Abstract
The RNA component of the signal recognition particle (SRP) is universally required for cotranslational protein targeting. Biochemical studies have shown that SRP RNA participates in the central step of protein targeting by catalyzing the interaction of the SRP with the SRP receptor (SR). SRP RNA also accelerates GTP hydrolysis in the SRP·SR complex once formed. Using a reverse-genetic and biochemical analysis, we identified mutations in the E. coli SRP protein, Ffh, that abrogate the activity of the SRP RNA and cause corresponding targeting defects in vivo. The mutations in Ffh that disrupt SRP RNA activity map to regions that undergo dramatic conformational changes during the targeting reaction, suggesting that the activity of the SRP RNA is linked to the major conformational changes in the signal sequence-binding subunit of the SRP. In this way, the SRP RNA may coordinate the interaction of the SRP and the SR with ribosome recruitment and transfer to the translocon, explaining why the SRP RNA is an indispensable component of the protein targeting machinery.
INTRODUCTION
Cotranslational protein targeting is a major route by which proteins are targeted to the membrane of the endoplasmic reticulum (or plasma membrane in prokaryotes). The machinery required for cotranslational protein targeting consists of the signal recognition particle (SRP), a protein/RNA complex that binds ribosomes translating secretory and membrane proteins, and the SRP receptor (SR), which resides at the membrane and binds to the SRP (Keenan et al., 2001). One of the highly conserved features of the targeting machinery is that SRP requires an RNA subunit to function (Walter and Blobel, 1982). This study elaborates the mechanism by which the SRP RNA subunit contributes to the protein targeting reaction.
In the first step of cotranslational protein targeting, the SRP binds to the signal sequence of a nascent polypeptide chain emerging from the ribosome (Walter et al., 1981; Keenan et al., 2001; Halic et al., 2004). The resulting SRP-ribosome-nascent chain complex then binds to the SRP receptor (SR), which resides at the target membrane (Gilmore et al., 1982a,b). The SRP and the SR associate with each other through related GTPase modules, but only when GTP-bound (Miller et al., 1993; Egea et al., 2004; Focia et al., 2004). After transfer of the ribosome to the protein translocation channel (translocon), the SRP and the SR hydrolyze their respective bound GTPs, which causes them to dissociate, and allows for a new round of targeting (Connolly et al., 1991). The SRP and the SR reciprocally activate the other's GTPase and are therefore GTPase-activating proteins (GAPs) for each other (Powers and Walter, 1995).
Although SRP-dependent protein targeting is conserved in all organisms, the prokaryotic system has the fewest components and is therefore the simplest (Poritz et al., 1990; Larsen and Zwieb, 1993). In Escherichia coli, the SRP consists of a single protein, Ffh, and a small RNA, the 4.5S RNA (Poritz et al., 1990). Ffh consists of two domains: the M domain, which contains both the binding site for signal sequences and the 4.5S RNA, and the NG domain, which includes the GTPase module that binds the SRP receptor FtsY. A flexible linker joins the two domains of Ffh (Keenan et al., 1998; Egea et al., 2005). In E. coli, Ffh, FtsY, and the 4.5S RNA are all essential genes.
The SRP RNA is an almost universally conserved component of the SRP targeting system. The only known exception is chloroplast SRP, which lacks an SRP RNA but also functions in a different, posttranslational mode by binding to proteins entering the chloroplast from the cytosol and targeting them to the thylakoid membrane. In E. coli, the 4.5S RNA has two known biochemical activities: to catalyze the interaction of Ffh and FtsY, accelerating both the on and off rate of complex formation by over two orders of magnitude without changing the KD, and to enhance the GTPase activity of the Ffh·FtsY complex by about sevenfold (this RNA-dependent stimulation of GTPase activity is distinct from the much larger GTPase stimulation caused directly by the interaction of Ffh and FtsY; Peluso et al., 2000, 2001). Thus, the 4.5S RNA represents an amazing example of an RNA that modulates the behavior of proteins. However, the relationship between the biochemically defined activities of the 4.5S RNA and the universal requirement for an RNA in cotranslational protein targeting is unknown.
A clue to the role of the 4.5S RNA comes from structural work, which shows that the signal sequence-binding pocket (a deep groove lined by hydrophobic amino acids) is spatially contiguous with the 4.5S RNA (Keenan et al., 1998; Batey et al., 2000). Moreover, the hydrophobic groove can assume multiple conformations, indicating that its open state may not be stable in an aqueous environment but closes so that hydrophobic residues can pack against each other and thus be shielded from water (Rosendal et al., 2003). Because this conformational variability occurs in close juxtaposition to the 4.5S RNA, it is plausible that the RNA might monitor the occupancy of the signal sequence-binding pocket and transmit that information to the NG domain, thereby modulating its interaction with FtsY. However, it has remained unresolved how the 4.5S RNA might be linked to these conformational dynamics.
Solution studies looking at the conformation of Ffh suggested that in the absence of the 4.5S RNA, the M domain may occlude the binding site for FtsY. 4.5S RNA binding to Ffh may relieve this inhibition to promote complex formation (Buskiewicz et al., 2005a,b; Halic et al., 2006a,b; Mainprize et al., 2006; Schaffitzel et al., 2006). Although this model provides a plausible mechanism for 4.5S RNA activity, it would suggest that the role of 4.5S RNA is limited to mediate a single Ffh activation step during SRP assembly. Once bound, it would be an inert bystander that would not contribute actively to regulation of the SRP cycle, as there is currently no evidence that 4.5S RNA dissociates from Ffh after binding.
Here, we describe a reverse-genetic and biochemical analysis designed to ask if and how the 4.5S RNA facilitates communication between the M and NG domains of Ffh. We describe mutants in Ffh that bind 4.5S RNA normally and interact normally with FtsY when the 4.5S RNA is absent, but impair the rate of association or GTP hydrolysis when the 4.5S RNA is present. These mutations map to regions of Ffh that undergo major conformational rearrangements during the targeting cycle, suggesting that the 4.5S RNA coordinates the steps of the targeting reaction.
