Abstract
Trans-splicing has been used to repair mutant RNA transcripts via competition for the spliceosome using pre-trans-splicing molecules, or “PTMs.” Previous studies have demonstrated that functional PTMs can be designed for either 3′- or 5′-exon replacement, with a vast majority of the work to date focusing on repair of mutations within internal exons and via 3′-exon replacement. Here, we describe the first use of trans-splicing to target the first exon and intron of a therapeutically relevant gene and repair the mutant RNA by 5′-exon replacement. Our results show that 5′-PTMs can be designed to repair mutations in the β-globin transcript involved in sickle cell anemia and β-thalassemia while providing insight into considerations for competition between trans- versus cis-splicing in mammalian cells. Target transcripts with impaired cis-splicing capabilities, like those produced in some forms of β-thalassemia, are more efficiently repaired via trans-splicing than targets in which cis-splicing is unaffected as with sickle β-globin. This study reveals desirable characteristics in substrate RNAs for trans-splicing therapeutics as well as provides an opportunity for further exploration into general splicing mechanisms via 5′-PTMs.
Keywords: Hemoglobinopathies, trans-splicing
INTRODUCTION
Trans-splicing is a naturally occurring phenomenon in some viruses, prokaryotes, lower eukaryotes, and possibly even in mammalian cells (Ziff and Evans 1978; Michel and Dujon 1983; Milhausen et al. 1984; Blumenthal and Thomas 1988; Kohchi et al. 1988; de Souza et al. 1991; Finta and Zaphiropoulos 2002). The mechanism varies from the self-catalyzed reaction of the discontinuous group II intron to the spliceosome-driven splice leader approach used by nematodes and trypanosomes (Kohchi et al. 1988; de Souza et al. 1991). Trans-splicing can also be artificially derived by either modifying a natural splicing process, as in the group I ribozyme, or by manipulation of the cis-splicing machinery (Sullenger and Cech 1994; Puttaraju et al. 1999). The latter is termed spliceosomal mediated RNA trans-splicing, or “SMaRT,” and it has been studied as a potential therapeutic for diseases including cystic fibrosis, hemophilia A, a form of epidermolysis bullosa, and tauopathies (Mansfield et al. 2000; Liu et al. 2002; Chao et al. 2003; Dallinger et al. 2003; Rodriguez-Martin et al. 2005). The general methodology has also been used as a possible cancer therapeutic via intra-tumoral reconstitution of a toxin RNA (Nakayama et al. 2005).
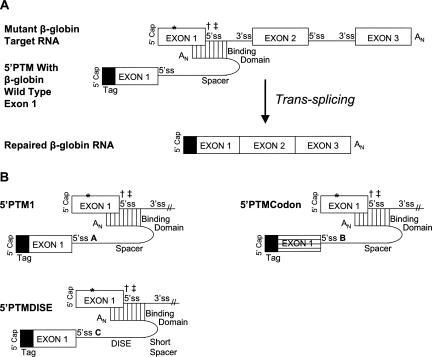
SMaRT technology uses a pre-trans-splicing molecule (PTM) to “trick” the spliceosome into using it as a substrate for splicing (Puttaraju et al. 1999). It achieves this by blocking the splicing signals on the nascent pre-mRNA transcript via base-pairing while concurrently providing the spliceosome with alternative splicing sequences (Fig. 1A). The goal is for the spliceosome to recognize the PTM's splice sites instead of the endogenous RNA's signals. Achieving this goal results in the trans-splicing of the PTM's cargo onto the target RNA and, in therapeutic applications, repair of the mutant endogenous RNA. The technology is desirable in that it preserves the endogenous regulatory milieu and requires only delivery of a portion of the transcript, the latter being important for some limited-capacity viral vectors.
FIGURE 1.
First exon replacement via trans-splicing using a 5′ pre-trans-splicing molecule (5′-PTM) and the 5′-PTM constructs. (A) Mechanism for 5′-trans-splicing RNA repair adapted for targeting β-globin mutations in the first exon/intron. A pre-trans-splicing molecule or “PTM” base pairs to the target RNA and conceals the endogenous 5′-splice site (5′-ss) while concurrently providing its own site that, if used by the spliceosome, introduces a therapeutic RNA sequence. The tag (black box) is sequence used to distinguish between trans- and cis-spliced products. The symbols “*,” “†,” and “‡” denote the location of point mutations used in this study that are involved in sickle cell anemia, β0-thalassemia, and β+-thalassemia, respectively. (B) 5′-PTM constructs. All 5′-PTMs were generated via RNA polymerase II expression plasmids within human embryonic kidney 293T cells. A 5′-tag sequence was added to each to improve distinction between cis and trans transcripts and alternative codons for exon 1 were used in 5′-PTMCodon for the same purpose. A 43-nt binding domain was used in all cases, and it bound the last 11 nt of exon 1 through the first 32 nt of IVS1 (binding domain sequence provided in Materials and Methods). The three 5′-PTMs each had a unique 5′-ss denoted here as A, B, and C, respectively (for details, see Materials and Methods). 5′-PTMDISE was further distinct in that the spacer length used was 12 nt rather than 84 nt as in the other constructs. This 5′-PTM also carried a 45-nt putative intronic splicing enhancer element called DISE (P. Seth and M. Garcia-Blanco, pers. comm.) downstream from its 5′-ss. The symbols “*,” “†,” and “‡” denote the location of point mutations used in this study that are involved in sickle cell anemia, β0-thalassemia, and β+-thalassemia, respectively.
PTMs can be crafted to replace RNA sequence either upstream of or downstream from its binding site on the endogenous pre-mRNA; these are described as 5′-PTM and 3′-PTM, respectively (Puttaraju et al. 1999; Mansfield et al. 2003). 3′-PTMs have been used to correct a number of different clinically relevant mutations using various delivery routes ranging from in vitro applications to mouse models of disease (Puttaraju et al. 1999, 2001; Mansfield et al. 2000; Kikumori et al. 2001; Liu et al. 2002, 2005; Chao et al. 2003; Dallinger et al. 2003; Tahara et al. 2004; Rodriguez-Martin et al. 2005). Liu and colleagues delivered a 3′-PTM to polarized human cystic fibrosis airway epithelial cells using an adeno-associated virus delivery vehicle (Liu et al. 2005). The group reported a 14% restoration of conductance through the CFTR channels in CF cells compared to that detected in normal cells.
Unlike 3′-PTMs, the study landscape for 5′-PTMs is quite barren. Mansfield and colleagues have published the only study using 5′-PTMs to date (Mansfield et al. 2003). They found that the location of binding by the PTM impacts the trans-splicing efficiency in mammalian cell culture, especially when dealing with a large target intron. Our study explores the effects of the targets themselves while also using the 5′-PTM in a novel context, that of repairing a natural first exon. Our models for this work are β-globin mutations in the first exon or intron that lead to either sickle cell anemia or β-thalassemia, respectively.
