Abstract
Surveys of nucleotide diversity in the wild ancestor of maize, Zea mays ssp. parviglumis, have revealed genomewide departures from the standard neutral equilibrium (NE) model. Here we investigate the degree to which population structure may account for the excess of rare polymorphisms frequently observed in species-wide samples. On the basis of sequence data from five nuclear and two chloroplast loci, we found significant population genetic structure among seven subpopulations from two geographic regions. Comparisons of estimates of population genetic parameters from species-wide samples and subpopulation-specific samples showed that population genetic subdivision influenced observed patterns of nucleotide polymorphism. In particular, Tajima's D was significantly higher (closer to zero) in subpopulation-specific samples relative to species-wide samples, and therefore more closely corresponded to NE expectations. In spite of these overall patterns, the extent to which levels and patterns of polymorphism within subpopulations differed from species-wide samples and NE expectations depended strongly on the geographic region (Jalisco vs. Balsas) from which subpopulations were sampled. This may be due to the demographic history of subpopulations in those regions. Overall, these results suggest that explicitly accounting for population structure may be important for studies examining the genetic basis of ecologically and agronomically important traits as well as for identifying loci that have been the targets of selection.
MOLECULAR population genetic approaches have been used increasingly to identify genes that have experienced adaptive evolution (e.g., Ford 2002; Wright et al. 2005; Voight et al. 2006). In a few model systems (e.g., Drosophila, humans, Arabidopsis, and maize), putative targets of selection have been identified as loci with extreme values of population genetic parameters relative to distributions of these statistics derived from a large number of loci (e.g., Parsch et al. 2001; Yamasaki et al. 2005; Toomajian et al. 2006). This approach does not require researchers to assume an explicit model of a population's demographic history. An alternative approach for identifying genes that have been subject to selection is to use likelihood to evaluate the fit of data to models that include specific demographic histories (e.g., Wall et al. 2002; Tenaillon et al. 2004; Wright et al. 2005). For most studies and for most study species, however, inferences of non-neutral evolution have been made by comparing the properties of a sample of DNA sequences to that expected under the standard neutral equilibrium (NE) model.
Because many species have complex demographic histories, central assumptions of the NE model—random mating and constant population size—are likely violated, leading to potentially unreliable inferences of non-neutral evolution (Andolfatto and Przeworski 2000; Akey et al. 2002) even when empirically derived distributions of statistics have been employed (Teshima et al. 2006). Violations of NE assumptions appear to be particularly common in plant species due to population subdivision, metapopulation dynamics, and shifts in patterns of geographic distribution (Innan and Stephan 2000; Wright et al. 2003; Nordborg et al. 2005; Schmid et al. 2005). Nevertheless, the effects of population subdivision on patterns of intraspecific nucleotide diversity remain unclear because most surveys of nucleotide diversity in plant species have used species-wide samples, in which one or a few individuals are selected from multiple geographically isolated populations (hereafter referred to as subpopulations) (reviewed in Wright and Gaut 2004).
Geographically structured subpopulations are expected to diverge as a result of neutral evolutionary processes as long as the effective number of migrants per generation is less than one (Wright 1951; Nagylaki 1980; Charlesworth et al. 2003). Theoretical studies indicate that population structure can also affect patterns of sequence variation. For example, population structure may produce excess linkage disequilibrium (LD) (Li and Nei 1974; Ohta 1982) and skew the frequency spectrum of polymorphism such that there is an excess of rare variants (Tajima 1989). These consequences of subdivision can mimic the effects of positive selection and therefore confound inferences about the role of adaptation in shaping nucleotide variation.
Population structure can also affect inferences about the evolutionary history of genes that have been shaped by natural selection (Charlesworth et al. 1997). When subpopulations are locally adapted to different environmental conditions, the signature of positive selection on ecologically important genes may differ among subpopulations. A sample of sequences taken from across these subpopulations with different evolutionary histories (as in species-wide samples) can produce patterns of nucleotide variation consistent with expectations under balancing selection, rather than under the positive selection that drove evolution (Nordborg and Innan 2003).
Sampling individuals from a single geographically distinct subpopulation can be problematic for different reasons. Wakeley and Aliacar (2001) have shown that the frequency distribution of polymorphism in samples drawn from a single subpopulation can be strongly affected by immigration and population extinction/recolonization. In particular, immigration from differentiated subpopulations and metapopulation dynamics can result in a pattern of diversity similar to that expected following an episode of strong selection in a panmictic stable population (see also Wright and Gaut 2004). Therefore, accurate inferences about whether and how natural selection has shaped sequence variation depend critically on an understanding of the extent and pattern of population structure.
Zea mays ssp. parviglumis (hereafter parviglumis), the closest wild relative of the domesticate, maize (Zea mays ssp. mays), is an important model for investigating the molecular population genetics of natural plant populations. The close relationship of parviglumis to maize has allowed for a wealth of sequence, genomic, and functional information to be applied to this nondomesticated taxon. parviglumis has also been a focus of attention because knowledge of the genomic diversity in parviglumis is needed to identify genes that were targets of artificial selection and to understand the demographic consequences of domestication (Doebley et al. 1997; Wang et al. 1999; Doebley 2004; Wright et al. 2005). Multiple surveys of nucleotide variation in parviglumis (reviewed in Wright and Gaut 2005), including a survey of 774 loci (Wright et al. 2005), have revealed that the majority of loci have negative values of Tajima's D, indicative of a genomewide excess of rare variants relative to NE expectations. As with most molecular population genetic studies in plants, these surveys have relied on species-wide samples drawn from multiple geographically distinct subpopulations. Therefore, population structure may be the reason for, or contribute to, the apparent excess of rare polymorphisms. It is also possible that there is little population structure within parviglumis and that the excess of rare variants is due to only recent population size changes.
Assessing the extent of population structure in parviglumis is important both for determining the forces that shape diversity within species and for correctly inferring the effects of domestication on genomic diversity. For example, if diversity in parviglumis is highly structured among subpopulations, sampling individuals from across the species' range may overestimate diversity in the progenitor population of parviglumis, leading to an overestimate of the strength of the genetic bottleneck associated with maize domestication (Hilton and Gaut 1998; Tenaillon et al. 2004). Similarly, allele frequencies in species-wide samples may not reflect allele frequencies within subpopulations, complicating the identification of targets of selection through the use of genome scans (Teshima et al. 2006). These potential problems would be particularly pronounced if maize were domesticated from one or a few genetically distinct subpopulations. The phylogenetic relationships among parviglumis subpopulations have been investigated using microsatellite diversity (Fukunaga et al. 2005), but the effects of population structure on levels and patterns of nucleotide diversity in parviglumis have not been previously characterized.