MATERIALS AND METHODS
Reagents
Proteins and 4.5S RNA were purified as described in Peluso et al. (2001). Mutations in Ffh were introduced using the QuickChange Mutagenesis protocol (Stratagene, La Jolla, CA). For all in vitro experiments, a truncated form of FtsY (aa 47–497) was used, as previously described (Powers and Walter, 1997). In all cases, assays were performed at 25°C in 50 mM HEPES, pH 7.5, 150 mM potassium acetate, 2 mM magnesium acetate, 0.01% Nikkol, and 2 mM dithiothreitol. The NG domain of Ffh was generated by limited digestion as described (Zopf et al., 1993). The M domain was removed by flowing the digested protein over SP Sepharose, and the NG domain was further purified by gel filtration on Superdex 75.
Fluorescence-binding Assays
Fluorescence-binding experiments were performed as described (Peluso et al., 2000). Rapid reactions [wild type (wt) and Ffh(L301P) + RNA] were performed on a stopped flow fluorimeter (KinTek). Slow reactions were performed on an SLM fluorimeter. For on rates, data were fit to a single exponential and observed rate constants were plotted as a function of concentration. Rate constants were calculated using the following equation: kobs = kon ∗ [Ffh] + koff. Off rates were calculated by preforming complexes of 2 μM of each Ffh (±4.5S RNA) and FtsY and then trapping dissociated components with an excess of GDP. Curves were fit to a single exponential function.
GTPase Assays
Assays were performed as described (Peluso et al., 2001) with slight modifications. To calculate the basal activities of Ffh mutants, trace amounts of [32P]GTP were added to varying concentrations of Ffh and reactions were followed to completion. The data were fit to a single exponential equation to calculate the kobs. In contrast, we used a multiple-turnover regime to measure the stimulated GTPase activity of the Ffh variants. A fixed concentration of Ffh (0.1 μM for Ffh(wt) + 4.5S RNA and other fast reactions and 0.5 μM for Ffh(wt) − 4.5S RNA and other slow reactions) was used with varying concentrations of FtsY. The initial linear portion of the reaction was followed, corrected for the contribution of basal hydrolysis from FtsY (<20% of total hydrolysis observed), and fit to the following equation: turnovers/complex = kobs ∗ time.
Gel Shift Assays
Gel shift analysis of binding of the 4.5S RNA to Ffh mutants was carried out by mixing trace amounts of 32P-end–labeled 4.5S RNA with 0.25 μM cold 4.5S RNA and 0.25 μM Ffh (cold RNA was important to help prevent aggregation of Ffh and RNA in the well) in assay buffer supplemented with 10% glycerol. This mixture was separated in TAE buffer (40 mM Tris-acetate, 20 mM sodium acetate, and 1 mM EDTA) supplemented with 2.5 mM magnesium acetate on a 7% (29:1 acrylamide:bisacrylamide) polyacrylamide gel.
Biotinylation Experiments
Wam121 cells [MC4100 ara+ ffh::kan attB::(OriR6K PBAD-ffh tet); Phillips and Silhavy, 1992] were transformed with plasmid pHP44 (pBR322-acrR′acrA acrB576-PSBT; Tian and Beckwith, 2002) and pHDB7 (pACYC184-Ffh; Lee and Bernstein, 2001) or variants in which point mutations were introduced. Transformants were selected on plates in the presence of arabinose and were then grown overnight at 37°C in liquid medium in the absence of arabinose. Cells were then diluted back and harvested during log phase as described (Tian and Beckwith, 2002). Western blots were performed using streptavidin-horseradish peroxidase (HRP) conjugate (Amersham). HRP was inactivated with azide, and the blots were reprobed with an antibody to Ffh (Poritz et al., 1990) and visualized using chemiluminescence.
RESULTS
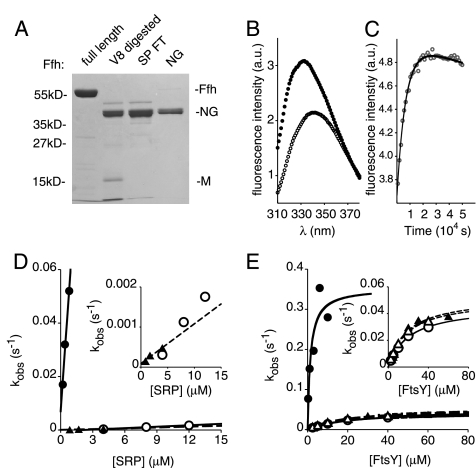
To assess how 4.5S RNA promotes Ffh·FtsY complex formation, we first asked whether the Ffh M domain inhibits the rate of binding of Ffh and FtsY. It has been proposed that 4.5S RNA may enhance the rate of complex formation by relieving this putative inhibition (Buskiewicz et al., 2005a,b). To test this possibility, we generated the NG domain of Ffh (Ffh-NG) by removal of the M domain by V8 protease digestion (Figure 1A). Next, we monitored the kinetics of complex formation by two independent assays: 1) measuring the change in intrinsic tryptophan fluorescence of FtsY (Jagath et al., 2000; Peluso et al., 2000) and 2) measuring the stimulation of GTP hydrolysis (Peluso et al., 2001). In both assays, Ffh-NG behaved as full-length Ffh in the absence of 4.5S RNA and by contrast to full-length Ffh could not be accelerated by addition of the RNA (Figure 1, B–E).
Figure 1.
