Abstract
The Ure2 protein of Saccharomyces cerevisiae can become a prion (infectious protein). At very low frequencies Ure2p forms an insoluble, infectious amyloid known as [URE3], which is efficiently transmitted to progeny cells or mating partners that consequently lose the normal Ure2p nitrogen regulatory function. The [URE3] prion causes yeast cells to grow slowly, has never been identified in the wild, and confers no obvious phenotypic advantage. An N-terminal asparagine-rich domain determines Ure2p prion-forming ability. Since ure2Δ strains are complemented by plasmids that overexpress truncated forms of Ure2p lacking the prion domain, the existence of the [URE3] prion and the evolutionary conservation of an N-terminal extension have remained mysteries. We find that Ure2p function is actually compromised in vivo by truncation of the prion domain. Moreover, Ure2p stability is diminished without the full-length prion domain. Mca1p, like Ure2p, has an N-terminal Q/N-rich domain whose deletion reduces its steady-state levels. Finally, we demonstrate that the prion domain may affect the interaction of Ure2p with other components of the nitrogen regulation system, specifically the negative regulator of nitrogen catabolic genes, Gzf3p.
“PRION” means infectious protein, a protein that transmits an infection without an essential accompanying nucleic acid, and generally a protein in an altered state that causes a polypeptide of similar primary sequence to convert to the same altered state (reviewed in Wickner et al. 2004; Ross et al. 2005). Prions can include any propagatable post-translational modification, most notably an altered protein fold. Indeed, most prions are infectious amyloid, which is a protein aggregate characterized by filamentous morphology, relative protease insensitivity, and a cross β-sheet content. The amyloid structural state is a thermodynamic and kinetic alternative to the soluble, globular state shared by many nonhomologous polypeptides (Chiti and Dobson 2006). However, in the case of prion amyloid, this conformation is both self-propagating and infectious.
Amyloid is of general concern since many human diseases are associated with a specific protein amyloid. One particular class of amyloid diseases is the collection of diseases known as the transmissible spongiform encephalopathies (TSEs), which are apparently caused by an infectious amyloid of PrP and are always fatal (Caughey and Baron 2006). A host organism must harbor a gene coding for PrP to be susceptible to infection by the prion, but deletion of the PrP gene has no demonstrable phenotypic effect in mice (Bueler et al. 1992, 1993). Presumably, any beneficial function of the prion protein must outweigh the penalty of prion susceptibility.
The prions [URE3], [PSI+], and [PIN+] of Saccharomyces cerevisiae are infectious amyloid forms of Ure2p, Sup35p, and Rnq1p, respectively (Wickner 1994; Derkatch et al. 2001; King and Diaz-Avalos 2004; Tanaka et al. 2004; Brachmann et al. 2005; Patel and Liebman 2007). Ure2p is a regulator of genes involved in nitrogen catabolite repression (NCR) (Cooper 2002) and Sup35p is a subunit of the translation termination factor (eRF3) (Frolova et al. 1994). Yeast cells carrying [URE3] or [PSI+] have reduced activity of Ure2p or Sup35p, respectively.
Normally, Ure2p exists as a soluble homodimer (Taylor et al. 1999), with the protein consisting of two domains: a carboxy-terminal NCR domain with a structure similar to glutathione-S-transferases (Bousset et al. 2001; Umland et al. 2001), and an unstructured amino-terminal Q/N-rich prion domain that enables the conversion and propagation of the [URE3] prion (Coschigano and Magasanik 1991; Masison and Wickner 1995; Pierce et al. 2005). When yeast cells are supplied with a good nitrogen source, such as ammonia or glutamine, Ure2p binds to the positive transcription factors Gln3p and Gat1p and prevents their entry into the nucleus (Beck and Hall 1999; Cardenas et al. 1999; Hardwick et al. 1999; Cox et al. 2000). The DAL5 gene encoding an allantoate permease is particularly sensitive to regulation by Ure2p (Rai et al. 1987). Among its fungal homologs, the amino-terminal Q/N-rich domain of Ure2p is a conserved feature and is ∼90 residues in length (Edskes and Wickner 2002; Baudin-Baillieu et al. 2003). In previous attempts to dissect Ure2p NCR function, a plasmid-based URE2 NCR domain was shown to complement ure2Δ cells as well as full-length URE2 (Masison and Wickner 1995).
In the laboratory setting with controlled conditions, the [URE3] prion occurs in a yeast population at a frequency of ∼1 per 1 million cells (Wickner 1994). [URE3] cells have relatively reduced growth rates and no obvious phenotypic advantages, although it is impossible to challenge yeast with all realistic environmental conditions. However, an extensive survey of wild and domestic yeast strains yielded no naturally occurring [URE3] strains, suggesting that the prion is not particularly advantageous since an infectious beneficial element should rapidly spread through natural populations (Nakayashiki et al. 2005).
Here we address the functional contribution of the Ure2p prion domain. We have integrated truncations of the URE2 gene lacking a complete prion domain into the endogenous URE2 locus. The strains lacking the entire prion domain of Ure2p have phenotypes that are intermediate between those containing the complete gene and those with the complete deletion. We observe that the prion domain contributes to the function and stability of the Ure2 protein and cannot be viewed solely as a prion-facilitating sequence, but may influence the interactions between Ure2p and other nitrogen regulatory proteins.
