Abstract
Studies of Vibrio cholerae diversity have focused primarily on pathogenic isolates of the O1 and O139 serotypes. However, autochthonous environmental isolates of this species routinely display more extensive genetic diversity than the primarily clonal pathogenic strains. In this study, genomic and metabolic profiles of 41 non-O1/O139 environmental isolates from central California coastal waters and four clinical strains are used to characterize the core genome and metabolome of V. cholerae. Comparative genome hybridization using microarrays constructed from the fully sequenced V. cholerae O1 El Tor N16961 genome identified 2,787 core genes that approximated the projected species core genome within 1.6%. Core genes are almost universally present in strains with widely different niches, suggesting that these genes are essential for persistence in diverse aquatic environments. In contrast, the dispensable genes and phenotypic traits identified in this study should provide increased fitness for certain niche environments. Environmental parameters, measured in situ during sample collection, are correlated to the presence of specific dispensable genes and metabolic capabilities, including utilization of mannose, sialic acid, citrate, and chitosan oligosaccharides. These results identify gene content and metabolic pathways that are likely selected for in certain coastal environments and may influence V. cholerae population structure in aquatic environments.
Vibrio cholerae encompasses more than 200 serotypes, two of which (O1 and O139) are causative agents of epidemic Asiatic cholera. Non-O1/O139 strains are autochthonous (indigenous) members of aquatic bacterial communities throughout the world and are also being implicated in some cases of mild gastroenteritis (20). The emergence of O139 infections among previously immune persons in India in 1992 is believed to have been enabled by the horizontal transfer of the lipopolysaccharide gene cluster from an O22 serotype strain to an O1 recipient (13). This event not only dramatically changed the dynamics of cholera in the Bay of Bengal region but also confirmed the potential for genetic exchange among virulent and avirulent strains in the environment. Cholera toxin genes were identified in non-O1/O139 strains in southern California, suggesting that boundaries to gene transfer lie at the species or genus level rather than at the strain level (18).
Lan and Reeves (22) introduced the species genome concept for bacteria in 2000, delineating the genome into a core that defines the species characteristics contained in ≥95% of strains and auxiliary or dispensable components that allow adaptation to individual niches. Recently, several studies using multiple sequenced bacterial genomes from the same species identified core and dispensable genes (including three strains of Listeria monocytogenes and eight strains of Streptococcus agalactiae) (30, 36). Nearly half of the sequenced bacterial genomes to date are from pathogenic strains, limiting our ability to study functions that facilitate adaptation to environments outside of animal hosts. Analysis of multiple environmental strains from diverse environments is needed to appreciate how genome content is distributed across a species.
It is generally agreed that a bacterial species should be defined by both its core genetic composition and the phenotypes encoded by this core that define its broad ecological niche (23, 40). Diagnostic phenotypic characteristics of Vibrio cholerae were determined in clinical laboratories, using pathogenic strains and enteric media. These characteristics may not hold true for environmental strains distantly related to the infectious strains. Phylogenetic studies have demonstrated the clonality of O1 biovars and O139 strains while uncovering broad genetic diversity in non-O1/O139 strains (5, 12). If the genetic diversity is any indication of the phenotypic diversity of the species, then the core phenotypic characteristics of V. cholerae should be reevaluated for collections of environmental strains.
Given the sampling limitations associated with complete genome sequencing, comparative genome hybridization (CGH) using DNA microarrays provides a relatively inexpensive and rapid method for probing the genomic diversity of a collection of closely related strains. CGH has been widely used to explore the genomic composition of clinical and environmental strains of several bacterial species (4, 8, 10, 11, 14, 17, 33, 34). In the present study, we use DNA microarrays from the sequenced V. cholerae O1 El Tor strain N16961 to profile the genomes of 41 environmental isolates from a wide range of central California coastal environments and four clinical strains from India and Bangladesh. The 45 strains were also assayed for growth on 190 different carbon sources, allowing the classification of core and dispensable metabolomes for V. cholerae strains. These combined analyses identify basic V. cholerae genomic and metabolic cores that contain genes and functions enabling persistence in the aquatic environment.
This study provides the first analysis of both comparative genome content and metabolic capabilities of a substantial collection of environmental V. cholerae strains from a region without epidemic cholera. Although the isolates are geographically segregated from areas where cholera is endemic, we show that their genomes are not substantially different from non-O1/O139 strains from endemic regions. Simultaneous strain isolation and measurement of environmental parameters allows the correlation of gene presence and carbon source utilization with in situ water temperature, salinity, turbidity, and inorganic nutrient concentrations. This analysis identifies genes and pathways that may allow adaptation to specific niches in the coastal marine environment and play a role in shaping the Vibrio cholerae population structure in these ecosystems.
MATERIALS AND METHODS
Site selection and sample collection.
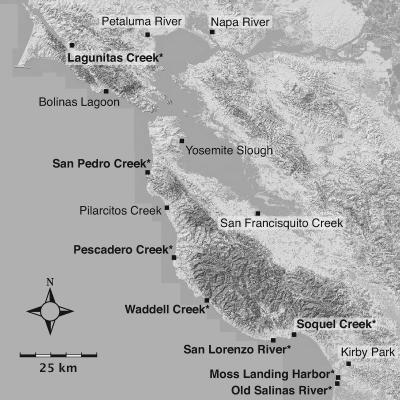
Fifteen sampling sites (Fig. 1) were chosen to span a broad range of coastal environments with different physical and chemical characteristics and land use distributions in the associated watersheds. Handler et al. (16) provide more detailed descriptions of the water quality and land use characteristics specific to each sampling site. Water column samples were collected monthly between January 2004 and June 2005 in triple-rinsed sterile high-density polyethylene bottles and transported on ice and processed within 7 h of collection.
FIG. 1.
Locations of sampling sites in and around San Francisco Bay, California. Sites where Vibrio cholerae strains were isolated are displayed in bold followed by an asterisk. Darker colored areas along the coast indicate dense vegetative cover. (Map background data available from U.S. Geological Survey/EROS, Sioux Falls, SD.)
Confirmation of isolates as Vibrio cholerae.
Up to 100 ml of sample water was membrane filtered on 0.45-μm-pore-sized nitrocellulose acetate filters and incubated on TCBS (thiosulfate citrate-bile salts-sucrose) agar. Presumptive colonies were screened biochemically, and confirmation of isolates as Vibrio cholerae was performed by PCR amplification of a 300-bp subset of the 16S-23S rRNA intergenic spacer using the clinical O1 strain N16961 as a positive control and Escherichia coli as a negative control (7). After minimal passage, isolates were stored at −80°C in LB broth with 15% glycerol. No effort was made to separate free-living V. cholerae strains from those that might be associated with particles, phytoplankton, or zooplankton in our water samples.
Environmental parameters.
Coincident with sample collection, water temperature, salinity, dissolved oxygen, pH, turbidity, and chlorophyll concentration were measured in situ using a calibrated water quality probe (model YSI 6600; Hydrolab, Yellow Springs, OH). Water samples were 0.2-μm-syringe filtered into acid-washed containers and stored at −20°C prior to analysis for dissolved nutrients. A five-channel, continuous flow analyzer was used to measure ammonium, soluble reactive phosphate, nitrate-nitrite, nitrite, and silicate following standard methods (3). Log transformations of salinity, ammonium, soluble reactive phosphate, nitrate, and nitrite, and inverse transformation of turbidity was performed to ensure the data were normally distributed. Selected environmental data are presented in Table S1 in the supplemental material.