RESULTS
Three 5′-PTM constructs were designed (Fig. 1B) and cloned into the RNA polymerase II expression vector pEGFPN1 (Clontech). The PTMs varied by the 5′-splice site (5′-ss) sequence used, spacer length, and inclusion of a potential intronic splice enhancer sequence (“DISE”) (P. Seth and M. Garcia-Blanco, pers. comm.). One construct, 5′-PTMCodon, was designed using the degeneracy of the genetic code to make the distinction between cis- and trans-spliced products clearer at the nucleotide level. A PTM-specific 5′-tag was also added to all PTM constructs for this same purpose.
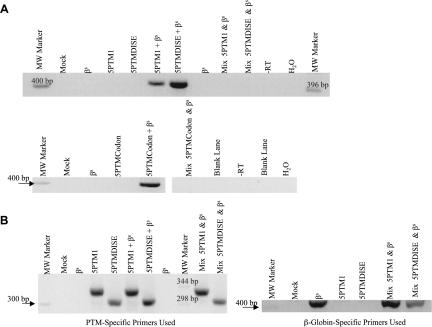
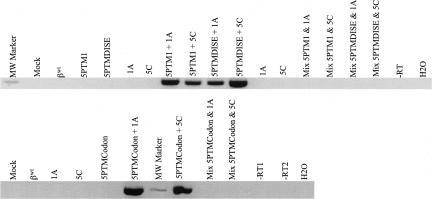
Each 5′-PTM construct was cotransfected along with genomic β-globin expression vectors into 293T human embryonic kidney cells, and trans-splicing was assessed 48 h later using total RNA in a one-step RT-PCR reaction. Using trans-splicing specific primers followed by sequence confirmation, we detected repair of the sickle β-globin transcript using each of the three 5′-PTMs (Fig. 2A). The “mix” control, comprised of PTM and β-globin transcripts expressed in separate cells then mixed before cell lysis, did not contain the amplification product, indicating that the trans-splicing event happened only within the cotransfected cells. Figure 2B shows that although the mix controls did not produce a trans-spliced RT-PCR product, the PTM and β-globin transcripts were both present and able to be independently amplified.
FIGURE 2.
Sickle β-globin transcripts are corrected via 5′-PTM-driven trans-splicing. (A) βs RNA is repaired via 5′-PTM trans-splicing in 293T cells. 5′-PTM (5′-PTM1, 5′-PTMDISE, and 5′-PTMCodon) and genomic βs-globin expression cassettes were cotransfected into 293T cells, and total RNA was harvested for analysis by RT-PCR. Primers specific for trans-spliced products were used to amplify the repaired RNA and yield a DNA product of 410 bp. The products were purified from an agarose gel and sequenced to confirm their identity. The transfection performed is listed above each lane in the gel. The term “mix” denotes mix controls that were used to ensure that trans-splicing events did not occur during RNA workup and analysis but rather occurred intracellularly. Mix controls were made by combining pre-lysis cells from PTM-alone and βs-target-alone transfections and then processing the mixed cells in the same way as the experimental samples from this point onward. (B) The mix samples contain both PTM and β-globin target RNAs. Total RNA from mix samples was reverse-transcribed and amplified using either β-globin or PTM-specific primers, and the products were run on agarose gels to confirm that both the 5′PTM and βs-globin target RNAs were present in the mix sample even though no trans-splicing products were present.
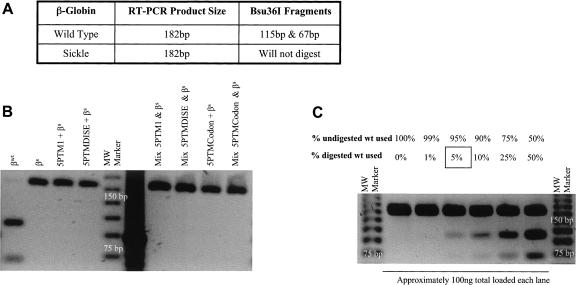
We then examined the efficiency of trans-splicing-mediated repair of the βs-globin RNA using a Bsu36I restriction enzyme digest assay. RT-PCR amplification using primers to the β-globin 5′-UTR and exon 2 was followed by an overnight Bsu36I digest. The RT-PCR design allowed for both the cis- (βs-globin encoding) and trans-spliced (wild-type β-globin encoding) products to be amplified equally, and the Bsu36I enzyme was used to distinguish between the two (Fig. 3A). We then attempted to quantify the levels of repair by densitometric analysis of the resultant restriction fragments. However, we could not detect any digestion fragments using any of the 5′-PTMs (Fig. 3B) and estimated that the level of repair would need to be 5% or more in order to be identified using this assay (Fig. 3C).
FIGURE 3.
Quantifying levels of sickle β-globin repair. (A) Bsu36I restriction enzyme digest used to quantify repair of sickle β-globin RNA. After RT-PCR using primers that bind both repaired and mutant targets, a Bsu36I restriction site exists only in the repaired β-globin product, thus allowing for distinction between the two amplification products. (B) Amplification products from cells cotransfected with 5′PTM and genomic βs-globin expression cassettes and digested with Bsu36I. The sickle transcript is in significant excess over the repaired RNA. Although trans-splicing is occurring (see Fig. 2), the levels are not high enough for detection via the Bsu36I restriction digest assay. (C) The detection limit of the Bsu36I assay is a 5% repair level. Different ratios of digested and undigested normal β-globin RT-PCR products were used to identify the lower detection limit of repaired product in the assay.
Since SMaRT relies on the PTM's ability to compete with cis-splicing for spliceosomal components, we hypothesized that perhaps the inability of the 5′-PTMs to repair more of the βs-globin RNA was due to the highly efficient cis-splicing of the sickle β-globin target itself. The sickle cell transcript has no aberrations in its splicing ability, and perhaps cis-splicing occurs so rapidly that the 5′-PTM constructs are not able to compete well against it. We therefore attempted to improve our trans-splicing levels by targeting mutations that disrupt normal cis-splicing of β-globin's exon 1.
