Abstract
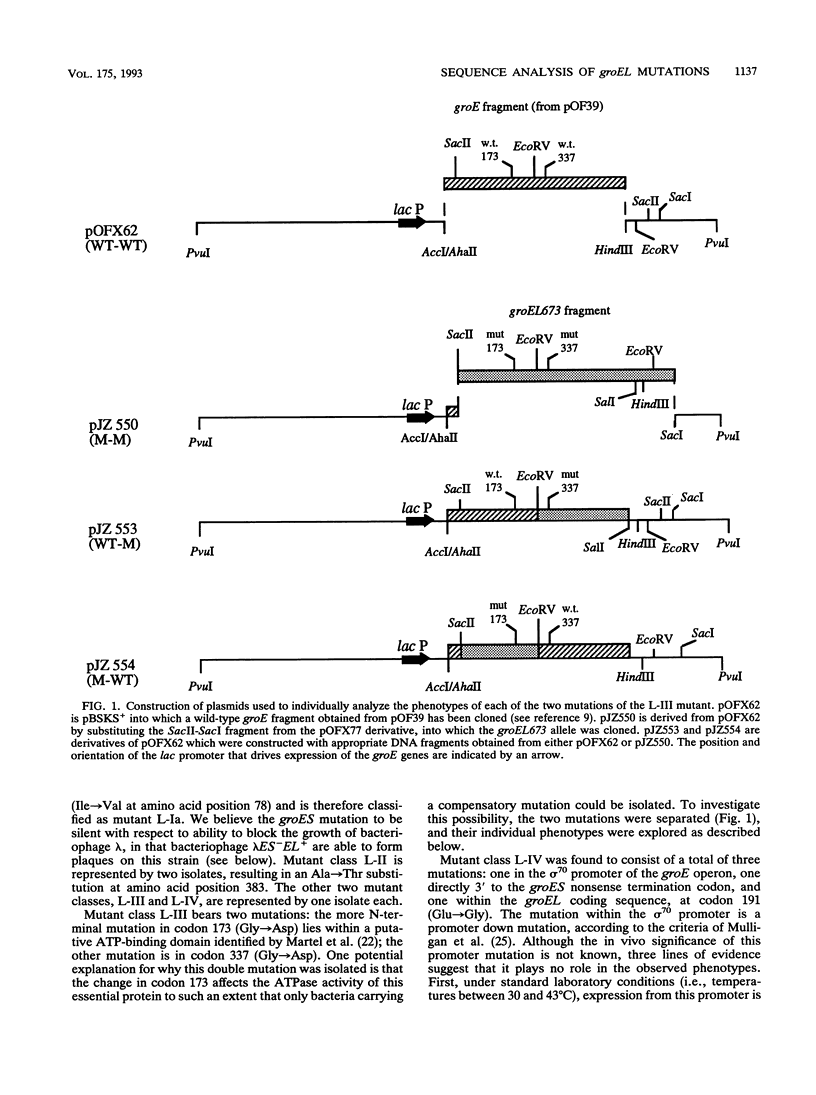
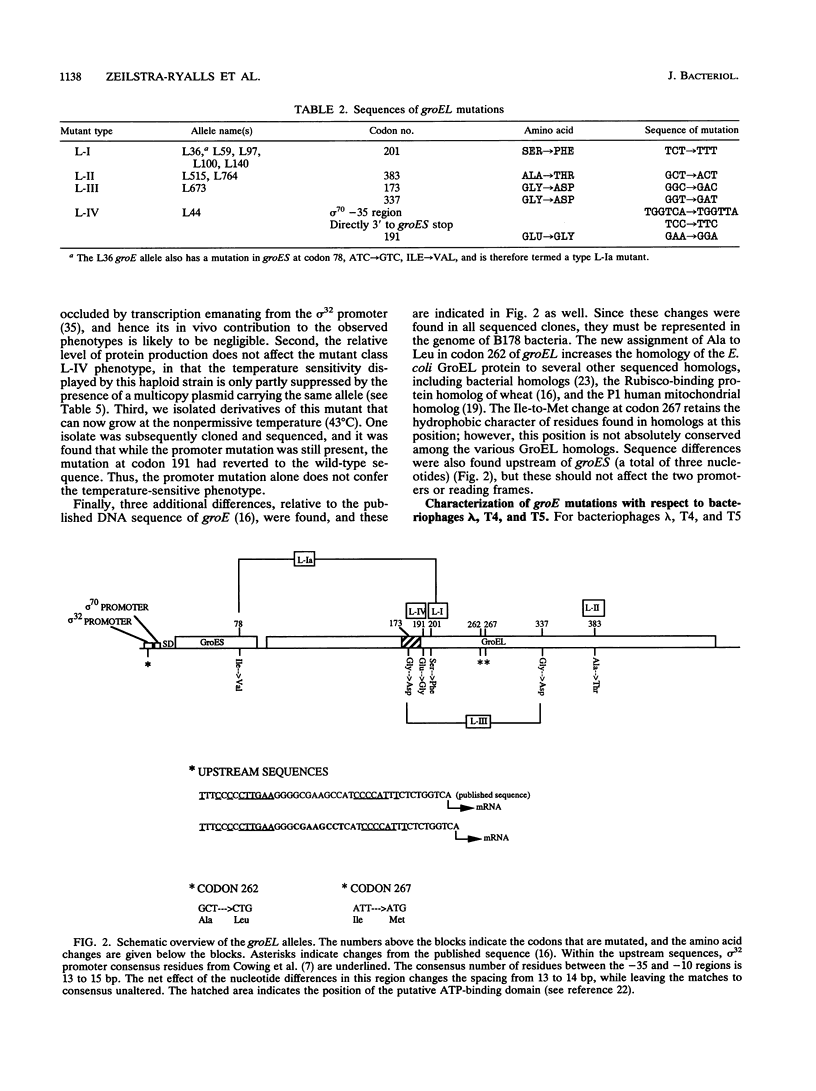
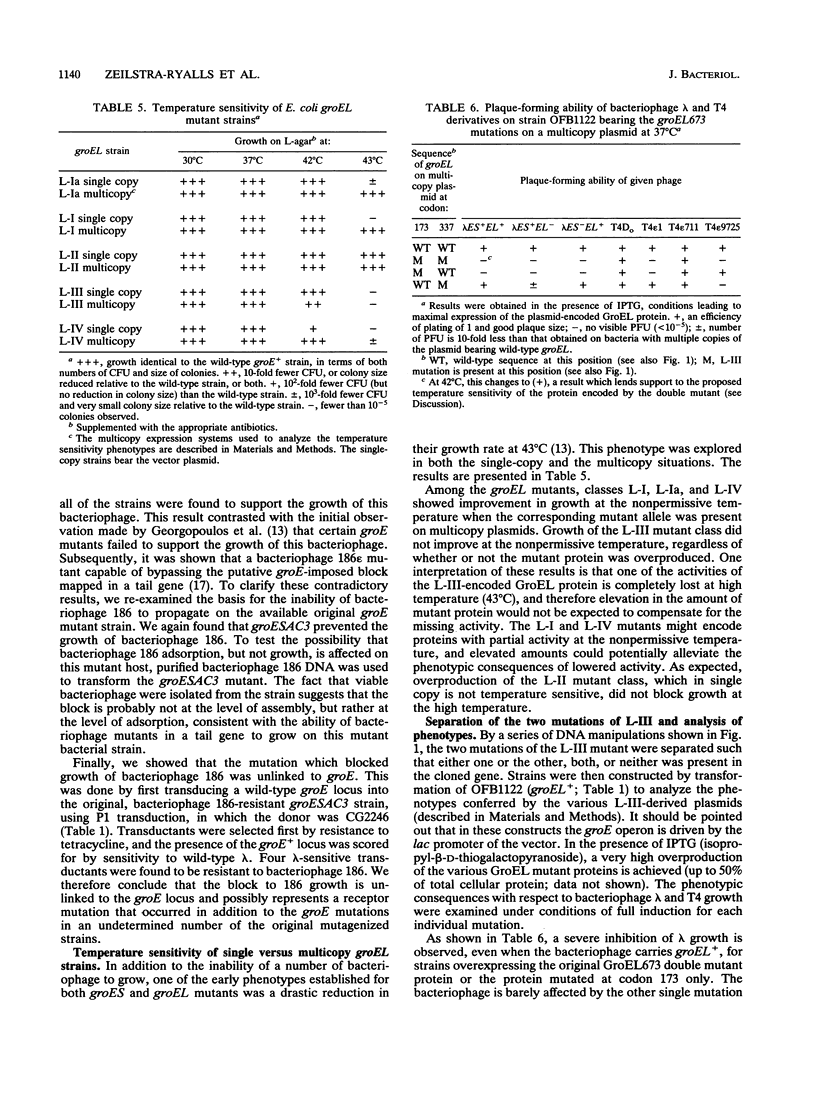
The groES and groEL genes of Escherichia coli have been shown previously to belong to a single operon under heat shock regulation. Both proteins have been universally conserved in nature, as judged by the presence of similar proteins throughout evolution. The GroEL protein has been shown to bind promiscuously to many unfolded proteins, thus preventing their aggregation. ATP hydrolysis by GroEL results in the release of the bound polypeptides, a process that often requires the action of GroES. In an effort to understand GroEL and GroES structure and function, we have determined the nucleotide changes of nine mutant alleles of groEL. All of these mutant alleles were isolated because they block bacteriophage lambda growth. Our sequencing results demonstrate that (i) many of these alleles are identical, in spite of the fact that they were independently isolated, and (ii) most of the different alleles are clustered in the same region of the gene. One of the mutant alleles was shown to possess two nucleotide alterations in the groEL coding phase, one of which is located in a putative ATP-binding domain. The two nucleotide changes were separated by genetic engineering, and each individual change was shown to exert an effect on bacteriophage growth. But, using genetic analyses, we demonstrate that the restriction on bacterial growth at elevated temperatures is conferred only by the mutation within the putative ATP-binding domain. We have cloned the mutant alleles on multicopy plasmids and overexpressed their products. By testing for the ability of bacteriophage either to propagate or to form colonies at 43 degrees C, we have been able to divide the mutant proteins into those with no activity and those with residual activity under the various conditions tested.
