Abstract
The p-nitroaromatic antibiotic chloramphenicol has been used extensively to treat life-threatening infections due to Haemophilus influenzae and Neisseria meningitidis; its mechanism of action is the inhibition of protein synthesis. We found that during incubation with H. influenzae cells and lysates, chloramphenicol is converted to a 4-aminophenyl allylic alcohol that lacks antibacterial activity. The allylic alcohol moiety undergoes facile re-addition of water to restore the 1,3-diol, as well as further dehydration driven by the aromatic amine to form the iminoquinone. Several Neisseria species and most chloramphenicol-susceptible Haemophilus species, but not Escherichia coli or other gram-negative or gram-positive bacteria we examined, were also found to metabolize chloramphenicol. The products of chloramphenicol metabolism by species other than H. influenzae have not yet been characterized. The strains reducing the antibiotic were chloramphenicol susceptible, indicating that the pathway does not appear to mediate chloramphenicol resistance. The role of this novel nitroreductase pathway in the physiology of H. influenzae and Neisseria species is unknown. Further understanding of the H. influenzae chloramphenicol reduction pathway will contribute to our knowledge of the diversity of prokaryotic nitroreductase mechanisms.
We previously reported (5) that chloramphenicol (Cm) was transported by the gram-negative bacterium Haemophilus influenzae in a saturable, energy-dependent mechanism with Michaelis-Menten kinetics. In these studies, high-pressure liquid chromatography (HPLC) was used to monitor the disappearance of Cm from the culture medium during incubation with H. influenzae. This process was time and temperature dependent and required incubation in a medium that would support growth. Apparent transport was abolished by drugs that collapse the proton motive force (dinitrophenol, CCCP [dicyclohexylcarbodiimide]) but was affected much less by compounds that block the final step in electron transport, azide and cyanide (5). Since CCCP inhibits the ATPase that is coupled to nutrient transport, we concluded that Cm is actively transported in H. influenzae.
In the years since this study was reported, no Cm import system has been identified experimentally in H. influenzae or in any other organism. Further consideration of the data led us to reevaluate our earlier conclusion. In a bacterial culture containing 5 × 108 CFU/ml, we observed a reduction in Cm concentration from 30 to 10 μM within an hour. If this loss of Cm was entirely due to bacterial internalization of Cm, the intracellular Cm concentration would be 40 mM, assuming that the volume of a single bacterial cell is 10−9 μl. Since the solubility of Cm in water is 7.7 mM, this seems unlikely unless Cm is stored at a high concentration in an intracellular compartment. We were not able to recover the lost Cm from bacteria by extraction. We therefore sought evidence that loss of Cm during incubation with H. influenzae was the result of metabolism of the drug.
Supporting this idea, we found that the disappearance of radiolabeled Cm from culture medium was associated with concurrent appearance of a modified, more hydrophilic radiolabeled compound. With cochromatography of two species of radiolabeled Cm (1,2-14C in the dichloroacetamido side chain and 3,5-3H in the ring), we found that the ratio of 3H to 14C was unchanged in the metabolite, indicating that the dichloroacetamido side chain was not cleaved from the parent antibiotic. The present study was undertaken to characterize the end product of Cm metabolism by H. influenzae and to determine whether other bacterial pathogens metabolize Cm similarly.
We present here the first description of Cm nitroreduction by H. influenzae. Two classes of chloramphenicol-transforming enzymes have been reported previously: (i) Cm acetyltransferases mediating Cm resistance and found in a wide range of bacterial species (32) and (ii) a Cm phosphotransferase that protects Streptomyces venezuelae from the antibacterial activity of the Cm that it produces (24). Both enzymes mediate resistance to the antibacterial activity of chloramphenicol. In contrast, the reaction we describe occurs in Cm-susceptible bacteria. Analysis of the products of metabolism identified three interconverting chemical species, including the p-amino allylic alcohol (Fig. 1). We found evidence of Cm metabolism in certain members of the Pasteurellaceae (including several other Haemophilus species) and in Neisseria species but not in Escherichia coli, Pseudomonas aeruginosa, Bacillus subtilis, or any of the other species we surveyed.
FIG. 1.
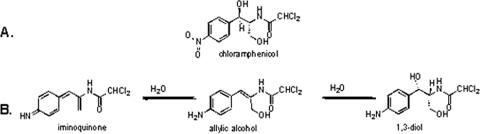
(A) Cm. C11H12Cl2N2O5, Mr = 322 Da. (B) Proposed structure of the proposed metabolites, showing the allylic alcohol (C11H12Cl2N2O2, Mr = 274 Da) gaining (C11H14N2O3, Mr = 292 Da) and losing (C11H10Cl2N2O, Mr = 256 Da) one molecule of water. Experimental evidence supporting the metabolites’ structures is described in Results.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The work presented here is based predominantly on H. influenzae strain Rd KW20, the strain whose genome sequence was published in 1995 (9). Additional strains used are listed in Table 1 (Haemophilus spp. and other Pasteurellaceae) and Table 2 (non-Pasteurellaceae). H. influenzae was cultivated in brain heart infusion medium supplemented with 10 μg of β-NAD and 10 μg of hemin per ml (sBHI) or in chemically defined medium (8). sBHI was also used for the culture of other bacteria with the exception of Neisseria species, which were cultivated in GC broth (Difco) containing Isovitalex (BD). The MIC of Cm and its metabolite were determined by a modification of the reference method (25) we described previously (16).
TABLE 1.
Detection of Cm metabolism by Pasteurellaceae strains
| Straina | Accession no.b | Cm metabolism detectedc | Source or reference |
|---|---|---|---|
| Haemophilus influenzae Rd KW20 (seq) | R652 | Y | 41 |
| Haemophilus influenzae Int1 (seq) | R2866 | Y | 26 |
| Haemophilus influenzae 12 (seq) | R2846 | Y | 3 |
| Haemophilus influenzae type a 9006 | R539 | Y | ATCCd |
| Haemophilus influenzae type b Eagan | E1 | Y | 34 |
| Haemophilus influenzae type c 9007 | R540 | Y | ATCC |
| Haemophilus influenzae type d 9008 | R541 | Y | ATCC |
| Haemophilus influenzae type e 8142 | R542 | Y | ATCC |
| Haemophilus influenzae type f 9833 | R543 | Y | ATCC |
| Haemophilus influenzae 86-0298 (seq) | R3642 | Y | 13 |
| Haemophilus influenzae U11 | R2859 | Y | 34 |
| Haemophilus influenzae 3224A (seq) | R3661 | Y | D. Dyere |
| Haemophilus influenzae 1128 | R3157 | Y | 2 |
| Haemophilus influenzae 49766 (ref) | R3253 | Y | ATCC |
| Haemophilus aegyptius 11116 | R1967 | Y | ATCC |
| Haemophilus aphrophilus 33389 | R1968 | Y | ATCC |
| Haemophilus avium 29546 | R1990 | Y | ATCC |
| Haemophilus equigenitalis 35865 | R1976 | Y | ATCC |
| Haemophilus haemolyticus 33390 | R1970 | Y | ATCC |
| Haemophilus paragallinarum 29545 | R1989 | Y | ATCC |
| Haemophilus parahaemolyticus 29237 | R1985 | Y | ATCC |
| Haemophilus parainfluenzae 33392 | R1966 | Y | ATCC |
| Haemophilus paraphrophilus 29241 | R1969 | Y | ATCC |
| Haemophilus parasuis 19417 | R1974 | N | ATCC |
| Haemophilus pleuropneumoniae 27099 | R1975 | Y | ATCC |
| Aggregatibacter segnis 33393 | R1973 | N | ATCC |
| Histophilus somni 8025 | R1959 | N | 15 |
| Pasteurella multocida Pm70 (seq) | R3687 | N | 21 |
Strains for which the genome sequence has been published are denoted by “(seq)”; strains recommended as reference isolates for clinical testing by the National Committee for Clinical Laboratory Standards (25) are denoted by “(ref).”