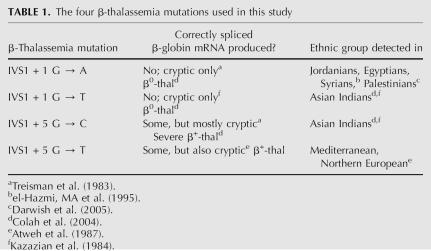
Four different point mutations within β-globin's intron 1 5′-splice site (IVS1 5′-ss) each result in another hematological disease called β-thalassemia. Unlike sickle cell anemia, β-thalassemia can be caused by various mutations in the β-globin gene, and often the mutation is population-specific. The four mutations we chose have all been documented in specific ethnic groups, and each causes impairment of normal β-globin cis-splicing (Table 1; Treisman et al. 1983; Kazazian et al. 1984; Atweh et al. 1987; el-Hazmi et al. 1995; Colah et al. 2004; Darwish et al. 2005). Specifically, in the absence of normal IVS-1 5′-ss usage, two different cryptic sites are substituted: E105 and E127. The former is found in β-globin exon 1 position 105 and the latter in exon 1 position 127 (Treisman et al. 1983; Atweh et al. 1987).
TABLE 1.
The four β-thalassemia mutations used in this study
Each 5′-PTM construct was cotransfected along with the four different genomic β-globin thalassemia expression vectors into 293T human embryonic kidney cells, and trans-splicing was assessed 48 h later using total RNA in a one-step RT-PCR reaction. Using trans-splicing-specific primers, we detected the expected product of 410 base pairs (bp), indicating that PTM-driven restoration of the normally spliced β-globin transcript using each of the three 5′-PTMs with each of the four β-thalassemia mutations had occurred (Fig. 4; only β-thalassemia mutants 1A and 5C are shown, but similar results were obtained for the 1T and 5T targets). The identity of the trans-spliced products was verified by sequence analysis. The mix controls for these experiments were derived in the same way as in the sickle β-globin studies and again were indicative of a true intracellular trans-splicing event.
FIGURE 4.
The β-thalassemia mutants are corrected via 5′-PTM-driven trans-splicing. βthal RNA is repaired via 5′PTM trans-splicing in 293T cells. 5′-PTM (5′-PTM1, 5′-PTMDISE, and 5′-PTMCodon) and genomic βthal-globin expression cassettes were cotransfected into 293T cells, and total RNA was harvested for analysis by RT-PCR. Primers specific for trans-spliced products were used to amplify the repaired RNA and yield a DNA product of 410 bp. The products were purified from an agarose gel and sequenced to confirm their identity. The figure shows data after testing β-thalassemia target mutants βthal + 1G → A and βthal + 5G → C (referred to as 1A and 5C in the figure), but similar results were obtained for βthal + 1G → T and βthal + 5G → T. The transfection performed is listed above each lane in the gel. Trans-splicing is expected to yield a product of 410 bp. The molecular weight marker band is 400 bp in all cases.
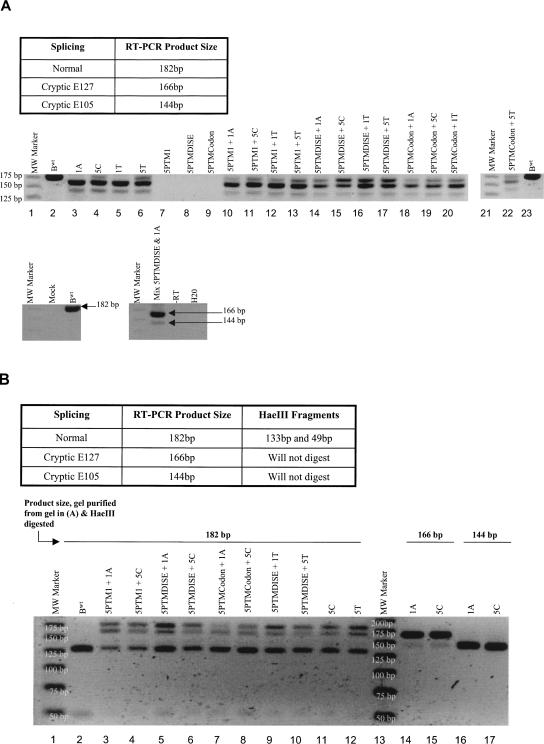
Total RNA was then amplified via one-step RT-PCR using a primer set that bound the 5′-UTR and exon 2 of β-globin. This experiment detected the effects of each β-thalassemia mutation alone as well as each 5′-PTM's ability to restore normal splice products in 293T cotransfection experiments since the assay allowed for amplification of both cryptic and normal RNAs (Fig. 5A). We found that the β-thalassemia point mutations produced splicing profiles similar to what has been previously described in the literature (Fig. 5A; Treisman et al. 1983; Atweh et al. 1987). The mutations within the core 5′-ss at + 1 (1A and 1T) produced no detectable 182-bp normal splice product, whereas mutations at position + 5 (5C and 5T) produced some, but still with the cryptic products predominating (Fig. 5A, lanes 3–6). Cotransfection of 5′-PTM1, 5′PTMDISE, or 5′-PTMCodon expression cassettes with any of the four β-thalassemia mutants resulted in the appearance or increase (relative to cryptic products within the same lane) of the 182-bp normal splice product (Fig. 5A). For example, compare lane 3 (1A mutant alone) with lane 14 (1A mutant with 5′-PTMDISE).
FIGURE 5.