Gene expression data.
We compared our CGH data with gene expression data from rice-water stool samples of cholera patients in Bangladesh (26) and fluid from the ileal loops of infected rabbits (41) to examine whether genes expressed while in the animal intestine also provide a selectable benefit for life in coastal ecosystems. Hybridization data were compiled as a log2 ratio with mid-log phase growth in LB used as the reference condition for the stool samples and genomic DNA used as reference for the ileal loop samples. Upregulated genes were defined as those greater than two standard deviations from the mean of the distribution.
Phenotype comparisons.
Isolates were assayed for nearly 190 different carbon substrates using proprietary phenotype microarrays (Biolog, Hayward, CA). Metabolic activity was quantified after a period of 48 h at 22°C. Isolates were scored as growth or no growth, using a procedure modified from the method for calling CGH data (27). Raw data for the phenotype microarrays consisted of intensity measurements of a chromogenic substrate at 15-min time points. The maximum intensity value was collected for all wells, and the maximum intensity in the negative control was removed from these values. Assays that were poorly reproducible between replicates were excluded. Substrates for strain N16961 with maximum intensity values greater than 175 and less than 175 were pooled into preliminary subsets A and B, respectively. Substrates with maximum intensities greater than median(A) − 2 × standard deviation(A) were designated “growth” and those with maximum intensities less than median(B) − 2 × standard deviation(B) were designated “no-growth.” Any substrates with maximum intensities between the two thresholds were designated “unknown.” Next, the maximum intensity for N16961 was subtracted from the maximum intensities for all other strains for each substrate. Substrates for all strains were called using the thresholds set for N16961, so that each assay was called growth or no growth relative to the score for N16961, based on the consensus of two replicates. Any substrates without a consensus were called unknown.
Statistical analysis.
All raw data management, scoring of data, and regression analysis for the CGH and phenotype microarrays were performed with MATLAB (Mathworks). Overrepresentation of functional role categories in a dispensable genome was verified with LACK 4.2 lexical analysis software (21). Nonmetric multidimensional scaling of microarray profiles was completed with Primer v.5 (PRIMER-E Ltd., Plymouth, United Kingdom). Canonical correspondence analysis was performed with PC ORD v4.0 (MjM Software). The unweighted-pair group method with arithmetic averages (UPGMA) tree of the binary CGH data was constructed using PAUP* 4.0b8 software with Jukes-Cantor distance and 1,000 bootstrap replicates. Logistic regression was accomplished with StatView 5.0.1 (SAS Institute, Inc).
RESULTS
Vibrio cholerae core genome as defined by comparative genome hybridization.
Water column samples were collected monthly from 15 water bodies along the central California coast between January 2004 and June 2005 (Fig. 1). Sampling sites were chosen to encompass a broad range of water quality characteristics and land use distributions in the associated watersheds (16). Isolates were collected in accordance with methods described elsewhere without enrichment (19) and identified on the basis of the 16S-23S rRNA intergenic spacer sequence (41). The concentration of V. cholerae in California coastal waters varies both spatially and temporally, with values ranging from 0.1 to 45 CFU/liter. 16S rRNA sequences for documented V. cholerae and Vibrio mimicus strains are very similar but easily distinguished from other Vibrio species. Phylogenetic analysis of 16S rRNA sequences from our environmental isolates and other Vibrio isolates confirms that all isolates are greater than 99% similar to strain N16961, by Jukes-Cantor distance and within the bootstrap-supported V. cholerae-V. mimicus cluster (data not shown). Our environmental strains were isolated from a region where cholera cases are virtually nonexistent, so there should be no fitness advantage for strains that harbor genes involved in causing cholera disease. Therefore, these environmental strains should contain genetic content that is adapted to fitness in aquatic environments without the influence of selection for genes involved in causing disease in humans.
Forty-one environmental and four clinical strains (see Table S2 in the supplemental material) were analyzed by CGH with the V. cholerae O1 El Tor strain N16961, using amplicon microarrays. The hybridization data and details of the CGH methods, including descriptions of probes printed on the microarray and explanations of the gene calls, are described in the report by Miller et al. (27). Briefly, the log2 ratio of the hybridization intensity of the query genomic DNA (gDNA) and that of the reference gDNA (strain N16961) was used to divide genes into three categories based on gene presence: positive, negative, and uncertain. Comparing the gene calls for each probed gene across the entire collection of isolates allowed the categorization of genes into biologically meaningful groups. Following the convention of Lan and Reeves, genes that were positive in 95% or more of independent strains were defined as the core genome for Vibrio cholerae, while genes that were negative in more than 5% of independent strains were defined as the dispensable genome (22). The core and dispensable gene sets contained 83.0 and 13.3% of the 3,357 probed genes. The remaining 3.6% of probed genes could not be determined to be (“called”) positive or negative in at least 70% of the strains and are hereafter referred to as the uncalled gene set. The uncalled gene set is identical to the one described by Miller et al. (27), but the core and dispensable gene sets differ slightly from the “conserved,” “absent,” and “variable” gene sets. The core gene set comprises the entire “conserved” gene set plus “variable” genes called present in ≥95% and <100% of the strains. Similarly, the dispensable gene set includes all “absent” genes and the “variable” genes called present in <95% of strains. The 95% cutoff was chosen to allow comparison with other studies and should minimize the miscategorization of core genes due to stochastic errors, resulting in a more biologically meaningful core gene set (6).
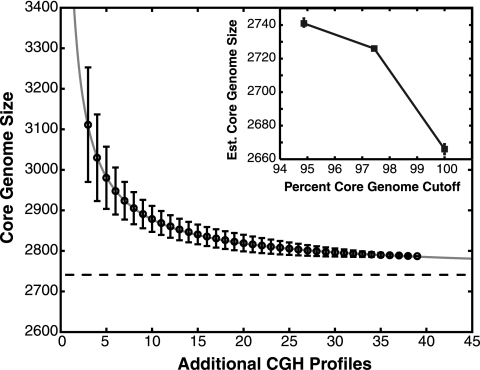
We used regression analysis to estimate the adequacy of our sample size for predicting core genome size (Fig. 2). Six strains that had a clonal genotype in the same sampling event were removed from the data set, and 10,000 random permutations of strain order for the remaining 39 strains were generated. A power law was fitted to a plot of the reduction in core genome size with each additional strain sampled (R-squared value, 0.9998). The predicted core genome size for an infinite number of sampled genomes is 2,741, indicating that we overestimated the core genome size by approximately 1.6%. Increasing the accuracy of the core genome size by an additional 27 genes (1%) would require the hybridization of 86 additional independent genomes. This regression analysis validates the use of this strain set in approximating the V. cholerae core genome. However, there are two notable sources of error in this analysis. First, there are 530 predicted protein-encoding genes that were not included on the amplicon microarrays. A substantial portion of these unprobed genes fall within the integron, so core genes will likely be underrepresented in the unprobed gene set. However, the exclusion of the unprobed genes will certainly result in a reduced core genome size. Second, there is likely to be a small percentage of core genes excluded due to their absence from N16961 but presence in most other V. cholerae strains.