Removal of the M domain does not alter the interaction kinetics of Ffh and FtsY in the absence of the 4.5S RNA. (A) The NG domain was severed from Ffh by limited proteolysis with V8 protease and purified as described in Materials and Methods. A Coomassie blue–stained SDS polyacrylamide gel displaying selected fractions from the purification procedure is shown. The lane labeled SP FT contains the flow through fraction from an SP Sepharose column, which binds to the liberated M domain, and the lane labeled NG contains purified Ffh-NG after gel filtration. This fraction was used in the subsequent assays. (B) Binding of the purified Ffh-NG fragment to FtsY produces a change of the tryptophan fluorescence of FtsY. •, the intensity of fluorescence from the complex of Ffh-NG and FtsY when excited with 290-nm light; ○, the fluorescence spectrum of unbound Ffh-NG and FtsY. Complex formation was initiated by addition of Mg2+ in excess over EDTA as previously described (Shan and Walter, 2003). (C) Representative data monitoring the increase in tryptophan fluorescence intensity as a function of time. The time course shown was obtained at 0.85 μM Ffh-NG and 0.1 μM FtsY and no 4.5S RNA. Data points represent intensity measurements taken when the sample was excited at 290 nm and emission was measured at 340 nm. (D) Ffh-NG binds to FtsY with rates indistinguishable from full-length Ffh in the absence of the 4.5S RNA. Observed rate constants from data such as in C are plotted as a function of concentration of Ffh (•: full-length Ffh + 4.5S RNA; ○: full-length Ffh − 4.5S RNA; ▴: Ffh NG − 4.5S RNA). The inset graph is magnified to show the slow reactions. (E) Stimulated GTP hydrolysis reactions for Ffh·FtsY and Ffh-NG·FtsY. Hydrolysis rates (per complex, per second) are plotted as a function of concentration of FtsY (•: full-length Ffh + 4.5S RNA, ○: full-length Ffh − 4.5S RNA, ▴: Ffh NG − 4.5S RNA, ▵: Ffh NG + 4.5S RNA).
As shown in Figure 1B, binding of Ffh-NG to FtsY caused an increase in fluorescence intensity and blue shift of the fluorescence emission peak similar to full-length Ffh (Jagath et al., 2000; Peluso et al., 2000). We used this fluorescence shift to measure binding of Ffh-NG to FtsY. The rate of Ffh-NG·FtsY complex formation (kon = 99 M−1s−1; Figure 1, C and D) was within error from that of full-length Ffh in the absence of the 4.5S RNA (kon = 180 M−1s−1) and over 500 times slower than full-length Ffh in the presence of the SRP RNA (kon = 5.7 × 104 M−1s−1; Peluso et al., 2000).
Monitoring stimulation of GTP hydrolysis after Ffh-NG·FtsY complex formation yielded similar results. The stimulated GTPase reaction measures the entire interaction cycle of Ffh and FtsY. At low concentrations of FtsY the GTP hydrolysis rate is primarily dependent on the rate of binding of Ffh and FtsY, whereas at higher concentrations the catalytic step of GTP hydrolysis becomes rate limiting (Peluso et al., 2001). Thus, the kmax is a direct measure of the catalytic rate, and the K1/2 depends on both the binding rate and the kmax. As the 4.5S RNA affects both the binding rate and the rate of catalysis, we observe an increase in the kmax as well as a decrease in the K1/2 upon addition of the 4.5S RNA (Figure 1E; cf. • and ○). When we compared the stimulated GTPase activity of Ffh-NG to that of full-length Ffh, we saw that both the kmax and the K1/2 were nearly identical to that measured for full-length Ffh in the absence of the 4.5S RNA (Figure 1E, Table 1). Furthermore, addition of 4.5S RNA in 10-fold excess of the 0.5 μM KD for the 4.5S RNA and Ffh-NG Buskiewicz et al., 2005a) had no affect on either parameter of the GTPase reaction for Ffh-NG. These results indicate that the Ffh M domain is required for 4.5S RNA to accelerate Ffh·FtsY complex formation. 4.5S RNA bound to the M domain therefore actively enhances the rate of complex formation, rather than the RNA-free M domain slowing it down.
Table 1.
Stimulated GTPase activity of Ffh variants in complex with FtsY
| kmax (s−1) |
K1/2 (μM) |
|||
|---|---|---|---|---|
| +4.5S RNA | −4.5S RNA | +4.5S RNA | −4.5S RNA | |
| Ffh(wt) | 0.354 | 0.034 | 1.39 | 16.3 |
| Ffh(NG) | 0.053 | 0.040 | 18.66 | 15.2 |
| Ffh(L301P) | 0.077 | 0.041 | 0.34 | 11.6 |
| Ffh(L303D) | 0.087 | 0.045 | 10.99 | 17.2 |
| Ffh(L350D) | 0.187 | 0.034 | 23.98 | 15.5 |
| Ffh(L354D) | 0.199 | 0.033 | 29.02 | 12.5 |
We next asked whether the M domain merely provides a binding site for the 4.5S RNA or if it actively participates in the Ffh·FtsY interaction. To answer this question, we sought mutations in Ffh that abrogate 4.5S RNA activity, without impairing 4.5S RNA binding to Ffh. We chose conserved residues in the M domain and in the linker that tethers the M and NG domains and changed these residues by site-directed mutagenesis. We then expressed and purified each of the Ffh mutants and assayed their ability to bind the 4.5S RNA and their basal GTPase activity. Mutants that behaved indistinguishably from wild-type Ffh were further tested for their ability to form a complex with FtsY and stimulate GTP hydrolysis in the complex.
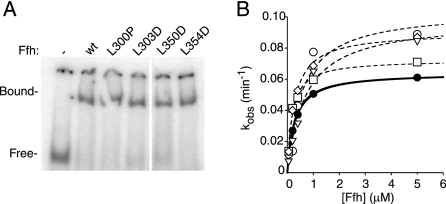
Through this analysis, we identified four point mutations in Ffh (L301P, L303D, L350D, and L354D) that impair the activity of 4.5S RNA in interesting ways. All four Ffh mutants bind to the 4.5S RNA (Figure 2A) and hydrolyze GTP in a manner indistinguishable from wild-type Ffh (the measurement refers to basal GTPase in the absence of FtsY; Figure 2B).
Figure 2.
