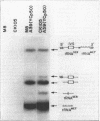
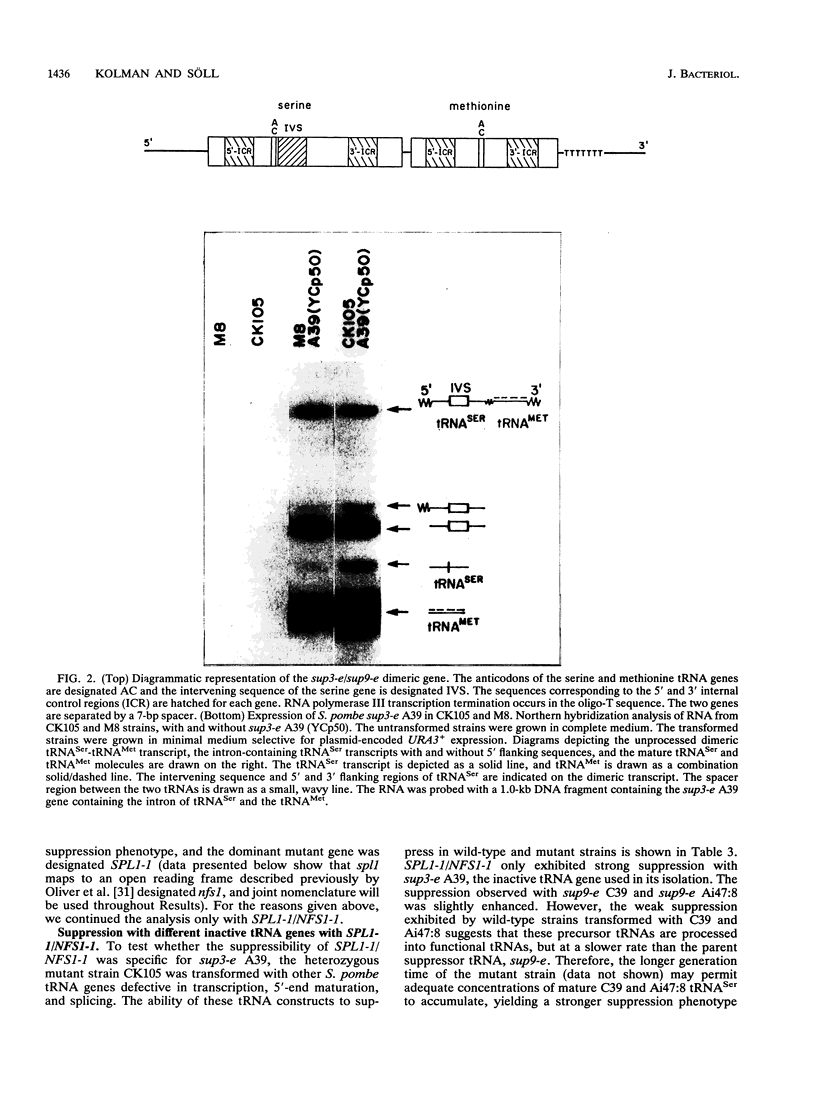
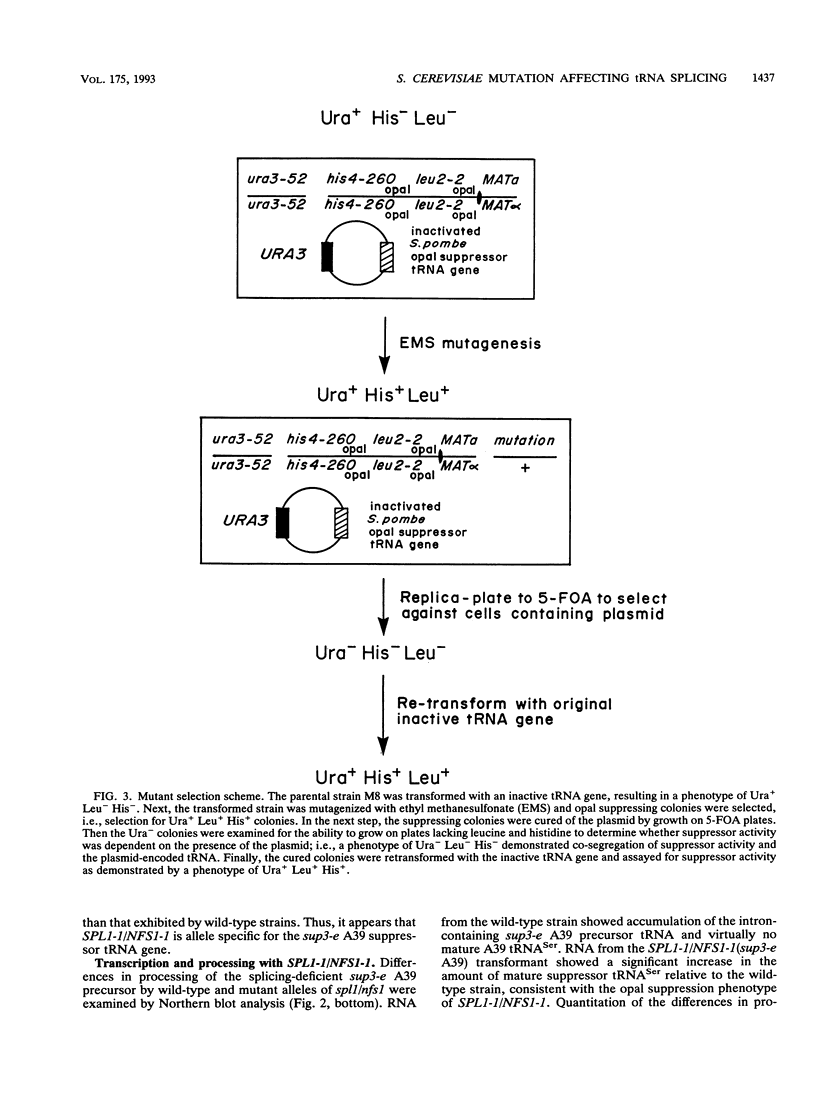
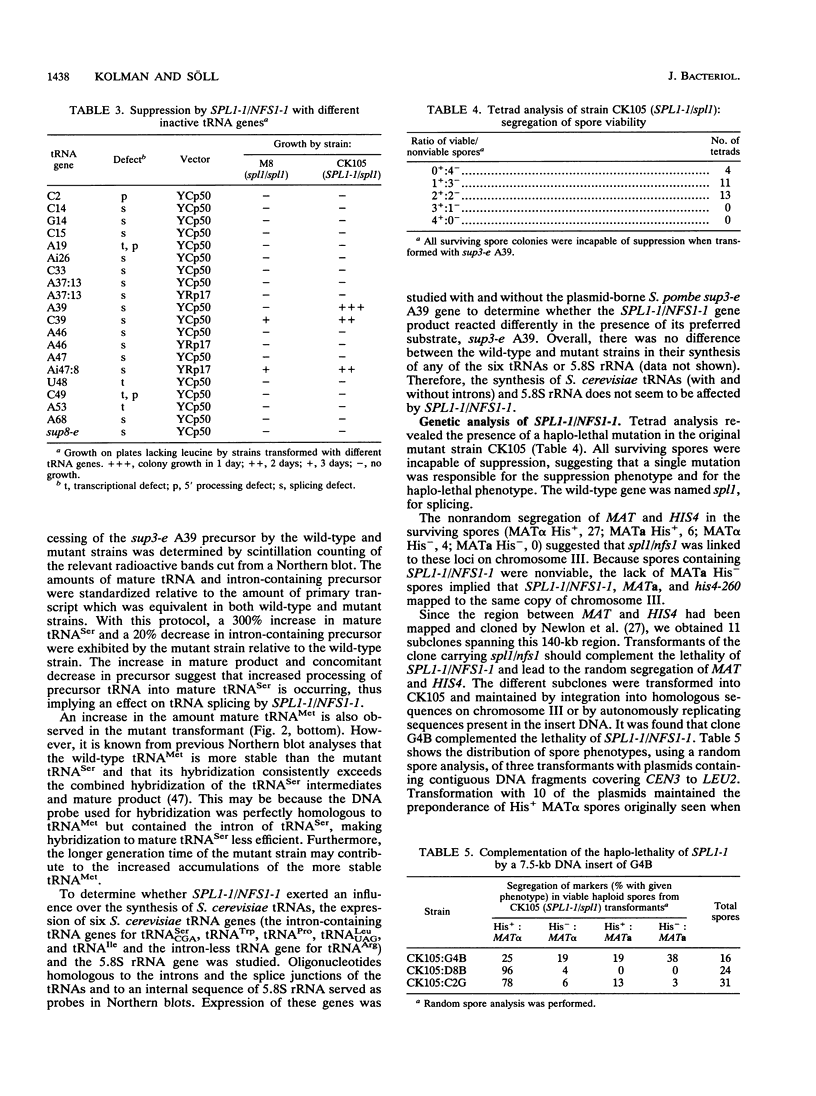
Abstract
A genetic approach was used to isolate and characterize Saccharomyces cerevisiae genes affecting tRNA processing. Three mutants were isolated which were able to process and utilize splicing-deficient transcripts from inactivated Schizosaccharomyces pombe suppressor tRNA genes. Extragenic recovery of suppressibility was verified by the suppression of nonsense mutations in LEU2, HIS4, and ADE1. One mutant, SPL1-1, was chosen for detailed analysis on the basis of its increased synthesis of mature suppressor tRNA over wild-type cell levels as determined by Northern (RNA) analysis. This mutant exhibited strong suppression exclusively with the defective tRNA gene used in the mutant selection. Genetic analysis revealed that a single, dominant, haplo-lethal mutation was responsible for the suppression phenotype. The mutation mapped on chromosome III to an essential 1.5-kb open reading frame (L. S. Symington and T. D. Petes, Mol. Cell. Biol. 8:595-604, 1988), recently named NFS1 (S. G. Oliver et al., Nature [London] 357:38-46, 1992), located adjacent (centromere proximal) to LEU2.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Atkinson N. S., Dunst R. W., Hopper A. K. Characterization of an essential Saccharomyces cerevisiae gene related to RNA processing: cloning of RNA1 and generation of a new allele with a novel phenotype. Mol Cell Biol. 1985 May;5(5):907–915. doi: 10.1128/mcb.5.5.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark M. W., Abelson J. The subnuclear localization of tRNA ligase in yeast. J Cell Biol. 1987 Oct;105(4):1515–1526. doi: 10.1083/jcb.105.4.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culbertson M. R., Winey M. Split tRNA genes and their products: a paradigm for the study of cell function and evolution. Yeast. 1989 Nov-Dec;5(6):405–427. doi: 10.1002/yea.320050602. [DOI] [PubMed] [Google Scholar]