A. L. Smith laboratory accession number.
Cm metabolism detected by TLC of ethyl extract after incubation of bacteria with Cm in culture medium. Y, yes; N, no.
ATCC, American Type Culture Collection.
Provided by D. Dyer, Laboratory for Genomics and Bioinformatics, University of Oklahoma Health Sciences Center.
TABLE 2.
Detection of Cm metabolism by strains other than Pasteurellaceae
| Straina | Accession no.b | Cm metabolism detectedc | Source or referenced |
|---|---|---|---|
| Neisseria meningitidis 5587 | R1904 | Y | 6 |
| Neisseria meningitidis 5591 | R1905 | Y | 6 |
| Neisseria meningitidis Z2491 (seq) | R3666 | Y | 27 |
| Neisseria gonorrhoeae FA19 | R3533 | Y | 29 |
| Neisseria gonorrhoeae F1090 (seq) | R3534 | Y | 29 |
| Neisseria gonorrhoeae F62 | R3535 | Y | 29 |
| Neisseria perflava | R3336 | Y | 20 |
| Neisseria cinerea | R3340 | Y | 19 |
| Neisseria kochii | R3345 | Y | 22 |
| Escherichia coli 25922 (ref) | R3662 | N | ATCC |
| Pseudomonas aeruginosa PAO1 (seq) | R3620 | N | 38 |
| Pseudomonas aeruginosa 27853 (ref) | R3663 | N | ATCC |
| Vibrio harveyi BB170 | R3395 | N | 39 |
| Bacillus subtilis NF 143 | R3716 | N | 10 |
| Listeria monocytogenes EGD | R3713 | N | 42 |
| Listeria innocua 33090 | R3714 | N | 42; ATCC |
| Staphylococcus aureus 25923 (ref) | R3356 | N | ATCC |
| Staphylococcus pneumoniae BAA-334 (seq) | R3668 | N | ATCC |
| Staphylococcus pneumoniae BAA-255 (seq) | R3667 | N | ATCC |
| Streptococcus agalactiae A909 (seq) | R3717 | N | 18 |
| Streptococcus agalactiae COH1 (seq) | R3718 | N | 30 |
Strains for which the genome sequence has been published are denoted by “(seq)”; strains recommended as reference isolates for clinical testing by the National Committee for Clinical Laboratory Standards (25) are denoted by “(ref).”
A. L. Smith laboratory accession number.
Cm metabolism detected by TLC of ethyl extract after incubation of bacteria with Cm in culture medium. Y, yes; N, no.
ATCC, American Type Culture Collection.
Detection of Cm metabolism in small-scale cultures.
Bacteria grown to mid-log phase were harvested by centrifugation, resuspended to approximately 5 × 108 CFU/ml in culture medium containing 0.15 mM Cm, and incubated at 37°C for various periods of time. For analysis by thin-layer chromatography (TLC) or UV absorbance spectroscopy, the bacteria were removed by centrifugation, and the supernatant was acidified by addition of an equal volume of 20 mM sodium acetate (pH 3.0) and extracted with two and one-half volumes of ethyl acetate. Whatman 20-by-20-cm PE SILG/UV plates (catalog no. 4410-222) containing a fluorescent indicator were used for TLC. In control experiments, ethyl(4-amino)cinnamate was used as a model aniline to verify that these conditions would allow extraction of the putative aniline metabolite. For initial tandem mass spectrometry (MS/MS) analysis, 109 CFU of H. influenzae Rd KW20 were incubated with 0.1 mg of Cm in 1 ml of defined medium. The culture supernatant was extracted as described to produce the material referred to below as the small-scale fermentation. Large-scale isolation of metabolic products is described below.
For study of Cm metabolism in cell-free systems, H. influenzae Rd KW20 cells were suspended in culture medium and disrupted by sonication. Unbroken cells were removed by centrifugation, and the cell-free lysate was incubated with Cm as described above and with NADH and NADPH (1.5 mM) added as indicated.
The UV absorbance spectrum of the ethyl acetate extracts was determined by using a Varian spectrophotometer. For TLC, the ethyl acetate extract was concentrated in a Speed-Vac and spotted onto silica gel plates (PE SIL G/UV; Whatman). The mobile phase was CHCl3-methanol-NH4OH at 80:20:2 (vol/vol/vol).
The Bratton-Marshall reaction, which detects diazotiable nitrogen species such as aromatic amines, was performed essentially as described previously (11). Each sample (bacterial suspension, lysate, or culture supernatant) was treated with an equal volume of 20% trichloroacetic acid and held on ice for 30 min before centrifugation (15 min at 4°C, 16,000 × g). For assay, 200 μl of a trichloroacetic acid supernatant was transferred to a well of a microtiter plate, to which were added successively 25 μl each of 0.1% sodium nitrite and 0.5% ammonium sulfamate, followed by 10 min of incubation at room temperature after each addition: 25 μl of 0.05% N-1-p-naphthylethylenediamine was added, and the plate was incubated for 20 min at room temperature to allow complete color development, followed by determination of the absorbance at 550 nm (A550).
Large-scale isolation of products.
H. influenzae Rd KW 20 was grown to late-log phase in 200 ml of defined medium (8) to a density of approximately 2.5 × 109 CFU/ml. A total of 20 mg of Cm was added, and the suspension was incubated for 10 h at 37°C. The progress of the reaction was monitored by UV absorbance. After 8 h, <10% of intact Cm remained, and the spectrum did not change during the last 2 h of incubation. Bacteria were removed by centrifugation, and the culture supernatant extracted for 20 min at 4°C and for 10 min at ambient temperature with an equal volume of ethyl acetate. The phases were separated, and the aqueous phase was reextracted with an additional 200 ml of ethyl acetate. The combined organic layers were concentrated to 20 ml by using a Büchi rotary evaporator, and a portion of this material was submitted for liquid chromatography coupled with mass spectroscopy (LC-MS) and for an accurate mass determination. The remainder was further concentrated to 2 ml and stored at −20°C for 2 weeks and then subjected to chromatography on a Biotage SP4 system using a 12S silica gel cartridge eluting at 12 ml/min. The mobile phase was a gradient of 2 to 50% methanol-NH4OH (99:1 [vol/vol]) in CHCl3-NH4OH (99:1 [vol/vol]) over 12 column volumes. Fractions (9 ml) were collected for evaluation by TLC. Aliquots were spotted onto TLC plates (Machery-Nagel 40 by 80 mm UV254), and the plates were developed by using a mobile phase of CHCl3-methanol-NH4OH at 89:10:1 (vol/vol/vol) and visualized with UV light. The fractions containing product (Rf = 0.08) were pooled and concentrated in vacuo to give 1.8 mg of residue.