Binding to 4.5S RNA and basal GTPase activity is not affected by mutations L301P, L303D, L350D, and L354D. (A) Gel shift analysis using 0.25 μM Ffh and 0.25 μM 32P-labeled 4.5S RNA shows that all mutant variants quantitatively shift the RNA. (B) Single-turnover GTP hydrolysis assays of wild-type (•), Ffh(L301P) (○), Ffh(L303D) (□), Ffh(L350D) (◇), and Ffh(L354D) (▿). Reactions were performed with trace [32P]GTP and varying amounts of Ffh. Curves (solid line Ffh(wt), dashed lines for Ffh mutants) were fit to the equation kobs = kcat ∗ [Ffh]/(KM + [Ffh]). Values of kcat are as follows: Ffh(wt), 0.070 min−1; Ffh(L301P), 0.077 min−1; Ffh(L303D), 0.092 min−1; Ffh(L350D), 0.10 min−1; and Ffh(L354D), 0.10 min−1.
Ffh Mutations L303D, L350D, and L354D Impair 4.5S RNA-catalyzed Ffh·FtsY Complex Formation
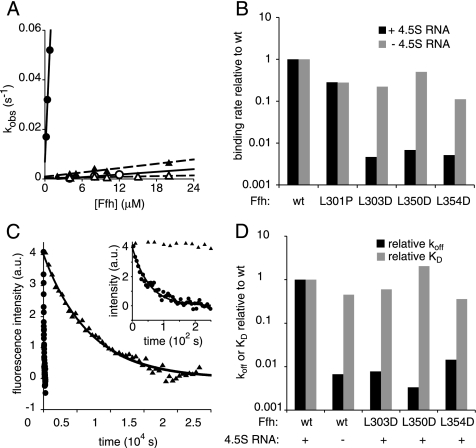
Three of the mutations (L303D, L350D, and L354D) led to dramatically reduced rates of Ffh·FtsY complex formation in the presence of the 4.5S RNA (Figure 3, A and B, Table 2). Compared with wild-type Ffh bound to 4.5S RNA, complex formation was slowed in each case by more than 100-fold (Figure 3B, ■). By contrast, the binding rates were only modestly affected in the absence of the 4.5S RNA (a 2- to 10-fold decrease; Figure 3B, ■).
Figure 3.
Ffh mutations of L303D, L350D, and L354D abrogate the activity of the 4.5S RNA to catalyze association of Ffh and FtsY. (A) Observed rate constants for binding of wild-type Ffh (•, ○) or Ffh(L303D) (▴, ▵) in the presence (filled symbols) or absence (open symbols) of 4.5S RNA are plotted as a function of Ffh concentration. Lines represent fits to the equation kobs = kon ∗ [Ffh] + koff [solid lines for wt Ffh, dashed lines for Ffh(L303D)]. (B) The mutations selectively affect the binding rate in the presence but not absence of the 4.5S RNA. The binding rates relative to wt Ffh are plotted (note log-scaled Y axis). (C) Dissociation of wild-type (•) or Ffh(L303D) (▴) from FtsY was measured in the presence of the 4.5S RNA by adding GDP to trap dissociated complexes and monitored by changes in tryptophan fluorescence. Samples were excited at 290 nm, and fluorescence emission at 340 nm was recorded. The x-axis in the inset is expanded to show the curve for wild-type Ffh. Data were fit to a single exponential equation to calculate the koff. (D) Plot of the koff or KD of Ffh mutants relative to wild-type Ffh in the presence of the 4.5S RNA. kon, koff, and KD values are summarized in Table 2.
Table 2.
Ffh·FtsY complex formation in the presence of the 4.5S RNA
| kon (M−1s−1) |
koff (s−1) | KDa (nM) | ||
|---|---|---|---|---|
| +4.5S RNA | −4.5S RNA | +4.5S RNA | +4.5S RNA | |
| Ffh(wt) | 57,000 | 180 | 0.0018 | 33 |
| Ffh(L301P) | 17,000 | 47 | n.d. | n.d. |
| Ffh(L303D) | 280 | 43 | 0.000014 | 20 |
| Ffh(L350D) | 410 | 99 | 0.000006 | 68 |
| Ffh(L354D) | 310 | 21 | 0.000026 | 120 |
a KD values are calculated from the ratio of the koff and the kon. Equilibrium binding experiments independently confirmed showed that the affinity of the interaction of Ffh and FtsY was unaffected by the mutations in Ffh (data not shown). n.d., not determined.
Because the KD of the wild-type Ffh·FtsY complex is the same in the presence or absence of 4.5S RNA (Peluso et al., 2000), mutations that affect the activity of the 4.5S RNA should affect the on and off rates of complex formation to the same degree. To test this prediction, we determined the off rates of the Ffh mutants in the presence of the 4.5S RNA by following tryptophan fluorescence after addition of GDP to trap Ffh and FtsY in their dissociated, GDP-bound states. As shown in Figure 3, C and D, all three Ffh mutants bound to 4.5S RNA showed dissociation rates comparable to those of complexes containing wild-type Ffh lacking 4.5S RNA (Figure 3D, ■). In summary, these data demonstrate that the mutations L303D, L350D, and L354D abrogate the activity of 4.5S RNA to catalyze complex formation between Ffh and FtsY but, importantly, do not impair the affinity with which Ffh and FtsY interact (Figure 3D, KDs ▩).