FIG. 2.
Estimation of Vibrio cholerae core genome size by regression analysis. Open circles with 95% confidence limits represent the mean number of core genes with increasing numbers of genomes sampled for 10,000 random permutations of sampling order. A power law regression fit [y = a × (◯b) + c] with an R-squared value of 0.9998 is included. Regression coefficients with 95% confidence limits (CL) are as follows: a, 906.1 (CL, 894.1, 918.0); b, −0.8215 (CL, −0.8348, −0.8083); and c, 2,741 (CL, 2,739, 2,744). The horizontal dashed line represents the extrapolated core genome size for Vibrio cholerae, which is equal to 2,741 genes for a threshold of genes shared among 95% of sampled genomes. (Inset) Closed squares show the reduction in projected core genome size with increased stringency for gene ubiquity from 95% to 100% of strains.
To analyze how our estimate of core genome content compares with V. cholerae strains from regions with endemic cholera, we compared our core genome to results of a CGH study of Bangladeshi strains. Dziejman et al. (11) used CGH to characterize “conserved” and “divergent” genes in four non-O1/non-O139 strains isolated from patients with diarrhea in Bangladesh and in four toxigenic O1 and O139 clinical strains. Among the eight V. cholerae strains profiled, 2,761 out of our 2,787 predicted core genes were called conserved in all eight strains. Of our 26 remaining core genes, only 5 were called divergent in more than one strain, and all of these genes were adjacent to genes classified as dispensable in our data set.
We mapped the core gene set onto the pathway-genome database VchoCyc, which was designed to predict 171 likely metabolic pathways in Vibrio cholerae (35). Nearly all predicted metabolic pathways are fully represented within the core genome, including glycolysis, the tricarboxylic acid cycle, and pentose phosphate pathways. Biosynthesis of metabolic intermediates, amino acids, cofactors, nucleotides, and cell building blocks is represented, except for ectoine, cysteine, and derivatives of mannose. Pathways involved in metabolism of carbon and nutrient sources are also represented, except for sialic acid assimilation, removal of superoxides, galactose degradation, glycogen degradation, and citrate fermentation. Because all strains of V. cholerae inhabit the aquatic environment, while only a subset of strains reside for any time in other environments, like the intestine, pathways encoded in the core genome should primarily provide for persistence in the aquatic environment.
Functional characteristics of the dispensable genome.
Gene ontology (GO) terms (2) were collected for all genes probed on the microarray, yielding 5,957 and 637 functional annotations for the entire and dispensable genomes, respectively. Lexical analysis was performed to identify GO terms that were overrepresented in the dispensable gene set relative to those of the entire genome and to compute a probability that the observed number of hits would be observed for a random sample of the same size using the cumulative binomial probability function. Results were deemed significant at a P value of <0.05 (Table 1). It should be noted that the results of this analysis only allow a glimpse into the functions encoded by the dispensable genome because our methods restrict our analysis to genes probed on the microarray. Pathogenesis, transposition, and prophage functions and O-antigen biosynthesis genes are highly enriched in the dispensable genome, as has been shown elsewhere (11, 29). Primary virulence determinants contained on the CTX phage and Vibrio pathogenicity island 1 were missing from all the environmental isolates. Overrepresented functions also included chemosensing, cell surface modification, and lipopolysaccharide transport that mediate interactions between the bacterium and the extracellular environment. Such functions are expected to vary between strains adapted to living in different niche environments and are preferentially absent from other bacterial genomes relative to other functional categories (29, 30). The transport of some metabolic substrates in the dispensable genome, including citrate, mannose, fructose, and chitosan oligosaccharide (nonacetylated glucosamine dimer), were also overrepresented. This is consistent with the postulation that gene alteration is most likely to occur in the peripheral metabolic network since modifications or loss of core enzymes would affect metabolism of many connected substrates that are funneled through the same enzyme (31, 37).
TABLE 1.
Overrepresented GO functions in the dispensable genomea
| GO term | No. of dispensable-genome hits | No. of entire-genome hits | P value | GO role |
|---|---|---|---|---|
| GO:0009405 | 29 | 63 | <0.0001 | Pathogenesis |
| GO:0004803 | 9 | 11 | <0.0001 | Transposase activity |
| GO:0006313 | 9 | 11 | <0.0001 | DNA transposition |
| GO:0019047 | 6 | 6 | 0.0001 | Provirus integration |
| GO:0008979 | 5 | 5 | 0.0002 | Prophage integrase activity |
| GO:0006935 | 19 | 72 | 0.0004 | Chemotaxis |
| GO:0004871 | 17 | 61 | 0.0004 | Signal transducer activity |
| GO:0009297 | 11 | 30 | 0.0005 | Fimbriae biogenesis |
| GO:0004872 | 15 | 51 | 0.0005 | Receptor activity |
| GO:0009401 | 9 | 24 | 0.0013 | Phosphoenolpyruvate-dependent sugar phosphotransferase system |
| GO:0016407 | 8 | 20 | 0.0016 | Acetyltransferase activity |
| GO:0008653 | 3 | 3 | 0.0043 | Lipopolysaccharide metabolism |
| GO:0008815 | 3 | 3 | 0.0043 | Citrate (pro-3S) lyase activity |
| GO:0009346 | 3 | 3 | 0.0043 | Citrate lyase complex |
| GO:0019038 | 3 | 3 | 0.0043 | Provirus |
| GO:0019190 | 3 | 3 | 0.0043 | Glucosamine-dimer permease activity |
| GO:0009289 | 5 | 10 | 0.0048 | Fimbrium |
| GO:0030030 | 5 | 10 | 0.0048 | Cell projection organization and biogenesis |
| GO:0015585 | 4 | 7 | 0.0072 | Fructose permease activity |
| GO:0015755 | 4 | 7 | 0.0072 | Fructose transport |
| GO:0006113 | 8 | 28 | 0.0116 | Fermentation |
| GO:0009243 | 3 | 5 | 0.0171 | O-antigen biosynthesis |
| GO:0004476 | 2 | 2 | 0.0198 | Mannose-6-phosphate isomerase activity |
| GO:0008219 | 2 | 2 | 0.0198 | Cell death |
| GO:0015437 | 2 | 2 | 0.0198 | Lipopolysaccharide-transporting ATPase activity |
| GO:0015920 | 2 | 2 | 0.0198 | Lipopolysaccharide transport |
| GO:0019491 | 2 | 2 | 0.0198 | Ectoine biosynthesis |
| GO:0003700 | 26 | 160 | 0.0253 | Transcription factor activity |
| GO:0005324 | 2 | 3 | 0.0416 | Long-chain fatty acid transporter activity |
| GO:0015288 | 2 | 3 | 0.0416 | Porin activity |
| GO:0015909 | 2 | 3 | 0.0416 | Long-chain fatty acid transport |
The entire genome and dispensable genome contained 5,957 and 656 assigned GO terms, respectively. Overrepresented GO terms in the dispensable genome were significant if the P value for the lexical analysis software LACK v4.2 binomial function was less than 0.05.