- DeMarini D. J., Winey M., Ursic D., Webb F., Culbertson M. R. SEN1, a positive effector of tRNA-splicing endonuclease in Saccharomyces cerevisiae. Mol Cell Biol. 1992 May;12(5):2154–2164. doi: 10.1128/mcb.12.5.2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher M. P. Processing of tRNA in prokaryotes and eukaryotes. CRC Crit Rev Biochem. 1984;17(1):45–71. doi: 10.3109/10409238409110269. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans D. J., Jones R., Woodley P. R., Wilborn J. R., Robson R. L. Nucleotide sequence and genetic analysis of the Azotobacter chroococcum nifUSVWZM gene cluster, including a new gene (nifP) which encodes a serine acetyltransferase. J Bacteriol. 1991 Sep;173(17):5457–5469. doi: 10.1128/jb.173.17.5457-5469.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer C. L., Peebles C. L., Gegenheimer P., Abelson J. Mechanism of action of a yeast RNA ligase in tRNA splicing. Cell. 1983 Feb;32(2):537–546. doi: 10.1016/0092-8674(83)90473-7. [DOI] [PubMed] [Google Scholar]
- Greer C. L., Söll D., Willis I. Substrate recognition and identification of splice sites by the tRNA-splicing endonuclease and ligase from Saccharomyces cerevisiae. Mol Cell Biol. 1987 Jan;7(1):76–84. doi: 10.1128/mcb.7.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho C. K., Rauhut R., Vijayraghavan U., Abelson J. Accumulation of pre-tRNA splicing '2/3' intermediates in a Saccharomyces cerevisiae mutant. EMBO J. 1990 Apr;9(4):1245–1252. doi: 10.1002/j.1460-2075.1990.tb08232.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hottinger H., Pearson D., Yamao F., Gamulin V., Cooley L., Cooper T., Söll D. Nonsense suppression in Schizosaccharomyces pombe: the S. pombe Sup3-e tRNASerUGA gene is active in S. cerevisiae. Mol Gen Genet. 1982;188(2):219–224. doi: 10.1007/BF00332678. [DOI] [PubMed] [Google Scholar]
- Hurt D. J., Wang S. S., Lin Y. H., Hopper A. K. Cloning and characterization of LOS1, a Saccharomyces cerevisiae gene that affects tRNA splicing. Mol Cell Biol. 1987 Mar;7(3):1208–1216. doi: 10.1128/mcb.7.3.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson M. R., Brigle K. E., Bennett L. T., Setterquist R. A., Wilson M. S., Cash V. L., Beynon J., Newton W. E., Dean D. R. Physical and genetic map of the major nif gene cluster from Azotobacter vinelandii. J Bacteriol. 