Abstract
During germ-band extension, Decapentaplegic (Dpp) signals from the dorsal ectoderm to maintain Tinman (Tin) expression in the underlying mesoderm. This signal specifies the cardiac field, and homologous genes (BMP2/4 and Nkx2.5) perform this function in mammals. We showed previously that a second Dpp signal from the dorsal ectoderm restricts the number of pericardial cells expressing the transcription factor Zfh1. Here we report that, via Zfh1, the second Dpp signal restricts the number of Odd-skipped-expressing and the number of Tin-expressing pericardial cells. Dpp also represses Tin expression independently of Zfh1, implicating a feed-forward mechanism in the regulation of Tin pericardial cell number. In the adjacent dorsal muscles, Dpp has the opposite effect. Dpp maintains Krüppel and Even-skipped expression required for muscle development. Our data show that Dpp refines the cardiac field by limiting the number of pericardial cells. This maintains the boundary between pericardial and dorsal muscle cells and defines the size of the heart. In the absence of the second Dpp signal, pericardial cells overgrow and this significantly reduces larval cardiac output. Our study suggests the existence of a second round of BMP signaling in mammalian heart development and that perhaps defects in this signal play a role in congenital heart defects.
BONE morphogenetic proteins (BMPs), a subfamily of the transforming growth factor-β (TGF-β) family of secreted cytokines, are critical for the induction of cardiac mesoderm in both flies and mammals (Cripps and Olson 2002). It has been proposed that BMP ligands signal to the cardiac field multiple times to regulate embryonic heart development in both groups (Zaffran and Frasch 2002). Consistent with this suggestion, we showed that the Drosophila BMP family member Decapentaplegic (Dpp) signals from the dorsal ectoderm to the dorsal mesoderm for a second time (Johnson et al. 2003). We reported that the second round of Dpp signaling represses the expression of the transcription factor Zfh1.
To date, three studies of Zfh1 activity in mesoderm development have been reported. The first study showed that Zfh1 positively regulates Even-skipped (Eve) expression in a subset of heart cells. Subsequently, a Zfh1-binding site was identified in the Eve mesodermal enhancer and a mutational analysis showed that the site was active specifically in heart cells (Su et al. 1999; Knirr and Frasch 2001). Another study showed that misexpression of Zfh1 throughout the mesoderm disrupts the development of dorsal somatic muscles (Postigo et al. 1999).
The Drosophila embryonic heart is composed of two major cell types: contractile cardial cells that form the heart tube and Zfh1-expressing pericardial cells that surround the cardial cells. Pericardial cells can be further divided into subpopulations on the basis of expression of specific genes, including Eve, Odd-skipped (Odd), and Tinman (Tin) (Su et al. 1999; Ward and Skeath 2000; Alvarez et al. 2003). Somatic dorsal muscle cells, positioned just ventral to the pericardial cells, express a unique set of genes and represent a third major cell type within the dorsal mesoderm.
A boundary exists between any two adjacent cell types with differing gene expression profiles (Irvine and Rauskolb 2001). Boundaries separate cells with differing fates, and proper boundary formation is a fundamental aspect of many developmental processes. The boundaries separating cardiac, pericardial, and dorsal muscle cells are established via a multistep process that initiates during germ-band extension. First, combinatorial Dpp, Wingless, and Hedgehog signaling from the dorsal ectoderm specifies the positions of heart precursors and dorsal muscle precursor cells (Xu et al. 1998; Halfon et al. 2000; Klinedinst and Bodmer 2003; Reim and Frasch 2005; Liu et al. 2006). Subsequent specification of heart vs. dorsal muscle fate requires the activity of Ras downstream of the epidermal growth factor and fibroblast growth factor receptors (Carmena et al. 1998a). Once specified, the precursor cells divide and populate the dorsal mesoderm. Notch-regulated asymmetric cell divisions as well as cross-repressive interactions ensure the appropriate segregation of daughter cells into the cardiac, pericardial, and dorsal muscle domains (Carmena et al. 1998b; Ward and Skeath 2000; Jagla et al. 2002; Han and Bodmer 2003). Although boundary-forming mechanisms in the dorsal mesoderm have been characterized, little is known about the mechanisms that maintain these boundaries.
Here we report that a second Dpp signal from the dorsal ectoderm to the mesoderm maintains the boundary between pericardial and dorsal muscle cells. Specifically, loss of the second round of Dpp signaling expands the number of Odd-expressing and the number of Tin-expressing pericardial cells while simultaneously reducing the number of dorsal muscle cells expressing Krüppel and Eve. We show that Dpp maintains the dorsal muscle–pericardial cell boundary via two mechanisms: the restriction of cell proliferation and the regulation of gene expression critical for cell fate. Finally, we show that embryonic pericardial cell overgrowth resulting from the loss of this Dpp signal has a detrimental effect on the function of the larval heart. Larvae without this signal have significantly reduced cardiac output in comparison to wild type.
MATERIALS AND METHODS
Drosophila genetics:
Fly stocks are as described: In(2L)dppd6 and In(2L)dppd12 (St. Johnston et al. 1990), Df(2L)dppd14 (Segal and Gelbart 1985), zfh12 (Lai et al. 1993), CycAC8 (Knoblich and Lehner 1993), lmd1 (Duan et al. 2001), 24B.Gal4 (Brand and Perrimon 1993), tinCΔ4.Gal4 (Lo and Frasch 2001), Prc.Gal4 (Chartier et al. 2002), LE.Gal4 (Glise and Noselli 1997), UAS.Dpp (Staehling-Hampton and Hoffmann 1994), UAS.CA-Tkv (Haerry et al. 1998), UAS.Zfh1.2B (FlyBase at http://flybase.bio.indiana.edu/), and HCH.GFP (Han and Olson 2005). All crosses were conducted at 25°. Standard methods were used to generate recombinant chromosomes when necessary and to identify homozygous mutant embryos (Johnson et al. 2003).
Immunohistochemistry and in situ hybridization:
Immunohistochemistry was performed essentially as described (Johnson et al. 2003). The following primary antibodies were utilized: rabbit α-dMef2 (Bour et al. 1995), guinea pig α-Kr, (Kosman et al. 1998), rabbit α-muscle myosin (Kiehart and Feghali 1986), rabbit α-Odd (Ward and Coulter 2000), mouse α-Prc (Developmental Studies Hybridoma Bank), rabbit α-phospho-histone 3 (Sigma, St. Louis), rabbit α-phospho-Smad1 (Persson et al. 1998), rabbit α-Tin for Figures 4 and 5 and Table 1 (Yin and Frasch 1998), rabbit α-Tin for Figure 6 (Venkatesh et al. 2000), mouse α-Zfh1b (Lai et al. 1991), and rabbit α-lacZ (Organon Teknika, Malvern, PA). Secondary antibodies include biotinylated goat α-rabbit, α-mouse, and α-guinea pig (Vector Laboratories, Burlingame, CA); Alexa Fluor 488- and 633-conjugated goat α-rabbit, α-mouse, and α-guinea pig (Molecular Probes, Eugene, OR); and horseradish peroxidase (HRP)-conjugated goat α-rabbit (Molecular Probes). The Vectastain Elite kit (Vector Laboratories) was employed to detect biotinylated secondary antibodies and the TSA Amplification kit (Molecular Probes) was utilized to detect HRP-conjugated secondary antibodies. A midline cDNA (RE27439), inserted in pFLC-1, was obtained from the Drosophila Genomics Resource Center. A riboprobe was generated, after linearization with XhoI, using the Stratagene (La Jolla, CA) in vitro transcription kit. In situ hybridization was performed essentially as described (Lockwood and Bodmer 2002).
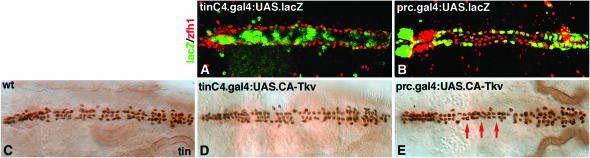
Figure 4.—
Expressing activated Tkv in pericardial cells, but not in cardiac cells, reduces the number of cells expressing Tin. Dorsal view of stage 17 embryos. (A) tinCΔ4.Gal4 drives lacZ expression (green) in a majority of cardiac cells but not in Zfh1-expressing pericardial cells (red). (B) prc.gal4 drives lacZ expression in a subset of Zfh1-expressing pericardial cells. (C) Wild type Tin expression. (D) tinCΔ4.Gal4:UAS.CA-Tkv embryos do not show a significant change in the number of Tin-expressing cells. (E) prc.Gal4:UAS.CA-Tkv embryos show a significant decrease in the number of Tin-expressing cells (see Table 1 for statistics). Note that, in the region indicated by red arrows, only two rows (of presumably cardiac cells) instead of three or four rows in the comparable region of wild-type embryos are present.
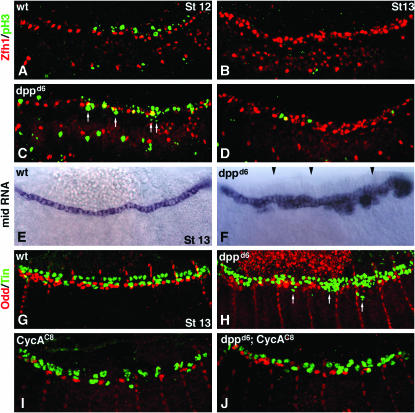
Figure 5.—
Dpp signaling reduces cell proliferation in the dorsal mesoderm by restricting mid expression. (A–D) Merged mesodermal scans of embryos double labeled for Zfh1 (red) and the mitosis marker phospho-histone3 (pH3, green). (A) Wild-type stage 12. Note the limited cell proliferation (pH3 staining) dorsal to the Zfh1-expressing pericardial cells. (B) Wild-type stage 13. No cell proliferation in the dorsal mesoderm. (C) dppd6 stage 12. Cell proliferation is apparent ventrally and medially to the Zfh1-expressing cells. White arrows identify ectopic pH3 staining. Note that apparent colocalization of Zfh1 and pH3 (yellow cells) is an artifact of merging scans. (D) dppd6 stage 13. Cell proliferation persists. (E and F) mid RNA expression stage 13. (E) Wild type. mid expression is restricted to cardiac cells. (F) dppd6. mid expression expands laterally, specifically in the posterior dorsal mesoderm. This image is a composite of four images. Black arrowheads denote points of overlay. The embryo shown is an extreme example. (G–I) Stage 13 embryos double labeled for Tin (green) and Odd (red). (G) Wild type. (H) dppd6. (I) CycAC8. (J) dppd6; CycAC8. The number of Odd-expressing cells in CycAC8 embryos is approximately half the number of wild type. However, the number of Odd-expressing cells in dppd6; CycAC8 double mutants is greater than that observed in CycAC8 embryos. In addition, none of these embryos co-express Tin and Odd. Numerous laterally displaced Tin-expressing cells (arrows in H) are observed in dppd6 embryos.