MATERIALS AND METHODS
Strains and media:
All full-length and truncated URE2 strains were derived from yeast strains FPS333 (MATα leu2 trp1 his3 kar1 ure2∷His3MX PDAL5∷ADE2 PDAL5∷CAN1) and FPS335 (MATα leu2 trp1 his3 kar1 ure2∷TRP1 PDAL5∷ADE2 PDAL5∷CAN1). These parental strains were created using PCR products derived from the template plasmid pFA6a-3HA-kanMX6 as described (Longtine et al. 1998). Full-length and truncated C-terminally HA-tagged URE2 genes were integrated into FPS333/335 at the endogenous locus using PCR products of the desired gene containing locus-specific flanking sequence. Beginning with strain FPS333, we constructed FPS337 (ure2∷URE21–354+3HA), FPS338 (ure2∷URE21–354+3HA), FPS340 (ure2∷URE2Δ2–64+3HA), FPS344 (ure2∷URE2Δ2–79+3HA), FPS345 (ure2∷URE2Δ2–79+3HA), FPS387 (ure2∷URE2Δ6–64+3HA), and FPS388 (ure2∷URE2Δ6–64+3HA). From strain FPS335 were prepared FPS352 (ure2∷URE2Δ2–64+3HA), FPS356 (ure2∷URE2Δ2–94+3HA), and FPS358 (ure2∷URE2Δ2–94+3HA). All integrations were confirmed by PCR and DNA sequencing and pairs with identical truncations were also confirmed to be phenotypically identical.
SUP35MC, which lacks the prion domain (residues 1–123), was PCR amplified with primers flanking the SUP35 locus using chromosomal template from strain 628-4B (MATα SUQ5 ade2-1 his3Δ202 kar1-1 ura2 SUP35∷SUP35Δ2–124). This SUP35MC PCR product was used to transform the [PSI+] strain 779-6A (MATa SUQ5 ade2-1 his3Δ202 leu2Δ1 trp1Δ63 ura3-52 [PSI+]), and transformants displaying a [psi−] phenotype were screened for the correct SUP35MC integrant by PCR.
Strains were cultured in the following media: YPAD (YPD supplemented with 40 mg/liter adenine), PRO medium [20 g/liter glucose; 1.7 g/liter yeast nitrogen base, lacking (NH4)2SO4 and amino acids; 10 g/liter proline, 24 mg/liter histidine, 24 mg/liter tryptophan, and 48 mg/liter leucine], NH4 medium [PRO medium supplemented with 50 mm (NH4)2SO4], and AA medium (PRO medium supplemented with 0.2 g/liter of each amino acid minus tyrosine).
Plasmids:
Plasmid-based complementation of ureΔ in strain FPS335 was performed with two different sets of plasmid constructs. The first set is derived from plasmid pH124 (Edskes et al. 1999) (CEN LEU2 PADH1) in which URE2 coding sequences with N-terminal tandem HA tags were inserted in the multiple cloning sites (MCS): pFPS56(URE2M,2HA,1–93), pFPS58(URE2M,2HA,94–354), and pFPS60(URE2M,2HA,1–354). The second set is derived from pH 316 (Moriyama et al. 2000) (CEN LEU2 PGAL1) in which coding sequences for full-length and truncated URE2 with N-terminal or C-terminal tandem HA tags have been inserted into the MCS: pFPS102(URE2M,2HA,Δ2–94), pFPS104(URE2M,2HA,1–354), pFPS110(URE2Δ2–94+2HA), and pFPS107(URE21–354+2HA).
Blotting methods:
Protein lysates for Western blotting were prepared by mechanically disrupting cells with glass beads, followed by heating in SDS–PAGE loading buffer. Immunoblotting was performed using rat anti-HA monoclonal antibody (Roche) and AP-conjugated anti-rat IgG (Promega) on proteins immobilized on PVDF membranes. Detection was performed using CDP-Star (Perkin Elmer) and CL-Xposure film (Pierce Biotechnology).
Detection of URE2 mRNA transcripts was performed by Northern blotting using 5–10 μg total RNA separated by electrophoresis and transferred to Hybond-N+ (GE Healthcare) membranes. URE2 transcripts were probed using the 550-bp restriction product produced by digesting URE2 DNA (pFPS55) with ScaI and NcoI. This restriction fragment does not overlap with the prion-domain coding region of URE2. The probe was labeled and detected using the alkaline-phosphatase AlkPhos Direct system (Amersham Biosciences).
Two-hybrid experiments:
Yeast two-hybrid interactions were observed in the haploid strain AH109 (MATa trp1 leu2 ura3 his3 gal4Δ gal80Δ LYS2∷GAL1UAS-GAL1TATA-HIS3 GAL2UAS-GAL2TATA-ADE2 URA3∷MEL1UAS-MEL1TATA-lacZ; CLONTECH) or in diploids resulting from the cross of strains PJ69-2A (MATa leu2-3,112 trp1-901 his3Δ200 ura3-52 gal4Δ gal80Δ GAL2∷ADE2 GAL1∷HIS3) and MaV204K (MATα leu2-3,112 trp1-901 his3Δ200 ade2Δ∷kanMX cyh2R can1R gal4Δ gal80Δ GAL1∷lacZ HIS3UASGAL1∷HIS3@LYS2 SPAL10 UASGAL1∷URA3) (Ito et al. 2000). Bait and prey constructs were prepared from pGBKT7 and pGADT7 (CLONTECH), respectively, as previously described (Pierce et al. 2005). In brief, bait and prey vectors coded fusions of Gal4p activation and binding domains with Ure2p1–80, Ure2pM,81–354, and Ure2p1–354. Screens for proteins that interact with Ure2p-binding-domain fusions were conducted in strain AH109 with a S. cerevisiae genomic library kindly provided by James et al. (1996). Screens for proteins that interact with Ure2p-activation domain fusions were conducted in diploids of PJ69-2A and MaV204K using a binding-domain ORF library generously provided by Takashi Ito (Ito et al. 2000). Positive two-hybrid interactions were selected on SC (−leu −trp −ade −his +3 mm 3-amino-triazole) medium. Using this system, we did not observe autoactivation caused by the Ure2p bait as was observed in a different two-hybrid system (Fernandez-Bellot et al. 1999; Kulkarni et al. 2001).
Protein stability:
The stabilities of the HA-tagged Ure2 proteins were monitored following the addition of a protein synthesis inhibitor, cycloheximide. Strains were grown in YPAD to OD550 ∼ 1, at which point 35 μg/ml cycloheximide was added. Cells were harvested and frozen at −80° every 15 min for 3 hr. Protein lysates were prepared as described above and equal amounts from all time points were gel separated, transferred, and blotted together. Relative protein values were quantified by densitometry.