Ffh Mutations L301P and L303D Diminish the Rate of Stimulated GTP Hydrolysis in the Ffh·FtsY Complex
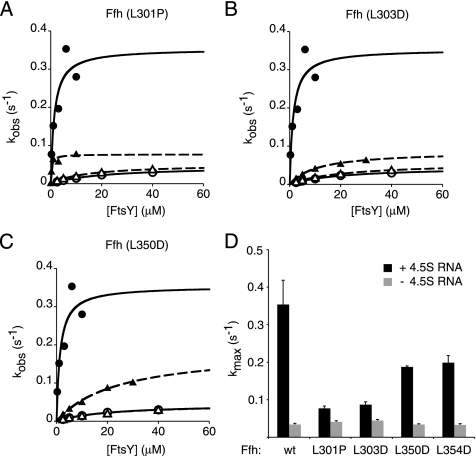
In addition to catalyzing Ffh·FtsY complex formation, the 4.5S RNA also stimulates the rate of GTP hydrolysis by the complex (kmax), albeit to a lesser extent (Peluso et al., 2001). All four Ffh mutants analyzed here showed some reduction of RNA stimulation of GTPase activity by the complex. Ffh(L350D) and Ffh(L354D) had less than a twofold reduction of maximal GTP hydrolysis activity in the presence of the 4.5S RNA (Figure 4, C and D, Table 1). The Ffh(L303D) and Ffh(L301P)·FtsY complex showed kmax levels that were reduced to near the level of the wild-type Ffh·FtsY complex lacking 4.5S RNA (Figure 4, A, B, and D, Table 1). In contrast to the other three mutants, Ffh(L301P) binds to FtsY with normal rates (Figure 3B and Table 2). Although the 4.5S RNA stimulation of Ffh·FtsY GTPase activity is small, the effects of the mutations were consistent across at least three independent experiments and two independent preparations of protein for each Ffh variant.
Figure 4.
Mutations L301P and L303D abrogate the activity of the 4.5S RNA to enhance the stimulated GTPase activity of the SRP and FtsY. Multiple turnover GTP hydrolysis reactions were carried out in which wild-type Ffh (•) or Ffh mutants (▴) were mixed with varying concentrations of FtsY in the presence (filled symbols) or absence (open symbols) of the 4.5S RNA. The Ffh mutants are shown in (A) Ffh(L301P), (B) Ffh(L303D), and (C) Ffh(L350D). Solid lines are curve fits to reactions containing wild-type Ffh and dashed lines to reactions containing Ffh mutants. (D) Plot of kmax in the presence and absence of 4.5S RNA. The values for kmax and K1/2 are summarized in Table 1. Error bars represent the accuracy of the fits to the data.
Taken together, our data show that the L301P mutation primarily impairs the activity of the 4.5S RNA to stimulate the GTPase activity of the complex, the L350D and L354D mutations primarily impair the activity of the 4.5S RNA to stimulate the rate of complex formation and GTP hydrolysis, and the L303D mutation impairs both.
Mutations That Impair Either Activity of the 4.5S RNA Lead to In Vivo Protein Targeting Defects
Previously it was unknown how the biochemical activities of the 4.5S RNA related to the cellular function of the 4.5S RNA in cotranslational protein targeting. Having identified Ffh mutants that specifically impair the activities of the 4.5S RNA, we were able to assess the importance of these activities for protein targeting in vivo without directly perturbing the 4.5S RNA. To do this, we expressed the Ffh mutants in cells that harbor a reporter for cotranslational protein targeting and conditionally express wild-type Ffh.
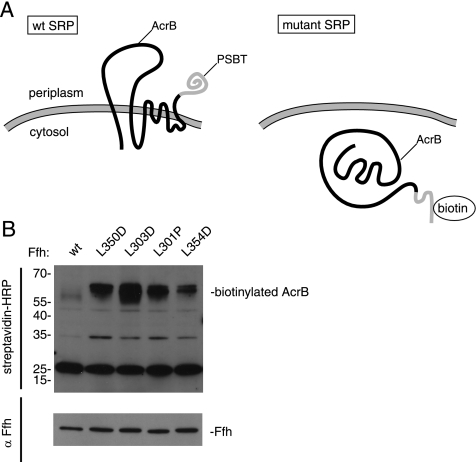
To monitor cotranslational protein targeting, we used the elegant system developed by Tian and Beckwith (2002) in which the multispanning membrane protein, AcrB is fused to the Proprionibacterium shermanii transcarboxylase (PSBT) biotinylation domain. When this fusion protein is targeted to the membrane by SRP, the PSBT is targeted to the periplasm, where it is not biotinylated (Figure 5A). However, if the SRP-targeting system is defective, the protein accumulates in the cytoplasm and is biotinylated. We introduced plasmids to direct the expression of either wild-type Ffh or Ffh bearing the L301P, L303D, L350D, and L354D mutations into E. coli cells harboring the AcrB-PSBT fusion and in which the sole genomic copy of Ffh was conditionally expressed by the presence of arabinose (Phillips and Silhavy, 1992).
Figure 5.
Ffh mutations L301P, L303D, L350D, and L354D show membrane protein integration defects in vivo. (A) The multispanning transmembrane protein AcrB fused to the PSBT biotinylation domain is efficiently targeted and integrated in cells expressing wild-type SRP, locating the PSBT biotinylation domain to the periplasmic space, where it is not biotinylated. If SRP-dependent protein targeting is impaired, AcrB accumulates in the cytoplasm, where it is biotinylated. (B) E. coli cells containing a genomic deletion of Ffh and a genomically inserted copy of wt Ffh that is exclusively expressed in the presence of arabinose and bearing two plasmids: one containing wild-type or mutant Ffh, and the other containing AcrB fused to the PSBT biotinylation domain were grown in the absence of arabinose. Cell extracts were fractionated by SDS polyacrylamide electrophoresis, and gels were blotted using a streptavidin-HRP conjugate (top panel) or anti-Ffh antibodies followed by HRP-conjugated secondary antibodies (bottom panel). HRP was visualized by chemiluminescence.