In this study, we analyzed sequence variation within and among seven subpopulations of parviglumis at five nuclear and two chloroplast loci. First, we present evidence for significant genetic structure among parviglumis subpopulations and describe patterns of gene flow among subpopulations. Second, we show that species-wide samples lead to estimates of population genetic parameters (π, θ, and Tajima's D) that are biased relative to NE expectations, consistent with previous studies. Third, through comparison of population genetic parameters estimated from subpopulation-specific samples vs. species-wide samples, we show that the genomewide excess of rare variants found in species-wide samples may be caused, in part, by population structure. Finally, we show that the consequences of subpopulation-specific sampling for estimation of population genetic parameters depends on the geographic region from which samples are taken, most likely due to different demographic histories.
MATERIALS AND METHODS
Population sampling:
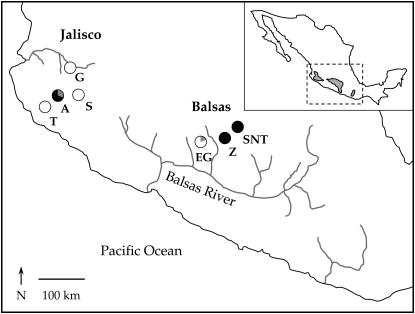
We sampled DNA sequences from seven subpopulations of the outcrossing annual Zea mays L. ssp. parviglumis Iltis and Doebley (supplemental Table 1 at http://www.genetics.org/supplemental/). We grew between 6 and 18 individuals from each population (84 total) with each individual from seed collected from separate maternal plants in natural populations. Seeds were collected in 2001 by Peter Tiffin, Jesus Sanchez (Universidad de Guadalajara), and Nicholas Lauter (University of Illinois). DNA was extracted from leaf material using DNeasy plant kits (QIAGEN, Valencia, CA). Four populations were from the Mexican state of Jalisco, the westernmost section of the species' range, and three populations were from the Balsas River region of the Mexican states of Mexico and Michoacán (Figure 1; supplemental Table 1 at http://www.genetics.org/supplemental/). These regions comprise two geographically distinct portions of the species' range and correspond with the two races of parviglumis distinguished by Wilkes (1967).
Figure 1.—
Geographic distribution of the seven subpopulations of Zea mays ssp. parviglumis included in this study. The four western subpopulations are found in the state of Jalisco and the three eastern subpopulations are found in the Balsas River region of the states of Mexico and Michoacán. The pie diagrams show the proportion of each of the three chloroplast haplotypes found in each subpopulation. The inset map of Mexico shows the entire geographic distribution of the taxon.
Five nuclear and two chloroplast loci, >6500 bases, were PCR amplified and sequenced from each of the 84 DNA samples (supplemental Table 2 at http://www.genetics.org/supplemental/). Three of the nuclear loci include coding regions (adh1 and glb1, chromosome 1; waxy, chromosome 9) and have been the subject of previous surveys of nucleotide diversity in smaller, species-wide samples of parviglumis (Eyre-Walker et al. 1998; Hilton and Gaut 1998; Zhang et al. 2002), and two of the loci are noncoding anonymous markers in maize: asg65 (Asgrow Seed maize clone, chromosome 2) and bnl7 (Brookhaven National Lab maize clone, core bin marker 7.06, probe p-umc168, chromosome 7). The two chloroplast loci (trnT-L, trnL-F) are intergenic spacers (Taberlet et al. 1991). Because teosinte is highly outcrossing, purified PCR products from nuclear genes were cloned into pGEM-T vectors (Promega, Madison, WI) and transformed into competent Escherichia coli cells. Plasmids were purified using a Qiaprep 8 Miniprep kit (QIAGEN). For each individual, one cloned DNA fragment was sequenced. To correct for Taq polymerase errors in cloned fragments, we identified individuals in the alignments that contained singletons and resequenced fragments from these individuals either directly from PCR products or by sequencing four or more clones from a second PCR. Sequences were assembled and aligned manually in BioEdit 7.0.4.1 (Hall 1999).
Population structure and patterns of migration:
We tested for evidence of population subdivision using an analysis of molecular variance (AMOVA; Excoffier et al. 1992) where sequence variation was hierarchically partitioned between the two geographic regions (Jalisco and Balsas; Figure 1), among subpopulations within regions, and among individuals within subpopulations. We also tested for genetic differentiation between pairs of populations using FST (ARLEQUIN; Schneider et al. 2000) and Snn (Hudson 2000). Statistical significance of covariance components (and Φ-statistics) from AMOVAs and pairwise FST's was determined on the basis of the distribution of values obtained from 10,000 permutations of the data under panmixis. The statistical significance of pairwise Snn values was determined by permuting the data 1000 times in DnaSP v 4.0.
Although FST is theorectially related to migration rates, estimating migration from FST is problematic because of biologically unrealistic assumptions, including equal population sizes and symmetric migration among populations (Whitlock and McCauley 1999). Examining pairwise FST's can also lead to unreliable inferences about patterns of migration because of interdependence among multiple populations (Fu et al. 2003). To avoid these problems, we estimated migration rates using the Bayesian version of LAMARC 2.02 (Kuhner et al. 2005), which accounts for the genealogical relationships among alleles and allows for asymmetrical migration between subpopulations, unequal population sizes, and population size changes (Beerli and Felsenstein 1999). The Bayesian approach may provide better estimates of parameters for sparse data sets, where the maximum-likelihood approach commonly fails to converge (Beerli 2006). We obtained asymmetric estimates of migration rates (effective number of migrants per generation) between populations (γ = 4Nemi) from the product of Mi = mi/μ and θi = 4Neμ on the basis of all five nuclear genes. We also simultaneously examined demographic history by obtaining estimates of the exponential population growth rate parameter, g [where θt = θpresentexp(−gt)], for each subpopulation. Default priors were used for recombination and migration; we adjusted priors for θ by specifying a linear density and a lower and upper limit of 0.001 and 0.1; these values encompass the range of estimates from previous studies of these genes in Zea, as well as estimates from this study.
We conducted two runs of LAMARC, each with one 1500-sample initial chain and one 100,000-sample final chain. The analysis was conducted with replication of chains and adaptive heating (Metropolis-coupled Markov chain Monte Carlo), where chains are repeated using different initial genealogies and where each chain is split into multiple searches, allowing for better sampling of parameter space. The results of different runs of LAMARC were very similar and therefore we present one set of results.
Patterns of nucleotide polymorphism:
We tested for the effects of population structure on patterns of nucleotide polymorphism by comparing estimates of population genetic parameters (π, θ, and Tajima's D) from species-wide samples and subpopulation-specific samples. Species-wide surveys are typically conducted by sampling one to a few individuals from multiple geographically isolated populations or ecotypes rather than from many individuals per population, as in our data set. Therefore, to simulate a species-wide survey using our data set, we resampled our entire data set by drawing two individuals from each of the seven subpopulations and estimating population genetic parameters. The resulting set of 14 sequences provides an adequate sample size for obtaining accurate estimates of population genetic parameters (Pluzhnikov and Donnelly 1996). This procedure was repeated for a total of 1000 iterations. Subpopulation-specific estimates of π, θ, and Tajima's D were obtained directly from our data using DnaSP v.4.0 (Rozas et al. 2003).
