Abstract
Carbapenem resistance due to KPC has rarely been observed outside the United States. We noticed a sharp increase in carbapenem-resistant Klebsiella pneumoniae strains possessing KPC in Tel Aviv Medical Center from 2004 to 2006. Sixty percent of the isolates belonged to a single clone susceptible only to gentamicin and colistin and carried the blaKPC-3 gene, while almost all other clones carried the blaKPC-2 gene. This rapid dissemination of KPC outside the United States is worrisome.
Carbapenem resistance in Klebsiella pneumoniae does not occur naturally and is due mainly to the presence of acquired carbapenem-hydrolyzing β-lactamases (16). KPC-type enzymes in carbapenem-resistant K. pneumoniae strains were first reported in 2001 in North Carolina (23), and until 2005, the geographical distribution of these enzymes in the family Enterobacteriaceae in general and in K. pneumoniae specifically was limited to the eastern United States (2, 4, 18, 22) where KPC-producing K. pneumoniae became a frequent nosocomial pathogen (3, 9). Outside of the United States, KPC-producing K. pneumoniae has been reported for only three patients; the first case was reported in 2005 in France and had a U.S. origin (14), and more recently, a case was reported in Colombia and an additional one in China (20, 21).
Carbapenemases KPC-2 and KPC-3 have been observed even more rarely among other gram-negative bacteria, including Enterobacter spp., Escherichia coli, and Serratia marcescens (9). Outside of the United States, KPC-2 was observed once from an S. marcescens isolate from China (25), from E. coli strains from our hospital (15), and during the same year, from an Enterobacter cloacae strain isolated from an outbreak in our neonatal intensive care unit (6). KPC-3 has never been reported outside the United States.
All the carbapenem-resistant K. pneumoniae isolates identified in the clinical laboratory of our hospital were collected from January 2004 to December 2006. In this study, all K. pneumoniae isolates manifesting carbapenem resistance were genotyped and analyzed for the presence of the blaKPC gene. The results presented suggest the rapid emergence of KPC in K. pneumoniae isolates, affecting multiple clones and leading to the emergence of carbapenem resistance in K. pneumoniae.
During the 3-year study period, from January 2004 to December 2006, a total of 4,149 single-patient K. pneumoniae isolates were identified in our hospital. Identification of strains and susceptibility testing were performed using a Vitek2 automated system (bioMerieux, Marcy l'Etoile, France) with an AST-GN09 card for the identification of gram-negative bacilli. Fifty-one isolates (1.2%) were carbapenem resistant, as defined by resistance to imipenem and/or meropenem. Sites of isolation included urine (n = 19), body fluids (n = 10), wounds (n = 9), catheter tips (n = 6), blood (n = 4), and respiratory tracts (n = 3). For all carbapenem-resistant isolates, resistance to imipenem, meropenem, and ertapenem was confirmed by using agar dilution according to the Clinical and Laboratory Standards Institute (8). Susceptibility testing for colistin and tigecycline was performed via Etest according to the manufacturer's instructions (AB Biodisk, Solna, Sweden). The genetic relatedness of all carbapenem-resistant K. pneumoniae strains was determined by pulsed-field gel electrophoresis (PFGE) analysis. DNA preparation and SpeI cleavage were performed as described previously (15), and chromosomal restriction fragments were documented and compared.
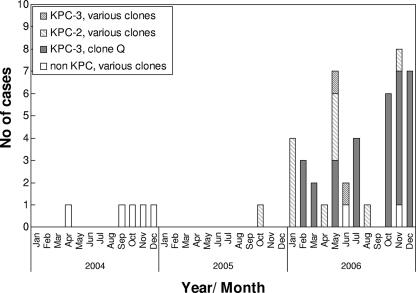
During 2004 and 2005, carbapenem-resistant K. pneumoniae was isolated from a total of six patients, while during 2006 this number increased dramatically to 45 single-patient isolates. The annual proportions of isolates resistant to carbapenems were 0.4%, 0.07%, and 3.1%, respectively, for the three years of the study. PFGE of all 51 resistant isolates indicated the presence of 12 different genetic clones, affecting one to three patients each, and a major clone (clone Q), affecting 31 (60%) cases (Fig. 1). Thus, 75% of the carbapenem-resistant K. pneumoniae isolates in our study represent a clonal transmission, while 25% represent different clones. All clones were resistant to all cephalosporins, aztreonam, ertapenem, imipenem, and/or meropenem, and to aminoglycosides. Resistance to aminoglycosides varied; eight clones were susceptible only to amikacin, and four clones, including the major clone (clone Q), were susceptible only to gentamicin. Two clones were susceptible to ciprofloxacin, but all were susceptible to colistin. Although clone Q was found to be the major clone isolated in the hospital, its isolation did not occur in clusters of space or time (with the exception of isolates from seven cases from one ward); it was isolated from 15 different wards over a span of 11 months.