Cells were grown to midlog phase in media lacking arabinose to shut off genomic Ffh expression. Cells were harvested, and extracts were electrophoresed and blotted for biotin using streptavidin-HRP. We found significant accumulation of biotinylated AcrB in all of the strains expressing mutant Ffh but not in control cells expressing wild-type Ffh (Figure 5B, top). The most dramatic accumulation of biotinylated AcrB was seen in cells expressing Ffh(L303D), which in vitro showed defects in both complex formation and stimulated GTPase activity. When the AcrB fusion protein was expressed, all four strains bearing mutant Ffh-expressing plasmids grew significantly slower than wild-type controls (data not shown). These results are consistent with previous studies, which demonstrate that SRP targeting may be significantly impaired without substantially affecting cell growth (Ulbrandt et al., 1997). Reduced levels of active SRP in combination with overexpression of an SRP substrate such as AcrB, however, can lead to pronounced synthetically toxic effects (Ulbrandt et al., 1997; Bernstein and Hyndman, 2001). Western blotting of cell extracts with an Ffh-specific antibody showed that all Ffh variants were expressed to comparable levels (Figure 5B, bottom).
Taken together, these data indicate that both biochemical activities of the 4.5S RNA—stimulating Ffh·FtsY complex formation and stimulating GTPase activity in the complex—are critical for protein targeting in vivo.
DISCUSSION
The 4.5S RNA accelerates the interaction of Ffh with FtsY and the GTPase activities of the two proteins in the complex. In this study, we have shown that the slow rates of complex formation and GTP hydrolysis that are observed in the absence of the 4.5S RNA are recapitulated with Ffh lacking the signal sequence-binding M domain, ruling out the possibility that the 4.5S RNA relieves an inhibition imposed by the RNA-free M domain. This finding led us to ask whether the 4.5S RNA requires specific features of Ffh beyond the binding site for its activity. We discovered point mutations in conserved Ffh residues that selectively abolish or diminish the catalytic effects of the 4.5S RNA on complex formation and on the simulated GTPase activity in the complex. These results demonstrate that the activity of the 4.5S RNA is intimately linked to features of the M domain and the linker that joins it to the NG domain, which interacts with the SR. Moreover, we show that mutations perturbing either activity of the 4.5S RNA significantly reduce the efficiency of the SRP-dependent protein targeting system.
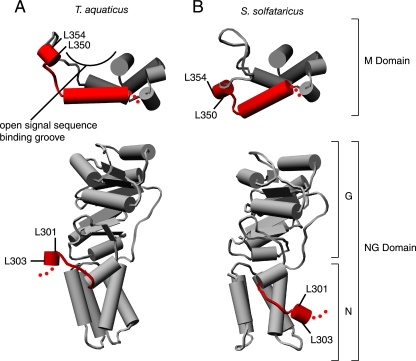
A striking feature of the mutations characterized here is that all four are found in positions of Ffh that are conformationally dynamic, based on comparison of available crystal structures (Figure 6). In particular, L350 and L354 are both part of a short helix in the finger loop of the M domain. In one crystal structure of the M domain (Thermus aquaticus; Keenan et al., 1998; Figure 6A, top), the residues line the top of one side of the signal sequence-binding groove. By contrast, in another crystal structure (Sulfolobus solfataricus; Rosendal et al., 2003), Figure 6B, top) the residues point away from the signal sequence-binding pocket. It is interesting to note that in the Keenan structure, two adjacent M domains in the crystal lattice intertwine, such that hydrophobic residues from one M domain partially occupy the signal sequence-binding groove of the other. Thus, the conformational differences in the two M domain structures may represent a change in conformation from a closed state (T. aquaticus structure) to an open state (S. solfataricus structure) that occurs when the signal sequence-binding pocket becomes occupied. This view suggests that the activity of the SRP RNA may be dependent on the occupancy of the signal sequence-binding groove. For example, binding of the SRP to a signal sequence may enhance the activity of the SRP RNA, giving cargo bound SRP a kinetic advantage to interact with FtsY.
Figure 6.
Mutations in Ffh that lead to defects in 4.5S RNA activity map to conformationally dynamic regions. Comparison of x-ray crystal structures from T. aquaticus (A) and S. solfataricus (B) reveals that the linker between the NG and M domains (bottom) and the finger loop of the M domain (top) are mobile. The corresponding amino acid positions of the mutations in E. coli Ffh described in this article are indicated: L301 (298 in T. aquaticus and 298 in S. solfataricus) and L303 (300,300) in the linker and L350(341,348) and L354(345,352) lead to defects in 4.5S RNA activity. Molecular graphics images were produced using the UCSF Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIH P41 RR-01081) (Pettersen et al., 2004).
Similarly, the other two mutated residues, L301 and L303, are part of a small helix, which together with the loop that connects it to the NG domain, are flipped across the surface of the compactly folded NG domain in the two structures (Figure 6, A and B, bottom). Conformational flexibility is also observed in single-particle reconstructions using cryo-electron microscopy techniques when the SRP is bound to the ribosome. In the absence of the SRP receptor, all of SRP54 (the metazoan ortholog of Ffh) could be fitted into defined density, indicating that the ribosome locks the M and NG domains into a fixed conformation relative to each other. By contrast, when SRP receptor is added, no density for the NG domains of either SRP54 or the NG domain of SRα (the metazoan ortholog of FtsY) was observed, indicating that binding to the SR induces a conformational change (Halic et al., 2006b). Solution studies further support the notion that complex formation between the SRP and SR induce a conformational change in this region (Buskiewicz et al., 2005a; Spanggord et al., 2005). Taken together with our results, these data indicate that a specific conformation of the linker is required for GTPase stimulation by the 4.5S RNA and that factors affecting the relative conformation of the M and NG domains of Ffh could affect the rate of Ffh·FtsY dissociation driven by GTP hydrolysis. For example, a ribosome bound to SRP may inhibit the conformation required for RNA-dependent GTPase stimulation and thus delay Ffh·FtsY dissociation until the ribosome has been transferred to the translocon.