To test whether species-wide and subpopulation-specific estimates of polymorphism differed from NE expectations, we compared our estimates of population genetic parameters to distributions obtained from coalescent simulations. Coalescent simulations were conducted using the standard Wright–Fisher neutral model, which assumes a large, panmictic population of constant size, and allowing for recombination. Simulations included 1000 replicates of 14 sequences drawn from our entire data set of sequences from seven populations (hereafter referred to as NE expectations). Our simulations were conditioned on empirical estimates of θ and recombination (4Nc) obtained from our entire data set. The simulations were conducted using the program described in Tenaillon et al. (2004), which is a modified version of the standard method described in Hudson (2002).
Linkage disequilibrium:
To examine the consequences of population structure for patterns of linkage disequilibrium, we obtained estimates of the population recombination rate (ρ), which is inversely proportional to linkage disequilibrium, for each subpopulation at each locus. Analyses were implemented in LDhat (http://www.stats.ox.ac.uk/∼mcvean/LDhat), which estimates ρ using the coalescent method of Hudson (2001). Because the ability to detect recombination was influenced by levels of nucleotide diversity—i.e., population recombination rate estimates were positively related to levels of nucleotide diversity (F = 22.3; P < 0.001; R2 = 0.40)—we scaled measures of ρ by θ (see Haddrill et al. 2005).
RESULTS
Population structure and patterns of migration:
Hierarchical AMOVAs provided no evidence that sequence variation was partitioned between the two geographic regions; covariance component estimates were close to zero (often slightly negative) for all five nuclear loci (Table 1). Despite a lack of regional structure, variation was significantly partitioned among subpopulations within geographic regions for all five loci, accounting for 6.0% (asg65) to 31.1% (waxy) of total variation. Individual subpopulations, however, harbored the majority of variation for each locus (74.2–91.5%).
TABLE 1.
Hierarchical analysis of molecular variance for the seven subpopulations from two geographic regions, Jalisco and Balsas
|
adh1
|
asg65
|
bnl7
|
glb1
|
waxy
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Population | % variancea | Φb | % variance | Φ | % variance | Φ | % variance | Φ | % variance | Φ |
| Among regions (Φct) | −5.33 | −0.053 | 2.42 | 0.024 | −2.31 | −0.023 | −2.04 | −0.020 | −5.29 | −0.053 |
| Among subpopulations within regions (Φsc) | 20.49*** | 0.195 | 6.04 | 0.062 | 23.92*** | 0.234 | 15.04*** | 0.147 | 31.13*** | 0.296 |
| Among individuals within subpopulations (Φst) | 84.85*** | 0.152 | 91.54 | 0.085 | 78.39*** | 0.216 | 87.00*** | 0.130 | 74.16*** | 0.258 |
P < 0.001.
The percentage of total variance explained by each hierarchical grouping, including the probability of having a more extreme variance component and Φ-statistic than the observed values assessed by permutation tests.
Fixation indices describing the correlation of haplotypes for each level of subdivision relative to a higher-level grouping: Φct, correlation within a region relative to the whole species; Φsc, correlation within populations relative to the region; Φst, correlation within populations relative to the whole species.
Consistent with the results from AMOVAs, pairwise FST's revealed strong genetic differentiation within the Jalisco region, even among closely situated populations (FST range: 0.0079–0.7024; mean: 0.2436; 25 of 30 estimates differed significantly from zero at P < 0.05), (supplemental Table 2 at http://www.genetics.org/supplemental/). By contrast, there was little evidence of differentiation between populations within the Balsas region (FST range: −0.0703–0.1642; mean: 0.0209; only 3 of 15 estimates differed significantly from zero). Genetic differentiation between geographic regions was intermediate between that found within the two regions (FST range: −0.0316–0.6586; mean: 0.1531). While some between-region population pairs were significantly differentiated, populations A and G (Jalisco) showed little or no differentiation from populations EG and Z (Balsas). The absence of differentiation between these population pairs appears to have contributed to the lack of regional structure indicated by AMOVAs. Patterns of population structure were largely consistent among the five loci, with adh1 and asg65 showing the weakest and waxy the strongest signal of population genetic subdivision (supplemental Table 2 at http://www.genetics.org/supplemental/). Analyses of population differentiation using Snn produced results similar to that from FST (supplemental Table 3 at http://www.genetics.org/supplemental/).
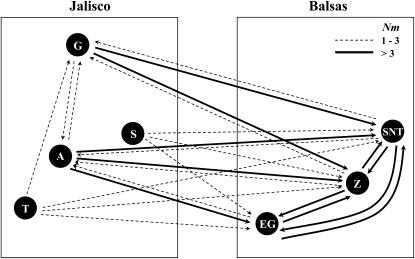
The coalescent-based analysis implemented in LAMARC suggested that migration between geographic regions was asymmetrical, with gene flow occurring principally from western Jalisco subpopulations (especially A and G) into the three eastern Balsas subpopulations. Rates of migration (4Nm) from population A or G to Balsas populations ranged from 3.6 to 10.6 and from 0.9 to 5.0 effective migrants/generation, respectively. Rates of migration from any Balsas population into any Jalisco population were generally much <1.0 and not >2.3. Consistent with the amount of genetic differentiation measured by pairwise FST and Snn (supplemental Tables 2 and 3 at http://www.genetics.org/supplemental/), rates of migration were very high among populations within the Balsas region and low among populations within the Jalisco region (Figure 2).
Figure 2.—
Asymmetric migration rates between pairs of subpopulations within and between the two geographic regions (indicated by boxes) estimated using the Bayesian version of LAMARC. Arrows indicate the direction of migration inferred from the analysis. Migration rates less than one are not shown.
Despite limited sequence variation, the chloroplast loci provided evidence of population subdivision and some suggestion of regional differentiation. Sequences from three Jalisco subpopulations (G, S, and T) were identical and differed from two Balsas populations (SNT and Z) at one SNP (in trnL-F) and a dinucleotide repeat motif (in trnT-L) (Figure 1). One subpopulation in each region, however, harbored haplotypes dominant in the other geographic region (A and EG). Jalisco subpopulation A was fixed for the trnT-L haplotype present in Balsas subpopulations SNT and Z and polymorphic for the trnL-F haplotype. Individuals from the Balsas population EG shared the trnL-F haplotype and five of six individuals shared the trnT-L haplotype with Jalisco populations (G, S, and T).