FIG. 1.
Molecular epidemiology and emergence of KPC in carbapenem-resistant K. pneumoniae strains in the Tel Aviv Medical Center from 2004 to 2006. In 2004, four panresistance carbapenem-resistant clones susceptible only to amikacin and colistin were identified. In 2005, KPC emerged in carbapenem-resistant K. pneumoniae isolates. In 2006, nine clones existed; two appeared previously in 2004 and 2005 and lacked KPC, and seven, including clone Q, the major clone that emerged in February 2006, possessed KPC. Four of the seven clones (including clone Q) were susceptible only to gentamicin and colistin, and three clones were susceptible only to amikacin and colistin.
In order to identify the molecular mechanism related to carbapenem resistance in the K. pneumoniae strains in our hospital, two isolates, 469 (clone P) and 490 (clone Q), representing two different antibiotic susceptibility profiles (Table 1), were selected for detailed molecular characterization. The presence of imipenem-hydrolyzing activity in cell extracts was demonstrated by streaking the tested strains away from an imipenem disk placed on a lawn inoculum of a susceptible E. coli strain, ATCC 25922, as described previously (24). An imipenem-susceptible K. pneumoniae clinical strain was used as a negative control for carbapenemase production. Imipenem-hydrolyzing activity measured spectrophotometrically at 299 nm showed specific activities of 44 and 46.5 mU/mg (where U = μmol imipenem/min) for isolates 469 and 490, respectively. Beta-lactamases in cell extracts from the two isolates examined by isoelectric focusing (IEF) demonstrated two nitrocefin-positive bands focusing at pI 6.7 and pI 7.5 for K. pneumoniae 469 and three bands focusing at pI 5.4, 6.7, and 7.6 for K. pneumoniae 490.
TABLE 1.
Antimicrobial susceptibility patterns of carbapenem-resistant K. pneumoniae strains 469 and 490 and their respective transformants
| Antimicrobial agent or enzyme | MIC (μg/ml)
|
||||
|---|---|---|---|---|---|
| K. pneumoniae 469 | Transformant 469 | K. pneumoniae 490 | Transformant 490 | E. coli GeneHogs | |
| Ampicillin/sulbactam | >32 | >32 | >32 | >32 | <4 |
| Ceftriaxone | >64 | >64 | >64 | 8 | <1 |
| Ceftazidime | >64 | >64 | >64 | 16 | <1 |
| Cefepime | >64 | 32 | >64 | 4 | <1 |
| Aztreonam | >64 | 32 | >64 | >64 | <1 |
| Piperacillin | >128 | >128 | >128 | >128 | <4 |
| Piperacillin/tazobactam | >128 | >128 | >128 | 64 | <4 |
| Ciprofloxacin | <0.25 | <0.25 | >4 | <0.25 | <0.25 |
| Amikacin | 4 | 16 | >64 | <2 | <2 |
| Gentamycin | >16 | >16 | 2 | <1 | <1 |
| Colistin | 0.25 | 0.064 | 0.25 | 0.064 | 0.064 |
| Imipenem | 16 | 4 | 32 | 4 | <0.25 |
| Meropenem | 8 | 4 | 32 | 2 | <0.25 |
| Ertapenem | 16 | 4 | 64 | 2 | <0.25 |
| Tigecycline | 2 | 0.5 | 2 | 0.5 | 0.016 |
| bla enzymes detected | KPC-2 OXA-4, CTX-M-10 | KPC-2 OXA-4, CTX-M-10 | KPC-3 TEM-1 | KPC-3 TEM-1 | None |
PCR screening was performed with cell lysates for identification of the carbapenemase genes using specific bla primers designed for identifying known β-lactamase genes including blaOXA (including OXA-23, -24, -40, and -58) (1, 10, 17), blaKPC, blaSME, blaIMI, blaNMC (5, 15), blaGES, blaIMP (23), blaVIM (13), blaSIM (12), blaGIM, and blaSPM (7). The two K. pneumoniae isolates were found to carry blaKPC. PCR products were cloned and sequenced as described previously (15). The nucleotide acid and deduced protein sequences of both isolates were analyzed and identified as KPC-2 in strain 469 and as KPC-3 in strain 490, corresponding to the beta-lactamase with the experimental pI of 6.7.