TABLE 1.
Odd-expressing and Tin-expressing cells in embryos with altered dpp or zfh1 activity
| Mean no. of Odd-skipped expressing cells (SD)
|
Mean no. of Tinman-expressing cells (SD)
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Genotype | Stage 13a | t-test vs. wild typeb | Stage 17a | t-test vs.wild typeb | Stage 13a | t-test vs. wild typeb | Stage 17c | t-test vs. wild typeb |
| Wild type | 44 (2.4) | — | 82.8 (4.4) | — | 73 (3.2) | — | 127 (4.2) | — |
| dppd6 | 52.8 (5.2) | 0.014 | 99.6 (15.3) | 0.021 | 85.4 (4.3) | 0.002 | 144.4 (7.7) | <0.001 |
| LE:Dpp | 43.4 (4.7) | 0.821 | 73.8 (2.2) | 0.002 | ||||
| zfh12 | 46 (5.9) | 0.447 | 71.9 (10.3) | 0.016 | 59.9 (0.7) | 0.005 | 106.8 (5.2) | <0.001 |
| 24B:zfh1; zfh12 | 60 (12.4) | 0.022 | 127.8 (7.3) | 0.002 | 92.0 (8.7) | 0.002 | 162.2 (15.3) | <0.001 |
| dppd6; zfh12 | 43.4 (2.3) | 0.706 | 70 (6.5) | 0.018 | 62.8 (4.5) | 0.006 | 104.0 (3.6) | <0.001 |
| prc:CA-Tkv | 76.3 (3.3) | 0.265 | 118.6 (12.7) | 0.027 | ||||
| tinCΔ4:CA-Tkv | 74.8 (1.2) | 0.347 | 131.4 (4.9) | 0.184 | ||||
Unilateral counts of a bilateral expression pattern.
P-value (numbers in italic are statistically significant).
Bilateral counts of a bilateral expression pattern.
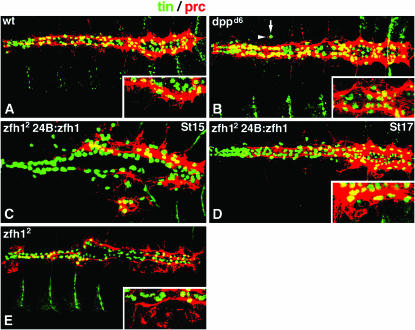
Figure 6.—
Modifying Zfh1 expression alters Prc expression. Embryos double labeled for Prc (red) and Tin (green). Insets are high-magnification merged scans from the posterior regions of the heart. (A) Wild type stage 17. Prc is broadly expressed throughout the pericardial cell domain, most prominently in the posterior. (B) dppd6 stage 17. Prc expression is expanded. Ectopic Tin-expressing cells (arrow) co-express Prc (arrowhead). (C) 24B.Gal4:UAS.Zfh1; zfh12 stage 15 and (D) stage 17. Ventrally positioned Tin-expressing cells co-express Prc. (E) zfh12 stage 17. Prc expression domain is greatly reduced.
Cell counts and statistics:
Cell identities were assigned on the basis of staining intensity and position along the dorsal ventral axis. Cell number quantification was performed as described (Johnson et al. 2003) except that embryos stage 13 and younger were viewed laterally to distinguish dorsal from lateral mesoderm. Statistical analysis of pericardial cell transcription factor expression utilized unilateral (stage 13) or bilateral (stage 15+) cell counts of the entire dorsal mesoderm with a minimum of five embryos assayed per genotype. For the quantification of dorsal muscle transcription factor expression, we assayed the number of nuclei per segment (minimum of 50 segments per genotype). Unpaired, two-tailed t-tests were used to determine whether the difference in the number of expressing cells between the two genotypes was statistically significant.
Larval heartbeat analysis:
First instar larvae containing the heart expressing transgene HCH.GFP were collected 24–28 hr after egg lay and mounted on 60-mm petri plates coated with 0.1% poly-l-lysine (Ted Pella). Larval heartbeats were captured at room temperature for 2 min at a rate of 12 frames/sec with a Roper Cool SNAP ES digital camera (Roper Scientific). Image acquisition and processing was performed with MetaMorph 6.0/6.1 software (Universal Imaging). Custom software was constructed using C++ Borland Builder 6 Enterprise edition that provides a user interface for computer-assisted tracking of individual cells. A pair of cardial cells two to three cell diameters from the anterior end of the heart (a region also known as the aorta; Lovato et al. 2002; Sellin et al. 2006) and a second pair of cardial cells two to three cell diameters from the posterior end of the heart (a region also referred to as the heart proper; Lovato et al. 2002; Sellin et al. 2006) were tracked for each animal. The heartbeat of first instar larvae is discontinuous and tracking was performed only during active contractions. The position of each tracked cell was recorded and the distance between each pair of cells was calculated to determine diastolic and systolic distances for each heartbeat. Heart rate was also determined. The heart rate and pulse distance reported is an average of every heartbeat tracked in all animals of each genotype.
RESULTS
Dpp signaling specifically restricts the number of pericardial cells:
Our previous study suggested that a second round of Dpp dorsal ectoderm-to-mesoderm signaling, stimulated by enhancers located in the dpp disk region, initiates during germ-band retraction (stage 12; Johnson et al. 2003). We refer to this as the second round of signaling because a distinct set of enhancers located in the dpp Haplo-insufficiency (Hin) region activates Dpp dorsal ectoderm-to-mesoderm signaling during germ-band extension (stage 8; Newfeld and Takaesu 2002). Further, our data and that of others (e.g., Kato et al. 2004) revealed that dpp dorsal ectoderm expression driven by the Hin region enhancers persists long after germ-band retraction. These studies showed that Hin-region-driven dpp expression is sufficient for Dpp ectodermal functions such as dorsal closure and dorsal branch migration.
Given these data, it appears that the dppd6 inversion (Figure 1A) prevents the augmentation of dpp expression in the dorsal ectoderm during germ-band retraction that is normally provided by disk region enhancers. The presence of numerous mesodermal phenotypes in dppd6 mutants (Johnson et al. 2003) suggests that the augmentation of dpp expression is necessary to boost Dpp dorsal ectoderm signals so that they can reach the underlying mesoderm. Perhaps there are barriers of distance or extracellular matrix density between these germ layers that must be overcome.
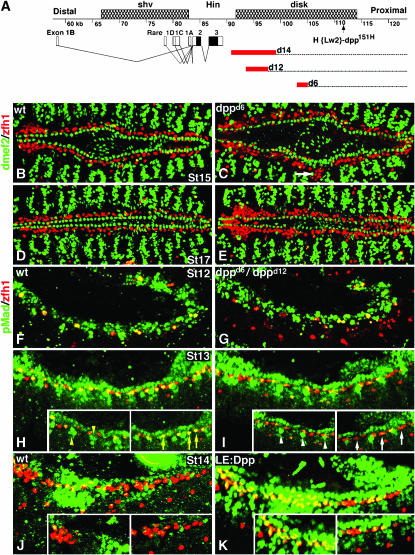
Figure 1.—
dpp mutant embryos display pericardial but not cardiac cell overgrowth and have reduced pMad accumulation in the dorsal mesoderm. (A) Map of the dpp locus showing three genetically defined regions: shortvein (shv), Hin, and disk. The structures of dpp transcripts are shown with black boxes representing the open reading frame and open boxes corresponding to untranslated regions. dpp disk region mutations used in this study are the deficiency dppd14 and the inversions dppd12 and dppd6. Red boxes represent the degree of uncertainty in the position of the distal breakpoint associated with each aberration. The dpp151H enhancer trap is also shown (Johnson et al. 2003). (B–E) Embryos double labeled for dMef2 (green) and Zfh1 (red). (B and C) Stage 15. (B) Wild type. (C) dppd6 embryos contain a significantly greater number of Zfh1-expressing pericardial cells (see Johnson et al. 2003 for statistics) but show no change in the number of dMef2-expressing cells. Note some Zfh1-expressing cells are visible in the dorsal muscle domain (white arrow). (D and E). Stage 17. (D) Wild type. (E) dppd6 embryos continue to display pericardial cell hyperplasia. (F–K) Merged scans of embryos double labeled for pMad (green) and Zfh1 (red). Insets are single mesodermal scans to exclude ectodermal pMad accumulation. (F and G) Stage 12. (F) Wild type. (G) dppd12/dppd6. pMad is detected in all Zfh1-expressing pericardial precursor cells in both genotypes. (H and I) Stage 13. (H) Wild type. pMad is detected in a majority of the Zfh1-expressing cells (yellow arrows) as well as in non-Zfh1-expressing dorsal mesoderm cells (yellow arrowheads). (I) dppd6/dppd12. pMad is not detected in a subset of Zfh1-expressing cells (white arrows). pMad is detected in fewer nonpericardial dorsal mesoderm cells (white arrowheads). (J and K) Stage 14. (J) Wild type. pMad is largely undetectable in Zfh1-expressing cells. (K) LE.Gal4:UAS.Dpp. Ectopic pMad accumulates in Zfh1-expressing and non-Zfh1-expressing dorsal mesoderm cells.
We continued our analysis of dppd6 mutant phenotypes by clearly documenting that Dpp signals from the dorsal ectoderm during germ-band retraction act specifically on pericardial but not on cardiac cells (Johnson et al. 2003). First, we double labeled wild-type and dppd6 embryos with anti-Zfh1 and anti-dMef2 antibodies. Zfh1 and dMef2 have mutually exclusive expression patterns: Zfh1 is expressed in all pericardial cells (Lai et al. 1991; Ward and Skeath 2000), while dMef2 is expressed in all muscle cell lineages, including cardiac cells and dorsal muscle cells (Lilly et al. 1994; Nguyen et al. 1994). dppd6 mutants lack Dpp signals from the dorsal ectoderm during germ-band retraction. In dppd6 embryos, the number of Zfh1-expressing pericardial cells is greater than that of wild type at stage 15 (Figure 1, B and C), indicating that Dpp normally represses Zfh1 expression. In Johnson et al. (2003), we showed utilizing t-tests that differences in Zfh1 cell number between wild-type and dppd6 embryos are statistically significant. Alternatively, the number of dMef2-expressing cardiac cells in dppd6 embryos is not different from wild type at stage 13 (t-test, P = 0.224) or stage 17 (t-test, P = 0.149).