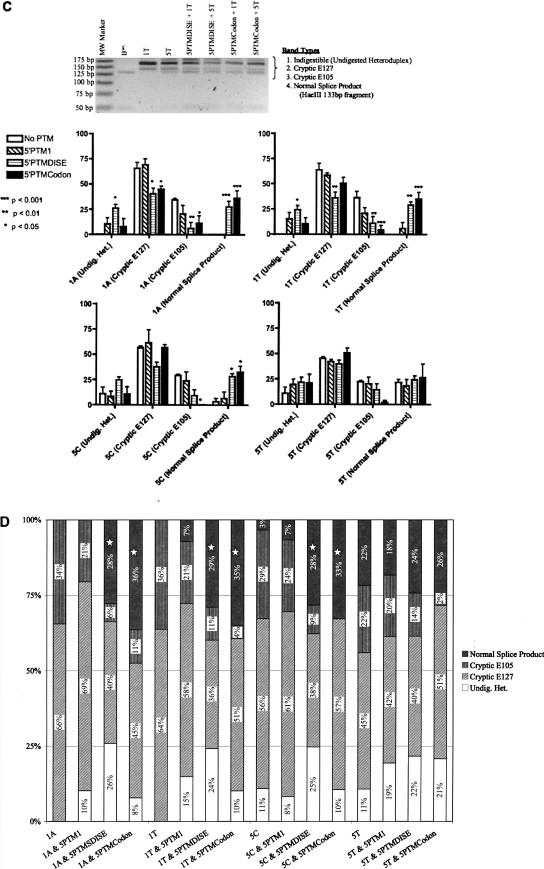
Quantification of repair levels of β-thalassemia RNA. (A) 5′-PTMs can restore the normal splice product in each β-thalassemia mutant. The two cryptic 5′-splice sites, E127 and E105, produce RT-PCR products that differ in size from that generated via normal splicing (144 bp and 166 bp versus 182 bp). When any of the three 5′PTMs are used, the normal splice product appears (for β-thal 1A and 1C) or increases relative to the cryptic products in the same lane (for β-thal 5C and 5T). This is illustrated by comparing the band types produced by each β-thalassemia mutant alone to the 5′PTM-treated counterpart in the figure. For example, compare lane 3 (1A mutant alone) with lane 14 (1A mutant with 5′-PTMDISE). (B) Testing an HaeIII restriction enzyme assay for potential use in quantifying β-thalassemia repair levels. After RT-PCR using primers that bind both normal and cryptic splice transcripts, an HaeIII restriction site exists only in the normal splice product, thus allowing for distinction between the possible outcomes. The three different RT-PCR products (normal, cryptic E127, cryptic E105) shown in A were excised individually from the agarose gel and purified. Each band was then digested overnight with HaeIII and run on an agarose gel as shown here. The two fragments resulting from HaeIII digestion of the normal 182-bp product are 133 bp and 49 bp, both of which can be seen in the figure. The normal splice product inherently produced by the 5C and 5T mutants is also cleaved as expected. The two cryptic RT-PCR products, 166 bp and 144 bp, do not produce the 133-bp and 49-bp HaeIII digestion fragments. This illustrates the assay's utility in quantifying repair levels via appearance of HaeIII digestion fragments (we used the 133-bp band specifically). The “182-bp” band remaining in some lanes is likely an indigestible heteroduplex product (explained in text). The 166-bp band seen in the 182-bp product digestion lanes is carryover from the gel slice excision process. The same is true for the 144-bp band seen in the 166-bp product digestion lanes. (C) Quantification of repair levels engendered by each 5′-PTM targeting the four β-thalassemia mutants. After harvesting RNA from transfected 293T cells, performing RT-PCR, and running the products on a gel, the normal (182 bp), cryptic E127 (166 bp), and cryptic E105 (144 bp) RT-PCR products were digested with HaeIII and run on a high-resolution agarose gel containing ethidium bromide (see gel, this figure). The 133-bp HaeIII digestion fragment was used as the indicator for the normal splice product. Quantification of each band (excluding the other HaeIII fragment, 49 bp) within each lane was performed using Quantity One densitometric software from the Bio-Rad company. The intensity values were normalized for nucleotide length, and each band type quantity was expressed as a fraction of the total of all bands quantified using a particular 5′-PTM/β-thalassemia mutant combination (for details, see Materials and Methods). The four β-thalassemia forms are graphed separately, and the label within each x-axis parentheses denotes the band type quantified. Each experiment was done in triplicate or greater, and the error bars shown here indicate the standard error of the mean. Significance testing for each β-thalassemia form was performed using the untreated (i.e., the β-thalassemia mutant alone) value for a particular band type (i.e., normal, cryptic E127, cryptic E105, or undigested heteroduplex) and comparing it to its PTM-containing counterpart. The different P-values are denoted as follows: (*) p < 0.05; (**) p < 0.01; (***) p < 0.001. (D) Percent repair of β-thalassemia mutants using various 5′-PTM constructs. The percent of each band type present for each 5′-PTM/β-thalassemia combination is shown here along with the untreated controls (each β-thalassemia mutant alone). The stars denote statistically significant increases in levels of normal splice product when treatment is used (i.e., 5′-PTM is cotransfected).
We then used an HaeIII restriction enzyme assay to verify and quantify the levels of normally spliced product. An HaeIII site exists between the locations of the cryptic splice sites and the normal 5′-splice site in β-globin exon 1, and thus only the normally spliced RT-PCR product will retain this restriction site after splicing (Fig. 5B). We used this unique HaeIII characteristic to verify the presence of the normal splice product by digesting some of the gel-purified 182-bp products seen in Figure 5A, and this result is shown in Figure 5B. The 133-bp and 49-bp HaeIII digestion fragments seen indicate the presence of the normally spliced product. However, a seemingly “182 bp” band is retained in the 182-bp digestion lanes. It is unlikely to be the result of incomplete digestion since it is not present in the βwt lane. A more likely possibility is that it is a heteroduplex formed between the normal 182-nucleotide (nt) and abundant cryptic 166-nt cDNAs that is unable to be digested by HaeIII. Attempts to sequence this fragment suggested the presence of more than one sequence in the band, further supporting the heteroduplex hypothesis. We chose to include this heteroduplex in the following quantification analysis since it may have acted as a “sink” for the cDNAs.
Finally, the gel-purified 166-bp and 144-bp bands were not cleaved by HaeIII. This illustrated the assay's ability to distinguish between cryptic and normal double-stranded DNA fragments (Fig. 5B, lanes 14–17). It should also be noted that due to the close similarity among product sizes, some carryover was expected during excision of the individual gel slices. This is seen by the presence of 166-bp and 144-bp bands in the 182-bp and 166-bp gel extract lanes, respectively. Since we extracted all band types in a lane as one single gel slice prior to HaeIII digestion, carryover was not an issue during the actual quantification.
We quantified the levels of normal splicing restored via each 5′-PTM using one of the HaeIII digestion fragments (133 bp) as the indicator for “normal” with this identity confirmed via sequencing of the fragment. Quantifying one of the HaeIII digestion products versus using the original 182-bp band allowed us to account for the indigestible heteroduplex while providing a conservative, yet true, assessment of repair efficiency. We used Quantity One software (Bio-Rad) to quantify the intensity of the 133-bp HaeIII fragment in each lane and compared this to the intensities of the other bands in the same lane (Fig. 5C, representative gel image).
As shown in Figure 5C, we found that 5′-PTMDISE and 5′-PTMCodon can restore the normal splice product in the two β0-thalassemia mutations (1A and 1T) up to levels reaching 36% and that these results are statistically significant (Fig. 5C, p < 0.05 in all four cases). Both PTMs also significantly increase the baseline level of normal splice product in the 5C β-thalassemia case (Fig. 5C, p < 0.05). The statistically significant decreases in cryptic products seen in the 1A, 1T, and 5C mutants perhaps reflect a 5′-PTM-driven increased conversion of pre-mRNA into normal rather than cryptic products (Fig. 5C, p < 0.05). The significant increases in heteroduplex product when comparing 5′-PTMDISE and “No PTM” in the 1A and 1T cases may also be caused by the significant increase in normal product, since we hypothesize that the heteroduplex is due to base-pairing between normal and E127 cryptic cDNAs. The E127 cryptic product is still in excess over the normal product even when repair efficiencies are at their highest; hence any increase in normal product may be redirected to forming the heteroduplex as well as the normal double-stranded DNA. Unlike 5′-PTMDISE, normal splice products stemming from 5′-PTMCodon may be less apt to form a heteroduplex with the cryptic product since this particular 5′-PTM's exon 1 nucleotide sequence is somewhat dissimilar to the endogenous sequence. This may be one reason why there is no statistically significant increase in heteroduplex levels when comparing 5′-PTMCodon to its “No PTM” counterpart in 1A and 1T.