Significantly, we found that the two activities of the 4.5S RNA, to promote complex formation and to promote GTP hydrolysis, are differentially sensitive to the mutations in Ffh that mimic the absence of 4.5S RNA in vitro. Although the two mutations in the signal sequence-binding domain, L350D and L354D, primarily affect 4.5S RNA stimulation of binding rate to FtsY, L301P primarily affects 4.5S RNA stimulation of GTPase activity, and L303D causes dramatic reductions in both binding rate and GTPase activity. Although these distinctions are not absolute, the differences contrast with previously reported mutations in the tetraloop region of the 4.5S RNA that act similarly to the L303D mutation and compromise both the rate of complex formation and the maximal rate of GTP hydrolysis in the complex (Jagath et al., 2001; Siu et al., 2006). Thus, the data presented here demonstrate that the two activities can be differentially affected. Therefore the activities of the 4.5S RNA to promote complex formation and disassembly could, in principle, be differentially regulated, consistent with the models presented above.
Taken together, the mutations identified in this study support the model that the SRP RNA links the major conformational changes in the signal sequence-binding subunit of the SRP to the interaction cycle of the SRP and the SR. Such molecular communication within SRP provides an attractive mechanism for coordination of the interaction of the SRP and the SR with ribosome recruitment and transfer to the translocon and an explanation for the centrality of the SRP RNA to efficient protein targeting.
ACKNOWLEDGMENTS
The authors thank Harris D. Bernstein, Julia R. Kardon, Saskia Neher, Beaker, and members of the Walter lab for helpful comments. Our special appreciation goes to Shu-ou Shan for her extensive mentoring of N.B. in the art of pre-steady state kinetic analyses. This work was supported by a grant from the National Institutes of Health to P.W. P.W. is an Investigator of the Howard Hughes Medical Institute. N.B. was supported by a predoctoral fellowship from the National Science Foundation.
Abbreviations used:
- SRP
signal recognition particle
- SR
SRP receptor.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-02-0117) on May 16, 2007.
REFERENCES
- Batey R. T., Rambo R. P., Lucast L., Rha B., Doudna J. A. Crystal structure of the ribonucleoprotein core of the signal recognition particle. Science. 2000;287:1232–1239. doi: 10.1126/science.287.5456.1232. [DOI] [PubMed] [Google Scholar]
- Bernstein H. D., Hyndman J. B. Physiological basis for conservation of the signal recognition particle targeting pathway in Escherichia coli. J. Bacteriol. 2001;183:2187–2197. doi: 10.1128/JB.183.7.2187-2197.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buskiewicz I., Kubarenko A., Peske F., Rodnina M. V., Wintermeyer W. Domain rearrangement of SRP protein Ffh upon binding 4.5S RNA and the SRP receptor FtsY. RNA. 2005a;11:947–957. doi: 10.1261/rna.7242305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buskiewicz I., Peske F., Wieden H. J., Gryczynski I., Rodnina M. V., Wintermeyer W. Conformations of the signal recognition particle protein Ffh from Escherichia coli as determined by FRET. J. Mol. Biol. 2005b;351:417–430. doi: 10.1016/j.jmb.2005.06.023. [DOI] [PubMed] [Google Scholar]
- Connolly T., Rapiejko P. J., Gilmore R. Requirement of GTP hydrolysis for dissociation of the signal recognition particle from its receptor. Science. 1991;252:1171–1173. doi: 10.1126/science.252.5009.1171. [DOI] [PubMed] [Google Scholar]
- Egea P. F., Shan S. O., Napetschnig J., Savage D. F., Walter P., Stroud R. M. Substrate twinning activates the signal recognition particle and its receptor. Nature. 2004;427:215–221. doi: 10.1038/nature02250. [DOI] [PubMed] [Google Scholar]
- Egea P. F., Stroud R. M., Walter P. Targeting proteins to membranes: structure of the signal recognition particle. Curr. Opin. Struct. Biol. 2005;15:213–220. doi: 10.1016/j.sbi.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Focia P. J., Shepotinovskaya I. V., Seidler J. A., Freymann D. M. Heterodimeric GTPase core of the SRP targeting complex. Science. 2004;303:373–377. doi: 10.1126/science.1090827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore R., Blobel G., Walter P. Protein translocation across the endoplasmic reticulum. I. Detection in the microsomal membrane of a receptor for the signal recognition particle. J. Cell Biol. 1982a;95:463–469. doi: 10.1083/jcb.95.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore R., Walter P., Blobel G. Protein translocation across the endoplasmic reticulum. II. Isolation and characterization of the signal recognition particle receptor. J. Cell Biol. 1982b;95:470–477. doi: 10.1083/jcb.95.2.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halic M., Becker T., Pool M. R., Spahn C. M., Grassucci R. A., Frank J., Beckmann R. Structure of the signal recognition particle interacting with the elongation-arrested ribosome. Nature. 2004;427:808–814. doi: 10.1038/nature02342. [DOI] [PubMed] [Google Scholar]
- Halic M., Blau M., Becker T., Mielke T., Pool M. R., Wild K., Sinning I., Beckmann R. Following the signal sequence from ribosomal tunnel exit to signal recognition particle. Nature. 2006a;444:507–511. doi: 10.1038/nature05326. [DOI] [PubMed] [Google Scholar]
- Halic M., Gartmann M., Schlenker O., Mielke T., Pool M. R., Sinning I., Beckmann R. Signal recognition particle receptor exposes the ribosomal translocon binding site. Science. 2006b;312:745–747. doi: 10.1126/science.1124864. [DOI] [PubMed] [Google Scholar]