We also noted that the expression domains of dMef2 and Zfh1 remain mutually exclusive in dppd6 embryos although ectopic Zfh1-expressing cells are observed as lateral to their typical location in a region of the dorsal mesoderm usually associated with dorsal muscle cells (Figure 1C, white arrow). In stage 17 embryos, the expression of dMef2 and Zfh1 is mutually exclusive in both wild-type and dppd6 embryos (Figure 1, D and E) although pericardial cell hyperplasia is still apparent in dppd6 embryos. Taken together, these results show that Dpp does not regulate a cell fate choice between cardiac and pericardial cells but that Dpp specifically restricts the number of pericardial cells.
To determine whether Dpp acts directly on pericardial cells or indirectly through an intermediate, we looked for the presence of the phosphorylated form of the Dpp signal transducer Mad (pMad) in Zfh1-expressing cells. During germ-band retraction (stage 12) pMad is widely visible in the ectoderm and in a majority of Zfh1-expressing pericardial precursor cells in both wild-type and dppd12/dppd6 embryos (Figure 1, F and G). In wild type, immediately following germ-band retraction (stage 13), the number of Zfh1-expressing cells increases and a majority of these cells continue to accumulate pMad (Figure 1H, yellow arrows). This observation is consistent with a previous report of pMad accumulation in the dorsal mesoderm (Knirr and Frasch 2001). In contrast, pMad is undetectable in a number of Zfh1-expressing cells in both the anterior and the posterior regions of the dorsal mesoderm in dppd12/dppd6 embryos immediately following germ-band retraction (Figure 1I, white arrows). These pMad data demonstrate that pericardial cells respond to Dpp signals during stages 12 and 13 and that dpp disk region mutations abrogate this aspect of Dpp signaling.
In our previous study, we showed that overexpressing Dpp in the dorsal ectoderm following germ-band retraction using the driver LE.Gal4 causes a loss of Zfh1-expressing cells. If Dpp directly signals to pericardial cells, as suggested above, we would expect to see enhanced pMad expression in LE.Gal4:UAS.Dpp embryos as compared with wild type. Following complete germ-band retraction (stage 14) in wild-type embryos, pMad is detectable in just a few Zfh1-expressing cells (Figure 1J). In LE.Gal4:UAS.Dpp embryos at the same stage pMad is detected in many Zfh1-expressing cells (Figure 1K). Moreover, ectopic pMad is detected in lateral regions of the dorsal mesoderm in LE.Gal4:UAS.Dpp embryos (Figure 1K insets). These results demonstrate that pericardial cells in the dorsal mesoderm are targets of Dpp signaling during germ-band retraction.
Dpp restricts the number of Odd pericardial cells in a zfh1-dependent manner:
Lineage tracing of pericardial-cell-specific transcription factors have identified three distinct cell types within the Zfh1-expressing pericardial cell population: Eve expressing, Odd expressing, and Tin expressing (Su et al. 1999; Ward and Skeath 2000; Alvarez et al. 2003). To identify which pericardial cells responded to Dpp signals during germ-band retraction, we examined the expression of these genes in both dpp and zfh1 mutant backgrounds. Our previous study showed that Eve-expressing pericardial cells are unaffected by dpp disk region mutations (Johnson et al. 2003). However, the number of Odd-expressing pericardial cells (OPCs) is significantly greater in dppd6 stage 13 embryos than in wild type and these supernumerary OPCs persist throughout development (compare Figure 2, A and B, with Figure 2, C and D; Table 1). This result indicates that Dpp normally represses Odd expression.
Figure 2.—
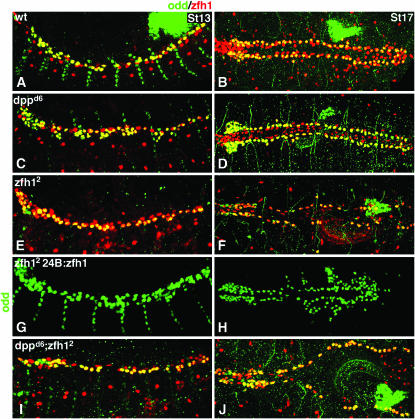
Dpp restricts the number of Odd-skipped pericardial cells via zfh1. Stage 13 embryos in lateral view (A, C, E, G, and I) and stage 17 in dorsal view (B, D, F, H, and J) double labeled for Odd (green) and Zfh1 (red). See Table 1 for statistics. (A and B) Wild type. Odd is expressed in a subset of Zfh1-expressing pericardial cells. (C and D) dppd6. The number of Odd-expressing pericardial cells is significantly increased. All Odd pericardial cells co-express Zfh1. (E and F) zfh12. The number of Odd-expressing cells is comparable to wild type at stage 13 (E) but is significantly less than wild type by stage 17 (F). (G and H) 24B.Gal4:UAS.Zfh1; zfh12. Pan-mesodermal Zfh1 expression in zfh12 mutants not only rescues Odd expression in pericardial cells but also induces ectopic Odd-expressing cells in lateral regions of the mesoderm. (I and J) dppd6; zfh12. The number of Odd-expressing cells is comparable to wild type at stage 13 (I) but is significantly decreased by stage 17 (J), a phenocopy of zfh12 single mutants.
Since Zfh1 is also expressed in all OPCs in dppd6 embryos, we hypothesized that Dpp could restrict Odd expression by restricting the number of cells expressing zfh1. To test this hypothesis, we first assessed the number of OPCs in zfh12 embryos. We chose zfh12 because it is widely utilized and has been described as a genetic and protein null allele (e.g., Lai et al. 1993). However, the exact nature of the zfh12 mutation is unknown. Homozygous zfh12 embryos show no staining with the anti-Zfh1-d antibody (Lai et al. 1993) generated against a fusion protein containing amino acids 648–775 of Zfh1 (Lai et al. 1991). Thus, it is possible that this allele encodes a truncated Zfh1 protein that is capable of partially fulfilling zfh1 functions. Supporting this possibility, the range of mutant phenotypes seen in zfh12 embryos is highly variable (Lai et al. 1993). Further, in our hands, other Zfh1 antibodies (e.g., Zfh1-a recognizing amino acids 1–561 and Zfh1-c recognizing amino acids 562–787; Lai et al. 1991) recognize a Zfh1 protein in zfh12 mutants (as shown in Figures 2 and 3). Nevertheless, zfh12 is the strongest mutant available and its use allows our data to be viewed in the context of other heart development studies.
Figure 3.—
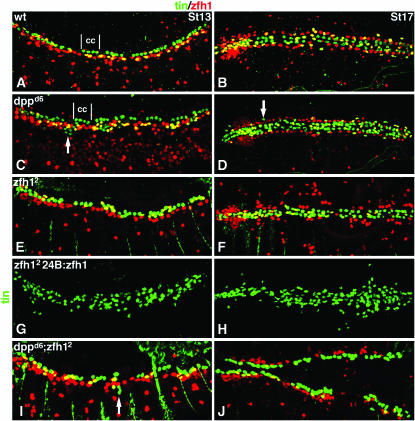
Dpp restricts the number of Tin pericardial cells via zfh1 and independently of zfh1. Stage and view as in Figure 2 for embryos double labeled for Tin (green) and Zfh1 (red). See Table 1 for statistics. (A and B) Wild type. Tin is expressed in a subset of Zfh1-expressing pericardial cells (yellow) and in a subset of cardial cells (cc). (C and D) dppd6. The number of Tin-expressing pericardial cells is significantly increased. Note that Tin-positive cells are located ventral to the Zfh1-expressing pericardial cells but they do not co-express Zfh1 (white arrows). (E and F) zfh12. The number of Tin-expressing cells is significantly fewer than wild type. (G and H) 24B.Gal4:UAS.Zfh1; zfh12. Pan-mesodermal Zfh1 expression in zfh12 mutants not only rescues Tin expression in pericardial cells but also induces ectopic Tin-expressing cells in lateral regions of the mesoderm. (I and J) dppd6; zfh12. The number of Tin-expressing cells is significantly decreased compared to wild type—a phenocopy of zfh12 single mutants. Some Tin-expressing cells positioned ventral to the Zfh1-expressing pericardial cells do not co-express Zfh1 in double-mutant embryos (white arrow in I).
Our analysis showed that zfh12 embryos contain the normal number of OPCs at stage 13 (Figure 2E), yet by stage 17 the number of OPCs is significantly decreased (Figure 2F), indicating that Zfh1 is a positively acting factor required to maintain the correct number of OPCs. This result is specific since the loss of OPCs in zfh12 mutants can be rescued by overexpressing Zfh1 using the pan-mesodermal driver 24B.Gal4 and ectopic OPCs are observed in lateral regions of the mesoderm when Zfh1 is misexpressed (Figure 2, G and H).
To determine if restriction of zfh1 expression is sufficient for Dpp to specify the correct number of OPCs, we then assayed Odd expression in dppd6; zfh12 embryos. The number of OPCs in dppd6; zfh12 embryos resembles that of zfh12 embryos at stages 13 and 17 (Figure 2, I and J). Taken together, these results demonstrate that zfh1 is epistatic to dpp in regulating OPC number and indicate that normally Dpp defines the number of OPCs by restricting zfh1 expression.