Population growth:
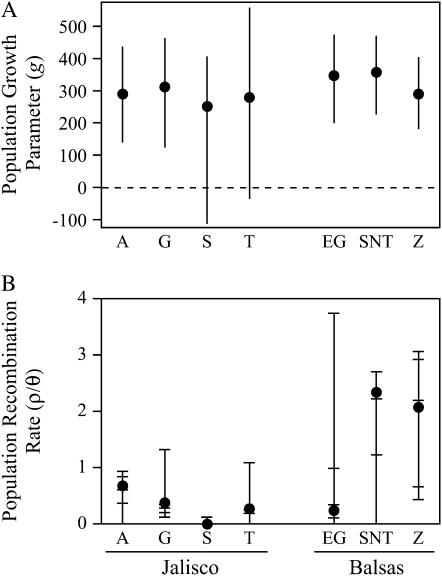
The LAMARC analysis suggested that all three Balsas subpopulations along with Jalisco subpopulations A and G have expanded in size (Figure 3). Large and positive values of the exponential growth parameter, g [where θt = θpresentexp(−gt)], indicate population expansion whereas negative values indicate population shrinkage; the scale of g is not symmetrical and thus relatively small positive values (g = 10) may indicate little or no growth while small negative values (g = −10) may indicate important population size declines (LAMARC documentation: http://evolution.gs.washington.edu/lamarc/). For all Balsas subpopulations and Jalisco subpopulations A and G, 95% C.I.'s of the population growth parameter, g, were >100, indicating population growth (Figure 3A). For subpopulations S and T, 95% C.I.'s for g encompassed positive and negative values, providing no evidence for recent population size changes. We also tested for evidence of population growth using Fs (Fu 1997) and R2 (Ramos-Onsins and Rozas 2002). These analyses also suggested population growth in Balsas subpopulations but no strong support for growth in any Jalisco subpopulation (supplemental Table 4 at http://www.genetics.org/supplemental/).
Figure 3.—
Comparison of population growth and the population recombination rate for each of the seven subpopulations from the Jalisco and Balsas regions. (A) The most probable estimates (±95% C.I.'s) of the population growth parameter, g, from the Bayesian LAMARC analysis. (B) The composite maximum-likelihood estimate of the population recombination rate (ρ/θ) from the five nuclear loci (solid circles). Estimates of ρ/θ from each of the five loci are shown for each subpopulation (horizontal bars) along with the full range of estimates (vertical bars).
Tests for deviations from NE expectations:
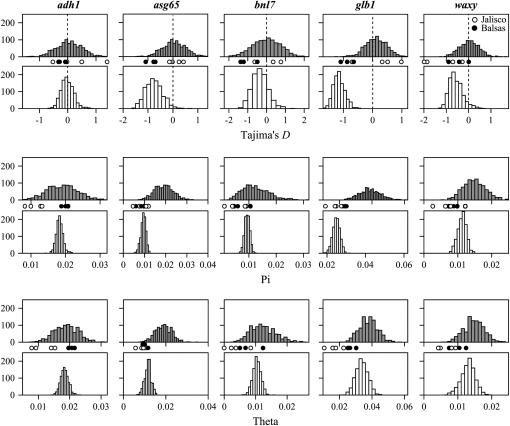
We tested for deviations from NE expectations by comparing our empirical estimates of population genetic parameters (from species-wide and subpopulation-specific samples) to expected distributions obtained from coalescent simulations. Overall, both species-wide and subpopulation-specific samples had lower levels of diversity than NE expectations from coalescent simulations (Table 2; supplemental Table 5 at http://www.genetics.org/supplemental/); however, only species-wide samples had consistently lower (more negative) Tajima's D values than coalescent distributions (Table 3; supplemental Table 6 at http://www.genetics.org/supplemental/). The distributions of species-wide θ and π, obtained from resampling the entire data set, had significantly lower means and variances than the distributions of these statistics obtained from coalescent simulations (NE expectations) (Figure 4; supplemental Table 6 at http://www.genetics.org/supplemental/). Subpopulation-specific estimates of θ and π were highly variable but tended to be lower than NE expectations, particularly for Jalisco populations (θ: 0–0.0302; π: 0–0.0235; Figure 4; Table 2). The difference between the empirical estimates and NE expectations suggests that nonequilibrium demographic processes or population structure have influenced levels of diversity.
TABLE 2.
Nucleotide polymorphism, measured by θ and π, for subpopulation-specific and species-wide samples of five nuclear genes
|
adh1
|
asg65
|
bnl7
|
glb1
|
waxy
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Population | θ | π | θ | π | θ | π | θ | π | θ | π |
| Jalisco subpopulations | ||||||||||
| A | 0.0158 | 0.0157 | 0.0105 | 0.0111 | 0.0023* | 0.0091 | 0.0227* | 0.0185* | 0.0094 | 0.0080* |
| G | 0.0141 | 0.0157 | 0.0090 | 0.0087 | 0.0086 | 0.0092 | 0.0189* | 0.0212* | 0.0085* | 0.0066* |
| S | 0.0093* | 0.0082* | 0.0092 | 0.0101 | 0.0000* | 0* | 0.0157* | 0.0189* | 0.0048* | 0.0028* |
| T | 0.0078* | 0.0100* | 0.0055* | 0.0054* | 0.0039* | 0.0032* | 0.0105* | 0.0112* | 0.0044* | 0.0025* |
| Balsas subpopulations | ||||||||||
| EG | 0.0211 | 0.0203 | 0.0098 | 0.0087 | 0.0074 | 0.0059 | 0.0252* | 0.0218* | 0.0079* | 0.0076* |
| SNT | 0.0205 | 0.0203 | 0.0094 | 0.0072 | 0.0050* | 0.0035* | 0.0302 | 0.0235* | 0.0104 | 0.0091 |
| Z | 0.0194 | 0.0186 | 0.0113 | 0.0096 | 0.0121 | 0.0107 | 0.0260* | 0.0216* | 0.0121 | 0.0098 |
| Species-wide | 0.0185 | 0.0173 | 0.0147 | 0.0097 | 0.0128 | 0.0094 | 0.0412 | 0.0214* | 0.0153 | 0.0091 |
Population-specific estimates shown in italics differed significantly from the resampled distribution using species-wide sampling. Asterisks denote values that differ significantly from the coalescent distributions (P < 0.05). For all significant differences, population-specific estimates were less than resampled and coalescent distributions.
TABLE 3.
Tajima's D for subpopulation-specific and species-wide samples of five nuclear genes
| Population | adh1 | asg65 | bnl7 | glb1 | waxy |
|---|---|---|---|---|---|
| Jalisco subpopulations | |||||
| A | −0.032 | 0.232 | 0.729 | −0.728* | −0.352 |
| G | 0.479 | −0.120 | 0.301 | 0.519 | −0.710 |
| S | −0.592 | 0.454 | — | 0.955* | −1.811* |
| T | 1.567* | −0.089 | −0.789 | 0.341 | −1.989* |
| Balsas subpopulations | |||||
| EG | −0.236 | −0.724 | −1.242 | −0.874* | 0.047 |
| SNT | −0.041 | −1.145 | −1.316 | −1.135* | −0.442 |
| Z | −0.221 | −0.706 | −0.524 | −0.702* | −0.858* |
| Species-wide | −0.213 | −1.114 | −0.830 | −1.568* | −1.202* |
Values in italics differ significantly from the resampled distributions based on species-wide sampling. Asterisks denote values that differ significantly from coalescent distributions (P < 0.05).
