Abstract
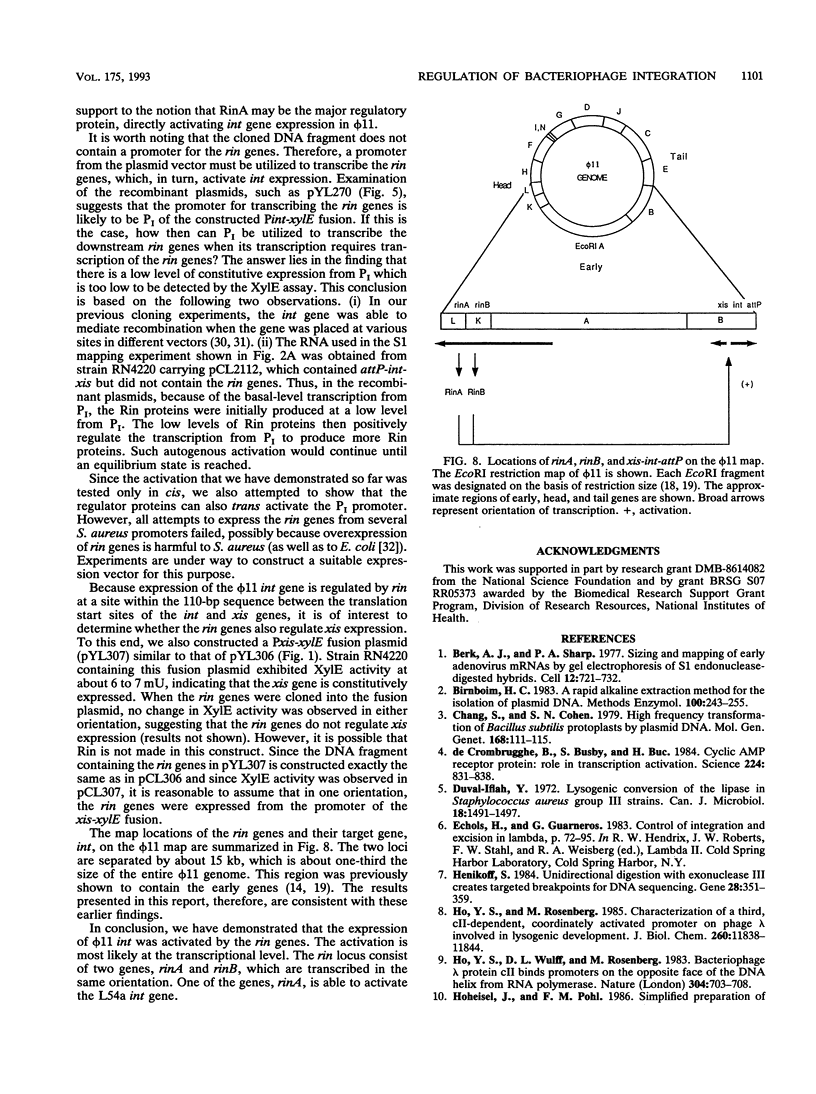
The int gene of staphylococcal bacteriophage phi 11 is the only viral gene responsible for the integrative recombination of phi 11. To study the regulation of int gene expression, we determined the 5' end of the transcript by S1 mapping. The presumed promoter is located just 22 nucleotides upstream of the int open reading frame in a region which is conserved between phi 11 and a closely related staphylococcal phage, L54a. To clone the possible regulatory gene, a vector which contained the reporter gene, xylE, of Pseudomonas putida under the control of the phi 11 int promoter was constructed. Subsequently, a 2-kb DNA fragment from the phi 11 genome, which mapped distal to the int gene, was shown to increase the XylE activity from the int promoter. Sequencing and subsequent deletion analysis of the 2-kb fragment revealed that two phi 11 regulatory genes, rinA and rinB, were both required to activate expression of the int gene. Northern (RNA) analysis suggested that the activation was, at least partly, at the transcriptional level. In addition, one of these regulatory genes, rinA, was capable of activating L54a int gene transcription.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C. A rapid alkaline extraction method for the isolation of plasmid DNA. Methods Enzymol. 1983;100:243–255. doi: 10.1016/0076-6879(83)00059-2. [DOI] [PubMed] [Google Scholar]
- Chang S., Cohen S. N. High frequency transformation of Bacillus subtilis protoplasts by plasmid DNA. Mol Gen Genet. 1979 Jan 5;168(1):111–115. doi: 10.1007/BF00267940. [DOI] [PubMed] [Google Scholar]
- Duval-Iflah Y. Lysogenic conversion of the lipase in Staphylococcus pyogenes group 3 strains. Can J Microbiol. 1972 Sep;18(9):1491–1497. doi: 10.1139/m72-228. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Ho Y. S., Rosenberg M. Characterization of a third, cII-dependent, coordinately activated promoter on phage lambda involved in lysogenic development. J Biol Chem. 1985 Sep 25;260(21):11838–11844. [PubMed] [Google Scholar]
- Ho Y. S., Wulff D. L., Rosenberg M. Bacteriophage lambda protein cII binds promoters on the opposite face of the DNA helix from RNA polymerase. Nature. 1983 Aug 25;304(5928):703–708. doi: 10.1038/304703a0. [DOI] [PubMed] [Google Scholar]
- Hoheisel J., Pohl F. M. Simplified preparation of unidirectional deletion clones. Nucleic Acids Res. 1986 Apr 25;14(8):3605–3605. doi: 10.1093/nar/14.8.3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes D. S., Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981 Jun;114(1):193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- Kreiswirth B. N., Löfdahl S., Betley M. J., O'Reilly M., Schlievert P. M., Bergdoll M. S., Novick R. P. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature. 1983 Oct 20;305(5936):709–712. doi: 10.1038/305709a0. [DOI] [PubMed] [Google Scholar]