California Vibrio cholerae isolates display extensive genomic diversity.
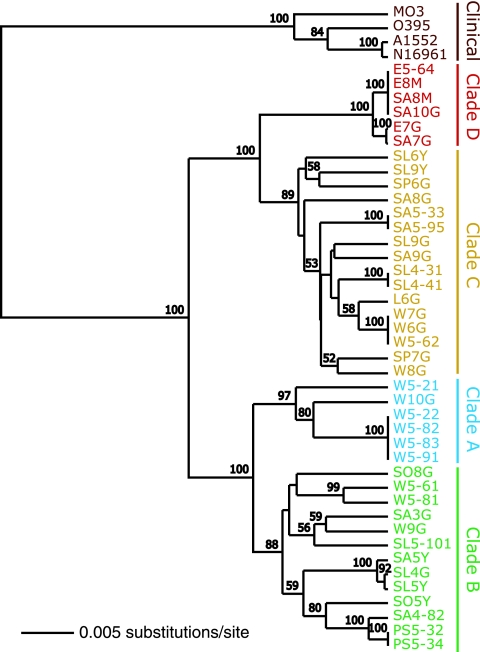
From the 41 environmental strains analyzed by CGH, there were 30 unique genome profiles with 24 genotypes sampled once, 3 sampled twice, 1 sampled three times, and 2 genotypes sampled four times (Fig. 3). One genotype sampled three times and one genotype sampled four times were composed of isolates from distinct sampling events up to 11 months apart. Grouping genotypes into clusters with similar patterns of positive and negative calls allowed analysis of temporal and geographical patterns in genotype distribution that would be otherwise impossible due to the rare resampling of identical genotypes. Hierarchical clustering (Fig. 3 and see binary CGH data used to construct Fig. 3 in Table S5 in the supplemental material) and nonmetric multidimensional scaling (data not shown) provided five distinct genotype clusters: one cluster of clinical strains and four clusters of environmental strains containing between 6 and 16 strains. One thousand bootstrap replicates of the UPGMA tree provided 100% support for the five genotype clusters (Fig. 3). The four environmental genotype clusters, hereafter referred to as clades A to D, represent useful groups of strains with similar genomes.
FIG. 3.
Division of strains into clades based on CGH profile. The UPGMA tree was generated using Jukes-Cantor distances and 1,000 bootstrap replicates, which provide 100% support for the five genotype clusters. Clades A, B, C, and D and clinical strains are shown in cyan, green, yellow, red, and brown, respectively. Bootstrap scores greater than 50 are displayed above the respective nodes.
Environmental factors influence distribution of genotypes.
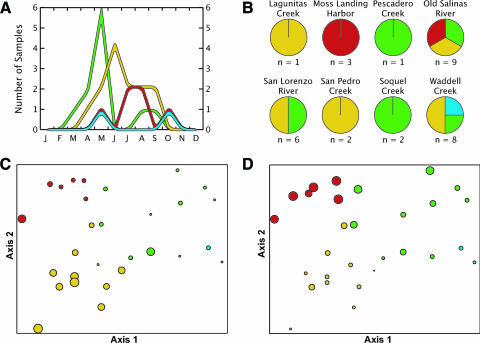
The temporal and geographical distribution of the four environmental genotype clusters are shown in Fig. 4. Clade B is the dominant group present in the spring months, while clades C and D are the most dominant groups during the summer. Clade A was detected only at one site (Waddell Creek) but remains culturable in October after clades B and C have declined. Clade C was evenly distributed geographically across the study area, while clade D was only found in the southernmost sites whose watersheds are dominated by agricultural land use (16). In contrast, clades A and B were found primarily in the more forested, less anthropogenically impacted watersheds to the north along the coast. Interestingly, while some sites support a broad diversity of genotypes (e.g., Old Salinas River), others are dominated by a single clade (e.g., Moss Landing Harbor).
FIG. 4.
Distribution of genotype groups illustrates relationship with changes in the environment. Clades A, B, C, and D are shown in cyan, green, yellow, and red, respectively. (A) Number of unique genotypes in each clade isolated, plotted for each month throughout the year. (B) Diversity of genotypes isolated from individual sampling sites over the entire sampling period listed, from north to south. Total numbers of strains sampled are listed below each pie graph. (Bottom) Canonical correspondence analysis ordination plots for (C) water temperature and (D) log ammonium. Each spot represents a genotype colored by clade, with the size of the spot proportional to the magnitude of the parameter when the strain was isolated. R values for axes 1 and 2 for water temperature are −0.742 and −0.294, respectively, confirming that clade C, followed by D, is most likely to be found in warmer water. R values for axes 1 and 2 for log ammonium are −0.470 and 0.779, respectively, confirming that clade D, followed by B, is the most likely to be found in nutrient-enriched waters.
We used canonical correspondence analysis to examine the relationship between environmental factors and the presence of specific clades. Clades A and B are more likely to be isolated from colder water, while clades C and D are more likely to be found in warmer water. Clades B and D are positively related to increased ammonium concentrations, and clades A and C are most likely to be cultivated from waters with depleted ammonium concentrations. Soluble reactive phosphate and nitrate-nitrite were positively correlated with ammonium, so these inorganic nutrients also correlate with the presence of clades B and D. All of these correlations were significant using a two-tailed t test (P < 0.04).
Dispensable genes whose presence correlates to specific environmental conditions may provide a selective advantage to Vibrio cholerae for survival and growth under those conditions. Isolates that cluster together in clades defined in Fig. 3 have similar dispensable genomes. Because membership in a specific clade is determined by only a subset of dispensable genes, many of the remaining genes that comprise the dispensable genome vary between strains within the same clade. Assuming that natural selection acts on the level of the gene, especially if gene loss and horizontal exchange are common, then we expect genes under selection in a subset of our sites or sampling events to correlate with environmental parameters that reflect the differences among samples. A two-tailed t test was used to identify specific dispensable genes whose presence was significantly correlated (P < 0.05) to environmental water parameters. Step-wise exclusion of water parameters from a logistic regression model was used to identify which combinations of parameters are the best predictors for the observed pattern of gene conservation (Table 2). Strains isolated from samples with colder water temperatures are more likely to contain genes annotated with functions in iron transport, transport and metabolism of chitosan oligosaccharides, chemotaxis, and chitin degradation. In contrast, strains from warmer waters are more likely to possess a gene cluster involved in exopolysaccharide production, another for protection against superoxide, and a fructose transporter. The presence of the gene cluster involved in fructose transport was shown to confer mannose metabolism due to an adjacent mannose-6-phosphate isomerase (27). Strains that are isolated from samples with lower inorganic nutrient concentrations are more likely to contain genes for ectoine biosynthesis, and strains from lower salinity samples more often contain genes encoding citrate transport and fermentation functions. Interestingly, conservation of a gene cluster enabling metabolism of sialic acid (N-acetylneuraminate) does not significantly correlate with water temperature but displays a strong seasonal signal, where 11 out of 12 strains that possess these genes were isolated from March through June, while only 60% of all strains were isolated during this period.