Nonsense suppression:
Translational readthrough in the SUP35MC strain was determined using the in vivo dual-luciferase assay system as described (Harger and Dinman 2003). CEN URA3 plasmids (a gift from Jonathan Dinman) pJ375 (wild-type control), pYDL-UAA, pYDL-UAG, and pYDL-UGA (containing premature stop codons in 5′ end of firefly gene) were transformed into the SUP35MC and parental strain, selected, and maintained on SD lacking uracil. Luciferase activity was assessed using the Dual-Luciferase reporter assay system (Promega E1910) protocol as described in the manual. Luciferase activity was monitored with a Zylux Femtomaster FB15. Translational readthrough is expressed as the ratio of the experimental values relative to the control values (average of firefly luciferase/average Renilla luciferase). Readthrough is expressed as a percentage of the wild-type ratio ±SD with n = 3 for each plasmid.
RESULTS
Two-hybrid screen for proteins that bind the prion domain of Ure2p:
One possible role of the Ure2p prion domain could be the facilitating of interactions between Ure2p and other proteins. A S. cerevisiae Gal4p activation-domain (AD) genomic library was screened with Ure2p1–80 fused to Gal4p binding domain (BD) as bait using the yeast two-hybrid binding assay (Fields and Song 1989). Several protein fragment fusions were isolated that positively interacted with the Ure2p prion domain in the two-hybrid assay, but interactions were weak and could never be reproduced between cloned full-length fusions and the Ure2p prion domain (data not shown).
In addition, full-length Ure2p was used as bait in two-hybrid screens. Full-length Ure2p, fused to the Gal4p BD, was screened against the James et al. (1996) genomic library and, fused to the Gal4p AD, against a Gal4p BD library (Ito et al. 2000). With full-length Ure2p as bait, several proteins were identified that reproducibly yielded positive interactions in the two-hybrid system (supplemental Table 1 at http://www.genetics.org/supplemental/). One of these, Gzf3p, was of particular interest because it belongs to the GATA family of transcription factors that are involved in nitrogen regulation (Soussi-Boudekou et al. 1997).
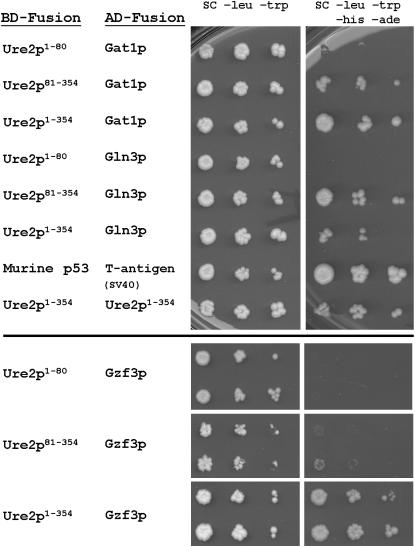
We examined further the two-hybrid interaction between GATA proteins and both full-length and truncated Ure2p. Full-length Gal4p AD fusions were constructed for Gzf3p, Ure2p, and two other GATA proteins: Gln3p and Gat1p. Gln3p has previously been shown to interact specifically with the C-terminal domain of Ure2p in two-hybrid assays (Kulkarni et al. 2001) and was used as a positive control, as was the Ure2p–Ure2p self interaction. The Gln3p and Gzf3p AD-fusion vectors were slightly toxic to the reporter strain, resulting in slower growth that could be slightly offset by the presence of the Ure2p BD-fusion vector, whereas the Gat1p AD-fusion vector had no observable detrimental effect upon growth (data not shown). The domain of Ure2p responsible for the two-hybrid interactions was confirmed for all pairs (Figure 1). In each case, the C-terminal NCR domain (residues 81–354) yielded a positive two-hybrid interaction, while the isolated N-terminal domain alone never produced a positive interaction.
Figure 1.—
Yeast two-hybrid binding interactions. Examples of two-hybrid interactions using reporter strain AH109 and Ure2p fragments fused to the Gal4p binding domain as bait. No autoactivation is caused by the Ure2p bait alone (data not shown).
As was observed with Gln3p, Gat1p has a specific two-hybrid interaction with the NCR domain of Ure2p. Gzf3p, however, proved to have a robust two-hybrid interaction with full-length Ure2p, but only a very weak interaction with the Ure2p NCR domain and no observable interaction specifically with the prion domain alone (Figure 1). In fact, there were no observed reproducible two-hybrid interactions between the isolated Ure2p prion domain and any other protein (Figure 1, supplemental Table 1 at http://www.genetics.org/supplemental/ and data not shown).
We considered the possibility that the failure of Gal4p-Ure281–354 or Gal4p-Ure21–80 fusions to interact with Gzf3 might be a result of altered stability. We consider this unlikely since each fusion is substantially overexpressed from the ADH1 promoter and Gal4p-Ure281–354 is stable enough to produce robust interactions with both Gln3p and Gat1p.
Phenotypic complementation by URE2 genes:
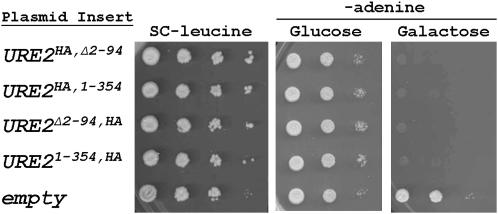
The C-terminal domain of Ure2p lacking the prion domain, when modestly overexpressed from a plasmid, can completely complement ure2Δ while the prion domain alone has no effect on nitrogen regulation (Masison and Wickner 1995; Figure 2 and data not shown). We critically tested whether the Ure2p prion domain affects nitrogen regulation when expressed at normal levels by constructing integrated URE2 genes lacking most or the entire prion domain and using as reporter the ADE2 gene linked to the DAL5 promoter (Brachmann et al. 2006). DAL5 encodes a permease for the uptake of allantoate (an alternative nitrogen source) and is normally transcriptionally repressed by Ure2p. With this reporting construct, a Ure2p activity gradient can be observed, such that high Ure2p activity completely inhibits growth in the absence of adenine in the medium while partial Ure2p activity results in some adenine prototrophy, but cells possess red coloration with intensity proportional to Ure2p activity.