In order to verify whether the carbapenem resistance phenotype of K. pneumoniae is plasmid encoded, plasmid DNA was purified using a NucleoBond PC 100 plasmid mini-kit (Macherey-Nagel, Germany) and E. coli GeneHogs (Invitrogene, United Kingdom), and transformants were selected on LB agar plates with ampicillin (100 μg/ml). Selected transformed colonies were subjected to antibiotic susceptibility testing (Table 1), IEF, and PCR screening for the identification of carbapenemases that were acquired upon transformation. Transfer of the blaKPC-encoding plasmids raised the MICs of extended-spectrum cephalosporins, aztreonam, and carbapenems compared to that of the susceptible E. coli GeneHogs recipient strain, but none of the transformants became resistant to imipenem or meropenem (Table 1). This observation suggests that the background of the strain is important for phenotypic resistance and that additional mechanisms, such as porin alterations that reduce the entry of carbapenems (11), are involved in carbapenem resistance in these strains. IEF confirmed by PCR and sequencing analysis using plasmid DNA from transformants supported cotransmission of blaOXA-4 and blaCTX-M-10 with blaKPC-2 in isolate 469 and cotransmission of blaTEM-1 and blaKPC-3 in isolate 490, suggesting that these genes identified in the clinical strains were encoded on the blaKPC-carrying plasmid in each isolate.
We screened all 51 carbapenem-resistant K. pneumoniae isolates for the presence of blaKPC. The blaKPC gene was not found in any carbapenem-resistant K. pneumoniae strains in 2004 (Fig. 1). Cell extracts from all the non-KPC-producing strains were assayed for their abilities to hydrolyze imipenem by using a spectrophotometric assay with imipenem as a substrate and gave negative results, suggesting that carbapenem resistance in these isolates did not involve a carbapenem-hydrolyzing enzyme. Ninety-three percent (43 of 46) of the isolates collected from 2005 to 2006 carried the blaKPC gene. All isolates belonging to clone Q (31 isolates) possessed blaKPC-3, while the other isolates belonging to six different pulsotypes possessed mainly blaKPC-2 (except for two isolates carrying blaKPC-3). KPC-3 and KPC-2 differ in only one amino acid (H272Y); thus, the coexistence of these two enzymes in our hospital is not surprising and may represent one mutational event followed by clonal spread. The emergence of KPC and its rapid spread after introduction to the hospital are worrisome findings, as therapeutic choices against these panresistance organisms are limited. It is possible that the prevalence of KPC-producing K. pneumoniae could have been underestimated in this study due to the fact that only carbapenem-resistant isolates (either imipenem or meropenem or both) were included and that KPC-harboring K. pneumoniae strains that did not exhibit carbapenem resistance MICs were missed in the Vitek2 assay, as has been shown previously (19). Nevertheless, this study has shown for the first time the rapid dissemination of carbapenem-resistant K. pneumoniae due to KPC-2 and KPC-3 outside the United States. Given reports of the presence of blaKPC from three continents, laboratories, clinicians, infection control personnel, and administrators alike should be alerted to design measures for early identification and control of the organisms bearing this resistance gene.
Acknowledgments
This work was supported by a grant from the Center for the Study of Emerging Diseases, Israel.
Footnotes
Published ahead of print on 11 June 2007.