Figure 4.—
Distributions of Tajima's D, π, and θ for the five nuclear loci from (i) coalescent simulations of a single panmictic population (shaded histograms) and (ii) resampled distributions generated by randomly selecting sequences from the species-wide pool of sequences (open histograms). Subpopulation-specific estimates of each parameter are denoted by open circles (Jalisco) and solid circles (Balsas).
Under mutation-drift equilibrium, both θ and π estimate 4Nμ, and Tajima's D is not expected to deviate significantly from zero. Similar to previous surveys of nucleotide diversity in parviglumis, estimates of D from our species-wide sample were consistently negative (D = −1.568 to −0.213; Table 3) and the mean D value for the five genes (−0.984) was significantly less than zero (t = 4.36; P = 0.0121). Although none of the species-wide estimates deviated significantly from zero under the conservative assumption of no recombination, three of five values (for asg65, glb1, waxy) were less than the lower bound of the 95% C.I. obtained from our coalescent simulations conducted with recombination (Figure 4; supplemental Table 6 at http://www.genetics.org/supplemental/). These results indicate that species-wide samples contain an excess of rare variants relative to NE expectations. Subpopulation-specific estimates of Tajima's D varied widely and included both negative and positive values (D = −1.989–1.567; Table 3). However, we did not observe a strong and consistent skew to D estimates as in species-wide samples. Of 19 D estimates from Jalisco subpopulations, 5 fell outside of the coalescent 95% C.I. (3 lower and 2 higher than expected) and 4 of 15 D estimates from Balsas subpopulations fell below the coalescent 95% C.I. (Figure 4; Table 3). Overall, subpopulation-specific estimates showed a less consistent deviation from NE expectations compared to species-wide samples.
Tests for differences between subpopulation-specific and species-wide samples:
We tested whether the empirical sampling approach influenced estimates of poplation genetic parameters by comparing each of our subpopulation-specific estimates to resampled distributions of species-wide samples. Overall, we found that estimates of nucleotide diversity from subpopulation-specific samples tended to be lower than species-wide samples, with 20 of 35 estimates falling below the 95% C.I.'s for species-wide samples (Figure 4; Table 2). However, this difference was driven mainly by subpopulations from Jalisco; Balsas subpopulations have levels of nucleotide diversity similar to those of species-wide samples. Subpopulation-specific estimates of θ fell below the 95% C.I.'s from species-wide resampled distributions for 15 of 20 estimates from Jalisco subpopulations but only for 5 of 15 estimates from Balsas subpopulations (Figure 4; Table 2). When directly comparing estimates from the two regions, we found that the mean of the population estimates of θ was significantly lower for Jalisco than Balsas populations for the two most variable loci, adh1 (Wilcoxon sign rank test; θ: P = 0.05; π: P < 0.05; n = 7) and glb1 (θ: P = 0.05; π: P = 0.05; n = 7). The same trend was observed for θ in the other three nuclear loci, but the differences between geographic regions were not significant. Levels of nucleotide variation measured using π showed similar patterns to those for θ (Figure 4; Table 2).
The frequency spectrum of polymorphism, as measured by Tajima's D, tended to be higher (closer to zero) in subpopulation-specific estimates than that found in species-wide samples (mean D: subpopulation specific = −0.37, species-wide = −0.99), with 14 of 35 estimates falling outside of the 95% C.I.'s for species-wide samples (Figure 4, Table 3). As with estimates of diversity, we found that the frequency spectrum of individual Jalisco subpopulations often deviated from that of species-wide samples, whereas spectra from Balsas subpopulations closely resembled those from species-wide samples. Estimates of D from Jalisco populations ranged widely from −1.989 to 1.567 with a mean near zero (mean = −0.09) with approximately half of the values less than zero and half greater than zero (Figure 4; Table 3). Of 19, 11 fell outside of the resampled 95% C.I., with approximately half of the significant values less than the lower bound and half greater than the upper bound. In contrast to Jalisco, estimates of D for Balsas subpopulations were strongly skewed to negative values (range: −1.32–0.05), with only 3 of 15 values falling below the resampled 95% C.I. A direct comparison of D values between the two geographic regions showed that the mean and variance in Tajima's D was greater for Jalisco than for Balsas subpopulations (Welch's ANOVA: F = 6.433; P = 0.017; Levene's test for unequal variance: F = 4.476; P = 0.042; n = 7).
Linkage disequilibrium:
We examined LD within subpopulations and in species-wide samples using estimates of the population recombination rate scaled by nucleotide diversity (ρ/θ). Overall, subpopulation-specific estimates tended to exhibit higher LD (lower ρ/θ) than species-wide samples across the five nuclear loci (supplemental Table 6 at http://www.genetics.org/supplemental/). Despite this overall pattern, we found regional differences in subpopulation-specific estimates of LD. Figure 3B shows composite likelihood scores for ρ/θ for each subpopulation along with the range of locus-specific estimates. Linkage disequilibrium was relatively high in Jalisco subpopulations, whereas Balsas subpopulations (especially SNT and Z) exhibited lower levels of LD, which were comparable to species-wide estimates (Figure 3B; supplemental Table 6 at http://www.genetics.org/supplemental/). Although the range of values for Balsas subpopulations overlaps with those of Jalisco subpopulations, the composite maximum-likelihood estimates across the five loci are well above the range of values observed for all four Jalisco subpopulations. This pattern of population and regional variation in LD remains the same even when ρ is not scaled by θ.
DISCUSSION
A basic assumption of most statistical tests in molecular population genetics is that sequences are randomly sampled from a panmictic population of constant size. Violating these assumptions can bias estimates of nucleotide diversity and lead to erroneous evidence for positive or balancing selection. Although it is well known that genetic variation in many plant species is structured among subpopulations and that the distribution and abundance of species has shifted repeatedly over time, the vast majority of surveys of sequence variation in plant species have relied on species-wide sampling strategies. In Zea mays ssp. parviglumis, species-wide sampling has revealed genomewide departures from a neutral equilibrium model, as manifest by an excess of uncommon polymorphic variants (Zhang et al. 2002; Tenaillon et al. 2004; Moeller and Tiffin 2005; Wright et al. 2005). Similarly in this study, comparisons of resampled distributions of our entire data set to coalescent distributions indicated that species-wide samples strongly deviated from NE expectations. Our analyses of patterns of nucleotide polymorphism in a geographically explicit context suggest that the excess of uncommon polymorphisms in species-wide samples is due in part to population subdivision.