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baneyx F., Gatenby A. A. A mutation in GroEL interferes with protein folding by reducing the rate of discharge of sequestered polypeptides. J Biol Chem. 1992 Jun 5;267(16):11637–11644. [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Churchward G., Belin D., Nagamine Y. A pSC101-derived plasmid which shows no sequence homology to other commonly used cloning vectors. Gene. 1984 Nov;31(1-3):165–171. doi: 10.1016/0378-1119(84)90207-5. [DOI] [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C., Hsu L. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc Natl Acad Sci U S A. 1972 Aug;69(8):2110–2114. doi: 10.1073/pnas.69.8.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppo A., Manzi A., Pulitzer J. F., Takahashi H. Abortive bacteriophage T4 head assembly in mutants of Escherichia coli. J Mol Biol. 1973 May 5;76(1):61–87. doi: 10.1016/0022-2836(73)90081-8. [DOI] [PubMed] [Google Scholar]
- Cowing D. W., Bardwell J. C., Craig E. A., Woolford C., Hendrix R. W., Gross C. A. Consensus sequence for Escherichia coli heat shock gene promoters. Proc Natl Acad Sci U S A. 1985 May;82(9):2679–2683. doi: 10.1073/pnas.82.9.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis R. J., van der Vies S. M. Molecular chaperones. Annu Rev Biochem. 1991;60:321–347. doi: 10.1146/annurev.bi.60.070191.001541. [DOI] [PubMed] [Google Scholar]
- Fayet O., Louarn J. M., Georgopoulos C. Suppression of the Escherichia coli dnaA46 mutation by amplification of the groES and groEL genes. Mol Gen Genet. 1986 Mar;202(3):435–445. doi: 10.1007/BF00333274. [DOI] [PubMed] [Google Scholar]
- Fayet O., Ziegelhoffer T., Georgopoulos C. The groES and groEL heat shock gene products of Escherichia coli are essential for bacterial growth at all temperatures. J Bacteriol. 1989 Mar;171(3):1379–1385. doi: 10.1128/jb.171.3.1379-1385.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman D. I., Olson E. R., Georgopoulos C., Tilly K., Herskowitz I., Banuett F. Interactions of bacteriophage and host macromolecules in the growth of bacteriophage lambda. Microbiol Rev. 1984 Dec;48(4):299–325. doi: 10.1128/mr.48.4.299-325.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgopoulos C. P., Hendrix R. W., Casjens S. R., Kaiser A. D. Host participation in bacteriophage lambda head assembly. J Mol Biol. 1973 May 5;76(1):45–60. doi: 10.1016/0022-2836(73)90080-6. [DOI] [PubMed] [Google Scholar]
- Georgopoulos C., Ang D. The Escherichia coli groE chaperonins. Semin Cell Biol. 1990 Feb;1(1):19–25. [PubMed] [Google Scholar]
- Goloubinoff P., Gatenby A. A., Lorimer G. H. GroE heat-shock proteins promote assembly of foreign prokaryotic ribulose bisphosphate carboxylase oligomers in Escherichia coli. Nature. 1989 Jan 5;337(6202):44–47. doi: 10.1038/337044a0. [DOI] [PubMed] [Google Scholar]
- Hemmingsen S. M., Woolford C., van der Vies S. M., Tilly K., Dennis D. T., Georgopoulos C. P., Hendrix R. W., Ellis R. J. Homologous plant and bacterial proteins chaperone oligomeric protein assembly. Nature. 1988 May 26;333(6171):330–334. doi: 10.1038/333330a0. [DOI] [PubMed] [Google Scholar]