REFERENCES
- 1.Afzal-Shah, M., N. Woodford, and D. M. Livermore. 2001. Characterization of OXA-25, OXA-26, and OXA-27, molecular class D β-lactamases associated with carbapenem resistance in clinical isolates of Acinetobacter baumannii. Antimicrob. Agents Chemother. 45:583-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bradford, P. A., S. Bratu, C. Urban, M. Visalli, N. Mariano, D. Landman, J. J. Rahal, S. Brooks, S. Cebular, and J. Quale. 2004. Emergence of carbapenem-resistant Klebsiella species possessing the class A carbapenem-hydrolyzing KPC-2 and inhibitor-resistant TEM-30 β-lactamases in New York City. Clin. Infect. Dis. 39:55-60. [DOI] [PubMed] [Google Scholar]
- 3.Bratu, S., D. Landman, R. Haag, R. Rocco, A. Eramo, M. Alam, and J. Quale. 2005. Rapid spread of carbapenem-resistant Klebsiella pneumoniae in New York City. Arch. Intern. Med. 165:1430-1435. [DOI] [PubMed] [Google Scholar]
- 4.Bratu, S., M. Mooty, S. Nichani, D. Landman, C. Gullans, B. Pettinato, U. Karumudi, P. Tolaney, and J. Quale. 2005. Emergence of KPC-possessing Klebsiella pneumoniae in Brooklyn, New York: epidemiology and recommendations for detection. Antimicrob. Agents Chemother. 49:3018-3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bratu, S., P. Tolaney, U. Karumudi, J. Quale, M. Mooty, S. Nichani, and D. Landman. 2005. Carbapenemase-producing Klebsiella pneumoniae in Brooklyn, NY: molecular epidemiology and in vitro activity of polymyxin B and other agents. J. Antimicrob. Chemother. 56:128-132. [DOI] [PubMed] [Google Scholar]
- 6.Carmeli, Y., G. Grisaru-Soen, A. Leavitt, M. J. Schwaber, S. Dolberg, and S. Navon-Venezia. 2006. Abstr. 46th Intersci. Conf. Antimicrob. Agents Chemother., abstr. C2-82.
- 7.Castanheira, M., M. A. Toleman, R. N. Jones, F. J. Schmidt, and T. R. Walsh. 2004. Molecular characterization of a β-lactamase gene, blaGIM-1, encoding a new subclass of metallo-β-lactamase. Antimicrob. Agents Chemother. 48:4654-4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clinical and Laboratory Standards Institute/NCCLS. 2005. Performance standards for antimicrobial susceptibility testing; 15th informational supplement. CLSI/NCCLS M100-S15. Clinical and Laboratory Standards Institute, Wayne, PA.
- 9.Desphande, L. M., R. N. Jones, T. R. Fritsche, and H. S. Sader. 2006. Occurrence and characterization of carbapenemase-producing Enterobacteriacea: report from the SENTRY Antimicrobial Surveillance Program (2000-2004). Microb. Drug Resist. 12:223-230. [DOI] [PubMed] [Google Scholar]
- 10.Heritier, C., L. Poirel, D. Aubert, and P. Nordmann. 2003. Genetic and functional analysis of the chromosome-encoded carbapenem-hydrolyzing oxacillinase OXA-40 of Acinetobacter baumannii. Antimicrob. Agents Chemother. 47:268-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaczmarek, F. M., F. Dib-Hajj, W. Shang, and T. D. Gootz. 2006. High-level carbapenem resistance in a Klebsiella pneumoniae clinical isolate is due to the combination of blaACT-1 β-lactamase production, porin OmpK35/36 insertional inactivation, and down-regulation of the phosphate transport porin PhoE. Antimicrob. Agents Chemother. 50:3396-3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee, K., J. H. Yum, D. Yong, H. M. Lee, H. D. Kim, J. D. Docquier, G. M. Rossolini, and Y. Chong. 2005. Novel acquired metallo-β-lactamase gene, blaSIM-1, in a class 1 integron from Acinetobacter baumannii clinical isolates from Korea. Antimicrob. Agents Chemother. 49:4485-4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Libisch, B., M. Gacs, K. Csiszar, M. Muzslay, L. Rokusz, and M. Fuzi. 2004. Isolation of an integron-borne blaVIM-4 type metallo-β-lactamase gene from a carbapenem-resistant Pseudomonas aeruginosa clinical isolate in Hungary. Antimicrob. Agents Chemother. 