1989 Feb;171(2):1017–1027. doi: 10.1128/jb.171.2.1017-1027.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson M. R., Cash V. L., Weiss M. C., Laird N. F., Newton W. E., Dean D. R. Biochemical and genetic analysis of the nifUSVWZM cluster from Azotobacter vinelandii. Mol Gen Genet. 1989 Oct;219(1-2):49–57. doi: 10.1007/BF00261156. [DOI] [PubMed] [Google Scholar]
- Kennedy C., Dean D. The nifU, nifS and nifV gene products are required for activity of all three nitrogenases of Azotobacter vinelandii. Mol Gen Genet. 1992 Feb;231(3):494–498. doi: 10.1007/BF00292722. [DOI] [PubMed] [Google Scholar]
- Lee M. C., Knapp G. Transfer RNA splicing in Saccharomyces cerevisiae. Secondary and tertiary structures of the substrates. J Biol Chem. 1985 Mar 10;260(5):3108–3115. [PubMed] [Google Scholar]
- McCraith S. M., Phizicky E. M. A highly specific phosphatase from Saccharomyces cerevisiae implicated in tRNA splicing. Mol Cell Biol. 1990 Mar;10(3):1049–1055. doi: 10.1128/mcb.10.3.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCraith S. M., Phizicky E. M. An enzyme from Saccharomyces cerevisiae uses NAD+ to transfer the splice junction 2'-phosphate from ligated tRNA to an acceptor molecule. J Biol Chem. 1991 Jun 25;266(18):11986–11992. [PubMed] [Google Scholar]
- Mulligan M. E., Buikema W. J., Haselkorn R. Bacterial-type ferredoxin genes in the nitrogen fixation regions of the cyanobacterium Anabaena sp. strain PCC 7120 and Rhizobium meliloti. J Bacteriol. 1988 Sep;170(9):4406–4410. doi: 10.1128/jb.170.9.4406-4410.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan M. E., Haselkorn R. Nitrogen fixation (nif) genes of the cyanobacterium Anabaena species strain PCC 7120. The nifB-fdxN-nifS-nifU operon. J Biol Chem. 1989 Nov 15;264(32):19200–19207. [PubMed] [Google Scholar]
- Nichols M., Bell J., Klekamp M. S., Weil P. A., Söll D. Multiple mutations of the first gene of a dimeric tRNA gene abolish in vitro tRNA gene transcription. J Biol Chem. 1989 Oct 15;264(29):17084–17090. [PubMed] [Google Scholar]