Figure 2.—
Plasmid-based complementation of ure2Δ. Galactose-induced overexpression of Ure2p and Ure2pΔ2–94 completely represses PDAL5-ADE2 expression. Cells were grown on PRO medium plus 5 mm (NH4)SO4 containing either glucose or galactose as the carbon source. Ten-fold dilutions were spotted on the indicated media. Neither N- nor C-terminal HA epitope tags on Ure2p have any apparent effect.
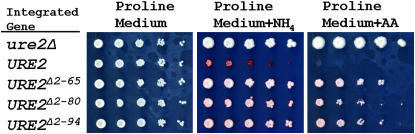
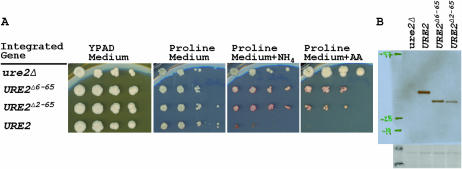
We find that, when expressed from the normal URE2 chromosomal site, deletion of part or all of the prion domain of Ure2p substantially affects nitrogen regulation (Figure 3). For each truncation, the repression of the DAL5 promoter is reduced relative to the full-length variant, but still maintains intermediate repressor activity. Thus, deletion of the prion domain compromises the activity of Ure2p.
Figure 3.—
Integrated URE2 gene truncations function incompletely. Ten-fold dilutions of strains carrying full-length and truncated chromosomal URE2 were plated on media with different primary nitrogen sources. Full-length Ure2p repressor activity is observed on a rich nitrogen source; however, Ure2p with a truncated prion domain only partially represses.
The prion domain and Ure2p steady-state protein levels:
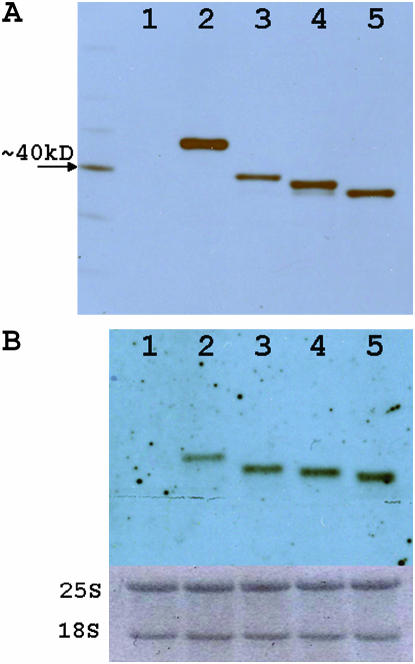
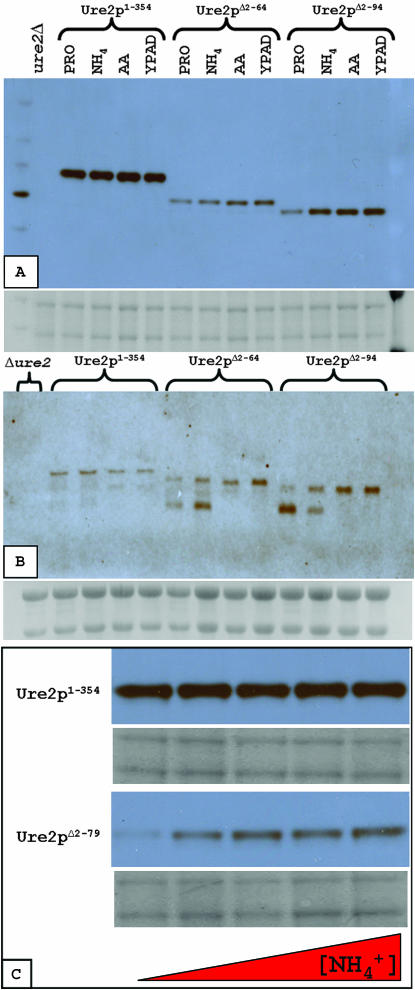
The relative expression of Ure2p deletions grown on YPAD medium was assayed by Western blotting (Figures 4A and 5C). All protein variants separated consistent with their predicted masses. The truncated Ure2 proteins were expressed at significantly lower levels relative to the full-length protein. This difference is not explained by transcript levels, as the truncated URE2 transcripts are conversely more abundant as judged by Northern analysis (Figure 4B).
Figure 4.—
Relative URE2 protein and transcript levels. Western blot (A) and Northern blot (B) analyses for isogenic strains differing at the URE2 locus grown on YPAD are as follows: (1) ure2Δ, (2) URE2, (3) URE2Δ2–65, (4) URE2Δ2–80, and (5) URE2Δ2–94.
Figure 5.—
Ure2 protein and transcript levels in various media. Strains were grown in the following media: YPAD, proline medium (PRO), PRO supplemented with 50 mm ammonium sulfate (NH4) or 2 g/liter amino acids (AA). (A) Western blot analysis of relative Ure2p levels. (B) Northern blot analysis of relative URE2 transcript levels. (C) Levels of truncated Ure2pΔ1–80, but not full-length Ure2p, are increased in response to supplemental ammonium (0–50 mm 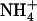 from left to right) in proline medium.
from left to right) in proline medium.
Relative protein and RNA transcript levels were also compared for strains grown on media containing different primary nitrogen sources (Figure 5). Unexpectedly, the truncated Ure2 protein levels were affected by the nitrogen source (Figure 5, A and C), while full-length Ure2p levels remained constant (Figure 5, A and C). Moreover, a smaller URE2 transcript species always appeared in the truncated strains when grown on poorer nitrogen sources (Figure 5B).