48:3576-3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Naas, T., P. Nordmann, G. Vedel, and C. Poyart. 2005. Plasmid-mediated carbapenem-hydrolyzing β-lactamase KPC in a Klebsiella pneumoniae isolate from France. Antimicrob. Agents Chemother. 49:4423-4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Navon-Venezia, S., I. Chmelnitsky, A. Leavitt, D. Schwartz, and Y. Carmeli. 2006. Plasmid-mediated imipenem-hydrolyzing enzyme KPC-2 among multiple carbapenem-resistant Escherichia coli clones in Israel. Antimicrob. Agents Chemother. 50:3098-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nordmann, P., and L. Poirel. 2002. Emerging carbapenemases in Gram-negative aerobes. Clin. Microbiol. Infect. 8:321-331. [DOI] [PubMed] [Google Scholar]
- 17.Poirel, L., S. Marque, C. Heritier, C. Segonds, G. Chabanon, and P. Nordmann. 2005. OXA-58, a novel class D β-lactamase involved in resistance to carbapenems in Acinetobacter baumannii. Antimicrob. Agents Chemother. 49:202-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith Moland, E., N. D. Hanson, V. L. Herrera, J. A. Black, T. J. Lockhart, A. Hossain, J. A. Johnson, R. V. Goering, and K. S. Thomson. 2003. Plasmid-mediated, carbapenem-hydrolysing β-lactamase, KPC-2, in Klebsiella pneumoniae. J. Antimicrob. Chemother. 51:711-714. [DOI] [PubMed] [Google Scholar]
- 19.Tenover, F. C., R. K. Kalsi, P. P. Williams, R. B. Carey, S. Stocker, D. Lonsway, J. K. Rasheed, J. W. Biddle, J. E. McGowan, and B. Hanna. 2006. Carbapenem resistance in Klebsiella pneumoniae not detected by automated susceptibility testing. Emerg. Infect. Dis. 12:1209-1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Villegas, M. V., K. Lolans, A. Correa, C. J. Suarez, J. A. Lopez, M. Vallejo, J. P. Quinn, and the Colombian Nosocomial Resistance Study Group. 2006. First detection of the plasmid-mediated class A carbapenemase KPC-2 in clinical isolates of Klebsiella pneumoniae from South America. Antimicrob. Agents Chemother. 50:2880-2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wei, Z. Q., X. X. Du, Y. S. Yu, P. Shen, Y. G. Chen, and L. J. Li. 2007. Plasmid-mediated KPC-2 in a Klebsiella pneumoniae isolate from China. Antimicrob. Agents Chemother. 51:763-765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Woodford, N., P. M. Tierno, Jr., K. Young, L. Tysall, M. F. I. Papelou, E. Ward, R. E. Painter, D. F. Suber, D. Shungu, L. L. Silver, K. Inglima, J. Kornblum, and D. M. Livermore. 2004. Outbreak of Klebsiella pneumoniae producing a new carbapenem-hydrolyzing class A β-lactamase, KPC-3, in a New York medical center. Antimicrob. Agents Chemother. 48:4793-4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yan, J. J., P. Hsueh, W. Ko, K. Luh, S. Tsai, H. Wu, and J. Wu. 2001. Metallo-β-lactamases in clinical Pseudomonas isolates in Taiwan and identification of VIM-3, a novel variant of the VIM-2 enzyme. Antimicrob. Agents Chemother. 45:2224-2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yigit, H., A. M. Queenan, G. J. Anderson, A. Domenech-Sanchez, J. W. Biddle, C. D. Steward, S. Ablerti, K. Bush, and F. C. Tenover. 2001. Novel carbapenem-hydrolyzing β-lactamase KPC-1 from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 45:1151-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang, R., H. W. Zhou, J. C. Cai, and G. X. Chen. 2007. Plasmid-mediated carbapenem-hydrolysing {beta}-lactamase KPC-2 in carbapenem-resistant Serratia marcescens isolates from Hangzhou, China. J. Antimicrob. Chemother. 59:574-576. [DOI] [PubMed] [Google Scholar]