LC-MS.
A Waters Micromass Quattro II tandem quadrupole mass spectrometer (Waters, Milford, MA) operated in positive electrospray ionization (ESI) mode and fitted with a Shimadzu LD-10AD solvent delivery system and SIL-10ADvp autoinjector (Shimadzu Scientific Instruments, Inc., Columbia, MD) was used to acquire all LC-MS/MS data. The instrument was operated at a source temperature of 150°C using nitrogen as a nebulizing and bath gas with an ESI probe voltage of 3.5 kV. The signal response for the protonated molecular species, [M + H]+, of Cm and its metabolites was optimized at a cone voltage of 30 V. MS/MS was performed with the collision cell (Q2) maintained at 10−3 mbar pressure of argon and 20 eV collision energy. The resolution of the instrument's first mass analyzer (Q1) was set to allow the transmission of the isotopic envelopes associated with the dichloro precursor ions, while that of the second mass analyzer (Q3) was adjusted to provide a product ion spectra at nominal mass resolution. The data acquisition was in the multichannel analysis mode using Micromass MassLynx 3.4 software (Micromass, Ltd., Manchester, United Kingdom).
Cm and its metabolites from the small-scale production were characterized by the flow injection analysis using a Javelin BetaBasic C8 filter column (2.0 mm inner diameter by 20 mm, 5-μm particle size; Keystone Scientific, Inc., Bellefonte, PA) under isocratic conditions (methanol-water, 40:60 [vol/vol]) at a flow rate of 0.10 ml min/min, enabling the selection and analysis of precursor ions without possible isobaric interference. The samples were taken up in 200 μl of methanol, vortex mixed for 30 s, and subjected to MS/MS analysis as described above using 20-μl aliquots of each.
Similarly, LC-MS and LC-MS/MS analyses were performed on the ethyl acetate extract obtained from the large-scale isolation experiment outlined above. Separations of the parent compound and its metabolites were achieved by using a Phenomenex Luna Phenyl-Hexyl HPLC column (150 mm by 2.0 mm inner diameter, 5 μm; Phenomenex, Inc., Torrance, CA) operating at a flow rate of 0.300 μl/min. Samples of 10 μl to 50 μl, diluted in methanol (1:9 [vol/vol]), were applied to the column, and the mobile phase (acetonitrile-water-0.1% formic acid) was linearly programmed from 5 to 95% acetonitrile over a 35-min period after a 2-min hold under initial conditions. Mass spectrometric data were acquired in the scan mode over a mass range from 50 to 1,000 Da.
High-resolution accurate mass measurements.
A Bruker APEX Qe 47 Fourier transformed ion cyclotron resonance mass spectrometer [FT(ICR)]MS/MS (Bruker Daltonics, Billerica, MA) equipped with a capillary flow injection system and operated in the positive ion ESI mode was used for high-resolution accurate mass measurement determinations.
NMR.
The 1H nuclear magnetic resonance (NMR) spectrum was obtained in 64 transients at 295°K on a Bruker AV500 in CDCl3. Chemical shifts (δ) are reported in ppm downfield from TMS = 0, and coupling constants (J) are reported in Hz.
Electrospray mass spectrometry of the NMR sample.
Positive ion electrospray MS and MS/MS spectra used to confirm the structural identity of the NMR sample were obtained on a Bruker Esquire ion trap mass spectrometer (Bruker/Hewlett-Packard). The NMR sample, diluted 1:100 into methanol, was infused at a rate of 1 μl/min via syringe pump (Cole Parmer model 74900). Mass assignments were determined from spectra by using Bruker Data Analysis software.
DNA sequence analysis.
The complete genome sequences of H. influenzae strains Rd KW20 and 86-028NP and the incomplete sequences of strains R2846 and R2866 were accessed through the Microbial Genomes Database of NCBI (http://www.ncbi.nlm.nih.gov/sites/entrez?db=genomeprj&cmd=Retrieve&dopt=Overview&list_uids=9621). BLAST (1) was used to search the H. influenzae genomes for homologs of known bacterial nitroreductase genes.
RESULTS
Survey of bacterial species for Cm metabolism using TLC.
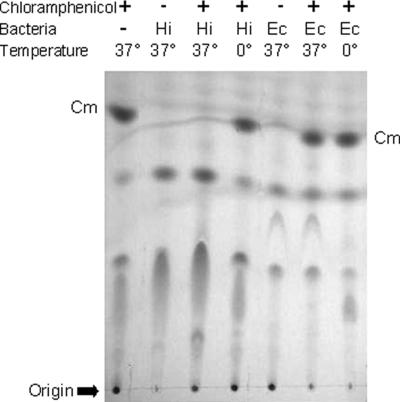
We tested a variety of gram-negative and gram-positive bacteria, with a particular emphasis on pathogens (Tables 1 and 2). Most of the bacterial strains used in the present study have been well characterized, and the genomes of some have been completely sequenced. Reactions were incubated for 1 h at 37°C and then extracted with ethyl acetate and analyzed by TLC. Control reactions contained bacteria but no Cm and a second control reaction containing both bacteria and Cm was maintained on ice. Metabolism of Cm was detected by the disappearance of a spot with Rf equivalent to that of pure Cm (Fig. 2). Cm-metabolizing activity was seen only in Haemophilus and Neisseria species; we did not detect evidence of Cm metabolism by Escherichia coli, Pseudomonas aeruginosa, Bacillus subtilis, or any bacteria other than Haemophilus and Neisseria spp. Notably, Pasteurella multocida, Histophilus somni (H. somnus), Aggregatibacter segnis (H. segnis), and H. parasuis did not metabolize Cm, indicating that this property is not shared by all Pasteurellaceae. The data below suggest that the product of Cm metabolism by the Neisseria species is not the same as that in H. influenzae.
FIG. 2.
H. influenzae Rd KW20 (Hi) or E. coli 25922 (Ec) were incubated with Cm for 1 h at either 37 or 0°C in sBHI. Ethyl acetate extracts of the culture supernatants were subjected to TLC.
Analysis of products of Cm metabolism by UV absorbance spectrum.
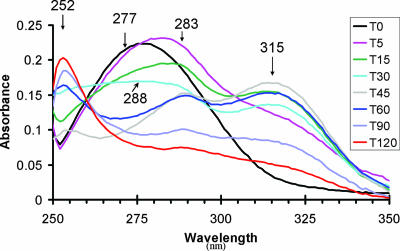
Examination of ethyl acetate extracts of culture supernatants collected during incubation of Cm with H. influenzae Rd KW20 indicated the conversion of Cm into at least two intermediate compounds before accumulation of the equilibrium-favored product (Fig. 3). The absorption spectrum of Cm has a single peak with maximum absorbance at 277 nm. Within 5 min, the peak shifted to 283 nm, and a shoulder was apparent. By 15 min, the shoulder developed into a peak with a maximum at 315 nm. The height of this peak was greatest at 45 min, and it disappeared over the next 75 min while the absorbance between 260 and 290 nm decreased. During the same period a new, sharper peak appeared at 252 nm. This was considered to be the final product since after 2 h of incubation the spectrum did not change further (data not shown). A transient peak at 288 nm was most apparent at 45 and 60 min. It is noteworthy that the samples were derived from culture supernatants, indicating that if the reaction occurred intracellularly, the intermediates diffused readily in and out of the cell, and the final product was extracellular.