TABLE 2.
Statistically significant correlations between gene presence and measured environmental variables for selected clusters of genesa
| Gene cluster or gene | Predicted functiond | Parameter (gene presence)e | Coefficient | P value | No. of strains per indicated cladef
|
|||
|---|---|---|---|---|---|---|---|---|
| A | B | C | D | |||||
| VC0198 to VC0203 | Iron(III) transport | Water temp (−) | −0.310 | 0.0350 | 6 | 10 | 3 | 0 |
| VC0790 to VC0801 | Citrate transport and fermentation | Salinity (−) | −3.723 | 0.0111 | 6 | 13 | 15 | 0 |
| VC0919 to VC0923 | Exopolysaccharide biosynthesis | Water temp (+) | 0.400 | 0.0172 | 0 | 1 | 15 | 6 |
| Silicate (−) | −0.010 | 0.0376 | ||||||
| VC1280 to VC1286 | PTS system, chitosan | Water temp (−) | −0.616 | 0.0023 | 6 | 13 | 0 | 0 |
| VC1394 to VC1403 | Chemotaxis | Water temp (−) | −0.287 | 0.0549 | 1 | 11 | 0 | 0 |
| VC1405 to VC1413b | Chemotaxis | Water temp (−) | −0.616 | 0.0023 | 6 | 13 | 0 | 0 |
| VC1583 to VC1587c | Superoxide dismutase | Water temp (+) | 0.302 | 0.0299 | 0 | 2 | 13 | 6 |
| VC1773 to VC1784 | Sialic acid assimilation | Month (−) | −0.674 | 0.0220 | 1 | 8 | 3 | 0 |
| VC1820 to VC1827 | PTS system, fructose | Water temp (+) | 0.331 | 0.0259 | 6 | 0 | 15 | 6 |
| VC1952 | Chitinase | Water temp (−) | −0.468 | 0.0061 | 6 | 10 | 4 | 0 |
| VCA0667 to VCA0673c | Sodium/solute symporter | Water temp (−) | −0.547 | 0.0057 | 6 | 10 | 0 | 0 |
| VCA0822 to VCA0825 | Ectoine biosynthesis | Ammonium (−) | −2.488 | 0.0065 | 6 | 13 | 13 | 0 |
| VCA0991 to VCA0993 | Glutaredoxin II | Turbidity (+) | −25.146 | 0.0061 | 2 | 5 | 10 | 6 |
| VCA1102 to VCA1111 | Mixed functions | Ammonium (−) | −6.094 | 0.0222 | 6 | 13 | 6 | 0 |
Statistically significant correlations (P < 0.05) between gene presence and measured environmental variables for selected clusters of genes as determined by logistic regression.
Deletion does not span the VC1407 gene.
No probes for the genes VC1586, VCA0668, VCA0670, and VCA0672.
Primary annotated functions of contiguous genes from the TIGR database.
Log transformations were used to normalize salinity and ammonium data, and inverse transformations were used to normalize turbidity data. Polarity of association with gene presence is indicated by a positive (+) or negative (−) change in the parameter.
Number of strains in each clade for which the gene cluster was called positive for the gene presence, out of 6 strains for clade A, 13 for clade B, 16 for clade C, and 6 for clade D.
Clinical and environmental Vibrio cholerae strains exhibit high phenotypic diversity.
Phenotypic characteristics of a particular strain determine its fitness in a given environment. By screening for metabolic capabilities, we are not constrained by limitations such as gene annotation that restrict our ability to interpret genome data. Novel functions that are underrepresented or misannotated in sequence databases are unlikely to be uncovered by genome comparison but can be elucidated through assays of phenotypic characteristics.
All 45 strains of Vibrio cholerae were tested for metabolic diversity using proprietary phenotype microarrays (Biolog, Hayward, CA). Each assay was scored as growth or no growth (see Table S3 and Table S4 in the supplemental material). Out of 190 carbon sources assayed, at least one strain grew on each of 47 substrates. Of these 47 substrates, 95% or more (at least 43 of 45) strains grew on 26 of the carbon sources. These 26 substrates (N-acetylglucosamine, succinate, d-galactose, d-trehalose, glycerol, d-gluconate, l-lactate, d-mannitol, d,l-malate, d-fructose, α-d-glucose, maltose, d,l-α-glycerolphosphate, maltotriose, adenosine, fumarate, inosine, l-serine, l-malate, pyruvate, dextrin, l-asparagine, l-glutamate, α-ketobutyrate, d-glucosamine, and sucrose) comprise functions residing in the V. cholerae core metabolome. The remaining 21 carbon sources were classified as the dispensable metabolome (Table 3). It should be noted that the metabolomes listed in Table 3 are undoubtedly incomplete because only a subset of possible metabolites were tested. Strains were grown in liquid culture in microtiter plates, so metabolic pathways involved in other growth conditions might not have been detected (i.e., surface-attached cells or eukaryote-associated growth).
TABLE 3.
Dispensable carbon sources for Vibrio cholerae metabolismd
| Dispensable metabolite | Parameter (gene presence)c | Coefficient | P value | No. of strains per indicated cladee
|
|||
|---|---|---|---|---|---|---|---|
| A | B | C | D | ||||
| l-Aspartate | 6 | 12 | 16 | 4 | |||
| d-Mannose | Water temp (−) | 0.452 | 0.0101 | 6 | 1 | 15 | 6 |
| d-Serine | 6 | 10 | 11 | 6 | |||
| d-Glucuronatea | Turbidity (+) | −16.237 | 0.0166 | 0 | 0 | 10 | 4 |
| d-Glucose-6-phosphate | 6 | 10 | 12 | 6 | |||
| α-Keto-glutarate | 6 | 9 | 12 | 5 | |||
| l-Glutamine | 6 | 10 | 15 | 6 | |||
| β-Methyl-d-glucoside | 5 | 12 | 8 | 4 | |||
| d-Fructose-6-phosphate | 6 | 10 | 12 | 6 | |||
| Glycyl-l-aspartate | 6 | 11 | 16 | 4 | |||
| Citric acid | Ammonium (−) | −2.903 | 0.0438 | 6 | 10 | 16 | 4 |
| l-Threoninea,b | 0 | 0 | 0 | 0 | |||
| d-Cellobiosea | 0 | 1 | 0 | 0 | |||
| l-Alanyl-glycine | 4 | 6 | 9 | 1 | |||
| p-Hydroxyphenylacetatea | Salinity (+) | 3.723 | 0.0111 | 0 | 0 | 1 | 6 |
| Tyraminea | Salinity (+) | 3.723 | 0.0111 | 0 | 0 | 1 | 5 |
| α-Cyclodextrina | 0 | 2 | 0 | 0 | |||
| Gelatina | 5 | 6 | 9 | 5 | |||
| Laminarina | 0 | 2 | 0 | 0 | |||
| N-Acetylgalactosaminea | 0 | 6 | 1 | 0 | |||
| Sialic acid | Month (−) | −0.674 | 0.0220 | 1 | 8 | 3 | 0 |
Dispensable substrates were not metabolized by the reference strain N16961; growth was observed for some isolates.
