Abstract
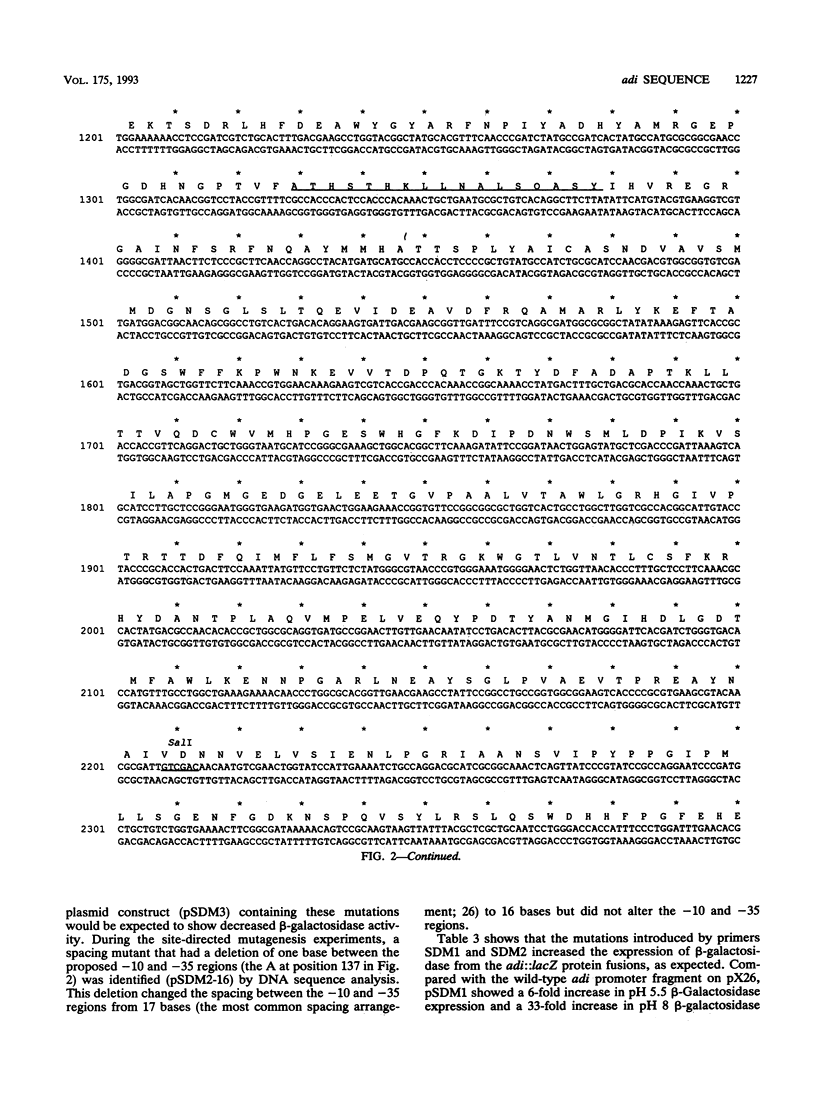
Arginine decarboxylase (encoded by adi) is induced under conditions of acidic pH, anaerobiosis, and rich medium. The DNA sequence of a 3-kb fragment of the Escherichia coli chromosome encoding biodegradative arginine decarboxylase was determined. This sequence encodes a protein of 755 amino acids with a molecular size of 84,420 daltons. The molecular weight and predicted Adi amino acid composition agree with those found in earlier work. The amino acid sequence of arginine decarboxylase showed homology to those of three other decarboxylases of E. coli: (i) CadA, encoding lysine decarboxylase; (ii) SpeC, encoding biosynthetic ornithine decarboxylase; and (iii) SpeF, encoding biodegradative ornithine decarboxylase and the lysine decarboxylase of Hafnia alvei. Unlike SpeC and SpeF, Adi is not similar to the biosynthetic arginine decarboxylase, SpeA. adi is also dissimilar to cadA and speF in that it does not appear to be part of an operon containing a metabolically related transport protein, indicating that it represents a new type of biodegradative decarboxylase regulation. Transcriptional fusions between fragments upstream of adi and lacZ, primer extension, and site-directed mutagenesis experiments defined the pH-regulated promoter. Deletion analysis of the upstream region and cloning of fragments to make adi::lacZ protein fusion implicated a region beyond an upstream SspI site in pH regulation. Induction of adi in the presence of sublethal concentrations of novobiocin or coumermycin A1, inhibitors of DNA gyrase, was dramatically decreased, indicating that DNA supercoiling is involved in adi expression. These results and those of promoter structure studies indicated that acid regulation of adi may involve a mechanism different from that of acid regulation of cad.