- Kretschmer P. J., Egan J. B. Genetic map of the Staphylococcal bacteriophage phi11. J Virol. 1975 Sep;16(3):642–651. doi: 10.1128/jvi.16.3.642-651.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. Y., Iandolo J. J. Integration of staphylococcal phage L54a occurs by site-specific recombination: structural analysis of the attachment sites. Proc Natl Acad Sci U S A. 1986 Aug;83(15):5474–5478. doi: 10.1073/pnas.83.15.5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. Y., Iandolo J. J. Mechanism of bacteriophage conversion of lipase activity in Staphylococcus aureus. J Bacteriol. 1985 Oct;164(1):288–293. doi: 10.1128/jb.164.1.288-293.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. Y., Iandolo J. J. Structural analysis of staphylococcal bacteriophage phi 11 attachment sites. J Bacteriol. 1988 May;170(5):2409–2411. doi: 10.1128/jb.170.5.2409-2411.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löfdahl S., Sjöström J. E., Philipson L. Cloning of restriction fragments of DNA from staphylococcal bacteriophage phi 11. J Virol. 1981 Feb;37(2):795–801. doi: 10.1128/jvi.37.2.795-801.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löfdahl S., Zabielski J., Philipson L. Structure and restriction enzyme maps of the circularly permuted DNA of staphylococcal bacteriophage phi 11. J Virol. 1981 Feb;37(2):784–794. doi: 10.1128/jvi.37.2.784-794.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick R. Properties of a cryptic high-frequency transducing phage in Staphylococcus aureus. Virology. 1967 Sep;33(1):155–166. doi: 10.1016/0042-6822(67)90105-5. [DOI] [PubMed] [Google Scholar]
- Oskouian B., Stewart G. C. Repression and catabolite repression of the lactose operon of Staphylococcus aureus. J Bacteriol. 1990 Jul;172(7):3804–3812. doi: 10.1128/jb.172.7.3804-3812.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattee P. A., Thompson N. E., Haubrich D., Novick R. P. Chromosomal map locations of integrated plasmids and related elements in Staphylococcus aureus. Plasmid. 1977 Nov;1(1):38–51. doi: 10.1016/0147-619x(77)90007-5. [DOI] [PubMed] [Google Scholar]
- Raibaud O., Schwartz M. Positive control of transcription initiation in bacteria. Annu Rev Genet. 1984;18:173–206. doi: 10.1146/annurev.ge.18.120184.001133. [DOI] [PubMed] [Google Scholar]
- Ray C., Hay R. E., Carter H. L., Moran C. P., Jr Mutations that affect utilization of a promoter in stationary-phase Bacillus subtilis. J Bacteriol. 1985 Aug;163(2):610–614. doi: 10.1128/jb.163.2.610-614.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Ye Z. H., Buranen S. L., Lee C. Y. Sequence analysis and comparison of int and xis genes from staphylococcal bacteriophages L54a and phi 11. J Bacteriol. 1990 May;172(5):2568–2575. doi: 10.1128/jb.172.5.2568-2575.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Z. H., Lee C. Y. Nucleotide sequence and genetic characterization of staphylococcal bacteriophage L54a int and xis genes. J Bacteriol. 1989 Aug;171(8):4146–4153. doi: 10.1128/jb.171.8.4146-4153.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zukowski M. M., Gaffney D. F., Speck D., Kauffmann M., Findeli A., Wisecup A., Lecocq J. P. Chromogenic identification of genetic regulatory signals in Bacillus subtilis based on expression of a cloned Pseudomonas gene. Proc Natl Acad Sci U S A. 1983 Feb;80(4):1101–1105. doi: 10.1073/pnas.80.4.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Crombrugghe B., Busby S., Buc H. Cyclic AMP receptor protein: role in transcription activation. Science. 1984 May 25;224(4651):831–838. doi: 10.1126/science.6372090. [DOI] [PubMed] [Google Scholar]