- O'Connor J. P., Peebles C. L. In vivo pre-tRNA processing in Saccharomyces cerevisiae. Mol Cell Biol. 1991 Jan;11(1):425–439. doi: 10.1128/mcb.11.1.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor J. P., Peebles C. L. PTA1, an essential gene of Saccharomyces cerevisiae affecting pre-tRNA processing. Mol Cell Biol. 1992 Sep;12(9):3843–3856. doi: 10.1128/mcb.12.9.3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver S. G., van der Aart Q. J., Agostoni-Carbone M. L., Aigle M., Alberghina L., Alexandraki D., Antoine G., Anwar R., Ballesta J. P., Benit P. The complete DNA sequence of yeast chromosome III. Nature. 1992 May 7;357(6373):38–46. doi: 10.1038/357038a0. [DOI] [PubMed] [Google Scholar]
- Pearson D., Willis I., Hottinger H., Bell J., Kumar A., Leupold U., Söll D. Mutations preventing expression of sup3 tRNASer nonsense suppressors of Schizosaccharomyces pombe. Mol Cell Biol. 1985 Apr;5(4):808–815. doi: 10.1128/mcb.5.4.808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phizicky E. M., Consaul S. A., Nehrke K. W., Abelson J. Yeast tRNA ligase mutants are nonviable and accumulate tRNA splicing intermediates. J Biol Chem. 1992 Mar 5;267(7):4577–4582. [PubMed] [Google Scholar]
- Phizicky E. M., Schwartz R. C., Abelson J. Saccharomyces cerevisiae tRNA ligase. Purification of the protein and isolation of the structural gene. J Biol Chem. 1986 Feb 25;261(6):2978–2986. [PubMed] [Google Scholar]
- Rockmill B., Lambie E. J., Roeder G. S. Spore enrichment. Methods Enzymol. 1991;194:146–149. doi: 10.1016/0076-6879(91)94012-2. [DOI] [PubMed] [Google Scholar]
- Rubin G. M. Preparation of RNA and ribosomes from yeast. Methods Cell Biol. 1975;12:45–64. doi: 10.1016/s0091-679x(08)60951-6. [DOI] [PubMed] [Google Scholar]
- Sharp S. J., Schaack J., Cooley L., Burke D. J., Söll D. Structure and transcription of eukaryotic tRNA genes. CRC Crit Rev Biochem. 1985;19(2):107–144. doi: 10.3109/10409238509082541. [DOI] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989 May;122(1):19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swerdlow H., Guthrie C. Structure of intron-containing tRNA precursors. Analysis of solution conformation using chemical and enzymatic probes. J Biol Chem. 1984 Apr 25;259(8):5197–5207. [PubMed] [Google Scholar]