Full text
PDF










Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aliabadi Z., Park Y. K., Slonczewski J. L., Foster J. W. Novel regulatory loci controlling oxygen- and pH-regulated gene expression in Salmonella typhimurium. J Bacteriol. 1988 Feb;170(2):842–851. doi: 10.1128/jb.170.2.842-851.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Applebaum D. M., Dunlap J. C., Morris D. R. Comparison of the biosynthetic and biodegradative ornithine decarboxylases of Escherichia coli. Biochemistry. 1977 Apr 19;16(8):1580–1584. doi: 10.1021/bi00627a008. [DOI] [PubMed] [Google Scholar]
- Auger E. A., Redding K. E., Plumb T., Childs L. C., Meng S. Y., Bennett G. N. Construction of lac fusions to the inducible arginine- and lysine decarboxylase genes of Escherichia coli K12. Mol Microbiol. 1989 May;3(5):609–620. doi: 10.1111/j.1365-2958.1989.tb00208.x. [DOI] [PubMed] [Google Scholar]
- Blethen S. L., Boeker E. A., Snell E. E. Argenine decarboxylase from Escherichia coli. I. Purification and specificity for substrates and coenzyme. J Biol Chem. 1968 Apr 25;243(8):1671–1677. [PubMed] [Google Scholar]
- Boeker E. A., Fischer E. H., Snell E. E. Arginine decarboxylase from Escherichia coli. 3. Subunit structure. J Biol Chem. 1969 Oct 10;244(19):5239–5245. [PubMed] [Google Scholar]
- Boeker E. A., Fischer E. H., Snell E. E. Arginine decarboxylase from Escherichia coli. IV. Structure of the pyridoxal phosphate binding site. J Biol Chem. 1971 Nov 25;246(22):6776–6781. [PubMed] [Google Scholar]
- Boeker E. A., Snell E. E. Arginine decarboxylase from Escherichia coli. II. Dissociation and reassociation of subunits. J Biol Chem. 1968 Apr 25;243(8):1678–1684. [PubMed] [Google Scholar]
- Buch J. K., Boyle S. M. Biosynthetic arginine decarboxylase in Escherichia coli is synthesized as a precursor and located in the cell envelope. J Bacteriol. 1985 Aug;163(2):522–527. doi: 10.1128/jb.163.2.522-527.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadaban M. J., Cohen S. N. Lactose genes fused to exogenous promoters in one step using a Mu-lac bacteriophage: in vivo probe for transcriptional control sequences. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4530–4533. doi: 10.1073/pnas.76.9.4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dente L., Cesareni G., Cortese R. pEMBL: a new family of single stranded plasmids. Nucleic Acids Res. 1983 Mar 25;11(6):1645–1655. doi: 10.1093/nar/11.6.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorman C. J., Barr G. C., Ni Bhriain N., Higgins C. F. DNA supercoiling and the anaerobic and growth phase regulation of tonB gene expression. J Bacteriol. 1988 Jun;170(6):2816–2826. doi: 10.1128/jb.170.6.2816-2826.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drlica K. Biology of bacterial deoxyribonucleic acid topoisomerases. Microbiol Rev. 1984 Dec;48(4):273–289. doi: 10.1128/mr.48.4.273-289.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drlica K. Control of bacterial DNA supercoiling. Mol Microbiol. 1992 Feb;6(4):425–433. doi: 10.1111/j.1365-2958.1992.tb01486.x. [DOI] [PubMed] [Google Scholar]
- Drlica K., Rouviere-Yaniv J. Histonelike proteins of bacteria. Microbiol Rev. 1987 Sep;51(3):301–319. doi: 10.1128/mr.51.3.301-319.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enea V., Zinder N. D. Interference resistant mutants of phage f1. Virology. 1982 Oct 15;122(1):222–226. doi: 10.1016/0042-6822(82)90395-6. [DOI] [PubMed] [Google Scholar]
- FALKOW S. Activity of lysine decarboxlase as an aid in the identification of Salmonellae and Shigellae. Am J Clin Pathol. 1958 Jun;29(6):598–600. doi: 10.1093/ajcp/29.6_ts.598. [DOI] [PubMed] [Google Scholar]