Dpp restricts the number of Tin pericardial cells via a feed-forward mechanism:
A second pericardial cell type, defined by Tin expression, is also responsive to Dpp signals during germ-band retraction. We noted that the number of Tin-expressing pericardial cells (TPC) in dppd6 embryos is greater than that of wild type at stages 13 and 17 (compare Figure 3, A and B, with Figure 3, C and D; Table 1). Since zfh1 is required downstream of Dpp to restrict the number of OPCs, we reasoned that a similar mechanism could be in place to restrict the number of TPCs. To test this hypothesis, we examined Tin expression in zfh12 embryos and found that the number of TPCs is significantly reduced (Figure 3, E and F). In addition, we found that this phenotype can be rescued by expressing Zfh1 in the mesoderm and that ectopic TPCs appear when Zfh1 is misexpressed in lateral regions of the mesoderm (Figure 3, G and H). These results indicate that Zfh1 is a positively acting factor required to maintain the correct number of TPCs. In dppd6; zfh12 embryos, the number of TPCs is significantly reduced by stage 13 (Figure 3, I and J), which phenocopies zfh12 embryos. These experiments demonstrate that zfh1 is also epistatic to dpp in regulating the number of TPCs. Thus, Dpp restricts zfh1 expression and, as a direct result, restricts the number of TPCs and OPCs.
The pattern of Tin-expressing cells in dppd6 embryos suggests that Dpp may also restrict TPC number independently of Zfh1. Tin is expressed in four of the six non-Zfh1-expressing cardiac cells per heart hemi-segment in wild type (Ward and Skeath 2000; Lo and Frasch 2001) and dppd6 embryos (Figure 3C). Interestingly, at stage 13, a subset of ectopic Tin-expressing cells in dppd6 and dppd6; zfh12 embryos is located ventral to the cardiac cells in a domain usually associated with pericardial or dorsal muscle cells, but these cells do not co-express Zfh1 (Figure 3, C and I). The position of the Tin-expressing cells in dppd6 embryos plus the fact that dMef2-expressing cardiac cells are unaffected in dppd6 embryos (Figure 1, D and E) indicate that the ectopic Tin-expressing cells in dppd6 mutants are pericardial cells. The absence of Zfh1 expression in the ectopic Tin-expressing cells in dppd6 and dppd6; zfh12 embryos further indicates that Dpp restricts the number of TPCs in a Zfh1-independent manner. Since dpp acts through Zfh1-dependent and Zfh1-independent mechanisms to restrict the number of TPCs, we conclude that dpp represses Tin expression via a feed-forward mechanism.
To further demonstrate that TPCs, and not Tin cardial cells, respond to Dpp signals, we expressed a constitutively active form of the Dpp type I receptor Thickveins (UAS.CA-Tkv) using two cell-type-specific drivers: the cardiac cell driver tinCΔ4.Gal4 (Figure 4A) and the pericardial cell driver prc.Gal4 (Figure 4B). Our experiments show that the wild-type Tin expression pattern (Figure 4C) is unaffected by expressing CA-Tkv specifically in cardiac cells (Figure 4D). In contrast, expressing CA-Tkv in pericardial cells reduces the number of Tin-expressing cells in medial regions of the heart (Figure 4E; Table 1). The mild reduction in Tin expression observed in prc.Gal4:UAS.CA-Tkv embryos is due to the fact that prc.Gal4 drives expression in only a subset of pericardial cells and does not initiate until late stages of embryogenesis. Nonetheless, these studies strengthen our hypothesis that Dpp signals initiating during germ-band retraction specifically restrict the number of TPCs.
Dpp restricts cell proliferation in the dorsal mesoderm:
The increases in pericardial cell number observed in dpp mutants could reflect changes in gene expression in a stable cell population or could be the result of changes in cell number. To distinguish between these possibilities, we assessed cell proliferation in the dorsal mesoderm during and after germ-band retraction. In wild-type embryos at stage 12, cell proliferation is very limited (Figure 5A) and completely absent by stage 13 (Figure 5B). In contrast, dppd6 embryos show expanded cell proliferation during stage 12 particularly in mesoderm cells positioned immediately ventral to Zfh1-expressing cells (Figure 5C, arrows). Cell division in the dorsal mesoderm persists in dppd6 embryos throughout stage 13 (Figure 5D). These results argue that Dpp restricts pericardial cell number by limiting cell proliferation in the dorsal mesoderm.
Overexpression of the Tbx family member midline (mid) induces cell proliferation and ectopic Tin expression in lateral regions of the dorsal mesoderm (Qian et al. 2005). Further, these ectopic Tin-expressing cells do not express the cardiac cell marker Toll and are therefore likely to be Tin pericardial cells (Reim et al. 2005). Thus we reasoned that Dpp could repress cell proliferation and Tin pericardial cell expression independently of Zfh1 by restricting mid expression. To test this, we investigated mid expression in dpp and zfh1 mutant embryos.
In wild-type embryos, mid is expressed solely in cardiac cells (Figure 5E) but in dppd6 embryos, mid expression expands into ventral regions of the dorsal mesoderm, presumably into the pericardial or dorsal muscle domain (Figure 5F). In contrast, mid expression is unaltered in zfh12 and 24B:Zfh1 embryos (data not shown), indicating that Dpp regulates mid expression independently of Zfh1. We propose that the ventrally positioned TPCs observed in dppd6 embryos (Figure 3C; white arrow) arise from misregulation of mid.
However, we were not yet fully convinced that cell proliferation could account for all ectopic pericardial cells observed in dpp mutants. We further tested this possibility in CycAC8 embryos. Mitosis does not occur after maximum germ-band extension in these embryos and the number of OPCs in CycAC8 embryos is approximately half the number of OPCs in wild-type embryos (Han and Bodmer 2003). We found that the number of OPCs in dppd6; CycAC8 double mutants is greater than that observed in CycAC8 embryos (Figure 5, G–J). Further cell counts and t-tests indicate that this difference is statistically significant (1.18 OPCs in CycAC8 vs. 1.90 OPCs in dppd6; CycAC8 per hemi-segment; t-test P < 0.001). This result indicates that Dpp inhibits cell proliferation and restricts pericardial-specific gene expression in dorsal mesoderm cells.
Lineage analyses have shown that TPCs and OPCs arise from separate precursor cells and that following germ-band retraction Tin and Odd are not co-expressed in dorsal mesoderm cells of wild-type embryos (Figure 5G). Since Dpp signals regulate cell proliferation and the number of TPCs and OPCs, we wanted to understand whether Dpp signals might play a role in maintaining the lineage identities of TPCs and OPCs. Therefore, we examined dppd6 embryos double labeled for Tin and Odd expression. We found that, although the number of TPCs and OPCs is increased, co-expression of these proteins was not observed (Figure 5H). Further, co-expression of Tin and Odd was not observed in dppd14 (not shown) or CycAC8 or dppd6; CycAC8 embryos (Figure 5, I and J). Thus, Dpp signals do not maintain TPC or OPC lineage identities.
zfh1 expression is necessary and sufficient to induce pericardial cell fate in wild-type embryos but zfh1 is bypassed in dpp and lame duck mutants:
Our experiments show that Zfh1 activity is required for the specification of two distinct pericardial cell types: OPCs and TPCs. Moreover, a previous study (Su et al. 1999) showed that Zfh1 also specifies a third cell type: Eve-expressing pericardial cells. Taken together, these findings suggested to us that in wild-type embryos Zfh1 expression in the dorsal mesoderm is sufficient to induce a pericardial cell fate. To test this hypothesis, we assayed the expression of the extracellular matrix protein Pericardin (Prc) in embryos misexpressing Zfh1. Prc is broadly expressed in, and then secreted from, pericardial cells, including OPCs and TPCs (Figure 6A), and appears to serve a pericardial-specific function (Chartier et al. 2002).
We found that ectopic expression of Zfh1 either in the excess pericardial cells of dppd6 embryos or artificially in 24B.Gal4:UAS.Zfh1; zfh12 embryos induced ectopic Prc expression (Figure 6, B–D). On the other hand, Prc expression is dramatically reduced, although not absent, in zfh12 embryos (Figure 6E), perhaps as a result of residual zfh1 activity in this mutant as discussed above. These results support the hypothesis that zfh1 is a key regulator of pericardial cell fate. Moreover, since alterations in zfh1 expression alter the expression of three pericardial cell proteins—Eve (Su et al. 1999), Odd, and Prc (this study)—we conclude that in wild-type embryos Zfh1 expression is necessary and sufficient to specify pericardial cell fates.
However, in dppd6 mutant embryos, ectopic Tin-expressing cells that do not express Zfh1 are visible in positions normally reserved for dorsal muscle cells (Figure 3, C and D). Further, these ectopic Tin-expressing cells also express Prc (Figure 6B), indicating that they are pericardial cells. This implies that in dppd6 mutants the requirement for Zfh1 in the specification of pericardial cell fate is bypassed. Our hypothesis regarding the origin of the ectopic pericardial cells, based on their location, is that they derive from the transformation of cells normally destined to become dorsal muscle cells—cells that do not normally express Zfh1.
To test this hypothesis, we examined lame duck (lmd) mutant embryos. lmd encodes a Gli-like transcription factor expressed in dorsal muscle precursors but not in cardiac or pericardial cells (Duan et al. 2001). In lmd1 mutants, dorsal muscle precursors fail to develop into fusion-competent myoblasts, leading to significant defects in dorsal muscle architecture (Duan et al. 2001; Ruiz-Gomez et al. 2002). We examined lmd mutants because of the possibility that the missing muscle precursors were transformed into adjacent cell types—e.g., into pericardial cells in dorsal regions.
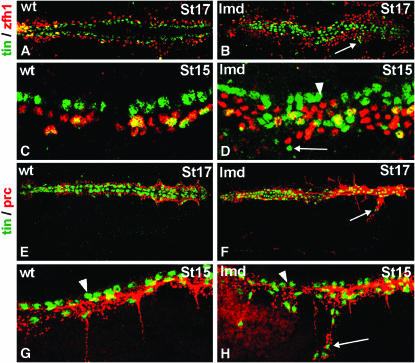
This hypothesis was validated by the observation that there is widespread pericardial cell hyperplasia in lmd mutants (Figure 7, A–D). This aspect of the lmd mutant phenotype is more pronounced than the hyperplasia seen in dpp mutants. Both display an excess of Zfh1-expressing pericardial cells (compare Figures 3D, 6B, and 7B). Importantly for our hypothesis that dpp mutants bypass the Zfh1 requirement in pericardial cell fate specification, lmd mutants also have ectopic Tin-expressing cells ventral to their normal position that do not express Zfh1 (Figure 3, C and D; Figure 7, B and D). Further, the ectopic Tin-expressing cells in dpp and lmd mutants also express Prc and thus they must be pericardial cells (Figure 6B, Figure 7, F and H).