Population structure and patterns of migration:
Both nuclear and chloroplast loci indicated that nucleotide polymorphism was structured among populations, but without clear geographic patterning. Although subpopulations were sampled from two disjunct regions, we found little evidence for differentiation between these regions. For nuclear loci, each of the subpopulations contained the majority of nuclear diversity (∼80%) and the remainder was harbored among subpopulations within geographic regions. For chloroplast loci, population subdivision was strong, with haplotypes commonly fixed within populations, and there was some suggestion of differentiation between regions although not complete. This partitioning of sequence variation among subpopulations can have important effects on statistical tests of the NE model, even when the majority of polymorphism is harbored within populations. In particular, species-wide samples may often contain an excess of rare variants because multiple subpopulations each contain singleton polymorphisms (Hammer et al. 2003). Thus, as an increasing number of subpopulations is pooled for analysis, the frequency spectrum of polymorphism is likely to show a greater skew away from NE expectations and make the identification of loci under selection increasingly difficult.
Beyond the partitioning of sequence variation among populations, the specific pattern of gene flow among subpopulations can influence levels and patterns of polymorphism found within subpopulations. Our coalescent-based analyses indicated that migration has occurred primarily from two western Jalisco populations (A and G) into eastern Balsas populations, but not the reverse. These results are surprising in light of evidence suggesting that Jalisco populations are derived from Balsas populations (Fukunaga et al. 2005) and that colonization of Jalisco may have occurred from refugia in the Balsas Valley following the most recent glacial maximum (Buckler et al. 2006). It is important to note that inferences about migration from LAMARC are made under the assumption that migration structure has been stable and that populations have persisted for a long time period (Kuhner 2006). Given that these assumptions may be unrealistic for parviglumis, it is possible that asymmetric migration estimates instead reflect shared ancestry (recent founding of populations A and G from Balsas populations) rather than asymmetrical patterns of gene flow.
Subpopulation-specific patterns of nucleotide polymorphism:
To test whether genomewide departures from NE expectations found in species-wide samples may be due to population structure, we compared levels and patterns of polymorphism within individual subpopulations to those from species-wide samples. Overall, we found that subpopulation-specific estimates of θ and π tended to be lower than those from species-wide samples. Moreover, frequency spectra of polymorphism from individual subpopulations were less likely to deviate from NE expectations. Specifically, the average of within-population Tajima's D values was greater (and closer to zero) than the mean of Tajima's D values estimated from our entire sample (mean within-population D = −0.37; mean species-wide D = −0.99); this difference was particularly evident for populations in the Jalisco region (mean D = −0.09). In general, these results suggest that species-wide samples tend to underestimate Tajima's D and to overestimate diversity relative to estimates obtained from local subpopulations. Although subpopulation-specific samples tend not to collectively show consistent deviations from NE expectations, we found high variance among estimates, including strongly positive and negative values. This high variance is consistent with predictions from theoretical studies that have examined the influence of evolutionary and demographic processes on sequence variation within subpopulations (Wakeley and Aliacar 2001; Wright and Gaut 2005).
Similar to the patterns that we identified in parviglumis, molecular population genetic studies in humans have suggested that sampling across multiple subpopulations influences estimates of the extent and pattern of nucleotide polymorphism. Specifically, studies of X-linked and autosomal loci that sampled a few individuals from a geographically diverse set of populations reported more negative values of Tajima's D compared to studies that sampled more intensively from a few populations (Ptak and Przeworski 2002). For loci on the Y chromosome, Hammer et al. (2003) found a negative correlation between Tajima's D and the number of populations pooled for analysis. In both cases, the global sampling approach recovered more rare alleles present in only one or a few subpopulations. Thus, when nucleotide variation is structured among subpopulations, sampling strategy clearly influences the estimation of population genetic parameters and inferences about natural selection and demographic history. The inconsistency in sampling strategy among studies will also limit the ability of researchers to compare loci within genomes and examine patterns across genomes.
Regional variation in patterns of nucleotide polymorphism:
Although sampling sequences from individual subpopulations tended to produce, on average, less consistent skew to the frequency spectrum of polymorphism, our data suggest that this result may depend upon the geographic region from which subpopulations were sampled. In particular, differences in demographic history between the Jalisco and Balsas regions may account for our observed differences in diversity, the frequency spectrum of polymorphism, and linkage disequilibrium.
Jalisco populations had relatively low levels of nucleotide variation, highly variable Tajima's D's, and higher linkage disequilibrium, all of which may reflect the action of random genetic drift during a population bottleneck. This scenario appears to be plausible in light of evidence suggesting that Jalisco populations have been recently founded from glacial refugia in the Balsas Valley (Buckler et al. 2006). In addition to reducing overall variation, a population bottleneck is expected to cause a shift in the frequency spectrum of segregating polymorphisms because rare variants are lost from populations at a higher rate than common variants. Depending on the composition of the ancestral population and the timing of the bottleneck, episodes of drift can result in an excess of either high- or low-frequency variants (Charlesworth et al. 2003). This pattern was evident in the Jalisco region where subpopulation-specific frequency spectra were often strongly skewed, as indicated by positive and negative D values. On the basis of our analysis, we cannot rule out the possibility that low rates of gene flow among differentiated Jalisco subpopulations contribute to the high variance in D and high levels of linkage disequilibrium; however, it does not appear that patterns of gene flow lead to a consistent direction of skew in the frequency spectra within subpopulations.
Unlike Jalisco populations, Balsas subpoplations tended to have high levels of nucleotide variation, consistently negative Tajima's D's, and low linkage disequilibrium, all of which were comparable to species-wide estimates. Given that Balsas populations exhibited little genetic differentiation from one another and that estimates of population genetic parameters from individual subpopulations did not differ significantly from that of species-wide samples, it seems unlikely that population structure can account for these results. Instead, our tests for population size changes from LAMARC and from two other statistical tests (Fs and R2) suggest recent population growth, which could be responsible for the excess of rare variants in Balsas subpopulations. These results, along with comparatively low linkage disequilibrium, also argue against recent bottlenecks or admixture as causes of skewed frequency spectra.
Species-wide samples of diversity from Arabidopsis thaliana (Schmid et al. 2005), Populus tremula (Ingvarsson 2005), and nondomesticated Helianthus annuus (Liu and Burke 2006) are also characterized by an excess of rare variants. In A. thaliana, this skew has been suggested to be the result of population expansion (Innan and Stephan 2000; Kuittinen and Aguadé 2000). More recent analyses have shown that the skew in the frequency spectrum may be more pronounced for nonsynonymous compared to synonymous sites, suggesting that both demographic events and purifying selection contribute to the excess of rare variants (Nordborg et al. 2005). Our results similarly indicate that rare variants were significantly more common for nonsynonymous sites relative to synonymous sites and that this pattern did not differ between the geographic regions (supplemental Figure 1 at http://www.genetics.org/supplemental/). Although purifying selection may contribute to a pattern of polymorphism, it does not appear to explain differences in frequency spectra between the Jalisco and Balsas regions.