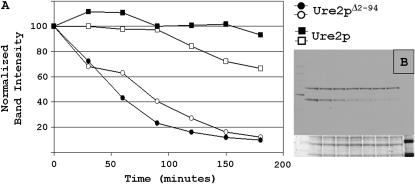
To address the lower steady-state levels of the truncated Ure2 proteins, protein levels were observed following the addition of cycloheximide, an inhibitor of protein synthesis. Ure2p1–354 and Ure2p94–354 levels were monitored over a 180-min period following the addition of cycloheximide to cultures in log-phase growth (Figure 6). The Ure2 protein lacking the prion domain is clearly less stable.
Figure 6.—
Ure2p turnover. Cycloheximide was added to strains with chromosomal full-length or truncated URE2. (A) Normalized Ure2p levels quantified by densitometry following Western blotting. (B) Example of data used for graph: full-length Ure2p is the top band and Ure2pΔ2–94 is the bottom band. Equal amounts of lysate protein from each strain were loaded in the same gel lane to ensure precise comparison.
The prion domain of Ure2p is amino terminal, so complete truncation of this domain necessarily results in removal of the beginning residues. Since the N-end residues can sometimes influence protein stability by signaling proteins for proteasomal degradation (Varshavsky 1996), the lower levels of prion domain-truncated Ure2p could have resulted from more degradation via the proteasome. We therefore constructed a strain with a minimally truncated URE2 gene that codes for Ure2p1–5,65–354, thus maintaining the first five amino acids. This strain similarly displays the partial repression observed in the other URE2-truncated strains (Figure 7A). Moreover, Ure2p1–5,65–354 has lower steady-state levels than the full-length Ure2p (Figure 7B), implying that the observed effects are not due to the N-end rule.
Figure 7.—
Retention of the Ure2p amino-terminal five amino acids does not restore function. (A) Isogenic strains differing at the URE2 locus were plated on various media. (B) Western blot was used to measure relative Ure2p levels.
Deletion of the prion domain of Sup35p:
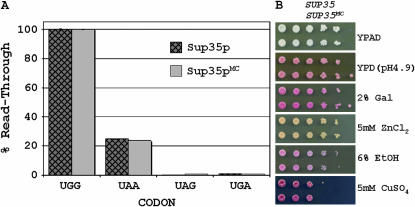
We were curious if the removal of the prion domain of another yeast prion protein would result in similar subtle differences in protein function and stability. The yeast prion protein Sup35p consists of three domains: an N-terminal prion domain, a highly charged middle domain, and a C-terminal eRF3 domain. Previously, it was reported that simultaneous deletion of the N and M domains (residues 1–253) of Sup35p resulted in phenotypic differences compared to wild type on several growth media (True and Lindquist 2000). We created yeast strains containing chromosomally integrated full-length and truncated SUP35 to similarly address functionality of the Sup35p prion domain. In our case, we limited our truncation to only the prion domain of Sup35p (residues 1–123). The strains coding full-length and truncated Sup35p were grown under conditions that were previously reported to reveal differences of Sup35p function (True and Lindquist 2000). In no case was there any observed difference in phenotype (Figure 8B). Likewise, steady-state protein levels of full-length and truncated Sup35p were observably equal for all growth conditions as judged by Western blotting (data not shown). Additionally, stop-codon readthrough assays revealed no intrinsic differences in translation termination activity between the full-length and truncated proteins (Figure 8A).
Figure 8.—
Integrated SUP35 gene truncations function completely in translational readthrough assay. (A) Nonsense suppression was assayed using a dual luciferase reporter with the different stop codons and using full-length Sup35p or a truncation lacking the prion domain. (B) Strains coding Sup35p with and without the prion domain were plated to selective media that were previously reported to reveal phenotypic differences for strains lacking the prion and middle domain of Sup35p. Top row, SUP35; bottom row, SUP35MC, 10-fold dilutions.
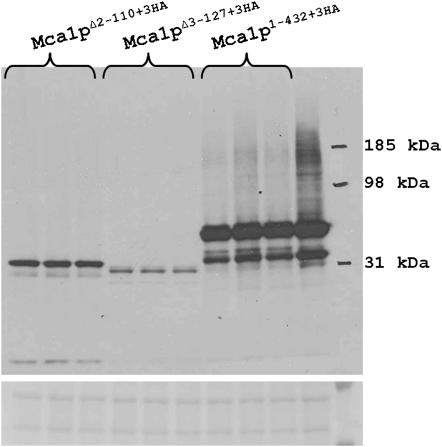
The prion domains of Sup35p and Ure2p are similar in that they are localized to the amino termini of their respective proteins, are rich in hydrophilic amino acids such as glutamine and asparagine, and diverge much more rapidly than their C-terminal domains. We selected a third protein with a similar amino-terminal domain, Mca1p, and asked how deletion of this domain affects protein steady-state levels. Relative Mca1p levels were compared from strains coding full-length and truncated (Mca1pΔ2–110 and Mca1pΔ3–127) proteins. As was observed with Ure2p, truncation of this region reduced steady-state protein levels (Figure 9).
Figure 9.—
Full-length and truncated Mca1p levels. Western blotting of Mca1p reveals that truncation of the prion-like domain of Mca1p dramatically reduces protein levels.
DISCUSSION
Here we show that removal of most or all of the prion domain dramatically impairs the normal in vivo nitrogen regulation activity of Ure2p. This effect may be explained by our finding that the prion domain plays a role in stabilizing Ure2p and by the requirement for the prion domain for Ure2p to interact with the GATA factor Gzf3.
The Ure2p C-terminal domain of a range of yeast species retains both high level sequence identity and ability to complement ure2Δ. The prion domain varies far more rapidly in evolution but most species retain a Q/N-rich N-terminal extension, suggesting that it has an important function (Edskes and Wickner 2002; Baudin-Baillieu et al. 2003). Residues 10–39 vary less rapidly than does the rest of the Ure2 prion domain (Edskes and Wickner 2002), and statistical analysis confirms that this region is selected for protein function (Harrison et al. 2007). However, those analyses do not tell us whether that function is prion formation or nitrogen regulation.