Figure 7.—
Mutations in lame duck also display pericardial cell hyperplasia. (A–D) Embryos double labeled for Tin (green) and Zfh1 (red) at the indicated stages. (A and B) Dorsal (bilateral) view of stage 17 embryonic hearts at low magnification. (C and D) Lateral (unilateral) view of stage 15 embryonic hearts at high magnification. (A and C) In wild-type embryos, all Tin-positive pericardial cells co-express Zfh1 (yellow cells lateral to the green Tin-expressing cardiac cells). (B and D) In lmd1 embryos, Tin-expressing cardiac cells look normal (cells indicated by an arrowhead in D). There are an excessive number of Zfh1-expressing cells and an excess of Tin-expressing cells, including many quite lateral to their normal position that do not co-express Zfh1 (cells indicated by an arrow in B and D). (E–H) Embryos double labeled for Tin (green) and Prc (red). (E and F) Dorsal (bilateral) view of stage 17 embryonic hearts at low magnification. (G and H) Lateral (unilateral) view of stage 15 embryonic hearts at intermediate magnification. (E and G) In wild-type embryos, all Tin-expressing pericardial cells express Prc but Tin-expressing cardiac cells do not (cardiac cells are indicated by an arrowhead in G and H). (F and H) In lmd1 embryos, all Tin-expressing cells, except for Tin cardiac cells (arrowhead), also express Prc (cells indicated by an arrow).
Overall, the data from dpp and lmd mutants suggest that the ectopic Tin- and Prc-expressing pericardial cells that do not express Zfh1 are derived from the cell fate transformation of dorsal muscle cells into pericardial cells at a relatively late stage of development.
Dpp maintains the boundary between pericardial cells and dorsal muscle cells:
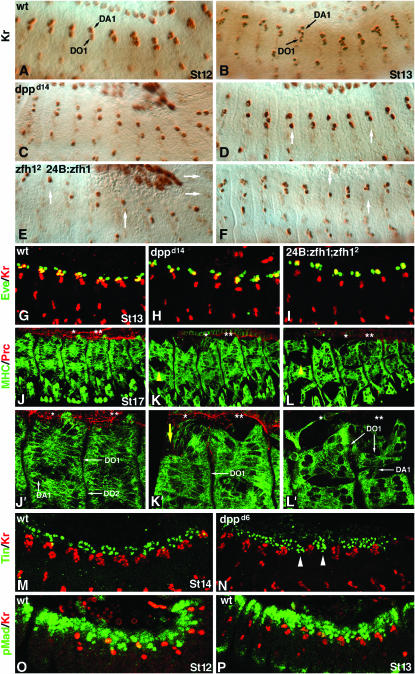
Our observation that loss of dpp or lmd results in an expanded pericardial cell domain led us to hypothesize that Dpp signals are specifically required to ensure that the dorsal muscle cells positioned just lateral to the pericardial cells are correctly specified. To test this hypothesis, we examined the expression of two dorsal muscle transcription factors, Kr and Eve, in dpp mutant embryos. In wild-type embryos, Kr expression becomes apparent in a pair of dorsal muscle precursors at the onset of germ-band retraction (Ruiz-Gomez et al. 1997). As the germ band retracts, Kr-expressing DA1 and DO1 founder cells fuse with neighboring cells to form multinucleate muscle precursors, and Kr expression concomitantly increases in the dorsal mesoderm (compare Figure 8A with 8B). Eve is expressed in DA1 founder cells and a subset of pericardial cells. Like Kr, Eve expression expands as DA1 founder cells begin to form multinucleate muscle precursors (Carmena et al. 1998b). In dpp mutant embryos, Kr- and Eve-expressing muscle founder cells are specified correctly but neither Kr nor Eve expression expands at wild-type levels as the germ band retracts or after germ-band retraction (Figure 8, C and D; Table 2). These results indicate that Dpp signals ensure proper development of the dorsal musculature.
Figure 8.—
Dpp indirectly maintains dorsal muscle cell fates. (A–F) Lateral views of embryos stained for Kr expression. See Table 2 for statistics. (A) Wild-type stage 12. Kr-expressing DA1 and DO1 muscle founder cells are shown. (B) Wild-type stage 13. An increased number of Kr-expressing nuclei are evident. (C) dppd14 stage12 and (D) stage 13. Roughly half of the Kr-expressing cells per segment are present. White arrows in D identify segments with reduced Kr expression. (E) 24B.Gal4:UAS.Zfh1; zfh12 stage 12 and (F) stage 13. Kr expression is dramatically reduced. (G–I) Stage 13 embryos double labeled for Kr (red) and Eve (green). (G) Wild type. (H) dppd14. (I) 24B.Gal4:UAS.Zfh1; zfh12. Eve pericardial cells, two per segment, are present in all genotypes, but the number of Kr and Eve co-expressing dorsal muscle cells (in yellow) is considerably reduced in dpp and zfh mutants. (J–L) Stage 17 embryos double labeled for myosin heavy chain (green) and Prc (red). The DO1, DO2, and DA1 muscles are indicated with white arrows. Asterisks indicate the two segments viewed at high magnification in J′–L′. (J and J′) Wild type. The DO1 and DO2 muscles are tightly associated. The DO1 and DA1 muscles abut the pericardial cells. (K and K′) dppd14. The DO1 and DO2 muscles are loosely associated (yellow arrowhead in K) and the size of the DO1 muscles is reduced (yellow arrow in K′). (L and L′) 24B.Gal4:UAS.Zfh1; zfh12. The DO1 and DO2 muscles are loosely associated (yellow arrowhead in L), a subset of DA1 muscles is absent, and the DO1 muscles are reduced. (M and N) Stage 14 embryos labeled for Tin (green) and Kr (red). (M) Wild type. Tin-expressing cells are exclusively positioned dorsal to the Kr-expressing cells. (N) dppd6. There are Tin-expressing cells positioned ventral to the dorsal-most Kr-expressing cells (white arrowheads). (O and P) Stage 12 and 13 embryos labeled for pMad (green) and Kr (red). (O) Wild type. (P) dppd6. Colabeling of Kr and pMad is not observed in either embryo, indicating that the effect of Dpp on Kr is indirect.
TABLE 2.
Kr and Eve expression in embryos with altered dpp or zfh1 activity
| Mean no. of Kr-expressing nuclei (SD)
|
Mean no. of Eve-expressing nuclei (SD)
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Genotype | Stage 12a | t-test vs. wild typeb | Stage 13a | t-test vs. wild typeb | Stage 12a | t-test vs. wild typeb | Stage 13a | t-test vs. wild typeb |
| Wild type | 2.78 (0.76) | — | 4.77 (1.22) | — | 3.32 (0.65) | — | 5.13 (0.97) | — |
| dppd14 | 2.43 (0.53) | 0.003 | 2.29 (0.51) | <0.001 | 2.92 (0.90) | 0.006 | 4.29 (1.13) | 0.001 |
| dppd6 | 2.38 (0.61) | 0.006 | 3.96 (1.31) | 0.011 | 3.10 (0.71) | 0.173 | 4.32 (0.98) | 0.004 |
| zfh12 | 2.69 (0.95) | 0.617 | 5.59 (1.06) | <0.001 | ||||
| 24B:zfh1; zfh12 | 1.60 (0.70) | <0.001 | 3.73 (2.09) | <0.001 | ||||
Number of nuclei per hemi-segment in lateral view.
P-value (numbers in italic are statistically significant).
Since the absence of Dpp signals results in an expansion of the pericardial cell domain and a concomitant reduction in the expression of dorsal muscle transcription factors, we suspected that Dpp functions to maintain the pericardial–dorsal muscle cell boundary. We hypothesized that Dpp signals pattern the dorsal musculature by preventing pericardial cells from occupying ventral regions of the dorsal mesoderm. If this hypothesis is correct, then the presence of ectopic pericardial cells in the dorsal mesoderm alone should phenocopy the loss of Dpp signals. Indeed, misexpression of Zfh1 throughout the dorsal mesoderm reduces Kr expression in a manner very similar to that observed in dpp mutants (Figure 8, E and F; Table 2). Further, double-labeling experiments with Kr and Eve show that the reduction in Eve expression in dpp mutant embryos is due to the loss of Eve-expressing muscle founder cells and not due to changes in Eve pericardial cell number (Figure 8, G–I). Thus, the presence of ectopic pericardial cells in the dorsal mesoderm, via Zfh1 misexpression, also prevents the expression of these two dorsal muscle cell transcription factors.
To better understand the role of Dpp signals in patterning the dorsal musculature, we examined the size and position of somatic muscle fibers in embryos stained for muscle myosin. In wild-type embryos, muscle myosin identifies the dorsal-most DO1 and DA1 muscles positioned proximal to the pericardial cells (Figure 8J). DO2 muscles are positioned just ventral to the DO1 muscles. The size of some DO1 muscles, particularly in the sixth row of somatic muscles, is reduced in dppd14 mutants, and the space between the DO1 and DO2 muscles is expanded (Figure 8K). Misexpressing Zfh1 in the dorsal mesoderm causes a similar phenotype: reduction in the size of DO1 muscles and dissociation of the DO1 and DO2 muscles (Figure 8L). In addition, the number and the size of the DA1 muscles are reduced in embryos misexpressing Zfh1. This result shows that increasing pericardial cell number simultaneously reduces the size and alters the pattern of the dorsal musculature.
We then reasoned that maintenance of the pericardial–dorsal muscle cell boundary might involve cross-repressive interactions between pericardial and dorsal muscle cells. Our data suggested that zfh1 defines the pericardial cell domain and that, in the absence of zfh1, the dorsal muscle domain would expand. Indeed, following germ-band retraction, Kr expression is expanded in zfh12 embryos as compared with wild type (Table 2). Moreover, in wild-type embryos, the pericardial cell domain is tightly restricted to the region dorsal of the Kr-expressing dorsal muscle domain (Figure 8M). However, in dppd6 embryos, pericardial cells are observed in the dorsal muscle domain (Figure 8N). These results argue that cross-repressive interactions between pericardial and dorsal muscle cells continue to maintain the pericardial–dorsal muscle cell boundary after the respective precursor cells have been specified.