No environmental isolates grew on l-threonine, but one clinical strain did.
Log transformations were used to normalize salinity and ammonium data, and inverse transformations were used to normalize turbidity data. Polarity of association with gene presence is indicated by a positive (+) or negative (−) change in the parameter.
Statistically significant (P < 0.05) correlations between dispensable substrate utilization and environmental parameters were verified by logistic regression and are displayed to the right of the dispensable metabolites to which they correspond.
Number of strains in each clade which grew on each sole carbon source out of 6 strains for clade A, 13 strains for clade B, 16 strains for clade C, and 6 strains for clade D.
The core metabolome was mapped to the VchoCyc database together with the core genome. We found all core metabolome substrates were accounted for by full complements of core genes in their degradation pathways. To examine the effect of in situ environmental parameters on the distribution of dispensable metabolic traits, we used logistic regression in the same manner as that used for linking dispensable gene presence with environmental parameters. The likelihood ratio and chi-square test was used to assess which parameters significantly (P < 0.05) predicted the observed phenotypic diversity (Table 3). Environmental strains isolated from warmer water and during spring months are more likely to grow on mannose and sialic acid, respectively. Strains isolated from high-turbidity and low-inorganic-nitrogen environments are more likely to grow on glucuronic acid and citric acid. Growth on p-hydroxyphenylacetate and tyramine is an attribute almost exclusively of clade D, which is found in higher-salinity environments.
The Vibrio cholerae core genome supports a generalist lifestyle.
Epidemic cholera has not been documented in central California for over 150 years, since 1850 when it claimed approximately 1,000 and 250 lives in Sacramento and San Francisco, respectively (15). Therefore, V. cholerae strains isolated from coastal waters here have likely not been in the human intestine for tens or hundreds of thousands of bacterial generations, if ever. These environmental isolates may even have diverged from ancestral pathogenic strains before the acquisition of the cholera toxin genes, so the isolates may never have contained genes under selection for causing cholera disease. If intestinal-specific genes were ever present in these isolates, then over time, gene loss and genome degradation would have periodically removed genes that were not selected for given the bacterium's lifestyle and ecology (28). If genes expressed while in the animal intestine provide no selectable benefit for life in coastal ecosystems, then these genes might be expected to be preferentially absent from the environmental isolates. To test whether this was the case, we compared our data with expression data from rice-water stool samples from three cholera patients in Bangladesh (26) and fluid from the ileal loops of infected rabbits (41). All strains of V. cholerae used in the expression studies were O1 El Tor biotype strains genetically similar to N16961. Genes with upregulated expression in each of the rice-water stool samples relative to that at mid-log phase in LB culture were mapped to the gene sets defined by CGH (Table 4). Remarkably, between 71 and 88% of the genes that were upregulated in stool samples from cholera patients belonged to the core gene set, while less than 7% were absent from all of the environmental isolates. For in vivo expression data from the rabbit ileal loop, approximately 90% of the upregulated genes mapped to the core gene set with either genomic DNA or mid-log phase in LB culture as a reference. Similar results were observed with additional ileal loop expression data using the O1 El Tor strain 92A1552 (N. Dolganov, personal communication). Comparison of these data with the those of the V. cholerae core and dispensable gene sets using a cumulative binomial distribution function indicates that genes upregulated in rabbit ileal loop samples are not preferentially absent from environmental isolates relative to those of the rest of the genome (P < 0.01). This trend is also true when looking at genes upregulated in stool from two or more of the cholera patients but is not statistically significant (P = 0.32). These data indicate that some of the genes in the core genome that are induced during in vivo growth are also found in the genomes of V. cholerae from California coastal waters. This finding likely indicates that such genes not only have a role during growth in vivo but are also required for survival of V. cholerae within one or more aquatic niches. Ultimately, the V. cholerae core genome contains genes useful for inhabiting multiple diverse habitats, including the animal intestine and coastal marine environments.
TABLE 4.
Distribution of upregulated genes in rice water stool and ileal loop samples in gene sets defined by comparative genome hybridization
| Samples | Upregulated genesa
|
|||
|---|---|---|---|---|
| Total genes | Core genes | Dispensable genes | Absent from California isolatesb | |
| Stool samplec | ||||
| Patient A | 119 | 105 | 7 | 0 |
| Patient B | 103 | 73 | 23 | 7 |
| Patient C | 244 | 181 | 49 | 15 |
| Two or more patientse | 77 | 62 | 11 | 2 |
| Ileal loop sampled | ||||
| gDNA (ref.) | 239 | 213 | 20 | 8 |
| LB (ref.) | 96 | 89 | 4 | 0 |
Upregulated genes have log2(intensity) values >2 standard deviations above the mean of all the probed genes.
Genes that were called absent in all environmental isolates but present in clinical strains. This is a subset of the dispensable gene set.
Rice water stool samples collected from three cholera patients in Bangladesh (26).
V. cholerae N16961 harvested from the rabbit ileal loop 8 h postinfection, using gDNA or RNA in mid-log LB culture as a reference (ref.) (41).
Compilation of genes scored as upregulated in stool samples of two or more of the three patients listed in the table.
DISCUSSION
Logistic regression of positive and negative gene calls identified several significant correlations (P < 0.05) with environmental parameters, particularly water temperature. Warm water is positively correlated with the presence of genes involved in exopolysaccharide production, superoxide dismutase, fructose transport, and mannose metabolism, while genes related to iron transport, chitin degradation, chemotaxis, and chitosan oligosaccharide transport and metabolism are more often found with strains isolated from colder water. No firm conclusions can be made based on the observed correlations, but we show below that they may prove useful in formulating hypotheses that then can be tested further to establish a causative link.
The coassociation of genes involved in mannose metabolism, superoxide protection, and exopolysaccharide production with warmer water temperature suggests that these genes may be selected for in environments with increased oxidative stress. Mannose was found to be a component of the exopolysaccharide produced by the V. cholerae O1 rugose variant, which provides increased resistance to osmotic and oxidative stress relative to that of smooth phenotype strains (39). During the summer months when water temperatures are higher, more solar radiation is absorbed by dissolved organic matter in coastal waters, leading to an increase in the production of reactive oxygen species (24). Strains containing genes for the production of protective exopolysaccharides and superoxide dismutase may be selectively isolated from warmer water samples due to their increased resistance to oxidative stress. Similarly, the coassociation of colder water temperature with genes encoding chitin degradation, chemotaxis, and chitosan oligosaccharide transport and metabolism functions suggests that these genes may all be under selection for chitin utilization. It is not surprising that chitin utilization is important in the coastal aquatic environment, and both particulate and soluble forms of chitin were shown to enhance the survival of V. cholerae at low temperature (1). Further exploration is required to assess whether the selection of chitin utilization genes in cold water also reflects the seasonal availability of chitin or other carbon sources at our sites.