- Jagath J. R., Matassova N. B., de Leeuw E., Warnecke J. M., Lentzen G., Rodnina M. V., Luirink J., Wintermeyer W. Important role of the tetraloop region of 4.5S RNA in SRP binding to its receptor FtsY. RNA. 2001;7:293–301. doi: 10.1017/s1355838201002205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagath J. R., Rodnina M. V., Wintermeyer W. Conformational changes in the bacterial SRP receptor FtsY upon binding of guanine nucleotides and SRP. J. Mol. Biol. 2000;295:745–753. doi: 10.1006/jmbi.1999.3427. [DOI] [PubMed] [Google Scholar]
- Keenan R. J., Freymann D. M., Stroud R. M., Walter P. The signal recognition particle. Annu. Rev. Biochem. 2001;70:755–775. doi: 10.1146/annurev.biochem.70.1.755. [DOI] [PubMed] [Google Scholar]
- Keenan R. J., Freymann D. M., Walter P., Stroud R. M. Crystal structure of the signal sequence binding subunit of the signal recognition particle. Cell. 1998;94:181–191. doi: 10.1016/s0092-8674(00)81418-x. [DOI] [PubMed] [Google Scholar]
- Larsen N., Zwieb C. The signal recognition particle database (SRPDB) Nucleic Acids Res. 1993;21:3019–3020. doi: 10.1093/nar/21.13.3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. C., Bernstein H. D. The targeting pathway of Escherichia coli presecretory and integral membrane proteins is specified by the hydrophobicity of the targeting signal. Proc. Natl. Acad Sci. USA. 2001;98:3471–3476. doi: 10.1073/pnas.051484198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mainprize I. L., Beniac D. R., Falkovskaia E., Cleverley R. M., Gierasch L. M., Ottensmeyer F. P., Andrews D. W. The structure of Escherichia coli signal recognition particle revealed by scanning transmission electron microscopy. Mol. Biol. Cell. 2006;17:5063–5074. doi: 10.1091/mbc.E06-05-0384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. D., Wilhelm H., Gierasch L., Gilmore R., Walter P. GTP binding and hydrolysis by the signal recognition particle during initiation of protein translocation. Nature. 1993;366:351–354. doi: 10.1038/366351a0. [DOI] [PubMed] [Google Scholar]
- Peluso P., Herschlag D., Nock S., Freymann D. M., Johnson A. E., Walter P. Role of 4.5S RNA in assembly of the bacterial signal recognition particle with its receptor. Science. 2000;288:1640–1643. doi: 10.1126/science.288.5471.1640. [DOI] [PubMed] [Google Scholar]
- Peluso P., Shan S. O., Nock S., Herschlag D., Walter P. Role of SRP RNA in the GTPase cycles of Ffh and FtsY. Biochemistry. 2001;40:15224–15233. doi: 10.1021/bi011639y. [DOI] [PubMed] [Google Scholar]
- Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. UCSF Chimera-a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Phillips G. J., Silhavy T. J. The E. coli ffh gene is necessary for viability and efficient protein export. Nature. 1992;359:744–746. doi: 10.1038/359744a0. [DOI] [PubMed] [Google Scholar]
- Poritz M. A., Bernstein H. D., Strub K., Zopf D., Wilhelm H., Walter P. An E. coli ribonucleoprotein containing 4.5S RNA resembles mammalian signal recognition particle. Science. 1990;250:1111–1117. doi: 10.1126/science.1701272. [DOI] [PubMed] [Google Scholar]
- Powers T., Walter P. Reciprocal stimulation of GTP hydrolysis by two directly interacting GTPases. Science. 1995;269:1422–1424. doi: 10.1126/science.7660124. [DOI] [PubMed] [Google Scholar]
- Powers T., Walter P. Co-translational protein targeting catalyzed by the Escherichia coli signal recognition particle and its receptor. EMBO J. 1997;16:4880–4886. doi: 10.1093/emboj/16.16.4880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosendal K. R., Wild K., Montoya G., Sinning I. Crystal structure of the complete core of archaeal signal recognition particle and implications for interdomain communication. Proc. Natl. Acad. Sci. USA. 2003;100:14701–14706. doi: 10.1073/pnas.2436132100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffitzel C., Oswald M., Berger I., Ishikawa T., Abrahams J. P., Koerten H. K., Koning R. I., Ban N. Structure of the E. coli signal recognition particle bound to a translating ribosome. Nature. 2006;444:503–506. doi: 10.1038/nature05182. [DOI] [PubMed] [Google Scholar]
- Shan S. O., Walter P. Induced nucleotide specificity in a GTPase. Proc. Natl. Acad. Sci. USA. 2003;100:4480–4485. doi: 10.1073/pnas.0737693100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu F. Y., Spanggord R. J., Doudna J. A. SRP RNA provides the physiologically essential GTPase activation function in cotranslational protein targeting. RNA. 2006;2:240–250. doi: 10.1261/rna.135407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanggord R. J., Siu F., Ke A., Doudna J. A. RNA-mediated interaction between the peptide-binding and GTPase domains of the signal recognition particle. Nat. Struct. Mol. Biol. 2005;12:1116–1122. doi: 10.1038/nsmb1025. [DOI] [PubMed] [Google Scholar]
- Tian H., Beckwith J. Genetic screen yields mutations in genes encoding all known components of the Escherichia coli signal recognition particle pathway. J. Bacteriol. 2002;184:111–118. doi: 10.1128/JB.184.1.111-118.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulbrandt N. D., Newitt J. A., Bernstein H. D. The E. coli signal recognition particle is required for the insertion of a subset of inner membrane proteins. Cell. 1997;88:187–196. doi: 10.1016/s0092-8674(00)81839-5. [DOI] [PubMed] [Google Scholar]
- Walter P., Blobel G. Signal recognition particle contains a 7S RNA essential for protein translocation across the endoplasmic reticulum. Nature. 1982;299:691–698. doi: 10.1038/299691a0. [DOI] [PubMed] [Google Scholar]
- Walter P., Ibrahimi I., Blobel G. Translocation of proteins across the endoplasmic reticulum. I. Signal recognition protein (SRP) binds to in-vitro-assembled polysomes synthesizing secretory protein. J. Cell Biol. 1981;91:545–550. doi: 10.1083/jcb.91.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zopf D., Bernstein H. D., Walter P. GTPase domain of the 54-kD subunit of the mammalian signal recognition particle is required for protein translocation but not for signal sequence binding. J. Cell Biol. 1993;120:1113–1121. doi: 10.1083/jcb.120.5.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]