FIG. 3.
H. influenzae Rd KW20 was incubated with Cm at 37°C for 120 min, and aliquots were taken, acidified, and extracted with ethyl acetate at the times indicated (T0 = zero time, T5 = 5 min, etc.). The absorbance between 250 and 350 nm was determined for each extract with the peak absorbance indicated.
When the same procedure was performed with N. meningitidis, we saw rapid loss of the Cm peak, a finding consistent with the TLC data. However, we did not detect new peaks which absorbed between 250 and 350 nm (data not shown). The product(s) of Neisseria Cm metabolism may not extract into ethyl acetate. For E. coli and H. somni, neither of which showed detectable Cm metabolism in the TLC assay, the Cm peak was gradually reduced in height over 24 h (data not shown).
Chemical assay of metabolite.
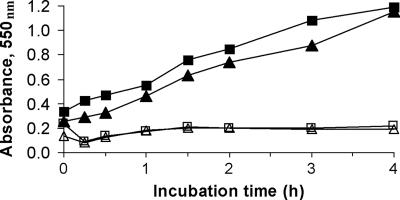
H. influenzae Rd KW20 was incubated with Cm in defined medium, and aliquots were taken at intervals up to 24 h for assay of primary aromatic amines using the Bratton-Marshall reaction. For H. influenzae, we saw chloramphenicol-dependent linear production of Bratton-Marshall-reactive material over the 6-h incubation (Fig. 4); material absorbing at 550 nm was not detected in the absence of Cm (Fig. 4) or if bacteria were maintained on ice (data not shown). For N. meningitidis, Bratton-Marshall-reactive material was produced at a similar rate as for H. influenzae; for E. coli, Bratton-Marshall-reactive material was detectable only after 24 h of incubation; and for H. somni, no Bratton-Marshall product was detectable even after 24 h of incubation (data not shown).
FIG. 4.
Production of primary amine-containing material during incubation of H. influenzae Rd KW20 with Cm. Bacteria (5 × 108 CFU/ml) were incubated at 37°C in defined medium with Cm at 100 μg/ml (or media alone), and aliquots were taken at the times indicated. Bacterial cell suspensions and culture supernatants were assayed with the Bratton-Marshall reaction. Symbols: □, cell suspension without Cm; ▵, culture supernatant without Cm; ▪, cell suspension incubated with Cm; ▴, culture supernatant after incubation with Cm.
Metabolism of Cm by H. influenzae lysates.
The data presented above were generated by incubating Cm with bacteria in media supporting viability. As a preliminary step in confirmation of the proposed pathway and characterization of the enzyme(s) involved in Cm metabolism, we sought evidence for metabolism in a cell-free system. Incubating Cm with bacterial sonicates free of intact cells, we demonstrated production of Bratton-Marshall-reactive material in the presence of NADPH to a greater extent than in the presence of NADH (Table 3). The addition of NADH to an NADPH-containing reaction further increased the production of Bratton-Marshall reactive metabolite (Table 3). It is possible that for the initial step in Cm metabolism, NADPH is the preferred cosubstrate, whereas NADH is a cosubstrate for a subsequent step.
TABLE 3.
Effect of pyridine nucleotides on the production of Bratton-Marshall reactive material by lysates of H. influenzae Rd KW20
| Sample condition | Mean A550a (SD) at:
|
A550 changeb | |
|---|---|---|---|
| Zero time | 2 h at 37°C | ||
| Medium | 0.185 (0.028) | 0.179 (0.017) | 0 |
| Lysate | 0.207 (0.009) | 0.207 (0.002) | 0 |
| + Cm | 0.199 (0.005) | 0.207 (0.005) | 0.008 |
| + Cm, NADPH | 0.173 (0.004) | 0.265 (0.004) | 0.092 |
| + Cm, NADH | 0.175 (0.010) | 0.205 (0.006) | 0.030 |
| + Cm, NADPH, NADH | 0.171 (0.007) | 0.340 (0.039) | 0.169 |
| Bacteria | |||
| Incubation at 0°C | 0.210 (0.006) | 0.212 (0.004) | 0.002 |
| Incubation at 37°C | 0.211 (0.006) | 0.443 (0.004) | 0.232 |
Mean of two assays performed in duplicate.
Initial mean A550 subtracted from the A550 determined after a 2-h incubation.
The Cm metabolite lacks antibacterial activity.
From the small-scale production we recovered the end product of Cm metabolism after resolution on a reversed-phase HPLC column; as a control, we chromatographed authentic Cm in the same system. The fractions shown to contain Cm and the principal metabolite were each lyophilized and then reconstituted in water, and the concentration was estimated from the absorbance at 278 nm (assuming that 20 ng of each compound/ml produces an optical density of 0.6). Using a microtiter format and an inoculum of 105 CFU of H. influenzae Rd KW20/ml, we determined the MIC of each fraction. For Cm it was 0.48 μg/ml, while for the metabolite it was >50 μg/ml.
Identification of H. influenzae metabolites by MS.
Similar incubation and extraction formats were used in both the small-scale and large-scale productions. As discussed below, the smaller-scale experiment yielded, almost exclusively, the allylic alcohol, whereas the larger-scale experiment provided a more complex mixture in which the hydrated 1,3-diol predominated in both the NMR and the mass spectral characterizations. Iminoquinone, the dehydration product of the allylic alcohol, was observed as a minor component in the large-scale experiment.
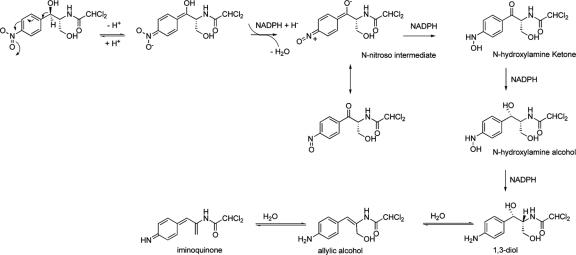
Initial small-scale studies compared authentic Cm with the final H. influenzae Rd KW20 metabolite (equivalent to the 252-nm peak detected in the 120-min sample in Fig. 3). As described below, the data were most consistent with reduction of the nitro group and α,β-dehydration to yield the allylic alcohol shown in Fig. 1B. The hypothesized pathway to this compound and to its hydration and dehydration products is depicted in Fig. 5.
FIG. 5.
Proposed pathway for the metabolic reduction of chloramphenicol. The nitroso intermediate and the final products were identified by MS analysis. The 1,3-diol was the predominant product observed in the 1H NMR analysis.