The Ure2p of S. paradoxus, closely related to S. cerevisiae, does not form a prion in S. paradoxus cells, despite possessing nearly identical primary structure (Talarek et al. 2005); nor could the Kluyveromyces lactis Ure2p form prions in S. cerevisiae (Baudin-Baillieu et al. 2003). If conservation of the prion domain were for facilitating prion formation, then this capability would be maintained. Thus, the “prion-domain”-like extension is apparently not preserved for forming prions.
Ure2p represses genes with GATA promoters when a good nitrogen source is abundant. In our reporter system, we observed the URE2-mediated repression of the ADE2 gene in the presence of various nitrogen sources. Full-length Ure2p functionally repressed the reporter in the presence of a good nitrogen source, whereas in the absence of Ure2p there was no observable repression. When Ure2p lacked a prion domain, there was neither complete repression nor total derepression, indicating an intermediate functionality.
The decrease in Ure2p function resulting from the truncation of the prion domain may be a result of diminished protein levels. Each truncation resulted in reduced Ure2p, while the overexpression of the Ure2p NCR domain resulted in a restoration of repressor function. The decrease in Ure2p levels is not merely a result of changing the amino-terminal residue. Of all the truncations, only one, Ure2pΔ2–64, would be predicted to have a higher turnover rate on the basis of the N-terminal residue if the initiating methionine were removed (Varshavsky 1996). Likewise, an earlier study with scrambled Ure2p prion domains, which also changed amino-terminal sequence, found no difference in protein levels (Ross et al. 2004). Most importantly, deletion of most of the prion domain, but leaving residues one to five intact, also resulted in defective function and low levels of Ure2 protein. Like Ure2p, Sup35p of Schizosaccharomyces pombe has a dispensable N-terminal domain whose deletion also destabilizes the protein (Kong et al. 2004). Further, we find that deletion of the Q/N-rich N-terminal domain of Mca1p, a yeast caspase homolog, destabilizes the protein. Our results suggest that some Q/N-rich N-terminal domains may function to protect the protein from degradation.
Changes in activity or stability of truncated Ure2p could result from loss of protein interactions that might normally occur through the prion domain. We find interactions between Ure2p and two GATA transcription factors, Gat1p and Gzf3p. The transcription factors regulating genes involved in nitrogen metabolism in yeast are members of the GATA family, named for their specific binding to GATAA promoter sequences (for review see Cooper 2002). Gat1p and Gln3p are positive activators of transcription that are regulated by Ure2p; Ure2p is known to interact with Gln3p (Kulkarni et al. 2001; Carvalho and Zheng 2003). Here we show that, like Gln3p, Gat1p also interacts with Ure2p in the two-hybrid system. Furthermore, Ure2p appears to interact with the transcriptional repressor, Gzf3p, and this interaction depends on the prion domain. Global regulation of nitrogen metabolism demands that Ure2p communicate with both activator and repressor transcription factors, and the Ure2p prion domain facilitates the interactions with Gzf3p.
Two-hybrid experiments revealed that the C-terminal domain of Ure2p is predominately responsible for interacting with other proteins. We found no proteins that bind to the Ure2p prion domain alone, but the prion domain is required for substantial binding to Gzf3p. Given the limitations of the two-hybrid assay, we are unable to distinguish between direct or indirect Ure2p-transcription factor interactions. The data are consistent with the Ure2p prion domain contributing to direct or indirect interactions with GATA transcription factors.
It is proposed that [PSI+] aids cells by helping them deal with stress (Eaglestone et al. 1999) or by helping them evolve (True and Lindquist 2000). One study reported that strains deleted for the prion domain (N) and the adjacent charged region (M) of Sup35 had fully 41 phenotypic differences compared to [psi−] cells, indicating a [PSI]-independent functional role for the prion domain (True and Lindquist 2000). Paradoxically, it was proposed that the prion-forming ability of the Q/N-rich N-terminal prion domains of Sup35p and Ure2p are an advantage to the cell, the former in aiding evolution of useful traits and the latter in promoting growth. Our Sup35 data do not reveal changes in phenotype on deletion of the prion domain reported by others, nor do we observe changes in the efficiency of readthrough of translation termination codons. Strain background or differences in the deletions studied may affect phenotypes, and slight functional contributions of the prion domain of Sup35p may be smaller than the sensitivity of our readthrough assays or have effects only in certain contexts of the termination codon.
However, there is mounting evidence that the Sup35 prion domain is functioning beyond its role in facilitating prion formation. In one study using C-terminal Sup35p mutants, deletion of the prion domain resulted in less nonsense codon suppression (Volkov et al. 2007). Likewise, in another study it was found that the Sup35p prion domain reduced translational termination when there were reduced levels of eRF1 (Urakov et al. 2006). Moreover, Urakov et al. contend that Sup35p may have an unidentified additional function that depends on the prion domain and is related neither to prion formation nor to translation termination.
Even deleterious infectious elements (viruses, plasmids, prions) may be widespread in nature, but an advantageous infectious element (e.g., mitochondria) is sure to be easily found in the wild. The absence of [URE3] and [PSI+] from wild strains, in spite of their infectivity and arising de novo at a much higher frequency than other infectious elements, proves that they are a substantial disadvantage to their hosts (Nakayashiki et al. 2005). Nor does this conclusion depend on the stability of the prion variant. The influenza virus does not stably propagate in any host. It, like most other viruses and bacteria, is purged from most hosts it infects. But because it is infectious, it is readily found in the wild. It is argued that the conservation of Q/N-rich N-terminal extensions across many species implies that prions are useful to yeast. But the inability of some “prion domains” to form prions vitiates this argument, and we have now provided a sufficient explanation of the conservation of these domains: the Ure2p prion domain facilitates the nitrogen regulation function by stabilizing the molecule and aiding its interaction with other factors. Like Ure2p, many human proteins can sometimes form amyloid in vivo, but the teleological explanation that they are therefore conserved for this function seems unlikely.