5′-PTM1 did produce the normal β-globin splice product (where there was none in the untreated sample) in the β0-thalassemia 1T case, but its level was not high enough to be statistically significant. This PTM's effect on normal splice levels was similarly not statistically significant in β-thalassemia 5C. Finally, none of the 5′-PTMs resulted in a clear increase of normal splice product in β-thalassemia 5T (Fig. 5C). The quantification data are summarized in Figure 5D. The means for each condition are plotted in bar graph form, and the six statistically significant increased normal splice product levels are starred with a maximum of 36% repair reached in the case of 5′-PTMCodon repairing the β-thalassemia 1A mutant.
DISCUSSION
We have shown that 5′-PTMs can be used to correct the first exon of a therapeutically relevant gene and that splicing-impaired targets may be well-suited for repair via SMaRT technology. Using mutations in the β-globin gene, we found that 5′-PTMs can trans-splice to repair defects causing sickle cell anemia and some forms of β-thalassemia at the RNA level (Figs. 2, 4). This is the first time a 5′-PTM has been targeted specifically to correct disease-causing mutations in a gene's first exon/intron, and it is only the second study of 5′-PTMs in general.
Our attempts to quantify repair efficiencies in the sickle cell case and subsequently in the β-thalassemia splice mutants revealed much about the nature of targets accessible to SMaRT. We estimated that our trans-splicing levels in the sickle cell experiments were below 5% when assessed by RT-PCR followed by restriction enzyme analysis (Fig. 3). However, repair efficiencies were as high as 36% when determined in a similar fashion but using a splicing-impaired target, namely, the β-thalassemia splice mutants (Fig. 5). These results suggest that perhaps either the PTM must out-compete the endogenous splice site for the splicing machinery or the target itself must be hindered in its splicing ability. The β-globin gene is normally spliced very efficiently (Audibert et al. 2002), and perhaps this is why, although we can detect the repaired transcript in the sickle cell case, it is not in high abundance. The sickle cell mutation does not impact the gene's ability to splice, but instead leads to a structurally compromised β-chain within the hemoglobin tetramer (Ingram 1956, 1959; Perutz 1960). Our 5′-PTM constructs may have been unable to compete effectively against this efficiently cis-spliced target.
However, when using β-thalassemia targets involved with splicing aberrations, 5′-PTMs (5′-PTMDISE and 5′-PTMCodon specifically) significantly increased the levels of normal β-globin RNA product made versus that in untreated samples (Fig. 5). Perhaps the β-thalassemia point mutations cause a “stall” in the formation of the spliceosomal complex during which the 5′-PTM can bind to the target and provide a viable alternative to the endogenous 5′-ss (Custodio et al. 1999). If this were the case, perhaps a 5′-PTM could be designed that artificially destabilizes the target endogenous RNA in sickle cell anemia to create the same “stalling” effect. The 5′-PTM binding domain could be extended to include binding of the 5′-cap region, and since the first exon is defined via interactions between the cap and IVS-1 5′-ss (Berget 1995; Lewis et al. 1996), PTM binding of both components on the target pre-mRNA could potentially further hide the endogenous sequence from the splicing machinery.
A second possibility for the evident increased effectiveness of 5′-PTMs in the β-thalassemia versus sickle cell anemia scenario involves the selective destabilization of certain transcripts and this impact on the repair calculation. Perhaps the β-thalassemia cryptic transcripts are undergoing nonsense-mediated decay (NMD), and hence the net level of cis-spliced RNA is lower here than in the sickle cell anemia case. This would decrease the denominator in the β-thalassemia repair efficiency ratio: (normal splice product)/(heteroduplex + cryptic E127 + cryptic E105 + normal splice products), and any trans-splicing would have a greater apparent effect compared to identical levels of trans-splicing in the sickle cell anemia example. Analysis of the mRNA sequences produced by the two cryptic 5′-splice sites reveals stop codons located further than 50–55 nt upstream of the nearby splice junction, and thus these cryptic transcripts may be susceptible to NMD (Maquat 2005). The cryptic E105 site results in a downstream premature stop codon that is 184 nt upstream of the exon 2/exon 3 splice junction. The E127 site leads to a downstream premature stop codon that is 131 nt upstream of the same junction. A 2002 study by Danckwardt and colleagues examined the possibility of NMD of cryptic transcripts produced from a β-thalassemia 5A mutation that are identical to those described here (Danckwardt et al. 2002). They found that the E127 cryptic RNA was subject to NMD. This study bolsters our suggestion that perhaps 5′-PTM repair is more detectable in our β-thalassemia examples than in the sickle cell case since the trans-spliced product potentially holds a selective advantage in stability over the former. However, they also found that the E105 transcript was not susceptible to NMD, and, in fact, its usage as a cryptic 5′-ss was dependent on E127 cryptic activity. This conclusion casts doubt on E105 transcripts skewing repair efficiency values in the β-thalassemia case, but it also poses an intriguing possibility for 5′-PTM design. Perhaps one might extend the PTM's binding domain so that it base pairs with the E127 cryptic region and, in doing so, prevents both cryptic sites (E105 and E127) from being used. The spliceosome might be even more apt to choose the 5′-ss provided by the 5′-PTM.
This study has shown that the first exon of a therapeutically relevant gene can be repaired via a 5′-PTM and SMaRT technology. It is also the first time SMaRT has been applied to either sickle cell anemia or β-thalassemia, and the different repair efficiencies achieved between the two illustrates important general considerations when applying this therapeutic approach. The target's endogenous splicing rate and the resulting transcript's stability play an important role in the success of the trans-splicing therapy and hence should be considered when selecting targets for SMaRT. Our diverse 5′-PTM designs also appear to influence the repair levels achieved since, among other differences, unique 5′-ss were used in each construct, and the PTMs repaired the targets to varying degrees. We are currently designing a 5′-PTM identical to 5′-PTMDISE save for the DISE sequence itself in hopes of revealing how this particular design characteristic may impact repair efficiencies.