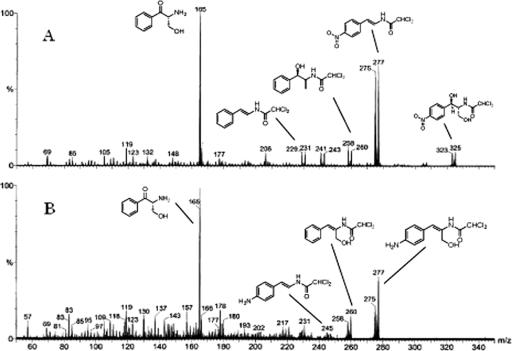
Table 4 depicts summarize the proposed neutral loss fragmentation pathways for the major ions observed in the product ion spectra of the [M + H]+ isotopic clusters for Cm at m/z 323 (Fig. 6A) and its allylic alcohol metabolite at m/z 275 (Fig. 6B), respectively. As may be seen, the tuning of the instrument with regard to resolution has made it possible to unambiguously determine chlorine containing fragment ions of their product ion spectra and provide neutral loss fragmentation pathways consistent with the structure of chloramphenicol (Fig. 1A) and its proposed allylic alcohol metabolite (Fig. 1B).
TABLE 4.
Comparison of the exact and measured masses for the [M + H]+ or [M + Na]+ ions of the three metabolite species
| Cm metabolite species | Calculated exact mass (Mr [Da]) | Measured accurate mass (Da) | Error (ppm) | Elemental composition |
|---|---|---|---|---|
| Iminoquinone | 257.0248 | 257.0243 | −2.1 | C11H11Cl2N2O |
| Allylic alcohol amine | 275.0354 | 275.0353 | −0.4 | C11H13Cl2N2O2 |
| 1,3-Diol | 315.0279 | 315.0275 | −1.3 | C11H14Cl2N2O3 Na |
FIG. 6.
Product ion spectra of Cm (A) and its metabolite (B) from the small-scale fermentation showing their [M + H]+ precursor ion clusters and proposed structures of major fragment ions (shown as the corresponding neutral species for clarity).
The m/z 275 isotope cluster selected as a precursor for the product ion spectra of the metabolite is clearly the protonated molecular species, [M + H]+, of the compound, since the presence of its sodium adduct, [M + Na]+, was also present during analysis (data not shown). Despite the predominate appearance of m/z 275 in both spectra, these ions differ in elemental composition and structure. In the Cm spectra, this fragment ion results from the consecutive loss of water (18 Da) and its primary alcohol side chain as formaldehyde (30 Da), yielding an overall loss of 48 Da from the initial 323 Da. However, the presence of a cluster at m/z 245 in the spectra of the metabolite, corresponding to this formaldehyde loss, clearly indicates the metabolite has retained this primary alcohol moiety. Further fragmentation pathways for Cm involve the combined neutral losses of its nitro group (as either nitrogen dioxide or nitrous acid), formaldehyde, and water to generate ions at m/z 258 and 229 (Fig. 6A). No such individual or combined neutral losses are observed in the spectrum of the metabolite, implying the biotransformation of Cm's benzylic secondary alcohol and p-nitro groups.
Of particular importance in the mass spectrum of the metabolite (Fig. 6B) is the appearance of the ion cluster at m/z 258. This loss of 17 Da is consistent with the presence of a primary amine under these conditions of ionization and product ion formation where the site-directed protonation of an amine leads to the elimination of ammonia and charge retention on the resultant fragment ion. Therefore, the occurrence of this cluster at m/z 258 is attributable to a [M + H - NH3]+ fragmentation pathway demonstrating the reduction of the nitro group to a primary amine. A second noteworthy feature is the conspicuous absence of a cluster at m/z 257 corresponding to the m/z 305 ion cluster, [M + H - H2O], observed in the Cm spectrum (Fig. 6A). This fragmentation is associated with the secondary benzylic alcohol in the Cm molecule, and thus its absence supports the allylic alcohol structure for the metabolite from the small scale fermentation. Finally, the spectra of both the parent Cm and allylic alcohol metabolite exhibit the loss of dichloroketene (Cl2C2O), as well as their respective aryl 4-substituents, resulting in a common m/z 165 base peak, illustrating their common backbone and further supporting the presence of the primary alcohol side chain in both compounds.
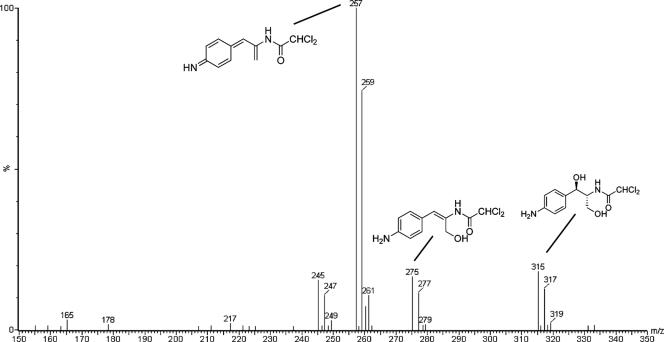
Mass spectral analysis of the larger scale fermentation products yielded the spectrum shown in Fig. 7. This spectrum exhibits the allylic alcohol amine species observed above at m/z 275, [M + H]+, in addition to its dehydrated iminoquione (m/z 257, [M + H]+) and hydrated 1,3-diol (m/z 315, [M + Na]+) forms. The heterogeneity of products and their relative abundance may reflect either differences between incubation conditions or the result of the manipulation of larger volumes during the extraction process. The identities of these three components were further confirmed by high-resolution accurate mass measurement. The calculated exact mass, the measured accurate mass, the absolute error (in ppm), and the elemental composition for the three species shown in Fig. 1B; the iminoquinone, the allylic alcohol, and the 1,3-diol, respectively, are shown in Table 4.
FIG. 7.
ESI spectrum of the ethyl acetate extract of the large-scale production showing the presence of the [M + H]+ for the iminoquinone (m/z 257), the allylic alcohol (m/z 275), and the [M + Na]+ for the 1,3-diol (m/z 315). For clarity, proposed structures are shown as the corresponding neutral species.
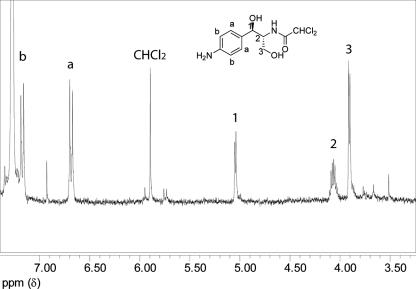
To provide further structural confirmation, we subjected the ethyl acetate extract to silica gel purification and obtained the one-dimensional 1H NMR structure of the major fraction isolated. The spectrum shown in Fig. 8 can be unambiguously assigned to the 1,3-diol. The aromatic pattern (δ 7.17, dd, J = 6.6, 1.7 Hz, δ 6.69, dd, J = 6.6, 1.8 Hz) strongly indicates a para-substituted aniline, while the upfield signals, particularly the benzylic methine which shows a clean doublet (δ 5.04, J = 3.6 Hz), and the C-2 methine (δ 4.06, ddd, J = 2.9, 4.0, 4.2 Hz) are consistent with the cis relationship of the two methines and with the coupling of the C-2 methine to the C-3 methylene (δ 3.91, J = 4.1 Hz). The NMR structure does not allow us to determine whether a single cis diastereomer is present. The NMR sample was diluted 1:100 into methanol and analyzed by MS. The positive ion mass spectra obtained from ion trap experiments gave the sodium adduct [M + Na]+, m/z 315 ion cluster, exclusively. MS/MS spectra of the mono-isotopic peak at 315 Da yielded fragment ions m/z 297, 279, 267, and 243 corresponding to the loss of H2O, 2H2O, H2O + H2CO, and 2H2O + HCl from the sodium adduct (data not shown). These data confirm the 1,3-diol as the observed species.