Acknowledgments
We thank Mike Pierce for his helpful advice and for providing URE2 two-hybrid plasmids. We are grateful to Philip James, Elizabeth Craig, and Takashi Ito for sharing their two-hybrid banks with us. This research was supported by the Intramural Research Program of the National Institutes of Health, National Institute of Diabetes Digestive and Kidney Diseases.
References
- Baudin-Baillieu, A., E. Fernandez-Bellot, F. Reine, E. Coissac and C. Cullin, 2003. Conservation of the prion properties of Ure2p through evolution. Mol. Biol. Cell. 14: 3449–3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck, T., and M. N. Hall, 1999. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature 402: 689–692. [DOI] [PubMed] [Google Scholar]
- Bousset, L., H. Belrhali, R. Melki and S. Morera, 2001. Crystal structures of the yeast prion Ure2p functional region in complex with glutathione and related compounds. Biochemistry 40: 13564–13573. [DOI] [PubMed] [Google Scholar]
- Brachmann, A., U. Baxa and R. B. Wickner, 2005. Prion generation in vitro: amyloid of Ure2p is infectious. EMBO J. 24: 3082–3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann, A., J. A. Toombs and E. D. Ross, 2006. Reporter assay systems for [URE3] detection and analysis. Methods 39: 35–42. [DOI] [PubMed] [Google Scholar]
- Bueler, H., M. Fischer, Y. Lang, H. Bluethmann, H. P. Lipp et al., 1992. Normal development and behavior of mice lacking the neuronal cell-surface PrP protein. Nature 356: 577–582. [DOI] [PubMed] [Google Scholar]
- Bueler, H., A. Aguzzi, A. Sailer, R.-A. Greiner, P. Autenried et al., 1993. Mice devoid of PrP are resistant to Scrapie. Cell 73: 1339–1347. [DOI] [PubMed] [Google Scholar]
- Cardenas, M. E., N. S. Cutler, M. C. Lorenz, C. J. Di Como and J. Heitman, 1999. The TOR signaling cascade regulates gene expression in response to nutrients. Genes Dev. 13: 3271–3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho, J., and X. F. Zheng, 2003. Domains of Gln3p interacting with karyopherins, Ure2p and the target of rapamycin protein. J. Biol. Chem. 278: 16878–16886. [DOI] [PubMed] [Google Scholar]
- Caughey, B., and G. S. Baron, 2006. Prions and their partners in crime. Nature 443: 803–810. [DOI] [PubMed] [Google Scholar]
- Chiti, F., and C. M. Dobson, 2006. Protein folding, functional amyloid and human disease. Annu. Rev. Biochem. 75: 333–366. [DOI] [PubMed] [Google Scholar]
- Cooper, T. G., 2002. Transmitting the signal of excess nitrogen in Saccharomyces cerevisiae from the Tor proteins to th GATA factors: connecting the dots. FEMS Microbiol. Rev. 26: 223–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coschigano, P. W., and B. Magasanik, 1991. The URE2 gene product of Saccharomyces cerevisiae plays an important role in the cellular response to the nitrogen source and has homology to glutathione S-transferases. Mol. Cell. Biol. 11: 822–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox, K. H., R. Rai, M. Distler, J. R. Daugherty, J. A. Coffman et al., 2000. Saccharomyces cerevisiae GATA sequences function as TATA elements during nitrogen catabolite repression and when Gln3p is excluded from the nucleus by overproduction of Ure2p. J. Biol. Chem. 275: 17611–17618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkatch, I. L., M. E. Bradley, J. Y. Hong and S. W. Liebman, 2001. Prions affect the appearance of other prions: the story of. [PIN] Cell 106: 171–182. [DOI] [PubMed] [Google Scholar]
- Eaglestone, S. S., B. S. Cox and M. F. Tuite, 1999. Translation termination efficiency can be regulated in Saccharomyces cerevisiae by environmental stress through a prion-mediated mechanism. EMBO J. 18: 1974–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edskes, H. K., V. T. Gray and R. B. Wickner, 1999. The [URE3] prion is an aggregated form of Ure2p that can be cured by overexpression of Ure2p fragments. Proc. Natl. Acad. Sci. USA 96: 1498–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edskes, H. K., and R. B. Wickner, 2002. Conservation of a portion of the S. cerevisiae Ure2p prion domain that interacts with the full-length protein. Proc. Natl. Acad. Sci. USA 99(Suppl. 4): 16384–16391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Bellot, E., E. Guillemet, A. Baudin-Baillieu, S. Gaumer, A. A. Komar et al., 1999. Characterization of the interaction domains of Ure2p, a prion-like protein of yeast. Biochem. J. 338: 403–407. [PMC free article] [PubMed] [Google Scholar]
- Fields, S., and O. Song, 1989. A novel genetic system to detect protein-protein interactions. Nature 340: 245–246. [DOI] [PubMed] [Google Scholar]
- Frolova, L., X. Legoff, H. H. Rasmussen, S. Cheperegin, G. Drugeon et al., 1994. A highly conserved eukaryotic protein family possessing properties of polypeptide chain release factor. Nature 372: 701–703. [DOI] [PubMed] [Google Scholar]
- Hardwick, J. S., F. G. Kuruvilla, J. K. Tong, A. F. Shamji and S. L. Schreiber, 1999. Rapamycin-modulated transcription defines the subset of nutrient- sensitive signaling pathways directly controlled by the tor proteins. Proc. Natl. Acad. Sci. USA 96: 14866–14870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harger, J. W., and J. D. Dinman, 2003. An in vivo dual-luciferase assay system for studying translational recoding in the yeast Saccharomyces cerevisiae. RNA 9: 1019–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison, L. B., Z. Yu, J. E. Stajich, F. S. Dietrich and P. M. Harrison, 2007. Evolution of budding yeast prion-determinant sequences across diverse fungi. J. Mol. Biol. 368: 273–282. [DOI] [PubMed] [Google Scholar]