- Foster J. W., Aliabadi Z. pH-regulated gene expression in Salmonella: genetic analysis of aniG and cloning of the earA regulator. Mol Microbiol. 1989 Nov;3(11):1605–1615. doi: 10.1111/j.1365-2958.1989.tb00146.x. [DOI] [PubMed] [Google Scholar]
- Gale E. F. The production of amines by bacteria: The decarboxylation of amino-acids by strains of Bacterium coli. Biochem J. 1940 Mar;34(3):392–413. doi: 10.1042/bj0340392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich J. A., Schwartz M. L., McClure W. R. Searching for and predicting the activity of sites for DNA binding proteins: compilation and analysis of the binding sites for Escherichia coli integration host factor (IHF). Nucleic Acids Res. 1990 Sep 11;18(17):4993–5000. doi: 10.1093/nar/18.17.4993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanatani M., Yazyu H., Shiota-Niiya S., Moriyama Y., Kanazawa H., Futai M., Tsuchiya T. Physical and genetic characterization of the melibiose operon and identification of the gene products in Escherichia coli. J Biol Chem. 1984 Feb 10;259(3):1807–1812. [PubMed] [Google Scholar]
- Harley C. B., Reynolds R. P. Analysis of E. coli promoter sequences. Nucleic Acids Res. 1987 Mar 11;15(5):2343–2361. doi: 10.1093/nar/15.5.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein J. Unified approach to alignment and phylogenies. Methods Enzymol. 1990;183:626–645. doi: 10.1016/0076-6879(90)83041-7. [DOI] [PubMed] [Google Scholar]
- Higgins C. F., Dorman C. J., Stirling D. A., Waddell L., Booth I. R., May G., Bremer E. A physiological role for DNA supercoiling in the osmotic regulation of gene expression in S. typhimurium and E. coli. Cell. 1988 Feb 26;52(4):569–584. doi: 10.1016/0092-8674(88)90470-9. [DOI] [PubMed] [Google Scholar]
- Kashiwagi K., Miyamoto S., Suzuki F., Kobayashi H., Igarashi K. Excretion of putrescine by the putrescine-ornithine antiporter encoded by the potE gene of Escherichia coli. Proc Natl Acad Sci U S A. 1992 May 15;89(10):4529–4533. doi: 10.1073/pnas.89.10.4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashiwagi K., Suzuki T., Suzuki F., Furuchi T., Kobayashi H., Igarashi K. Coexistence of the genes for putrescine transport protein and ornithine decarboxylase at 16 min on Escherichia coli chromosome. J Biol Chem. 1991 Nov 5;266(31):20922–20927. [PubMed] [Google Scholar]
- Kohara Y., Akiyama K., Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987 Jul 31;50(3):495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- MELNYKOVYCH G., SNELL E. E. Nutritional requirements for the formation of arginine decarboxylase in Escherichia coli. J Bacteriol. 1958 Nov;76(5):518–523. doi: 10.1128/jb.76.5.518-523.1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama S., Mizushima S. Novel rpoA mutation that interferes with the function of OmpR and EnvZ, positive regulators of the ompF and ompC genes that code for outer-membrane proteins in Escherichia coli K12. J Mol Biol. 1987 Jun 20;195(4):847–853. doi: 10.1016/0022-2836(87)90489-x. [DOI] [PubMed] [Google Scholar]
- Meng S. Y., Bennett G. N. Nucleotide sequence of the Escherichia coli cad operon: a system for neutralization of low extracellular pH. J Bacteriol. 1992 Apr;174(8):2659–2669. doi: 10.1128/jb.174.8.2659-2669.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng S. Y., Bennett G. N. Regulation of the Escherichia coli cad operon: location of a site required for acid induction. J Bacteriol. 1992 Apr;174(8):2670–2678. doi: 10.1128/jb.174.8.2670-2678.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore R. C., Boyle S. M. Nucleotide sequence and analysis of the speA gene encoding biosynthetic arginine decarboxylase in Escherichia coli. J Bacteriol. 1990 Aug;172(8):4631–4640. doi: 10.1128/jb.172.8.4631-4640.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris D. R., Boeker E. A. Biosynthetic and biodegradative ornithine and arginine decarboxylases from Escherichia coli. Methods Enzymol. 1983;94:125–134. doi: 10.1016/s0076-6879(83)94020-x. [DOI] [PubMed] [Google Scholar]