From the dispensable metabolome, we identified six phenotypes with significant (P < 0.05) correlations with environmental parameters, including water temperature, salinity, turbidity, ammonium concentration, and season. The ability of strains to grow on d-glucuronate was positively correlated with turbidity, while increased salinity was correlated with growth on p-hydroxyphenylacetate and tyramine. The latter two substrates are products of the anaerobic deamination and decarboxylation of the amino acid tyrosine, respectively. In the environment, these transformations could occur in animal waste or anaerobic sediment in water bodies receiving high nutrient inputs (9, 25). Six out of seven strains that grow on p-hydroxyphenylacetate and tyramine were isolated from the Old Salinas River and Moss Landing Harbor sites, in which agriculture makes up 68% of watershed land use, as opposed to 16% for other sites where V. cholerae strains were isolated (16). These sites receive runoff from agricultural fields and grazing areas where water can mix with raw or composted manure (L. Crawford-Miksza [California Department of Health Services, Food and Drug Laboratory Branch], personal communication). The ability of some strains to grow on p-hydroxyphenylacetate and tyramine suggests the presence of agricultural runoff that stimulates anaerobic activity in the surrounding sediments or provides a rich supply of amines and volatile fatty acids. The coassociation of growth on these substrates with increased salinity reflects higher salinities at sites in agricultural areas relative to the that of other sampling sites.
An obvious drawback to CGH is the restriction of data sets to the sequences printed on the microarrays. We cannot use CGH to explore expanded genome content or the phenotypes that those genes might encode. In the present study this means we are unable to assess the genomic basis for the ability of certain strains to metabolize 43% of the carbon substrates in the dispensable metabolome (d-glucuronate, l-threonine, p-hydroxyphenylacetate, tyramine, alpha-cyclodextrin, gelatin, laminarin, and N-acetyl-d-galactosamine). Despite the limited scope of the CGH sequence coverage, the trends observed in functional bias for the dispensable genome are corroborated by the expanded genome content in other environmental V. cholerae strains uncovered by subtractive hybridization. Unique sequence fragments found in two environmental V. cholerae strains from southern California were enriched in mobile elements and genes involved in cell surface modification, bioluminescence, transport, carbohydrate metabolism, virulence, stress resistance, and signal transduction (32).
Despite the high phenotypic diversity among environmental strains of Vibrio cholerae in central California, little is known about how various phenotypic traits might affect the fitness of these strains in coastal waters. More work is required to analyze the relative importance of individual carbon and nutrient sources to overall metabolic requirements in coastal aquatic systems. In studying a Vibrio splendidus population in Plum Island Sound, Thompson et al. uncovered extremely high genotypic diversity that appears to be neutral, revealing no population structure in time or space (38). While statistically significant correlations suggest that our genotype clusters align themselves with different environmental conditions, further characterization of the sampling sites and the relative fitness of genotypes is needed to assess the importance of the observed genomic diversity. Finally, this analysis only uses isolates we were able to cultivate on selective media, so molecular probing of environmental samples will be needed to understand how these patterns apply to the nonculturable population of Vibrio cholerae strains.
Supplementary Material
Acknowledgments
We thank Chris Francis, Alfred Spormann, and Alyson Santoro for their comments on the manuscript. Lilian Lam provided support in the laboratory, and Nadia Dolganov generously supplied in vivo expression data.
This work was funded by the Woods Institute for the Environment (to D.P.K., A.B.B., and G.K.S.), by the NIH (to G.K.S.), and by the Giannini Family Foundation (to M.C.M.).
Footnotes
Published ahead of print on 20 April 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Amako, K., S. Shimodori, T. Imoto, S. Miake, and A. Umeda. 1987. Effects of chitin and its soluble derivatives on survival of Vibrio cholerae O1 at low temperature. Appl. Environ. Microbiol. 53:603-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ashburner, M., C. A. Ball, J. A. Blake, D. Botstein, H. Butler, J. M. Cherry, A. P. Davis, K. Dolinski, S. S. Dwight, J. T. Eppig, M. A. Harris, D. P. Hill, L. Issel-Tarver, A. Kasarskis, S. Lewis, J. C. Matese, J. E. Richardson, M. Ringwald, G. M. Rubin, and G. Sherlock. 2000. Gene ontology: tool for the unification of biology. Nat. Genet. 25:25-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atlas, E. L., S. W. Hager, L. I. Gordon, and P. K. Park. 1971. A practical manual for the use of the Technicon AutoAnalyzer in seawater nutrient analysis (revised). Technical report 215. Oregon State University, Department of Oceanography, Corvallis, OR.
- 4.Behr, M. A., M. A. Wilson, W. P. Gill, H. Salamon, G. K. Schoolnik, S. Rane, and P. M. Small. 1999. Comparative genomics of BCG vaccines by whole-genome DNA microarray. Science 284:1520-1523. [DOI] [PubMed] [Google Scholar]
- 5.Byun, R., L. D. Elbourne, R. Lan, and P. R. Reeves. 1999. Evolutionary relationships of pathogenic clones of Vibrio cholerae by sequence analysis of four housekeeping genes. Infect. Immun. 67:1116-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Charlebois, R. L., and W. F. Doolittle. 2004. Computing prokaryotic gene ubiquity: rescuing the core from extinction. Genome Res. 14:2469-2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chun, J., A. Huq, and R. R. Colwell. 1999. Analysis of 16S-23S rRNA intergenic spacer regions of Vibrio cholerae and Vibrio mimicus. Appl. Environ. Microbiol. 65:2202-2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dorrell, N., J. A. Mangan, K. G. Laing, J. Hinds, D. Linton, H. Al-Ghusein, B. G. Barrell, J. Parkhill, N. G. Stoker, A. V. Karlyshev, P. D. Butcher, and B. W. Wren. 2001. Whole genome comparison of Campylobacter jejuni human isolates using a low-cost microarray reveals extensive genetic diversity. Genome Res. 11:1706-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drake, H. L., N. G. Aumen, C. Kuhner, C. Wagner, A. Griesshammer, and M. Schmittroth. 1996. Anaerobic microflora of everglades sediments: effects of nutrients on population profiles and activities. Appl. Environ. Microbiol. 62:486-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dziejman, M., E. Balon, D. Boyd, C. M. Fraser, J. F. Heidelberg, and J. J. Mekalanos. 2002. Comparative genomic analysis of Vibrio cholerae: genes that correlate with cholera endemic and pandemic disease. Proc. Natl. Acad. Sci. USA 99:1556-1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dziejman, M., D. Serruto, V. C. Tam, D. Sturtevant, P. Diraphat, S. M. Faruque, M. H. Rahman, J. F. Heidelberg, J. Decker, L. Li, K. T. Montgomery, G. Grills, R. Kucherlapati, and J. J. Mekalanos. 2005. Genomic characterization of non-O1, non-O139 Vibrio cholerae reveals genes for a type III secretion system. Proc. Natl. Acad. Sci. USA 102:3465-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Farfán, M., D. Minana-Galbis, M. C. Fuste, and J. G. Loren. 2002. Allelic diversity and population structure in Vibrio cholerae O139 Bengal based on nucleotide sequence analysis. J. Bacteriol. 184:1304-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Faruque, S. M., D. A. Sack, R. B. Sack, R. R. Colwell, Y. Takeda, and G. B. Nair. 2003. Emergence and evolution of Vibrio cholerae O139. Proc. Natl. Acad. Sci. USA 100:1304-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fitzgerald, J. R., D. E. Sturdevant, S. M. Mackie, S. R. Gill, and J. M. Musser. 2001. Evolutionary genomics of Staphylococcus aureus: insights into the origin of methicillin-resistant strains and the toxic shock syndrome epidemic. Proc. Natl. Acad. Sci. USA 98:8821-8826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gibbons, H. 1865. Malignant cholera in California. Pac. Med. Surg. J. Press VIII:191-197. [Google Scholar]