- Ito, T., K. Tahiro, S. Muta, R. Ozawa, T. Chiba et al., 2000. Toward a protein-protein interaction map of the budding yeast: a comprehensive system to examine two-hybrid interactions in all possible combinations between the yeast proteins. Proc. Natl. Acad. Sci. USA 97: 1143–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James, P., J. Halladay and E. A. Craig, 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144: 1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, C. Y., and R. Diaz-Avalos, 2004. Protein-only transmission of three yeast prion strains. Nature 428: 319–323. [DOI] [PubMed] [Google Scholar]
- Kong, C., K. Ito, M. A. Walsh, M. Wada, Y. Liu et al., 2004. Crystal structure and functional analysis of the eukaryotic class II release factor eRF3 from S. pombe. Mol. Cell 14: 233–245. [DOI] [PubMed] [Google Scholar]
- Kulkarni, A. A., A. T. Abul-Hamd, R. Rai, H. El Berry and T. G. Cooper, 2001. Gln3p nuclear localization and interaction with Ure2p in Saccharomyces cerevisiae. J. Biol. Chem. 276: 32136–32144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine, M. S., A. R. Mckenzie, D. J. Demarini, N. G. Shah, A. Wach et al., 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961. [DOI] [PubMed] [Google Scholar]
- Masison, D. C., and R. B. Wickner, 1995. Prion-inducing domain of yeast Ure2p and protease resistance of Ure2p in prion-containing cells. Science 270: 93–95. [DOI] [PubMed] [Google Scholar]
- Moriyama, H., H. K. Edskes and R. B. Wickner, 2000. [URE3] prion propagation in Saccharomyces cerevisiae: requirement for chaperone Hsp104 and curing by overexpressed chaperone Ydj1p. Mol. Cell. Biol. 20: 8916–8922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayashiki, T., C. P. Kurtzman, H. K. Edskes and R. B. Wickner, 2005. Yeast prions [URE3] and [PSI+] are diseases. Proc. Natl. Acad. Sci. USA 102: 10575–10580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel, B. K., and S. W. Liebman, 2007. “Prion proof” for [PIN+]: infection with in vitro-made amyloid aggregates of Rnq1p-(132-405) induces [PIN+]. J. Mol. Biol. 365: 773–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce, M. M., U. Baxa, A. C. Steven, A. Bax and R. B. Wickner, 2005. Is the prion domain of soluble Ure2p unstructured? Biochemistry 44: 321–328. [DOI] [PubMed] [Google Scholar]
- Rai, R., F. Genbauffe, H. Z. Lea and T. G. Cooper, 1987. Transcriptional regulation of the DAL5 gene in Saccharomyces cerevisiae. J. Bacteriol. 169: 3521–3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross, E. D., U. Baxa and R. B. Wickner, 2004. Scrambled prion domains form prions and amyloid. Mol. Cell Biol. 24: 7206–7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross, E. D., A. P. Minton and R. B. Wickner, 2005. Prion domains: sequences, structures and interactions. Nat. Cell Biol. 7: 1039–1044. [DOI] [PubMed] [Google Scholar]
- Soussi-Boudekou, S., S. Vissers, A. Urrestarazu, J. C. Jauniaux and B. Andre, 1997. Gzf3p, a fourth GATA factor involved in nitrogen-regulated transcription in Saccharomyces cerevisiae. Mol. Microbiol. 23: 1157–1168. [DOI] [PubMed] [Google Scholar]
- Talarek, N., L. Maillet, C. Cullin and M. Aigle, 2005. The [URE3] prion is not conserved among Saccharomyces species. Genetics 171: 23–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, M., P. Chien, N. Naber, R. Cooke and J. S. Weissman, 2004. Conformational variations in an infectious protein determine prion strain differences. Nature 428: 323–328. [DOI] [PubMed] [Google Scholar]
- Taylor, K. L., N. Cheng, R. W. Williams, A. C. Steven and R. B. Wickner, 1999. Prion domain initiation of amyloid formation in vitro from native Ure2p. Science 283: 1339–1343. [DOI] [PubMed] [Google Scholar]
- True, H. L., and S. L. Lindquist, 2000. A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature 407: 477–483. [DOI] [PubMed] [Google Scholar]
- Umland, T. C., K. L. Taylor, S. Rhee, R. B. Wickner and D. R. Davies, 2001. The crystal structure of the nitrogen catabolite regulatory fragment of the yeast prion protein Ure2p. Proc. Natl. Acad. Sci. USA 98: 1459–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urakov, V. N., I. A. Valouev, N. V. Kochneva-Pervukhova, A. N. Pakeiser, A. Y. Vishnevsky et al., 2006. N-terminal region of Saccharomyces cerevisiae eRF3 is essential for the functioning of the eRF1/eRF3 complex beyond translation termination. BMC Mol. Biol. 7: 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky, A., 1996. The N-end rule: functions, mysteries, uses. Proc. Natl. Acad. Sci. USA 93: 12142–12149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkov, K., K. Osipov, I. Valouev, S. Inge-Vechtomov and L. Mironova, 2007. N-terminal extension of Saccharomyces cerevisiae translation termination factor eRF3 influences the suppression efficiency of sup35 mutations. FEMS Yeast Res. 7: 357–365. [DOI] [PubMed] [Google Scholar]
- Wickner, R. B., 1994. [URE3] as an altered URE2 protein: evidence for a prion analog in S. cerevisiae. Science 264: 566–569. [DOI] [PubMed] [Google Scholar]
- Wickner, R. B., H. K. Edskes, B. T. Roberts, U. Baxa, M. M. Pierce et al., 2004. Prions: proteins as genes and infectious entities. Genes Dev. 18: 470–485. [DOI] [PubMed] [Google Scholar]