Since alleviation of symptoms in either sickle cell anemia or β-thalassemia does not require that all mutant transcripts be repaired, our trans-splicing efficiency levels may need only modest improvement for a potential therapeutic impact. Ho and colleagues found that β-globin transcript levels between 28% and 41% of that in unaffected individuals were associated with a clinically asymptomatic state in β-thalassemia family studies (Ho et al. 1996). Sickle cell symptoms are diminished by the presence of only 20%–25% fetal hemoglobin (Powars et al. 1984). Levasseur and colleagues designed a modified β-globin gene that they estimated would similarly inhibit hemoglobin S polymerization when incorporated into just 25% of circulating hemoglobin (Levasseur et al. 2004). The anti-sickling modifications could also be made to the 5′-PTM's β-globin cargo sequence, and RNA repair efficiencies < 25% would likely produce therapeutic hemoglobin levels since a single trans-spliced mRNA would produce multiple copies of the modified β-globin peptide.
An RNA repair approach like that used in SMaRT technology also provides the added benefit of preserving the target gene's endogenous regulatory mechanisms. This may be important for targets that are subject to complex transcriptional control processes like β-globin with its locus control region. The trans-splicing technology's safety profile may also improve since only those cells naturally producing the target transcript should be affected.
MATERIALS AND METHODS
Plasmids
Three 5′-PTM constructs were designed (Fig. 1B) and cloned into the RNA polymerase II expression vector pEGFPN1 (Clontech) using SacI and AgeI restriction enzymes (NEB). The EGFPN1 ORF was then removed using AgeI and NotI restriction enzymes (NEB), gel-extracting the desired EGFPN1-free vector (Qiaquick Gel Extraction Kit; QIAGEN), and filled in using the Klenow enzyme (large fragment from Fisher Biosciences). T4 DNA ligase (NEB) was then used to bluntly ligate the ends. The 43-nt binding domain sequence in the PTM was as follows: 5′-ccttaaacctgtcttgtaaccttgataccaacctgcccagggc. The 5′-ss used for each 5′-PTM were as follows: 5′-PTM1, gtgagt (canonical); 5′-PTMCodon, gaggtgagt (sequence found to be highly efficient in β-globin IVS-1 by Roca et al. 2005); and 5′-PTMDISE, gtgggt (hybrid of canonical and endogenous β-globin IVS-1 5′-ss).
The β-globin genomic sequences were cloned into the RNA polymerase II expression vector pcDNA3.1(+)Hyg (Invitrogen) using NheI and BamHI restriction enzymes (NEB). Six genomic β-globin constructs were made, and these included wild-type, sickle cell (A-to-T mutation at nucleotide 70), and β-thalassemia varieties. Four different β-thalassemia IVS-1 point mutations were used: + 1 (G to A), + 1 (G to T), + 5 (G to C), and + 5 (G to T). All four mutations were characterized via RT-PCR (Superscript III/ Platinum Taq One-Step RT-PCR; Invitrogen) and shown to activate two different cryptic 5′-splice sites (E105 and E127) leading to shortened amplimers.
Transfections
Human embryonic kidney (HEK) 293T cells were plated onto either six-well plates or 100 mm dishes and allowed to grow overnight to achieve 40%–80% confluency. The media (DMEM [Invitrogen], 10% fetal bovine serum [HyClone], 1× penicillin/streptomycin [Invitrogen]) was replaced the next morning immediately before transfection. A 2:1 ratio of PTM to β-globin expression plasmid was used for all cotransfected samples. Four micrograms (six-well plates) or 10 μg (100 mm dishes) of total DNA was suspended in 150 μL of Opti-MemI (Invitrogen) and allowed to interact with Superfect lipid transfection reagent (QIAGEN) for 10 min at room temperature. The DNA to Superfect ratio was 1:3 for all transfections. The DNA/Superfect complexes were then dripped onto the cells and placed in a humidified 37°C incubator with 5% CO2 for 48 h.
RNA harvest and RT-PCR
Forty-eight hours after transfection, cells were trypsinized and total RNA was harvested using QIAGEN's RNeasy Plus Miniprep kit. The mix controls were made after trypsinization but before cell lysis and contained one-third of the cells from each relevant well. The total RNAs were quantified using a NanoDrop spectrophotometer (NanoDrop Technologies).
Equal amounts of each total RNA were used in a one-step RT-PCR reaction with appropriate controls. The −RT control used total RNA from a cotransfection sample in a reaction containing only Platinum Taq (Invitrogen). The water control was a “no template” control for the RT-PCR. We used Invitrogen's Superscript III reverse transcriptase/Platinum Taq polymerase and the following conditions for specifically detecting the trans-spliced product: 30 min at 56°C, 2 min at 94°C, 25 cycles of (94°C for 30 sec/60°C for 30 sec/72°C for 30 sec), then 7 min at 72°C with a final hold at 4°C. The primers used to specifically amplify trans-spliced products bound to the 5′PTM-specific tag and β-globin exon 2. The RT-PCR products were then gel-purified using high-purity agarose-1000 (Invitrogen) and sequenced.
Bsu36I assay
Total RNA from βs/5′-PTM cotransfection experiments was isolated as previously described and amplified via one-step RT-PCR using primers that would amplify both cis- and trans-spliced products (primers bound 5′-UTR and exon 2 of β-globin) using the following conditions: 30 min at 56°C, 2 min at 94°C, 25 cycles of (94°C for 30 sec/57°C for 30 sec/72°C for 15 sec), then 7 min at 72°C with a final hold at 4°C. The RT-PCR products were gel-purified, digested overnight with Bsu36I (NEB), and then run on a 3% agarose-1000 (Invitrogen) gel to resolve the fragments.
HaeIII assay
Total RNA from β-thalassemia/5′-PTM cotransfection experiments was isolated as previously described and amplified via one-step RT-PCR using primers that would amplify both cryptic and normally spliced transcripts (primers bound 5′-UTR and exon 2 of β-globin) using the following conditions: 30 min at 56°C, 2 min at 94°C, 25 cycles of (94°C for 30 sec/55°C for 30 sec/72°C for 15 sec), then 7 min at 72°C with a final hold at 4°C. RT-PCR products resulting from both cryptic and normal splicing were isolated as single gel slices (one per lane) and purified. The purified products were then digested overnight with HaeIII (NEB). The fragments were resolved on a 3.5% agarose-1000 gel (Invitrogen) containing ethidium bromide, and the intensities of normal (133-bp HaeIII fragment) and abnormal (cryptic 166-bp/144-bp fragments or indigestible heteroduplex fragment present after complete HaeIII digestion) splice products were quantified using Quantity One densitometry software (Bio-Rad). Experiments were replicated three to five times, and band density (intensity) was normalized for nucleotide length. The means of the replicates were used to calculate the relative amounts of each band type (per PTM/β-thal mutant combination) and graphed in Figure 5C. One of these ratios was used in calculating the repair efficiency: (normal splice product)/(heteroduplex + cryptic E127 + cryptic E105 + normal splice products). Statistical significance testing was performed on band nucleotide length normalized relative intensity data using Bonferroni pairwise comparison testing with a 95% confidence interval using GraphPad Prism (GraphPad Software Inc.). Each band type/PTM/β-thal mutant combination was tested against the comparable “No PTM” combination to determine if there was a significant difference in relative mean intensity.