Conclusions:
We found that the importance of population structure to nucleotide diversity within parviglumis is dependent upon the geographic region that is sampled. In contrast to the Jalisco region, Balsas subpopulations harbored an excess of rare variants, similar to species-wide samples. Given that maize appears to have been domesticated from parviglumis populations growing in the Balsas region (Doebley 1990; Matsuoka et al. 2002), species-wide samples may provide accurate estimates of levels and patterns of diversity in the progenitor population of maize. Species-wide samples are also likely to be appropriate for characterizing the effects of domestication on nucleotide diversity (e.g., Hilton and Gaut 1998; Tenaillon et al. 2004) and the effects of artificial selection on individual genes (Wang et al. 1999; Whitt et al. 2002; Yamasaki et al. 2005), as well as for conducting genomewide scans to identify genes that evolved in response to artificial selection during domestication (Vigouroux et al. 2002; Wright et al. 2005).
Although species-wide samples may be useful for understanding aspects of maize domestication, surveys sampling multiple subpopulations may provide a more complete view of the processes shaping sequence variation. In addition to population subdivision and neutral evolutionary processes alone, patterns of sequence variation may be influenced by geographically variable selection. In the case of adaptive differentiation, a signature of selection at the molecular level may be undetectable when sampling across populations with different evolutionary histories. Local adaptation is remarkably common in plants (reviewed in Linhart and Grant 1996), and patterns of selection frequently differ among populations and over environmental gradients (Endler 1977, 1986), suggesting that a geographic perspective on molecular population genetic studies may be necessary for understanding the role of demographic and selective factors in shaping nucleotide diversity.
Acknowledgments
We thank Andy Muncaski and Nicholas Lauter for their help collecting parviglumis seeds, N. Lauter and Jesus Sanchez for generously providing us with seeds, Jesse Stringer for help in collecting sequence data, Eric Rynes for help implementing and interpreting the LAMARC analyses, and three anonymous reviewers for valuable comments on the manuscript. Financial support for this work was provided by National Science Foundation grant DEB 0235027 to P.T.
References
- Akey, J. M., G. Zhang, K. Zhang, L. Jin and M. D. Shriver, 2002. Interrogating a high-density SNP map for signatures of natural selection. Genome Res. 12: 1805–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andolfatto, P., and M. Przeworski, 2000. A genome-wide departure from the standard neutral model in natural populations of Drosophila. Genetics 156: 257–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerli, P., 2006. Comparison of Bayesian and maximum-likelihood inference of population genetic parameters. Bioinformatics 22: 341–345. [DOI] [PubMed] [Google Scholar]
- Beerli, P., and J. Felsenstein, 1999. Maximum-likelihood estimation of migration rates and effective population numbers in two populations using a coalescent approach. Genetics 152: 763–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckler, E. S., IV, M. M. Goodman, T. P. Holtsford, J. F. Doebley, and J. Sanchez G., 2006. Phylogeography of the wild subspecies of Zea mays. Maydica 51: 123–134. [Google Scholar]
- Charlesworth, B., M. Nordborg and D. Charlesworth, 1997. The effects of local selection, balanced polymorphism and background selection on equilibrium patterns of genetic diversity in subdivided populations. Genet. Res. 70: 155–174. [DOI] [PubMed] [Google Scholar]
- Charlesworth, B., D. Charlesworth and N. H. Barton, 2003. The effects of genetic and geographic structure on neutral variation. Annu. Rev. Ecol. Syst. 34: 99–125. [Google Scholar]
- Doebley, J., 1990. Molecular evidence and the evolution of maize. Econ. Bot. 44(3, Suppl.): 6–27. [Google Scholar]
- Doebley, J., 2004. The genetics of maize evolution. Annu. Rev. Genet. 38: 37–59. [DOI] [PubMed] [Google Scholar]
- Doebley, J., A. Stec and L. Hubbard, 1997. The evolution of apical dominance in maize. Nature 386: 485–488. [DOI] [PubMed] [Google Scholar]
- Endler, J. A., 1977. Geographic Variation, Speciation, and Clines. Princeton University Press, Princeton, NJ. [PubMed]
- Endler, J. A., 1986. Natural Selection in the Wild. Princeton University Press, Princeton, NJ.
- Excoffier, L., P. E. Smouse and J. M. Quattro, 1992. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131: 479–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre-Walker, A., R. L. Gaut, H. Hilton, D. L. Feldman and B. S. Gaut, 1998. Investigation of the bottleneck leading to the domestication of maize. Proc. Natl. Acad. Sci. USA 95: 4441–4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford, M. J., 2002. Applications of selective neutrality tests to molecular ecology. Mol. Ecol. 11: 1245–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, R. W., A. E. Gelfand and K. E. Holsinger, 2003. Exact moment calculations for genetic models with migration, mutation, and drift. Theor. Popul. Biol. 63: 231–243. [DOI] [PubMed] [Google Scholar]
- Fu, Y.-X., 1997. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147: 915–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukunaga, K., J. Hill, Y. Vigouroux, Y. Matsuoka, G. J. Sanchez et al. 2005. Genetic diversity and population structure of teosinte. Genetics 160: 2241–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddrill, P. R., K. R. Thornton, B. Charlesworth and P. Andolfattto, 2005. Multilocus patterns of nucleotide variability and the demographic and selection history of Drosophila melanogaster populations. Genome Res. 15: 790–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, T. A., 1999. BioEdit: a user-friendly biological sequence alignment editor and program for windows 95/98/NT. Nucleic Acids Symp. Ser. 41: 95–98. [Google Scholar]
- Hammer, M. F., F. Blackmer, D. Garrigan, M. W. Nachman and J. A. Wilder, 2003. Human population structure and its effects on sampling Y chromosome sequence variation. Genetics 164: 1495–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton, H., and B. S. Gaut, 1998. Speciation and domestication in maize and its wild relatives: evidence from the globulin-1 gene. Genetics 150: 863–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, R. R., 2000. A new statistic for detecting genetic differentiation. Genetics 155: 2011–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, R. R., 2001. Two-locus sampling distributions and their application. Genetics 159: 1805–1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, R. R., 2002. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics 18: 337–338. [DOI] [PubMed] [Google Scholar]
- Ingvarsson, P. K., 2005. Nucleotide polymorphism and linkage disequilibrium within and among natural populations of European aspen (Populus tremula L., Salicaceae). Genetics 169: 945–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innan, H., and W. Stephan, 2000. The coalescent in an exponentially growing metapopulation and its application to Arabidopsis thaliana. Genetics 155: 2015–2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhner, M. K., 2006. LAMARC 2.0: maximum likelihood and Bayesian estimation of population parameters. Bioinformatics 22: 768–770. [DOI] [PubMed] [Google Scholar]
- Kuhner, M. K., J. Yamato, P. Beerli, L. P. Smith, E. Rynes et al., 2005. LAMARC. University of Washington, Seattle.