- 16.Handler, N., A. Paytan, C. Higgins, R. Luthy, and A. Boehm. 2006. Human development is linked to multiple water body impairments along the California coast. Estuaries Coasts 29:860-870. [Google Scholar]
- 17.Hinchliffe, S. J., K. E. Isherwood, R. A. Stabler, M. B. Prentice, A. Rakin, R. A. Nichols, P. C. Oyston, J. Hinds, R. W. Titball, and B. W. Wren. 2003. Application of DNA microarrays to study the evolutionary genomics of Yersinia pestis and Yersinia pseudotuberculosis. Genome Res. 13:2018-2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang, S., W. Chu, and W. Fu. 2003. Prevalence of cholera toxin genes (ctxA and zot) among non-O1/O139 Vibrio cholerae strains from Newport Bay, California. Appl. Environ. Microbiol. 69:7541-7544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang, S. C., V. Louis, N. Choopun, A. Sharma, A. Huq, and R. R. Colwell. 2000. Genetic diversity of Vibrio cholerae in Chesapeake Bay determined by amplified fragment length polymorphism fingerprinting. Appl. Environ. Microbiol. 66:140-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaper, J. B., J. G. Morris, Jr., and M. M. Levine. 1995. Cholera. Clin. Microbiol. Rev. 8:48-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim, C. C., and S. Falkow. 2003. Significance analysis of lexical bias in microarray data. BMC Bioinformatics 4:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lan, R., and P. R. Reeves. 2000. Intraspecies variation in bacterial genomes: the need for a species genome concept. Trends Microbiol. 8:396-401. [DOI] [PubMed] [Google Scholar]
- 23.Lan, R., and P. R. Reeves. 2001. When does a clone deserve a name? A perspective on bacterial species based on population genetics. Trends Microbiol. 9:419-424. [DOI] [PubMed] [Google Scholar]
- 24.Lesser, M. P. 2006. Oxidative stress in marine environments: biochemistry and physiological ecology. Annu. Rev. Physiol. 68:253-278. [DOI] [PubMed] [Google Scholar]
- 25.Mackie, R. I., P. G. Stroot, and V. H. Varel. 1998. Biochemical identification and biological origin of key odor components in livestock waste. J. Anim. Sci. 76:1331-1342. [DOI] [PubMed] [Google Scholar]
- 26.Merrell, D. S., S. M. Butler, F. Qadri, N. A. Dolganov, A. Alam, M. B. Cohen, S. B. Calderwood, G. K. Schoolnik, and A. Camilli. 2002. Host-induced epidemic spread of the cholera bacterium. Nature 417:642-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller, M., D. Keymer, A. Avelar, A. Boehm, and G. Schoolnik. 2007. Detection and transformation of genome segments that differ within a coastal population of Vibrio cholerae strains. Appl. Environ. Microbiol. 73:3695-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mira, A., H. Ochman, and N. A. Moran. 2001. Deletional bias and the evolution of bacterial genomes. Trends Genet. 17:589-596. [DOI] [PubMed] [Google Scholar]
- 29.Nakamura, Y., T. Itoh, H. Matsuda, and T. Gojobori. 2004. Biased biological functions of horizontally transferred genes in prokaryotic genomes. Nat. Genet. 36:760-766. [DOI] [PubMed] [Google Scholar]
- 30.Nelson, K. E., D. E. Fouts, E. F. Mongodin, J. Ravel, R. T. DeBoy, J. F. Kolonay, D. A. Rasko, S. V. Angiuoli, S. R. Gill, I. T. Paulsen, et al. 2004. Whole genome comparisons of serotype 4b and 1/2a strains of the food-borne pathogen Listeria monocytogenes reveal new insights into the core genome components of this species. Nucleic Acids Res. 32:2386-2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pal, C., B. Papp, and M. J. Lercher. 2005. Adaptive evolution of bacterial metabolic networks by horizontal gene transfer. Nat. Genet. 37:1372-1375. [DOI] [PubMed] [Google Scholar]
- 32.Purdy, A., F. Rohwer, R. Edwards, F. Azam, and D. H. Bartlett. 2005. A glimpse into the expanded genome content of Vibrio cholerae through identification of genes present in environmental strains. J. Bacteriol. 187:2992-3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reen, F. J., E. F. Boyd, S. Porwollik, B. P. Murphy, D. Gilroy, S. Fanning, and M. McClelland. 2005. Genomic comparisons of Salmonella enterica serovar Dublin, Agona, and Typhimurium strains recently isolated from milk filters and bovine samples from Ireland, using a Salmonella microarray. Appl. Environ. Microbiol. 71:1616-1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salama, N., K. Guillemin, T. K. McDaniel, G. Sherlock, L. Tompkins, and S. Falkow. 2000. A whole-genome microarray reveals genetic diversity among Helicobacter pylori strains. Proc. Natl. Acad. Sci. USA 97:14668-14673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shi, J., P. R. Romero, G. K. Schoolnik, A. M. Spormann, and P. D. Karp. 2006. Evidence supporting predicted metabolic pathways for Vibrio cholerae: gene expression data and clinical tests. Nucleic Acids Res. 34:2438-2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tettelin, H., V. Masignani, M. J. Cieslewicz, C. Donati, D. Medini, N. L. Ward, S. V. Angiuoli, J. Crabtree, A. L. Jones, A. S. Durkin, et al. 2005. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome.” Proc. Natl. Acad. Sci. USA 102:13950-13955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thomason, B., and T. D. Read. 2006. Shuffling bacterial metabolomes. Genome Biol. 7:204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thompson, J. R., S. Pacocha, C. Pharino, V. Klepac-Ceraj, D. E. Hunt, J. Benoit, R. Sarma-Rupavtarm, D. L. Distel, and M. F. Polz. 2005. Genotypic diversity within a natural coastal bacterioplankton population. Science 307:1311-1313. [DOI] [PubMed] [Google Scholar]
- 39.Wai, S. N., Y. Mizunoe, A. Takade, S. I. Kawabata, and S. I. Yoshida. 1998. Vibrio cholerae O1 strain TSI-4 produces the exopolysaccharide materials that determine colony morphology, stress resistance, and biofilm formation. Appl. Environ. Microbiol. 64:3648-3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wertz, J. E., C. Goldstone, D. M. Gordon, and M. A. Riley. 2003. A molecular phylogeny of enteric bacteria and implications for a bacterial species concept. J. Evol. Biol. 16:1236-1248. [DOI] [PubMed] [Google Scholar]
- 41.Xu, Q., M. Dziejman, and J. J. Mekalanos. 2003. Determination of the transcriptome of Vibrio cholerae during intraintestinal growth and midexponential phase in vitro. Proc. Natl. Acad. Sci. USA 100:1286-1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.