- Ni Bhriain N., Dorman C. J., Higgins C. F. An overlap between osmotic and anaerobic stress responses: a potential role for DNA supercoiling in the coordinate regulation of gene expression. Mol Microbiol. 1989 Jul;3(7):933–942. doi: 10.1111/j.1365-2958.1989.tb00243.x. [DOI] [PubMed] [Google Scholar]
- Pettijohn D. E. Histone-like proteins and bacterial chromosome structure. J Biol Chem. 1988 Sep 15;263(26):12793–12796. [PubMed] [Google Scholar]
- Popkin P. S., Maas W. K. Escherichia coli regulatory mutation affecting lysine transport and lysine decarboxylase. J Bacteriol. 1980 Feb;141(2):485–492. doi: 10.1128/jb.141.2.485-492.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruss G. J., Drlica K. DNA supercoiling and prokaryotic transcription. Cell. 1989 Feb 24;56(4):521–523. doi: 10.1016/0092-8674(89)90574-6. [DOI] [PubMed] [Google Scholar]
- Recsei P. A., Snell E. E. Histidine decarboxylaseless mutants of Lactobacillus 30a: isolation and growth properties. J Bacteriol. 1972 Oct;112(1):624–626. doi: 10.1128/jb.112.1.624-626.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland G. C., Giffard P. M., Booth I. R. phs Locus of Escherichia coli, a mutation causing pleiotropic lesions in metabolism, is an rpoA allele. J Bacteriol. 1985 Nov;164(2):972–975. doi: 10.1128/jb.164.2.972-975.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabo D. L., Fischer E. H. Chemical properties of Escherichia coli lysine decarboxylase including a segment of its pyridoxal 5'-phosphate binding site. Biochemistry. 1974 Feb 12;13(4):670–676. doi: 10.1021/bi00701a006. [DOI] [PubMed] [Google Scholar]
- Shi X., Waasdorp B. C., Bennett G. N. Modulation of acid-induced amino acid decarboxylase gene expression by hns in Escherichia coli. J Bacteriol. 1993 Feb;175(4):1182–1186. doi: 10.1128/jb.175.4.1182-1186.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons R. W., Houman F., Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53(1):85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- Slonczewski J. L., Gonzalez T. N., Bartholomew F. M., Holt N. J. Mu d-directed lacZ fusions regulated by low pH in Escherichia coli. J Bacteriol. 1987 Jul;169(7):3001–3006. doi: 10.1128/jb.169.7.3001-3006.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R. F., Smith T. F. Automatic generation of primary sequence patterns from sets of related protein sequences. Proc Natl Acad Sci U S A. 1990 Jan;87(1):118–122. doi: 10.1073/pnas.87.1.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor C. W., Tabor H. Polyamines in microorganisms. Microbiol Rev. 1985 Mar;49(1):81–99. doi: 10.1128/mr.49.1.81-99.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson N., Dunyak D. S., Rosey E. L., Slonczewski J. L., Olson E. R. Identification of elements involved in transcriptional regulation of the Escherichia coli cad operon by external pH. J Bacteriol. 1992 Jan;174(2):530–540. doi: 10.1128/jb.174.2.530-540.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster C., Kempsell K., Booth I., Busby S. Organisation of the regulatory region of the Escherichia coli melibiose operon. Gene. 1987;59(2-3):253–263. doi: 10.1016/0378-1119(87)90333-7. [DOI] [PubMed] [Google Scholar]