- Kuittinen, H., and M. Aguadé, 2000. Nucleotide variation at the CHALCONE ISOMERASE locus in Arabidopsis thaliana. Genetics 155: 863–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W. H., and M. Nei, 1974. Stable linkage disequilibrium without epistasis in subdivided populations. Theor. Popul. Biol. 6: 173–183. [DOI] [PubMed] [Google Scholar]
- Linhart, Y. B., and M. C. Grant, 1996. Evolutionary significance of local genetic differentiation in plants. Annu. Rev. Ecol. Syst. 27: 237–277. [Google Scholar]
- Liu, A., and J. M. Burke, 2006. Patterns of nucleotide diversity in wild and cultivated sunflower. Genetics 173: 321–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka, Y., Y. Vigouroux, M. M. Goodman, J. Sanchez G., E. S. Buckler et al. 2002. A single domestication for maize shown by multilocus microsatellite genotyping. Proc. Natl. Acad. Sci. USA 99: 6080–6084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeller, D. A., and P. Tiffin, 2005. Genetic diversity and the evolutionary history of plant immunity genes in two species of Zea. Mol. Biol. Evol. 22: 2480–2490. [DOI] [PubMed] [Google Scholar]
- Nagylaki, T., 1980. The strong migration limit in geographically structured populations. J. Math. Biol. 9: 101–114. [DOI] [PubMed] [Google Scholar]
- Nordborg, M., and H. Innan, 2003. The genealogy of sequences containing multiple sites subject to strong selection in a subdivided population. Genetics 163: 1201–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordborg, M., T. T. Hu, Y. Ishino, J. Jhaveri, C. Toomajian et al., 2005. The pattern of polymorphism in Arabidopsis thaliana. PLoS Biol. 3: 1289–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta, T., 1982. Linkage disequilibrium due to random genetic drift in finite subdivided populations. Proc. Natl. Acad. Sci. USA 79: 1940–1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsch, J., C. D. Meiklejohn and D. L. Hartl, 2001. Patterns of DNA sequence variation suggest the recent action of positive selection in the janus-ocnus region of Drosophila simulans. Genetics 159: 647–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluzhnikov, A., and P. Donnelly, 1996. Optimal sequencing strategies for surveying molecular genetic diversity. Genetics 144: 1247–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptak, S. E., and M. Przeworski, 2002. Evidence for population growth in humans is confounded by fine-scale population structure. Trends Genet. 18: 559–563. [DOI] [PubMed] [Google Scholar]
- Ramos-Onsins, S. E., and J. Rozas, 2002. Statistical properties of new neutrality tests against population growth. Mol. Biol. Evol. 19: 2092–2100. [DOI] [PubMed] [Google Scholar]
- Rozas, J., J. C. Sanchez-DelBarrio, X. Messeguer and R. Rozas, 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19: 2496–2497. [DOI] [PubMed] [Google Scholar]
- Schmid, K. J., S. Ramos-Onsins, H. Ringys-Beckstein, B. Weisshaar and T. Mitchell-Olds, 2005. A multilocus sequence survey in Arabidopsis thaliana reveals a genome-wide departure from a neutral model of DNA sequence polymorphism. Genetics 169: 1601–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, S., D. Roessli and L. Excoffier, 2000. Arlequin ver 2.000: a software package for population genetics data analysis. University of Geneva, Geneva. [PMC free article] [PubMed]
- Taberlet, P., L. Gielly, G. Pautou and J. Bouvet, 1991. Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Mol. Biol. 17: 1105–1109. [DOI] [PubMed] [Google Scholar]
- Tajima, F., 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenaillon, M. I., J. U'Ren, O. Tenaillon, and B. S. Gaut, 2004. Selection versus demography: a multi-locus investigation of the domestication process in maize. Mol. Biol. Evol. 21: 1214–1225. [DOI] [PubMed] [Google Scholar]
- Teshima, K. M., G. Coop and M. Przeworski, 2006. How reliable are empirical genomic scans for selective sweeps? Genome Res. 16: 702–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toomajian, C., T. T. Hu, M. J. Aranzana, C. Lister, C. L. Tang et al., 2006. A nonparametric test reveals selection for rapid flowering in the Arabidopsis genome. PLoS Biol. 4: 732–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigouroux, Y., M. McMullen, C. T. Hittinger, K. Houchins, L. Schulz et al., 2002. Identifying genes of agronomic importance in maize by screening microsatellites for evidence of selection during domestication. Proc. Natl. Acad. Sci. USA 99: 9650–9655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voight, B. F., S. Kudaravalli, X. Wen and J. K. Pritchard, 2006. A map of recent positive selection in the human genome. PLoS Biol. 4: 446–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakeley, J., and N. Aliacar, 2001. Gene genealogies in a metapopulation. Genetics 159: 893–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall, J. D., P. Andolfatto and M. Przeworski, 2002. Testing models of selection and demography in Drosophila simulans. Genetics 162: 203–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, R.-L., A. Stec, J. Hey, L. Lukens and J. Doebley, 1999. The limits of selection during maize domestication. Nature 398: 236–239. [DOI] [PubMed] [Google Scholar]
- Whitlock, M. C., and D. D. McCauley, 1999. Indirect measures of gene flow and migration: Fst not equal to 1/(4Nm + 1). Heredity 82: 117–125. [DOI] [PubMed] [Google Scholar]
- Whitt, S. R., L. M. Wilson, M. I. Tenaillon, B. S. Gaut and E. S. Buckler, 2002. Genetic diversity and selection in the maize starch pathway. Proc. Natl. Acad. Sci. USA 99: 12959–12962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkes, H. G., 1967. Teosinte: The Closest Relative of Maize. Bussey Institute, Harvard University, Cambridge, MA.
- Wright, S., 1951. The genetical structure of populations. Ann. Eugen. 15: 323–354. [DOI] [PubMed] [Google Scholar]
- Wright, S. I., and B. S. Gaut, 2005. Molecular population genetics and the search for adaptive evolution in plants. Mol. Biol. Evol. 22: 506–519. [DOI] [PubMed] [Google Scholar]
- Wright, S.I., B. Lauga and D. Charlesworth, 2003. Subdivision and haplotype structure in natural populations of Arabidopsis lyrata. Mol. Ecol. 12: 1247–1263. [DOI] [PubMed] [Google Scholar]
- Wright, S. I., I. V. Bi, S. G. Schroeder, M. Yamasaki, J. F. Doebley et al., 2005. The effects of artificial selection on the maize genome. Science 308: 1310–1314. [DOI] [PubMed] [Google Scholar]
- Yamasaki, M., M. I. Tenaillon, I. V. Bi, S. G. Schroeder, H. Sanchez-Villeda et al., 2005. A large-scale screen for artificial selection in maize identifies candidate agronomic loci for domestication and crop improvement. Plant Cell 17: 2859–2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L., A. S. Peek, D. Dunams and B. S. Gaut, 2002. Population genetics of duplicated disease-defense genes, hm1 and hm2, in maize (Zea mays ssp. mays L.) and its wild ancestor (Zea mays ssp. parviglumis). Genetics 162: 851–860. [DOI] [PMC free article] [PubMed] [Google Scholar]