To further characterize the mechanism by which Dpp maintains the pericardial–dorsal muscle cell boundary, we double labeled embryos for pMad and Kr. We found that in wild type pMad is barely detectable in Kr-expressing dorsal muscle cells during germ-band retraction (Figure 8O). Following germ-band retraction, pMad accumulation is absent from all Kr-expressing cells although high levels of pMad can be seen in cells adjacent to the Kr-expressing cells (Figure 8P). This demonstrates that Dpp does not signal to Kr-expressing dorsal muscle founder cells during germ-band retraction although Dpp might signal to the adjacent cells to restrict zfh1 expression.
Embryonic pericardial cell overgrowth reduces larval cardiac output:
To determine what physiological effect pericardial hyperplasia might have on cardiac function, we analyzed the heartbeat of wild-type and dppd6 first instar larvae with live videomicroscopy. We studied the pulse distance traveled by pairs of cardiac cells each beat (the difference between the maximum separation at diastole and the minimum separation at systole) and the number of beats per second. In the anterior region of the heart (the aorta), we observed no difference in heart rate or pulse distance between wild-type and dppd6 larvae (data not shown).
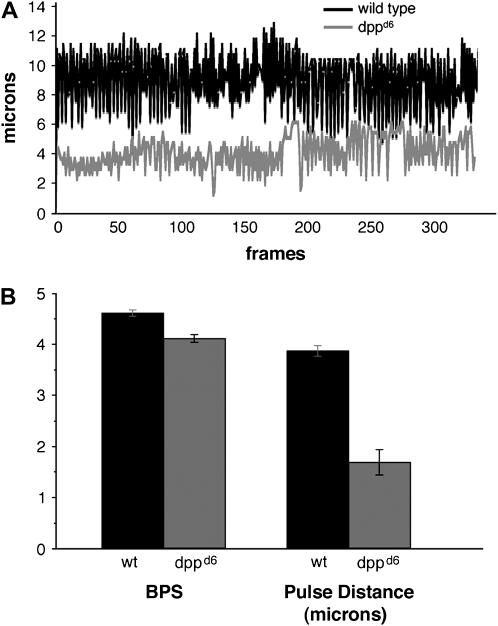
In the posterior region (the heart proper; Lovato et al. 2002, Sellin et al. 2006), dppd6 larvae show a considerable reduction in the systolic distance (mutant cells do not approach each other as closely as wild type do) and the diastolic distance (mutant cells do not separate from each other as far as wild type do). The disparity is so pronounced that heartbeat tracings for wild-type and dppd6 larvae are essentially nonoverlapping (Figure 9A). Quantifying the data, we found that dppd6 larvae have a 44% reduction in the average pulse distance per beat (Figure 9B), an indication that the total amount of fluid moved per beat (beat volume) is significantly reduced. Alternatively, the heart rate of dppd6 larvae in the posterior region is comparable to wild type (Figure 9B). We conclude that overall cardiac output, a function of beat volume and beat rate, is significantly reduced in dppd6 larvae.
Figure 9.—
Reduced cardiac output in dpp mutant larvae. (A) Representative traces showing the distance between a pair of cardiac cells in the posterior region of the heart over time in wild type (top) and dppd6 (bottom) first instar larvae. One second corresponds to 12 frames. (B) Average number of beats per second (BPS) and pulse distance for wild type (n = 3) and dppd6 (n = 4) first instar larvae. Pulse distance is the difference between the maximum diastolic position and the minimum systolic position for each heartbeat. Error bars signify the standard error of the mean.
DISCUSSION
Our data are wholly consistent with the hypothesis that the dppd6 inversion prevents the augmentation of dpp expression provided by disk region enhancers during germ-band retraction. The data further suggest that the augmentation of dpp expression is necessary to boost Dpp dorsal ectoderm signals such that they can reach the underlying mesoderm. Finally, we have shown that during germ-band retraction Dpp signals maintain the boundary between pericardial cells and dorsal muscle cells via two distinct mechanisms: the regulation of gene expression and the restriction of cell proliferation (see Figure 10 for a model). To regulate gene expression, Dpp signals directly to pericardial cells and restricts Odd and Tin expression in a zfh1-dependent manner. Dpp also limits Tin expression, independently of zfh1, by repressing the expression of mid, a stimulator of proliferation.
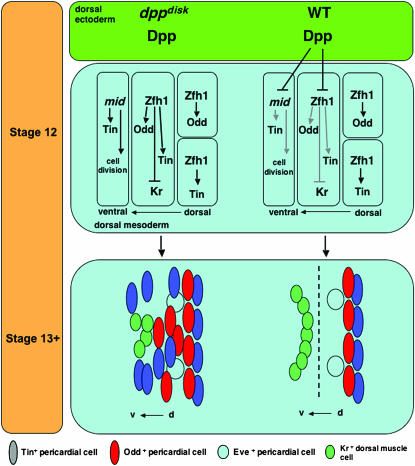
Figure 10.—
Dpp signals pattern the embryonic dorsal mesoderm during germ-band retraction. dppdisk at stage 12: in the absence of Dpp, we observed that (1) mid expression expands ventrally, drives cell proliferation, and induces ectopic Tin expression; (2) Zfh1 expression expands ventrally, inducing ectopic Tin and Odd expression and repressing Kr (and Eve) expression; and (3) pericardial cell specification of Odd and Tin by Zfh is unaffected. Wild type at stage 12: Dpp signals from the dorsal ectoderm restrict the expression of mid and Zfh1. dppdisk at stage 13+: in the absence of Dpp, we observed that (1) pericardial cells populate ventral regions of the dorsal mesoderm normally occupied exclusively by dorsal muscle cells and (2) the inappropriate presence of pericardial cells reduces the expression of the dorsal muscle genes Kr (and Eve). It appears that there is a failure to maintain the pericardial cell–dorsal muscle cell boundary and that this failure results in an increase in heart size. Wild type (WT): Dpp maintains the boundary between pericardial and dorsal muscle cells (dashed line) by restricting the number of Tin- and Odd-expressing pericardial cells and by reducing cell division via the repression of mid.
Dpp restricts the number of cells derived from symmetrically dividing lineages:
With respect to zfh1-dependent regulation, our data support the hypothesis that Dpp restricts Zfh1 expression to regulate the number of pericardial cells derived solely from symmetrically dividing lineages. Lineage analyses have identified both symmetric and asymmetric cell divisions of myogenic and pericardial precursor cells. Pericardial cells are derived from four separate lineages that arise from four distinct precursor cells. Asymmetric precursor cell divisions initiating between stages 8 and 10 give rise to the Odd-positive/Seven up (Svp)-positive pericardial cells and the Eve-positive/Tin-positive pericardial cells (EPCs) (Ward and Skeath 2000; Alvarez et al. 2003; Han and Bodmer 2003). On the other hand, symmetric division, initiating at the same stage, establishes the Odd-positive/Svp-negative pericardial cells (OPCs) and the Tin-positive/Eve-negative pericardial cells (TPCs). We show that dpp mutations do not affect the number of EPCs (Figure 8H) or the number of Odd-positive/Svp-positive cells (Johnson et al. 2003). However, embryos bearing dpp mutations show an increase in the number of OPCs (Figure 2) and TPCs (Figures 3 and 4). Therefore, the ectopic pericardial cells seen in dpp mutants derive from symmetrically dividing lineages.
Previous reports have shown that regulation of asymmetric cell division is a key mechanism in establishing boundaries among the various cell types in the dorsal mesoderm. For instance, in the absence of Numb, a Notch pathway antagonist, asymmetric progenitor cell division is abrogated and the number of Odd-positive/Svp-positive cells (Ward and Skeath 2000) and EPCs (Carmena et al. 1998b) increases at the expense of the Svp-expressing cardial cells and Eve-expressing dorsal muscle cells, respectively. Our study extends these observations by showing that pericardial cell types derived from symmetrically dividing lineages are also under strict regulatory control.
Dpp limits cell proliferation and the reactivation of Tin in a subset of pericardial cells:
With respect to zfh1-dependent regulation of pericardial cell number, Dpp restricts cell proliferation and, in turn, Tin expression by limiting mid expression. In wild-type embryos, cell division in the dorsal mesoderm is largely complete by the early stages of germ-band retraction (stage 11), whereas in dppd6 embryos cell proliferation in the dorsal mesoderm continues through stage 13 (Figure 5, C and D). Interestingly, the number of cells expressing Zfh1 increases from stage 12 to stage 13 in wild-type embryos in the absence of cell division (compare Figure 5A with 5B), demonstrating that patterning events subsequent to cell division regulate cell fate choices in the dorsal mesoderm. This hypothesis is supported by the fact that tracing pericardial cell lineages requires inducing mitotic clones by stage 8 (Alavarez et al. 2003). Therefore, the ectopically dividing mesoderm cells observed in dppd6 embryos are derived from cells with the potential to become Tin-expressing cells.
During stage 12, tin expression is reactivated in a subset of cardiac cells in a mid-dependent fashion, suggesting that tin expression in precursor cells alone is not sufficient for specifying the ultimate fate of their daughter cells (Reim et al. 2005). Moreover, misexpression of mid results in both ectopic cell division and expanded tin expression (Qian et al. 2005). Lineage studies support the necessity of reactivating Tin by showing that a single precursor cell gives rise to two Tin-positive/Eve-negative pericardial cells and two siblings that do not express Tin (Alvarez et al. 2003). Thus tin is not reactivated in all subpopulations of pericardial cells. Our data suggest that, during stage12, Dpp prevents tin reactivation in cells occupying lateral regions of the dorsal mesoderm by limiting mid expression.
A pericardial cell–dorsal muscle cell boundary may be essential for myoblast fusion:
Development of the dorsal musculature initiates when founder cells are specified in the mesoderm. These founder cells then fuse with neighboring cells to form syncitial myofibers. We found that, in the absence of Dpp, the pericardial cell domain expands into the dorsal muscle domain and reduces expression from the dorsal muscle genes Kr and Eve (Table 2). Since the separation between pericardial and dorsal muscle cells is lost in dpp mutant embryos, we conclude that Dpp maintains the pericardial–dorsal muscle cell boundary after it is established. Moreover, we find that reducing pericardial cell number increases Kr expression after germ-band retraction, suggesting that cross-repressive interactions between pericardial and dorsal muscle cells contribute to patterning of the dorsal mesoderm. The presence of ectopic pericardial cells in the dorsal mesoderm reduces the number of myofibers comprising the dorsal muscles (Figure 8, H and I) even though the dorsal muscle founder cells are, for the most part, correctly specified (Figure 8C). pMad does not accumulate in Kr-expressing founder cells yet Kr expression is significantly reduced in dpp mutant embryos. Therefore, changes in Kr and Eve expression observed in embryos with altered dpp or zfh1 activity reflect alterations in the number of myoblast fusion events in the dorsal mesoderm.