FIG. 8.
One-dimensional 1H NMR analysis of the 1,3-diol metabolite. The 500 MHz spectrum was obtained in CDCl3 at a concentration of approximately 6 mM. Peak assignments are as shown; small unassigned peaks are from impurities in the media. See Results for details.
Figure 1 shows the structure of Cm and the proposed metabolites based on these observations. The proposed structures are consistent with their observed protonated and sodiated molecular adducts and the fragmentation patterns observed in the positive ion spectra of the metabolites; these data reflect the biotransformation of Cm's aromatic nitro group to an amine. Elimination of water from the molecule to give the allylic alcohol occurred in the smaller-scale fermentation to a greater degree than in the larger scale. Absent stereochemical determination, we cannot distinguish between lack of dehydration and re-addition of water across the styryl system as the source of 1,3-diol observed in the larger-scale fermentation. With regard to the different proportion of side chain metabolites observed in the two sets of incubations and extractions, the silica gel purification used in the large-scale preparation may have facilitated addition of water across the allylic double bond, thereby producing only 1,3-diol as the isolated pure metabolite for NMR studies.
The most significant aspect of these studies is the biotransformation of the aromatic nitro group to the amino group. We propose the reaction pathway shown in Fig. 5. The intermediate collected after 45 min of incubation is likely to be the predicted nitroso intermediate (Mr = 306 Da). The spectra exhibit [M + H]+, [M + Na]+, and [M + H - H2O]+ at m/z 307, 329, and 189, respectively, with isotopic clusters indicating the presence of two chlorine atoms in each species.
DISCUSSION
These data indicate that H. influenzae metabolize Cm rapidly, converting it to a reduced form that lacks antibiotic activity. The reduction of Cm by bacteria was studied by scientists at Parke-Davis in the 1950s, when the antibiotic was first introduced (35-37). Based on the observation that after prolonged incubation of E. coli with Cm, the strain would eventually start to grow, these scientists hypothesized that E. coli metabolized Cm to an inactive form and that growth resumed when the concentration of active Cm was reduced to below the MIC. Smith and Worrel showed that several species of bacteria, including E. coli, were able to reduce the nitro group, forming an arylamine; this was accompanied by the hydrolysis of the amide linkage yielding Cm base and the oxidation of the hydroxyl groups to produce a series of compounds (37). Metabolism of Cm by E. coli was slow, requiring days of incubation. As far as we are aware these early studies on Cm metabolism in E. coli have not been pursued, and the enzymes responsible have not been characterized. Consistent with the reports of Smith and Worrel, we observed production of primary amine-containing material from Cm after 24 h of incubation with E. coli, but not within 1 to 2 h. Our data are consistent with these early reports, in that we saw slow conversion of Cm by E. coli to a product(s) likely to include an aromatic amine.
In contrast, we have shown here that H. influenzae Rd KW20 metabolizes Cm much more rapidly, yielding products that unlike the E. coli metabolites are extractable into ethyl acetate. These observations suggest that H. influenzae contains a nitroreductase pathway that is not present in E. coli or is not expressed under the conditions of our assay.
We searched for evidence that similar pathways have been previously described. Bacterial enzymes that catalyze the reduction of nitroaromatic compounds have been studied primarily in the context of degradation of 2-,4-dinitrophenol and related compounds by environmental organisms. In genera such as Pseudomonas and Ralstonia spp. that grow on nitrobenzene or nitrobenzoate, the reduction of the nitro group to hydroxylamine is followed by a mutase reaction that produces an o-aminophenol derivative. This compound then undergoes ring cleavage for metabolic utilization (reviewed in reference 17). Since these previously studied pathways do not lead to production of anilines, it appears that these pathways differ from the H. influenzae pathway.
We considered the possibility that the H. influenzae Cm metabolic pathway was similar to the reactions used by other bacteria and certain anaerobic parasites to activate nitroaromatic antimicrobial agents. Metronidazole and other nitroimidazole are prodrugs, activated within the microbial cell by oxidoreductases (reviewed in references 23 and 31). For the parasites Entamoeba histolytica, Giardia lamblia, and Trichomonas vaginalis, as well as for anaerobic bacteria susceptible to this drug, activation of metronidazole is primarily performed by pyruvate oxidoreductase, with the immediate electron donor being ferredoxin or flavodoxin. This enzyme is inactivated by oxygen, so the activity of metronidazole against the microaerophilic bacterium Helicobacter pylori was a puzzle until it was determined that in this organism the drug can also be reduced by oxygen-insensitive NADPH nitroreductases, encoded by rdxA and frxA. Both rdxA and frxA have a role in the resistance of H. pylori to metronidazole (12), but the natural substrates for these enzymes are not known. E. coli also possesses oxygen-insensitive nitroreductases, encoded by nfsA and nfsB (a homolog of rdxA), that reduce 5-nitrofurans (40). These enzymes may be responsible in part for the Cm-reducing activity described previously in E. coli (35).
We searched the H. influenzae genome sequences for genes that may encode enzymes in the proposed Cm metabolic pathway. These are likely to be oxidoreductases for which Cm is an incidental substrate and may be similar to nitroreducing enzymes described in other organisms. However, the nitrobenzene and nitrarene nitroreductases in P. pseudoalcaligenes, P. putida, and R. pickettii (17) have no homologs in H. influenzae. The first step in the Cm-reducing pathway may be catalyzed by an oxygen-tolerant nitroreductase similar to the H. pylori enzyme FrxA or to the E. coli enzymes NfsA and NfsB. Two candidate genes within the H. influenzae Rd KW20 genome sequence are HI1278 and HI1542, both of which are listed in the pfam database (http://www.sanger.ac.uk/Software/Pfam/) as members of the nitroreductase family PF00081. The predicted amino acid sequence of HI1278 is 75% similar to that of H. pylori FrxA (HP0642) and 49% similar to E. coli NfsB (equivalent to H. pylori RdxA). HI1542 is a gene of unknown function that is similar in sequence to genes in many gram-negative bacteria.
For subsequent steps in the pathway, we cannot identify candidate genes by homology since, to our knowledge, the reactions have not been previously described. Based on our survey of bacterial strains, candidates may be identified among the H. influenzae genes that lack homologs in the sequenced genomes of the closely related species H. somni and P. multocida, as well as lacking homologs in the genomes of the E. coli and P. aeruginosa. It is also possible that the Cm reduction pathway in H. influenzae might be catalyzed by enzymes that are shared with E. coli but regulated differently.