- Symington L. S., Petes T. D. Expansions and contractions of the genetic map relative to the physical map of yeast chromosome III. Mol Cell Biol. 1988 Feb;8(2):595–604. doi: 10.1128/mcb.8.2.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szekely E., Belford H. G., Greer C. L. Intron sequence and structure requirements for tRNA splicing in Saccharomyces cerevisiae. J Biol Chem. 1988 Sep 25;263(27):13839–13847. [PubMed] [Google Scholar]
- Takagi M., Kobayashi N., Sugimoto M., Fujii T., Watari J., Yano K. Nucleotide sequencing analysis of a LEU gene of Candida maltosa which complements leuB mutation of Escherichia coli and leu2 mutation of Saccharomyces cerevisiae. Curr Genet. 1987;11(6-7):451–457. doi: 10.1007/BF00384606. [DOI] [PubMed] [Google Scholar]
- Wang S. S., Hopper A. K. Isolation of a yeast gene involved in species-specific pre-tRNA processing. Mol Cell Biol. 1988 Dec;8(12):5140–5149. doi: 10.1128/mcb.8.12.5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis I., Hottinger H., Pearson D., Chisholm V., Leupold U., Söll D. Mutations affecting excision of the intron from a eukaryotic dimeric tRNA precursor. EMBO J. 1984 Jul;3(7):1573–1580. doi: 10.1002/j.1460-2075.1984.tb02013.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M., Culbertson M. R. Mutations affecting the tRNA-splicing endonuclease activity of Saccharomyces cerevisiae. Genetics. 1988 Apr;118(4):609–617. doi: 10.1093/genetics/118.4.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zillmann M., Gorovsky M. A., Phizicky E. M. Conserved mechanism of tRNA splicing in eukaryotes. Mol Cell Biol. 1991 Nov;11(11):5410–5416. doi: 10.1128/mcb.11.11.5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zyl W. H., Wills N., Broach J. R. A general screen for mutant of Saccharomyces cerevisiae deficient in tRNA biosynthesis. Genetics. 1989 Sep;123(1):55–68. doi: 10.1093/genetics/123.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]