ACKNOWLEDGMENTS
We thank Drs. Puneet Seth and Mariano Garcia-Blanco for the DISE sequence used in this study. We also are grateful to Drs. Marilyn Telen and Murat Arcasoy for their insightful suggestions and guidance. We thank David Boczkowski and Claudia Dollins for their critical reading of this manuscript.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.525607.
REFERENCES
- Atweh, G.F., Wong, C., Reed, R., Antonarakis, S.E., Zhu, D., Ghosh, P.K., Maniatis, T., Forget, B.G., Kazazian H.H., Jr A new mutation in IVS-1 of the human β globin gene causing β thalassemia due to abnormal splicing. Blood. 1987;70:147–151. [PubMed] [Google Scholar]
- Audibert, A., Weil, D., Dautry, F. In vivo kinetics of mRNA splicing and transport in mammalian cells. Mol. Cell. Biol. 2002;22:6706–6718. doi: 10.1128/MCB.22.19.6706-6718.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berget, S.M. Exon recognition in vertebrate splicing. J. Biol. Chem. 1995;270:2411–2414. doi: 10.1074/jbc.270.6.2411. [DOI] [PubMed] [Google Scholar]
- Blumenthal, T., Thomas, J. Cis and trans mRNA splicing in C. elegans . Trends Genet. 1988;4:305–308. doi: 10.1016/0168-9525(88)90107-2. [DOI] [PubMed] [Google Scholar]
- Chao, H., Mansfield, S.G., Bartel, R.C., Hiriyanna, S., Mitchell, L.G., Garcia-Blanco, M.A., Walsh, C.E. Phenotype correction of hemophilia A mice by spliceosome-mediated RNA trans-splicing. Nat. Med. 2003;9:1015–1019. doi: 10.1038/nm900. [DOI] [PubMed] [Google Scholar]
- Colah, R., Nadkarni, A., Gorakshakar, A., Phanasgaonkar, S., Surve, R., Subramaniam, P.G., Bondge, N., Pujari, K., Ghosh, K., Mohanty, D. Impact of β globin gene mutations on the clinical phenotype of β thalassemia in India. Blood Cells Mol. Dis. 2004;33:153–157. doi: 10.1016/j.bcmd.2004.05.002. [DOI] [PubMed] [Google Scholar]
- Custodio, N., Carmo-Fonseca, M., Geraghty, F., Pereira, H.S., Grosveld, F., Antoniou, M. Inefficient processing impairs release of RNA from the site of transcription. EMBO J. 1999;18:2855–2866. doi: 10.1093/emboj/18.10.2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallinger, G., Puttaraju, M., Mitchell, L.G., Yancey, K.B., Yee, C., Klausegger, A., Hintner, H., Bauer, J.W. Development of spliceosome-mediated RNA trans-splicing (SMaRT trade mark) for the correction of inherited skin diseases. Exp. Dermatol. 2003;12:37–46. doi: 10.1034/j.1600-0625.2003.120105.x. [DOI] [PubMed] [Google Scholar]
- Danckwardt, S., Neu-Yilik, G., Thermann, R., Frede, U., Hentze, M.W., Kulozik, A.E. Abnormally spliced β-globin mRNAs: A single point mutation generates transcripts sensitive and insensitive to nonsense-mediated mRNA decay. Blood. 2002;99:1811–1816. doi: 10.1182/blood.v99.5.1811. [DOI] [PubMed] [Google Scholar]
- Darwish, H.M., El-Khatib, F.F., Ayesh, S. Spectrum of β-globin gene mutations among thalassemia patients in the West Bank region of Palestine. Hemoglobin. 2005;29:119–132. [PubMed] [Google Scholar]
- de Souza, A.P., Jubier, M.F., Delcher, E., Lancelin, D., Lejeune, B. A trans-splicing model for the expression of the tripartite nad5 gene in wheat and maize mitochondria. Plant Cell. 1991;3:1363–1378. doi: 10.1105/tpc.3.12.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- el-Hazmi, M.A., Warsy, A.S., al-Swailem, A.R. The frequency of 14 β-thalassemia mutations in the Arab populations. Hemoglobin. 1995;19:353–360. doi: 10.3109/03630269509005827. [DOI] [PubMed] [Google Scholar]
- Finta, C., Zaphiropoulos, P.G. Intergenic mRNA molecules resulting from trans-splicing. J. Biol. Chem. 2002;277:5882–5890. doi: 10.1074/jbc.M109175200. [DOI] [PubMed] [Google Scholar]
- Ho, P.J., Rochette, J., Fisher, C.A., Wonke, B., Jarvis, M.K., Yardumian, A., Thein, S.L. Moderate reduction of β-globin gene transcript by a novel mutation in the 5′-untranslated region: A study of its interaction with other genotypes in two families. Blood. 1996;87:1170–1178. [PubMed] [Google Scholar]
- Ingram, V.M. A specific chemical difference between the globins of normal human and sickle-cell anaemia haemoglobin. Nature. 1956;178:792–794. doi: 10.1038/178792a0. [DOI] [PubMed] [Google Scholar]
- Ingram, V.M. Abnormal human haemoglobins. III. The chemical difference between normal and sickle cell haemoglobins. Biochim. Biophys. Acta. 1959;36:402–411. doi: 10.1016/0006-3002(59)90183-0. [DOI] [PubMed] [Google Scholar]
- Kazazian H.H., Jr, Orkin, S.H., Antonarakis, S.E., Sexton, J.P., Boehm, C.D., Goff, S.C., Waber, P.G. Molecular characterization of seven β-thalassemia mutations in Asian Indians. EMBO J. 1984;3:593–596. doi: 10.1002/j.1460-2075.1984.tb01853.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikumori, T., Cote, G.J., Gagel, R.F. Promiscuity of pre-mRNA spliceosome-mediated trans-splicing: A problem for gene therapy? Hum. Gene Ther. 2001;12:1429–1441. doi: 10.1089/104303401750298580. [DOI] [PubMed] [Google Scholar]