The role of this pathway in H. influenzae physiology is not known. Since we have not yet identified mutants in which Cm metabolism does not occur, we do not know whether this pathway affects the susceptibility of the organism to the antibacterial activity of Cm. There are H. influenzae strains in which the loss of an outer membrane protein confers Cm resistance (4). In such isolates the rapid metabolism of the reduced intracellular concentration of Cm by nitroreduction could contribute to resistance. The Haemophilus and Neisseria species that we found to metabolize Cm much more rapidly than E. coli are fully susceptible to Cm. In fact, although Cm is bacteriostatic for E. coli and most other species, both H. influenzae and N. meningitidis rapidly lose viability during incubation with Cm (7, 28). It is possible that Cm metabolism has an overall detrimental effect on these species, independent of Cm's inhibition of protein synthesis. If that is the case, one potential mechanism is the generation of nitrogen radicals. This is thought to be the major antimicrobial mechanism of rdxA-activated metronidazole. A second potential mechanism of toxicity mediated by Cm reduction is the consumption of reducing equivalents. If our proposed reaction scheme is correct, three NADPH or NADH molecules are oxidized for each molecule of Cm that is reduced. In an organism that is already paralyzed by having protein synthesis halted, such a drain of energy may be lethal. Consistent with this idea, it was recently reported that nitazoxanide, a nitrothiazolyl-salicylamide derivative with activity against H. pylori, is activated by pyruvate oxidoreductase, and that no associated DNA damage could be detected. It was hypothesized that the toxicity of nitazoxanide for H. pylori might result primarily from depletion of cellular NADPH pools (33). It is also possible that the two Michael acceptors, the iminoquinone and allylic alcohol, may be toxic if not adequately quenched by water and thus may be active metabolites that contribute to Cm toxicity in vivo by mechanisms distinct from inhibition of protein synthesis. Identification and mutagenesis of genes encoding the Cm-reducing enzymes would enable further pursuit of these hypotheses.
In summary, we have evidence that H. influenzae reduce Cm to a p-amino derivative or interconverting family of derivatives that have not previously been described. Further study of this novel pathway, and of a possibly related one in Neisseria species, may yield a better understanding of how the oxidative metabolism of these mucosal pathogens differs from that of better-understood organisms such as E. coli. Characterization of the enzymes involved may also be of practical importance: bacterial nitroreductases have attracted interest as potential anticancer prodrug-activating enzymes useful in antibody-directed enzyme prodrug therapy (ADEPT) or gene-directed enzyme prodrug therapy (GDEPT) (14). The observation of a novel prokaryotic reductase pathway that we describe in the present study should encourage further investigations in this direction.
Acknowledgments
We thank Martin Sadilek of the University of Washington for help with the Electrospray mass spectrometry experiments, Stona Jackson for assistance with the figures, and Steven Benkovic for invaluable discussions.
This study was supported by grants from National Institutes of Health (AI46512 and DC05833), the Hood Foundation, and the M. J. Murdock Charitable Trust.
Footnotes
Published ahead of print on 25 May 2007.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Bakaletz, L. O., and S. J. Barenkamp. 1994. Localization of high-molecular-weight adhesion proteins of nontypeable Haemophilus influenzae by immunoelectron microscopy. Infect. Immun. 62:4460-4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barenkamp, S. J., and E. Leininger. 1992. Cloning, expression, and DNA sequence analysis of genes encoding nontypeable Haemophilus influenzae high-molecular-weight surface-exposed proteins related to filamentous hemagglutinin of Bordetella pertussis. Infect. Immun. 60:1302-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burns, J. L., P. M. Mendelman, J. Levy, T. L. Stull, and A. L. Smith. 1985. A permeability barrier as a mechanism of chloramphenicol resistance in Haemophilus influenzae. Antimicrob. Agents Chemother. 27:46-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burns, J. L., and A. L. Smith. 1987. Chloramphenicol accumulation by Haemophilus influenzae. Antimicrob. Agents Chemother. 31:686-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campos, J., G. Trujillo, T. Seuba, and A. Rodriguez. 1992. Discriminative criteria for Neisseria meningitidis isolates that are moderately susceptible to penicillin and ampicillin. Antimicrob. Agents Chemother. 36:1028-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cole, F. S., R. S. Daum, L. Teller, D. A. Goldmann, and A. L. Smith. 1979. Effect of ampicillin and chloramphenicol alone and in combination on ampicillin-susceptible and -resistant Haemophilus influenzae type B. Antimicrob. Agents Chemother. 15:415-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coleman, H. N., D. A. Daines, J. Jarisch, and A. L. Smith. 2003. Chemically defined media for growth of Haemophilus influenzae strains. J. Clin. Microbiol. 41:4408-4410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fleischmann, R. D., M. D. Adams, O. White, R. A. Clayton, E. F. Kirkness, A. R. Kerlavage, C. J. Bult, J. F. Tomb, B. A. Dougherty, J. M. Merrick, et al. 1995. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269:496-512. [DOI] [PubMed] [Google Scholar]
- 10.Freitag, N. E., P. Youngman, and D. A. Portnoy. 1992. Transcriptional activation of the Listeria monocytogenes hemolysin gene in Bacillus subtilis. J. Bacteriol. 174:1293-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.German, D. P., M. H. Horn, and A. Gawlicka. 2004. Digestive enzyme activities in herbivorous and carnivorous prickleback fishes (Teleostei: Stichaeidae): ontogenetic, dietary, and phylogenetic effects. Physiol. Biochem. Zool. 77:789-804. [DOI] [PubMed] [Google Scholar]
- 12.Gerrits, M. M., E. J. van der Wouden, D. A. Bax, A. A. van Zwet, A. H. van Vliet, A. de Jong, J. G. Kusters, J. C. Thijs, and E. J. Kuipers. 2004. Role of the rdxA and frxA genes in oxygen-dependent metronidazole resistance of Helicobacter pylori. J. Med. Microbiol. 53:1123-1128. [DOI] [PubMed] [Google Scholar]
- 13.Harrison, A., D. W. Dyer, A. Gillaspy, W. C. Ray, R. Mungur, M. B. Carson, H. Zhong, J. Gipson, M. Gipson, L. S. Johnson, L. Lewis, L. O. Bakaletz, and R. S. Munson, Jr. 2005. Genomic sequence of an otitis media isolate of nontypeable Haemophilus influenzae: comparative study with H. influenzae serotype days, strain KW20. J. Bacteriol. 187:4627-4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hay, M. P., R. F. Anderson, D. M. Ferry, W. R. Wilson, and W. A. Denny. 2003. Synthesis and evaluation of nitroheterocyclic carbamate prodrugs for use with nitroreductase-mediated gene-directed enzyme prodrug therapy. J. Med. Chem. 46:5533-5545. [DOI] [PubMed] [Google Scholar]