Our data extend a previous study showing that misexpressing Zfh1 reduces dMef2 expression in somatic muscles (Postigo et al. 1999). Our study demonstrates that misexpression of Zfh1 induces ectopic pericardial cells and that the presence of pericardial cells in the dorsal muscle domain reduces myoblast fusion. Therefore, reduced dMef2 expression in embryos misexpressing Zfh1 is likely the result of reduced myoblast fusion and not of direct repression of dMef2 expression by Zfh1. Further, our analysis of lmd mutants that have reduced numbers of myoblasts revealed that they also contain an excessive number of pericardial cells. Together, these results suggest that maintaining the pericardial–dorsal muscle cell boundary requires Dpp-mediated cross-repressive interactions between these cell types. Thus, in the absence of Dpp, the transformation of dorsal muscle cells into pericardial cells reduces the number of myoblasts available for fusion.
Pericardial cell function:
Experiments in the larvae of Drosophila and other insects suggested that pericardial cells act as nephrocytes that filter the hemolymph (Crossley 1972). These studies also showed that pericardial cells secrete proteins into the hemolymph, suggesting that one pericardial cell function may be to provide short- or long-range signals. Consistent with this, reducing pericardial cell number reduces heart rate and increases the cardiac failure rate, suggesting that pericardial cells influence the development of cardiac cells (Fujioka et al. 2005).
Our study shows that pericardial cell hyperplasia reduces the luminal distance of the heart during systole as well as diastole, resulting in an overall decrease in average pulse distance of each contraction. However, pericardial overgrowth does not alter heart rate, indicating that cardiac cells develop appropriately in the presence of ectopic pericardial cells. Our luminal measurements suggest a role for pericardial cells in the mechanics of heart function. One hypothesis for this is based on the fact that pericardial cell hyperplasia results in excess levels of Prc in the extracellular matrix (ECM) surrounding the heart (Figure 6). Prc is a collagen IV-like ECM protein secreted at high levels from pericardial cells (Chartier et al. 2002). In dpp mutants, excess Prc is seen predominantly in the posterior of the heart where we observe the pulse-distance reduction. We propose that Prc secreted by pericardial cells limits the width of the dorsal vessel at diastole and thus modulates the pulse distance of each heart contraction. Pericardial cell overgrowth would increase Prc deposition, thereby reducing the size of the diastolic heart and the pulse distance. Consistent with this hypothesis, excessive expression of ECM proteins, including collagen IV, was correlated with heart failure in patients presenting with end-stage cardiomyopathy (Schaper and Speiser 1992).
Functional conservation of BMP signaling during heart development:
It is well documented that many of the early events driving Drosophila embryonic heart development have been conserved in vertebrates (Cripps and Olson 2002; Zaffran and Frasch 2002). Our data provide the first basis upon which to determine if Dpp regulation of Zfh1 or Tin late in heart development is also conserved.
Two orthologs of zfh1, Sip1 and Kheper, have been identified in vertebrates. Zebrafish embryos injected with the Dpp homolog BMP4 show reduced Kheper expression (Muraoka et al. 2000) while Xenopus embryos injected with the BMP antagonist Chordin display elevated Sip1 expression (Nitta et al. 2004). These results suggest the possibility that Dpp repression of zfh1 expression may be conserved in vertebrates. In addition, mammalian Sip1 plays an essential role in heart development. In mice, Sip1 is expressed in neural crest cells (NCCs), paraxial mesoderm, and neuroectoderm. The subset of NCCs that express Sip1 give rise to the septum and large arteries of the heart. Sip1 knockout mice fail to form these NCCs (Van de Putte et al. 2003) and these mice die midway through gestation with numerous heart defects. Mice lacking the BMP receptors BMPRIA or ALK2 specifically in NCCs also display numerous cardiac phenotypes (Kaartinen et al. 2004; Stottmann et al. 2004). In conditional knockout of ALK2 in NCCs, abnormalities are seen in the heart's outflow tract, and conditional knockout of BMPRIA in NCCs results in heart failure and early embryonic lethality similar to Sip1 knockout mice. Thus BMP signals are required for proper specification of NCCs, and loss of BMP signaling in NCCs phenocopies Sip1 knockout mice to an extent. It is tempting to speculate that, as in Drosophila, BMP signals regulate the Zfh1 ortholog Sip1 to correctly specify NCCs and, in turn, to properly pattern the mammalian heart.
With regard to the conservation of late-stage Dpp regulation of Tin, a recent article describing a study of mice with a conditional knockout of Nkx2.5 where expression is missing only during late stages of heart development (post E14.5) is highly relevant (Prall et al. 2007). Utilizing rescue of Nkx2.5 mutant embryos with BMP-signaling-pathway components, the study identified a direct connection among BMP4 signaling, Nkx2.5 activity, and heart cell proliferation. As Nkx2.5 is the Tin homolog, BMP4 is the Dpp homolog, and the mutant phenotype (heart cell hyperplasia) is the same in both species, this suggests that this aspect of Dpp signaling is conserved in mammals. Together with our study, these results suggest that defects in late-stage BMP signaling may play a role in congenital heart defects.
Acknowledgments
We thank Carlos Alonso, Rolf Bodmer, Manfred Frasch, Satoshi Goto, Carl-Henrik Heldin, Dan Kiehart, Dave Kosman, Ed Laufer, Hanh Nguyen, Eric Olson, Bruce Paterson, John Reinitz, Jim Skeath, the Bloomington Stock Center, and the Developmental Studies Hybridoma Bank for antibodies and flies. Zhi-Chun Lai was very generous with aliquots of Zfh1 antibodies and provided a zfh1 cDNA. We thank the Drosophila Genomics Resource Center for the mid cDNA and Doug Chandler, Maik Drechsler, Joel Frandsen, and Sudhir Kumar for support and technical assistance. This study was funded by the European Network of Excellence MYORES (A.P.) and the National Institutes of Health (S.J.N.; CA095875).
References
- Alvarez, A., W. Shi, B. Wilson and J. Skeath, 2003. pannier and pointedP2 act sequentially to regulate Drosophila heart development. Development 130: 3015–3026. [DOI] [PubMed] [Google Scholar]
- Bour, B., M. O'Brien, W. Lockwood, E. Goldstein, R. Bodmer et al., 1995. dMef2, a transcription factor that is essential for myogenesis. Genes Dev. 9: 730–741. [DOI] [PubMed] [Google Scholar]
- Brand, A., and N. Perrimon, 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415. [DOI] [PubMed] [Google Scholar]
- Carmena, A., S. Gisselbrecht, J. Harrison, F. Jimenez and A. Michelson, 1998. a Combinatorial signaling codes for the progressive determination of cell fates in the Drosophila embryonic mesoderm. Genes Dev. 12: 3910–3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmena, A., B. Murugasu-Oei, D. Menon, F. Jimenez and W. Chia, 1998. b Inscuteable and Numb mediate asymmetric muscle progenitor cell divisions during Drosophila myogenesis. Genes Dev. 12: 304–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartier, A., S. Zaffran, M. Astier, M. Semeriva and D. Gratecos, 2002. Pericardin, a Drosophila type IV collagen-like protein is involved in the morphogenesis and maintenance of the heart epithelium during dorsal ectoderm closure. Development 129: 3241–3253. [DOI] [PubMed] [Google Scholar]
- Cripps, R., and E. Olson, 2002. Control of cardiac development by an evolutionarily conserved transcriptional network. Dev. Biol. 246: 14–28. [DOI] [PubMed] [Google Scholar]
- Crossley, A., 1972. The ultrastructure and function of pericardial cells and other nephrocytes in an insect: Calliphora erythrocephala. Tissue Cell 4: 529–560. [DOI] [PubMed] [Google Scholar]
- Duan, H., J. Skeath and H. Nguyen, 2001. Drosophila Lame duck, a novel member of the Gli superfamily, acts as a key regulator of myogenesis by controlling fusion-competent myoblast development. Development 128: 4489–4500. [DOI] [PubMed] [Google Scholar]
- Fujioka, M., R. Wessells, Z. Han, J. Liu, K. Fitzgerald et al., 2005. Embryonic even skipped-dependent muscle and heart cell fates are required for normal adult activity, heart function, and lifespan. Circ. Res. 97: 1108–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glise, B., and S. Noselli, 1997. Coupling of Jun amino-terminal kinase and Decapentaplegic signaling pathways in Drosophila morphogenesis. Genes Dev. 11: 1738–1747. [DOI] [PubMed] [Google Scholar]
- Haerry, T., O. Khalsa, M. O'Connor and K. Wharton, 1998. Synergistic signaling by two BMP ligands through the SAX and TKV receptors controls wing growth and patterning in Drosophila. Development 125: 3977–3987. [DOI] [PubMed] [Google Scholar]
- Halfon, M., A. Carmena, S. Gisselbrecht, C. Sackerson, F. Jimenez et al., 2000. Ras pathway specificity is determined by the integration of multiple signal-activated and tissue-restricted transcription factors. Cell 103: 63–74. [DOI] [PubMed] [Google Scholar]
- Han, Z., and R. Bodmer, 2003. Myogenic cells fates are antagonized by Notch only in asymmetric lineages of the Drosophila heart, with or without cell division. Development 130: 3039–3051. [DOI] [PubMed] [Google Scholar]
- Han, Z., and E. Olson, 2005. Hand is a direct target of Tinman and GATA factors during Drosophila cardiogenesis and hematopoiesis. Development 132: 3525–3536. [DOI] [PubMed] [Google Scholar]