- Kohchi, T., Umesono, K., Ogura, Y., Komine, Y., Nakahigashi, K., Komano, T., Yamada, Y., Ozeki, H., Ohyama, K. A nicked group II intron and trans-splicing in liverwort, Marchantia polymorpha, chloroplasts. Nucleic Acids Res. 1988;16:10025–10036. doi: 10.1093/nar/16.21.10025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levasseur, D.N., Ryan, T.M., Reilly, M.P., McCune, S.L., Asakura, T., Townes, T.M. A recombinant human hemoglobin with anti-sickling properties greater than fetal hemoglobin. J. Biol. Chem. 2004;279:27518–27524. doi: 10.1074/jbc.M402578200. [DOI] [PubMed] [Google Scholar]
- Lewis, J.D., Izaurralde, E., Jarmolowski, A., McGuigan, C., Mattaj, I.W. A nuclear cap-binding complex facilitates association of U1 snRNP with the cap-proximal 5′-splice site. Genes & Dev. 1996;10:1683–1698. doi: 10.1101/gad.10.13.1683. [DOI] [PubMed] [Google Scholar]
- Liu, X., Jiang, Q., Mansfield, S.G., Puttaraju, M., Zhang, Y., Zhou, W., Cohn, J.A., Garcia-Blanco, M.A., Mitchell, L.G., Engelhardt, J.F. Partial correction of endogenous ΔF508 CFTR in human cystic fibrosis airway epithelia by spliceosome-mediated RNA trans-splicing. Nat. Biotechnol. 2002;20:47–52. doi: 10.1038/nbt0102-47. [DOI] [PubMed] [Google Scholar]
- Liu, X., Luo, M., Zhang, L.N., Yan, Z., Zak, R., Ding, W., Mansfield, S.G., Mitchell, L.G., Engelhardt, J.F. Spliceosome-mediated RNA trans-splicing with recombinant adeno-associated virus partially restores cystic fibrosis transmembrane conductance regulator function to polarized human cystic fibrosis airway epithelial cells. Hum. Gene Ther. 2005;16:1116–1123. doi: 10.1089/hum.2005.16.1116. [DOI] [PubMed] [Google Scholar]
- Mansfield, S.G., Kole, J., Puttaraju, M., Yang, C.C., Garcia-Blanco, M.A., Cohn, J.A., Mitchell, L.G. Repair of CFTR mRNA by spliceosome-mediated RNA trans-splicing. Gene Ther. 2000;7:1885–1895. doi: 10.1038/sj.gt.3301307. [DOI] [PubMed] [Google Scholar]
- Mansfield, S.G., Clark, R.H., Puttaraju, M., Kole, J., Cohn, J.A., Mitchell, L.G., Garcia-Blanco, M.A. 5′-exon replacement and repair by spliceosome-mediated RNA trans-splicing. RNA. 2003;9:1290–1297. doi: 10.1261/rna.5101903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maquat, L.E. Nonsense-mediated mRNA decay in mammals. J. Cell Sci. 2005;118:1773–1776. doi: 10.1242/jcs.01701. [DOI] [PubMed] [Google Scholar]
- Michel, F., Dujon, B. Conservation of RNA secondary structures in two intron families including mitochondrial-, chloroplast- and nuclear-encoded members. EMBO J. 1983;2:33–38. doi: 10.1002/j.1460-2075.1983.tb01376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milhausen, M., Nelson, R.G., Sather, S., Selkirk, M., Agabian, N. Identification of a small RNA containing the trypanosome spliced leader: A donor of shared 5′ sequences of trypanosomatid mRNAs? Cell. 1984;38:721–729. doi: 10.1016/0092-8674(84)90267-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama, K., Pergolizzi, R.G., Crystal, R.G. Gene transfer-mediated pre-mRNA segmental trans-splicing as a strategy to deliver intracellular toxins for cancer therapy. Cancer Res. 2005;65:254–263. [PubMed] [Google Scholar]
- Perutz, M.F. Structure of hemoglobin. Brookhaven Symp. Biol. 1960;13:165–183. [PubMed] [Google Scholar]
- Powars, D.R., Weiss, J.N., Chan, L.S., Schroeder, W.A. Is there a threshold level of fetal hemoglobin that ameliorates morbidity in sickle cell anemia? Blood. 1984;63:921–926. [PubMed] [Google Scholar]
- Puttaraju, M., Jamison, S.F., Mansfield, S.G., Garcia-Blanco, M.A., Mitchell, L.G. Spliceosome-mediated RNA trans-splicing as a tool for gene therapy. Nat. Biotechnol. 1999;17:246–252. doi: 10.1038/6986. [DOI] [PubMed] [Google Scholar]
- Puttaraju, M., DiPasquale, J., Baker, C.C., Mitchell, L.G., Garcia-Blanco, M.A. Messenger RNA repair and restoration of protein function by spliceosome-mediated RNA trans-splicing. Mol. Ther. 2001;4:105–114. doi: 10.1006/mthe.2001.0426. [DOI] [PubMed] [Google Scholar]
- Roca, X., Sachidanandam, R., Krainer, A.R. Determinants of the inherent strength of human 5′ splice sites. RNA. 2005;11:683–698. doi: 10.1261/rna.2040605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Martin, T., Garcia-Blanco, M.A., Mansfield, S.G., Grover, A.C., Hutton, M., Yu, Q., Zhou, J., Anderton, B.H., Gallo, J.M. Reprogramming of tau alternative splicing by spliceosome-mediated RNA trans-splicing: Implications for tauopathies. Proc. Natl. Acad. Sci. 2005;102:15659–15664. doi: 10.1073/pnas.0503150102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullenger, B.A., Cech, T.R. Ribozyme-mediated repair of defective mRNA by targeted, trans-splicing. Nature. 1994;371:619–622. doi: 10.1038/371619a0. [DOI] [PubMed] [Google Scholar]
- Tahara, M., Pergolizzi, R.G., Kobayashi, H., Krause, A., Crystal, R.G. Trans-splicing correction of CD40 ligand deficiency resulting in naturally regulated correction of a murine model of hyper-IgM X-linked immunodeficiency. Nat. Med. 2004;10:835–841. doi: 10.1038/nm1086. [DOI] [PubMed] [Google Scholar]
- Treisman, R., Orkin, S.H., Maniatis, T. Specific transcription and RNA splicing defects in five cloned β-thalassaemia genes. Nature. 1983;302:591–596. doi: 10.1038/302591a0. [DOI] [PubMed] [Google Scholar]
- Ziff, E.B., Evans, R.M. Coincidence of the promoter and capped 5′ terminus of RNA from the adenovirus 2 major late transcription unit. Cell. 1978;15:1463–1475. doi: 10.1016/0092-8674(78)90070-3. [DOI] [PubMed] [Google Scholar]