed] [Google Scholar]
- Saintigny Y, Makienko K, Swanson C, Emond MJ, Monnat RJ., Jr. Homologous recombination resolution defect in werner syndrome. Mol.Cell Biol. 2002;22:6971–6978. doi: 10.1128/MCB.22.20.6971-6978.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu.Rev.Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- Schuler M, Green DR. Transcription, apoptosis and p53: catch-22. Trends Genet. 2005;21:182–187. doi: 10.1016/j.tig.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Seki S, Hatsushika M, Watanabe S, Akiyama K, Nagao K, Tsutsui K. cDNA cloning, sequencing, expression and possible domain structure of human APEX nuclease homologous to Escherichia coli exonuclease III. Biochim.Biophys.Acta. 1992;1131:287–299. doi: 10.1016/0167-4781(92)90027-w. [DOI] [PubMed] [Google Scholar]
- Skalski V, Lin ZY, Choi BY, Brown KR. Substrate specificity of the p53-associated 3′-5′ exonuclease. Oncogene. 2000;19:3321–3329. doi: 10.1038/sj.onc.1203649. [DOI] [PubMed] [Google Scholar]
- Suh D, Wilson DM, Povirk LF. 3′-phosphodiesterase activity of human apurinic/apyrimidinic endonuclease at DNA double-strand break ends. Nucleic Acids Res. 1997;25:2495–2500. doi: 10.1093/nar/25.12.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun XZ, Nguyen J, Momand J. Purification of recombinant p53 from Sf9 insect cells. Methods Mol.Biol. 2003;234:17–28. doi: 10.1385/1-59259-408-5:17. [DOI] [PubMed] [Google Scholar]
- Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nat.Rev.Mol.Cell Biol. 2002;3:430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- Wang JC. DNA topoisomerases. Annu.Rev.Biochem. 1996;65:635–692. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- Ward JF. DNA damage produced by ionizing radiation in mammalian cells: identities, mechanisms of formation, and reparability. Prog.Nucleic Acid Res.Mol.Biol. 1988;35:95–125. doi: 10.1016/s0079-6603(08)60611-x. [DOI] [PubMed] [Google Scholar]
- Wiederhold L, Leppard JB, Kedar P, Karimi-Busheri F, Rasouli-Nia A, Weinfeld M, Tomkinson AE, Izumi T, Prasad R, Wilson SH, Mitra S, Hazra TK. AP endonuclease-independent DNA base excision repair in human cells. Mol.Cell. 2004;15:209–220. doi: 10.1016/j.molcel.2004.06.003. [DOI] [PubMed] [Google Scholar]
- Wilson DM., III Properties of and substrate determinants for the exonuclease activity of human apurinic endonuclease Ape1. J.Mol.Biol. 2003;330:1027–1037. doi: 10.1016/s0022-2836(03)00712-5. [DOI] [PubMed] [Google Scholar]
- Wilson DM, III, Sofinowski TM, McNeill DR. Repair mechanisms for oxidative DNA damage. Front Biosci. 2003;8:d963–d981. doi: 10.2741/1109. [DOI] [PubMed] [Google Scholar]
- Winters TA, Henner WD, Russell PS, McCullough A, Jorgensen TJ. Removal of 3′-phosphoglycolate from DNA strand-break damage in an oligonucleotide substrate by recombinant human apurinic/apyrimidinic endonuclease 1. Nucleic Acids Res. 1994;22:1866–1873. doi: 10.1093/nar/22.10.1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SW, Burgin AB, Jr., Huizenga BN, Robertson CA, Yao KC, Nash HA. A eukaryotic enzyme that can disjoin dead-end covalent complexes between DNA and type I topoisomerases. Proc Natl Acad Sci USA. 1996;93:11534–11539. doi: 10.1073/pnas.93.21.11534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yannone SM, Roy S, Chan DW, Murphy MB, Huang S, Campisi J, Chen DJ. Werner syndrome protein is regulated and phosphorylated by dna- dependent protein kinase. J.Biol.Chem. 2001;276:38242–38248. doi: 10.1074/jbc.M101913200. [DOI] [PubMed] [Google Scholar]
- Zhou T, Lee JW, Tatavarthi H, Lupski JR, Valerie K, Povirk LF. Deficiency in 3′-phosphoglycolate processing in human cells with a hereditary mutation in tyrosyl-DNA phosphodiesterase (TDP1) Nucleic Acids Res. 2005;33:289–297. doi: 10.1093/nar/gki170. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