- Irvine, K., and C. Rauskolb, 2001. Boundaries in development: formation and function. Ann. Rev. Cell Dev. Biol. 17: 189–214. [DOI] [PubMed] [Google Scholar]
- Jagla, T., Y. Bidet, J. Da Ponte, B. Dastugue and K. Jagla, 2002. Cross-repressive interactions of identity genes are essential for proper specification of cardiac and muscular fates in Drosophila. Development 129: 1037–1047. [DOI] [PubMed] [Google Scholar]
- Johnson, A., C. Bergman, M. Kreitman and S. Newfeld, 2003. Embryonic enhancers in the dpp disk region regulate a second round of Dpp signaling from the dorsal ectoderm to the mesoderm that represses Zfh-1 expression in a subset of pericardial cells. Dev. Biol. 262: 137–151. [DOI] [PubMed] [Google Scholar]
- Kaartinen, V., M. Dudas, A. Nagy, S. Sridurongrit, M. Lu et al., 2004. Cardiac outflow tract defects in mice lacking ALK2 in neural crest cells. Development 131: 3481–3490. [DOI] [PubMed] [Google Scholar]
- Kato, K., T. Chihara and S. Hayashi, 2004. Hedgehog and Dpp instruct polarized growth of cell extensions in the Drosophila trachea. Development 131: 5253–5261. [DOI] [PubMed] [Google Scholar]
- Kiehart, D., and R. Feghali, 1986. Cytoplasmic myosin from Drosophila. J. Cell Biol. 103: 1517–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinedinst, S., and R. Bodmer, 2003. Gata factor Pannier is required to establish competence for heart progenitor formation. Development 130: 3027–3038. [DOI] [PubMed] [Google Scholar]
- Knirr, S., and M. Frasch, 2001. Molecular integration of inductive and mesoderm-intrinsic inputs governs even-skipped enhancer activity in a subset of pericardial and dorsal muscle progenitors. Dev. Biol. 238: 13–26. [DOI] [PubMed] [Google Scholar]
- Knoblich, J., and C. Lehner, 1993. Synergistic action of Drosophila cyclins A and B during the G2-M transition. EMBO J. 12: 65–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosman, D., S. Small and J. Reinitz, 1998. Rapid preparation of a panel of polyclonal antibodies to Drosophila segmentation proteins. Dev. Genes Evol. 208: 290–294. [DOI] [PubMed] [Google Scholar]
- Lai, Z., M. Fortini and G. Rubin, 1991. The embryonic expression patterns of zfh1 and zfh2, two Drosophila genes encoding novel zinc-finger homeodomain proteins. Mech. Dev. 34: 123–134. [DOI] [PubMed] [Google Scholar]
- Lai, Z., E. Rushton, M. Bate and G. Rubin, 1993. Loss of function of the Drosophila zfh1 gene results in abnormal development of mesodermally derived tissues. Proc. Natl. Acad. Sci. USA 90: 4122–4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilly, B., S. Galewsky, A. Firulli, R. Schulz and E. Olson, 1994. dMef2: a MADS box transcription factor expressed in differentiating mesoderm and muscle cell lineages during Drosophila embryogenesis. Proc. Natl. Acad. Sci. USA 91: 5662–5666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J., L. Qian, R. Wessells, Y. Bidet, K. Jagla et al., 2006. Hedgehog and RAS pathways cooperate in the anterior-posterior specification and positioning of cardiac progenitor cells. Dev. Biol. 290: 373–385. [DOI] [PubMed] [Google Scholar]
- Lo, P., and M. Frasch, 2001. A role for the COUP-TF-related gene seven-up in the diversification of cardioblast identities in the dorsal vessel of Drosophila. Mech. Dev. 104: 49–60. [DOI] [PubMed] [Google Scholar]
- Lockwood, W., and R. Bodmer, 2002. The patterns of wingless, decapentaplegic, and tinman position the Drosophila heart. Mech. Dev. 114: 13–26. [DOI] [PubMed] [Google Scholar]
- Lovato, T., T. Nguyen, M. Molina and R. Cripps, 2002. The Hox gene abdominal-A specifies heart cell fate in the Drosophila dorsal vessel. Development 129: 5019–5027. [DOI] [PubMed] [Google Scholar]
- Muraoka, O., H. Ichikawa, H. Shi, S. Okumura, E. Taira et al., 2000. Kheper, a novel ZFH/deltaEF1 family member, regulates the development of the neuroectoderm of zebrafish (Danio rerio). Dev. Biol. 228: 29–40. [DOI] [PubMed] [Google Scholar]
- Newfeld, S., and N. Takaesu, 2002. An analysis using the hobo genetic system reveals that combinatorial signaling by the Dpp and Wg pathways regulates dpp expression in leading edge cells of the dorsal ectoderm in Drosophila. Genetics 161: 685–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, H., R. Bodmer, S. Abmayr, J. McDermott and N. Spoerel, 1994. dMef2: a Drosophila mesoderm-specific MADS box-containing gene with a biphasic expression profile during embryogenesis. Proc. Natl. Acad. Sci. USA 91: 7520–7524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitta, K., K. Tanegashima, S. Takahashi and M. Asashima, 2004. XSIP1 is essential for early neural gene expression and neural differentiation by suppression of BMP signaling. Dev. Biol. 275: 258–267. [DOI] [PubMed] [Google Scholar]
- Persson, U., H. Izumi, S. Souchelnytskyi, S. Itoh, S. Grimsby et al., 1998. The L45 loop in type I receptors for TGF-β family members is a critical determinant in specifying Smad isoform activation. FEBS Lett. 434: 83–87. [DOI] [PubMed] [Google Scholar]
- Postigo, A., E. Ward, J. Skeath and D. Dean, 1999. zfh-1, the Drosophila homologue of ZEB, is a transcriptional repressor that regulates somatic myogenesis. Mol. Cell. Biol. 19: 7255–7263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prall, O., M. Menon, M. Solloway, Y. Watanabe, S. Zaffran et al., 2007. An Nkx2.5/Bmp2/Smad1 negative feedback loop controls heart progenitor specification and proliferation. Cell 128: 947–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian, L., J. Liu and R. Bodmer, 2005. Neuromancer Tbx20-related genes (H15/midline) promote cell fate specification and morphogenesis of the Drosophila heart. Dev. Biol. 279: 509–524. [DOI] [PubMed] [Google Scholar]
- Reim, I., and M. Frasch, 2005. The Dorsocross T-box genes are key components of the regulatory network controlling early cardiogenesis in Drosophila. Development 132: 4911–4925. [DOI] [PubMed] [Google Scholar]
- Reim, I., J. Mohler and M. Frasch, 2005. Tbx20-related genes, mid and H15, are required for tinman expression, proper patterning, and normal differentiation of cardioblasts in Drosophila. Mech. Dev. 122: 1056–1069. [DOI] [PubMed] [Google Scholar]
- Ruiz-Gomez, M., S. Romani, C. Hartmann, H. Jackle and M. Bate, 1997. Specific muscle identities are regulated by Krüppel during Drosophila embryogenesis. Development 124: 3407–3414. [DOI] [PubMed] [Google Scholar]
- Ruiz-Gomez, M., N. Coutts, M. Suster, M. Landgraf and M. Bate, 2002. myoblasts incompetent encodes a zinc finger transcription factor required to specify fusion-competent myoblasts in Drosophila. Development 129: 133–141. [DOI] [PubMed] [Google Scholar]
- Schaper, J. and B. Speiser, 1992. The extracellular matrix in the failing human heart. Basic Res. Cardiol. 87(Supp1.): 303–309. [DOI] [PubMed] [Google Scholar]
- Segal, D., and W. Gelbart, 1985. Shortvein, a new component of the decapentaplegic gene complex in Drosophila. Genetics 109: 119–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellin, J., S. Albrecht, V. Kölsch and A. Paululat, 2006. Dynamics of heart differentiation, visualized utilizing heart enhancer elements of the Drosophila melanogaster bHLH transcription factor Hand. Gene Expr. Patterns 6: 360–375. [DOI] [PubMed] [Google Scholar]
- Staehling-Hampton, K., and F. Hoffmann, 1994. Ectopic Decapentaplegic in the Drosophila midgut alters the expression of five homeotic genes, dpp, and wingless, causing specific morphological defects. Dev. Biol. 164: 502–512. [DOI] [PubMed] [Google Scholar]
- St. Johnston, R., F. Hoffmann, R. Blackman, D. Segal, R. Grimaila et al., 1990. Molecular organization of the decapentaplegic gene in Drosophila. Genes Dev. 4: 1114–1127. [DOI] [PubMed] [Google Scholar]
- Stottmann, R., M. Choi, Y. Mishina, E. Meyers and J. Klingensmith, 2004. BMP receptor IA is required in mammalian neural crest cells for development of the cardiac outflow tract and ventricular myocardium. Development 131: 2205–2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, M., M. Fujioka, T. Goto and R. Bodmer, 1999. The Drosophila homeobox genes zfh-1 and even-skipped are required for cardiac-specific differentiation of a numb-dependent lineage decision. Development 126: 3241–3251. [DOI] [PubMed] [Google Scholar]
- Van de Putte, T., M. Maruhashi, A. Francis, L. Nelles, H. Kondoh et al., 2003. Mice lacking ZFHX1B, the gene that codes for Smad-interacting protein-1, reveal a role for multiple neural crest cell defects in the etiology of Hirschsprung disease-mental retardation syndrome. Am. J. Hum. Genet. 72: 465–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh, T., M. Park, K. Ocorr, J. Nemaceck, K. Golden et al., 2000. Cardiac enhancer activity of the homeobox gene tinman depends on CREB consensus binding sites in Drosophila. Genesis 26: 55–66. [PubMed] [Google Scholar]
- Ward, E., and D. Coulter, 2000. odd-skipped is expressed in multiple tissues during Drosophila embryogenesis. Mech. Dev. 96: 233–236. [DOI] [PubMed] [Google Scholar]
- Ward, E., and J. Skeath, 2000. Characterization of a novel subset of cardiac cells and their progenitors in the Drosophila embryo. Development 127: 4959–4969. [DOI] [PubMed] [Google Scholar]
- Xu, X., Z. Yin, J. Hudson, E. Ferguson and M. Frasch, 1998. Smad proteins act in combination with synergistic and antagonistic regulators to target Dpp responses to the Drosophila mesoderm. Genes Dev. 12: 2354–2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, Z., and M. Frasch, 1998. Regulation and function of tinman during dorsal mesoderm induction and heart specification in Drosophila. Dev. Genet. 22: 187–200. [DOI] [PubMed] [Google Scholar]
- Zaffran, S., and M. Frasch, 2002. Early signals in cardiac development. Circ. Res. 91: 457–469. [DOI] [PubMed] [Google Scholar]