- 15.Inzana, T. J., G. Glindemann, A. D. Cox, W. Wakarchuk, and M. D. Howard. 2002. Incorporation of N-acetylneuraminic acid into Haemophilus somnus lipooligosaccharide (LOS): enhancement of resistance to serum and reduction of LOS antibody binding. Infect. Immun. 70:4870-4879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jacobs, R. F., C. B. Wilson, J. G. Laxton, J. E. Haas, and A. L. Smith. 1982. Cellular uptake and intracellular activity of antibiotics against Haemophilus influenzae type b. J. Infect. Dis. 145:152-159. [DOI] [PubMed] [Google Scholar]
- 17.Johnson, G. R., and J. C. Spain. 2003. Evolution of catabolic pathways for synthetic compounds: bacterial pathways for degradation of 2,4-dinitrotoluene and nitrobenzene. Appl. Microbiol. Biotechnol. 62:110-123. [DOI] [PubMed] [Google Scholar]
- 18.Jones, A. L., K. M. Knoll, and C. E. Rubens. 2000. Identification of Streptococcus agalactiae virulence genes in the neonatal rat sepsis model using signature-tagged mutagenesis. Mol. Microbiol. 37:1444-1455. [DOI] [PubMed] [Google Scholar]
- 19.Knapp, J. S., P. A. Totten, M. H. Mulks, and B. H. Minshew. 1984. Characterization of Neisseria cinerea, a nonpathogenic species isolated on Martin-Lewis medium selective for pathogenic Neisseria spp. J. Clin. Microbiol. 19:63-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leng, Z., D. E. Riley, R. E. Berger, J. N. Krieger, and M. C. Roberts. 1997. Distribution and mobility of the tetracycline resistance determinant tetQ. J. Antimicrob. Chemother. 40:551-559. [DOI] [PubMed] [Google Scholar]
- 21.May, B. J., Q. Zhang, L. L. Li, M. L. Paustian, T. S. Whittam, and V. Kapur. 2001. Complete genomic sequence of Pasteurella multocida, Pm70. Proc. Natl. Acad. Sci. USA 98:3460-3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mazloum, H., P. A. Totten, G. F. Brooks, C. R. Dawson, S. Falkow, J. F. James, J. S. Knapp, J. M. Koomey, C. J. Lammel, and D. Peters. 1986. An unusual Neisseria isolated from conjunctival cultures in rural Egypt. J. Infect. Dis. 154:212-224. [DOI] [PubMed] [Google Scholar]
- 23.Mendz, G. L., and F. Megraud. 2002. Is the molecular basis of metronidazole resistance in microaerophilic organisms understood? Trends Microbiol. 10:370-375. [DOI] [PubMed] [Google Scholar]
- 24.Mosher, R. H., D. J. Camp, K. Yang, M. P. Brown, W. V. Shaw, and L. C. Vining. 1995. Inactivation of chloramphenicol by O-phosphorylation. A novel resistance mechanism in Streptomyces venezuelae ISP5230, a chloramphenicol producer. J. Biol. Chem. 270:27000-27006. [DOI] [PubMed] [Google Scholar]
- 25.National Committee for Clinical Laboratory Standards. 2004. Standards for antimicrobial susceptibility testing, 13th informational supplement. NCCLS publication no. M100-S14. National Committee for Clinical Laboratory Standards, Wayne, PA.
- 26.Nizet, V., K. F. Colina, J. R. Almquist, C. E. Rubens, and A. L. Smith. 1996. A virulent nonencapsulated Haemophilus influenzae. J. Infect. Dis. 173:180-186. [DOI] [PubMed] [Google Scholar]
- 27.Parkhill, J., M. Achtman, K. D. James, S. D. Bentley, C. Churcher, S. R. Klee, G. Morelli, D. Basham, D. Brown, T. Chillingworth, R. M. Davies, P. Davis, K. Devlin, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, S. Leather, S. Moule, K. Mungall, M. A. Quail, M. A. Rajandream, K. M. Rutherford, M. Simmonds, J. Skelton, S. Whitehead, B. G. Spratt, and B. G. Barrell. 2000. Complete DNA sequence of a serogroup A strain of Neisseria meningitidis Z2491. Nature 404:502-506. [DOI] [PubMed] [Google Scholar]
- 28.Rahal, J. J., Jr., and M. S. Simberkoff. 1979. Bactericidal and bacteriostatic action of chloramphenicol against meningeal pathogens. Antimicrob. Agents Chemother. 16:13-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ram, S., M. Cullinane, A. M. Blom, S. Gulati, D. P. McQuillen, B. G. Monks, C. O'Connell, R. Boden, C. Elkins, M. K. Pangburn, B. Dahlback, and P. A. Rice. 2001. Binding of C4b-binding protein to porin: a molecular mechanism of serum resistance of Neisseria gonorrhoeae. J. Exp. Med. 193:281-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rubens, C. E., L. M. Heggen, R. F. Haft, and M. R. Wessels. 1993. Identification of cpsD, a gene essential for type III capsule expression in group B streptococci. Mol. Microbiol. 8:843-855. [DOI] [PubMed] [Google Scholar]
- 31.Samuelson, J. 1999. Why metronidazole is active against both bacteria and parasites. Antimicrob. Agents Chemother. 43:1533-1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schwarz, S., C. Kehrenberg, B. Doublet, and A. Cloeckaert. 2004. Molecular basis of bacterial resistance to chloramphenicol and florfenicol. FEMS Microbiol. Rev. 28:519-542. [DOI] [PubMed] [Google Scholar]
- 33.Sisson, G., A. Goodwin, A. Raudonikiene, N. J. Hughes, A. K. Mukhopadhyay, D. E. Berg, and P. S. Hoffman. 2002. Enzymes associated with reductive activation and action of nitazoxanide, nitrofurans, and metronidazole in Helicobacter pylori. Antimicrob. Agents Chemother. 46:2116-2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith, A. L., D. H. Smith, D. R. Averill, Jr., J. Marino, and E. R. Moxon. 1973. Production of Haemophilus influenzae b meningitis in infant rats by intraperitoneal inoculation. Infect. Immun. 8:278-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith, G. N., and C. S. Worrel. 1949. Enzymatic reduction of chloramphenicol. Arch. Biochem. 24:216-223. [PubMed] [Google Scholar]
- 36.Smith, G. N., and C. S. Worrel. 1950. The decomposition of chloromycetin (chloramphenicol) by microorganisms. Arch. Biochem. 28:232-241. [PubMed] [Google Scholar]
- 37.Smith, G. N., and C. S. Worrel. 1953. Reduction of chloromycetin and related compounds by Escherichia coli. J. Bacteriol. 65:313-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 39.Surette, M. G., and B. L. Bassler. 1998. Quorum sensing in Escherichia coli and Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 95:7046-7050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Whiteway, J., P. Koziarz, J. Veall, N. Sandhu, P. Kumar, B. Hoecher, and I. B. Lambert. 1998. Oxygen-insensitive nitroreductases: analysis of the roles of nfsA and nfsB in development of resistance to 5-nitrofuran derivatives in Escherichia coli. J. Bacteriol. 180:5529-5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilcox, K. W., and H. O. Smith. 1975. Isolation and characterization of mutants of Haemophilus influenzae deficient in an adenosine 5′-triphosphate-dependent deoxyribonuclease activity. J. Bacteriol. 122:443-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wong, K. K., H. G. Bouwer, and N. E. Freitag. 2004. Evidence implicating the 5′ untranslated region of Listeria monocytogenes actA in the regulation of bacterial actin-based motility. Cell Microbiol. 6:155-166. [DOI] [PubMed] [Google Scholar]