- Hocking S. M., Egan J. B. Genetic studies of coliphage 186. I. Genes associated with phage morphogenesis. J Virol. 1982 Dec;44(3):1056–1067. doi: 10.1128/jvi.44.3.1056-1067.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ish-Horowicz D., Burke J. F. Rapid and efficient cosmid cloning. Nucleic Acids Res. 1981 Jul 10;9(13):2989–2998. doi: 10.1093/nar/9.13.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jindal S., Dudani A. K., Singh B., Harley C. B., Gupta R. S. Primary structure of a human mitochondrial protein homologous to the bacterial and plant chaperonins and to the 65-kilodalton mycobacterial antigen. Mol Cell Biol. 1989 May;9(5):2279–2283. doi: 10.1128/mcb.9.5.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keppel F., Lipinska B., Ang D., Georgopoulos C. Mutational analysis of the phage T4 morphogenetic 31 gene, whose product interacts with the Escherichia coli GroEL protein. Gene. 1990 Jan 31;86(1):19–25. doi: 10.1016/0378-1119(90)90109-5. [DOI] [PubMed] [Google Scholar]
- Mehra V., Sweetser D., Young R. A. Efficient mapping of protein antigenic determinants. Proc Natl Acad Sci U S A. 1986 Sep;83(18):7013–7017. doi: 10.1073/pnas.83.18.7013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan M. E., Hawley D. K., Entriken R., McClure W. R. Escherichia coli promoter sequences predict in vitro RNA polymerase selectivity. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):789–800. doi: 10.1093/nar/12.1part2.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reading D. S., Hallberg R. L., Myers A. M. Characterization of the yeast HSP60 gene coding for a mitochondrial assembly factor. Nature. 1989 Feb 16;337(6208):655–659. doi: 10.1038/337655a0. [DOI] [PubMed] [Google Scholar]
- Saraste M., Sibbald P. R., Wittinghofer A. The P-loop--a common motif in ATP- and GTP-binding proteins. Trends Biochem Sci. 1990 Nov;15(11):430–434. doi: 10.1016/0968-0004(90)90281-f. [DOI] [PubMed] [Google Scholar]
- Sternberg N. Properties of a mutant of Escherichia coli defective in bacteriophage lambda head formation (groE). II. The propagation of phage lambda. J Mol Biol. 1973 May 5;76(1):25–44. doi: 10.1016/0022-2836(73)90079-x. [DOI] [PubMed] [Google Scholar]
- Takano T., Kakefuda T. Involvement of a bacterial factor in morphogenesis of bacteriophage capsid. Nat New Biol. 1972 Sep 13;239(89):34–37. doi: 10.1038/newbio239034a0. [DOI] [PubMed] [Google Scholar]
- Tilly K., Murialdo H., Georgopoulos C. Identification of a second Escherichia coli groE gene whose product is necessary for bacteriophage morphogenesis. Proc Natl Acad Sci U S A. 1981 Mar;78(3):1629–1633. doi: 10.1073/pnas.78.3.1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeilstra-Ryalls J., Fayet O., Georgopoulos C. The universally conserved GroE (Hsp60) chaperonins. Annu Rev Microbiol. 1991;45:301–325. doi: 10.1146/annurev.mi.45.100191.001505. [DOI] [PubMed] [Google Scholar]
- Zhou C., Abaigar L., Jong A. Y. A protocol for using T7 DNA polymerase in oligonucleotide site-directed mutagenesis. Biotechniques. 1990 May;8(5):503–503. [PubMed] [Google Scholar]
- Zhou Y. N., Kusukawa N., Erickson J. W., Gross C. A., Yura T. Isolation and characterization of Escherichia coli mutants that lack the heat shock sigma factor sigma 32. J Bacteriol. 1988 Aug;170(8):3640–3649. doi: 10.1128/jb.170.8.3640-3649.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zweig M., Cummings D. J. Cleavage of head and tail proteins during bacteriophage T5 assembly: selective host involvement in the cleavage of a tail protein. J Mol Biol. 1973 Nov 5;80(3):505–518. doi: 10.1016/0022-2836(73)90418-x. [DOI] [PubMed] [Google Scholar]



