Abstract
Plant cell wall degradation is a premier event when Bacillus subtilis, a typical saprophytic bacterium, invades plants. Here we show the degradation system of rhamnogalacturonan type I (RG-I), a component of pectin from the plant cell wall, in B. subtilis strain 168. Strain 168 cells showed a significant growth on plant cell wall polysaccharides such as pectin, polygalacturonan, and RG-I as a carbon source. DNA microarray analysis indicated that three gene clusters (yesOPQRSTUVWXYZ, ytePQRST, and ybcMOPST-ybdABDE) are inducibly expressed in strain 168 cells grown on RG-I. Cells of an industrially important bacterium, B. subtilis strain natto, fermenting soybeans also express the gene cluster including the yes series during the assimilation of soybean used as a carbon source. Among proteins encoded in the yes cluster, YesW and YesX were found to be novel types of RG lyases releasing disaccharide from RG-I. Genetic and enzymatic properties of YesW and YesX suggest that strain 168 cells secrete YesW, which catalyzes the initial cleavage of the RG-I main chain, and the resultant oligosaccharides are converted to disaccharides through the extracellular exotype YesX reaction. The disaccharide is finally degraded into its constituent monosaccharides through the reaction of intracellular unsaturated galacturonyl hydrolases YesR and YteR. This enzymatic route for RG-I degradation in strain 168 differs significantly from that in plant-pathogenic fungus Aspergillus aculeatus. This is, to our knowledge, the first report on the bacterial system for complete RG-I main chain degradation.
Bacillus subtilis, the best-characterized gram-positive bacterium, is widely distributed and has various high levels of potential to incorporate external DNA; produce amino acids, antibacterial agents, and industrially important enzymes; secrete large amounts of proteins; form endospores; and associate with plants. The complete genome sequence for the type strain, B. subtilis 168, was determined in 1997, and about 4,000 genes were found contained in the bacterial genome (26). The clarification of function-unknown genes is now being focused on, and essential genes in the bacterium have been identified through the construction of knockout mutants for each gene (25). Although mechanisms for DNA uptake (10), amino acid fermentation (20), antibiotic production (49), protein secretion (49), and endospore formation (44) are well analyzed, little is known about the interaction between bacilli and plants.
B. subtilis is important for producing fermented foods and for composting plants (41). In Japan, this bacterium is called “Kosoukin,” which means “bacteria present in dead grass,” and a type of B. subtilis, strain natto, has been used for the production of “natto” for over 1,000 years. Natto is produced through the fermentation of boiled soybeans by strain natto. Natto-like fermented soybean foods such as tempe and Thua nao are also produced in Asia and Africa. In natto production, cells of strain natto sprayed on boiled soybeans degrade their cell walls and excrete large amounts of γ-polyglutamic acid through the assimilation of soybean nutrients (23, 24). Natto is thus considered a mimic ecosystem model for the interaction between bacilli and plants. In strain 168, this bacterium excretes little γ-polyglutamic acid due to mutations in two genes, degQ and swrA. Incorporation of corresponding wild-type genes into strain 168 cells restores production of the polymer (51). Since DegQ has been identified as an activator of gene transcription and SwrA as a regulator of swarming motility, strain 168 probably invades soybeans by degrading their cell walls.
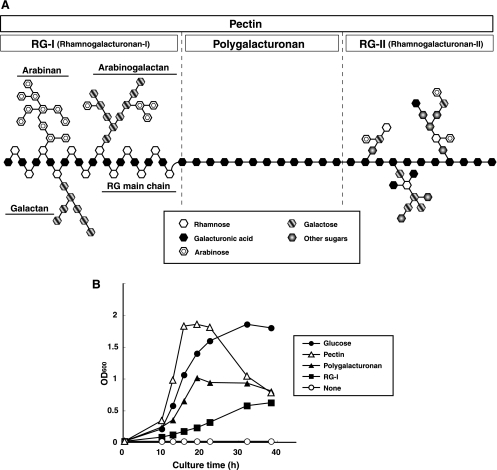
Plant cell wall degradation is a major event when saprophytic bacteria invade plants. The plant cell wall consists of macromolecule polysaccharides such as pectin, cellulose, and hemicellulose (3). Pectin contains polygalacturonan as a linear component and rhamnogalacturonan type I (RG-I) and rhamnogalacturonan type II (RG-II) as branched chains (6, 31, 52) (Fig. 1A). RG-I consists of alternating l-rhamnopyranose and d-galactopyranouronic acid as a backbone (RG chain) with additions of arabinans and galactans as side chains (30). RG-II contains polygalacturonan as a backbone and a complex of approximately 30 monosaccharides, including rare sugars such as apiose and aceric acid, as a side chain (42). Many plant-pathogenic fungi and bacteria produce enzymes responsible for plant cell wall degradation. Glycosidases and polysaccharide lyases, for example, cleave glycosidic bonds in pectin via hydrolysis and β-elimination reactions, respectively (7, 28). These polysaccharide-degrading enzymes function as a virulence factor in microbial plant pathogens.
FIG. 1.
Assimilation of pectin-related polysaccharides by B. subtilis. (A) Structure of pectin. Symbols of sugars are indicated in the box. (B) Growth of strain 168 cells on different carbon sources: glucose (closed circles), pectin (open triangles), polygalacturonan (closed triangles), and RG-I (closed squares). The nonaddition of saccharides as a carbon source was used as the negative control (open circle). Bacterial growth was determined by measuring turbidity (optical density at 600 nm [OD600]).
The degradation pathway for the polygalacturonan region in pectin has been extensively characterized in plant-pathogenic microbes such as Erwinia chrysanthemi (15) and Aspergillus species (9). A large number of polygalacturonan-degrading enzymes (i.e., pectin lyase, pectate lyase, and polygalacturonase) have also been identified from many fungi and bacteria, including B. subtilis (11, 45). Reports on the microbial degradation system for the RG region in pectin have been limited, however, except for A. aculeatus strain KSM 510 (33-37), possibly due to the complex structure of the heteropolysaccharide, although a few RG lyases from fungi and bacteria have been identified (14, 29, 36, 43). The analysis of RG degradation by B. subtilis is important for clarification of its saprophytic interaction with plants. RG is also known to be easily extracted as a water-soluble fraction from soybeans (38). This indicates that the polysaccharide is bared on the soybean surface during the preparation of boiled soybeans and that RG degradation by strain natto may be crucial for triggering bacterial growth for fermentation of boiled soybean.
This article deals with molecular identification of the RG-I degradation system in B. subtilis via transcriptome, genetic, and enzymatic analyses.
MATERIALS AND METHODS
Materials.
RG-I (from potatoes), azo-galactan (from potatoes), and polygalacturonan (from citrus pectin) were purchased from Megazyme. Pectin was purchased from Wako Pure Chemical and pectic acid from Lancaster Synthesis. Silicagel 60/Kieselguhr F254 thin-layer chromatography (TLC) plates were obtained from E. Merck. Ni2+-chelating Sepharose Fast Flow, HiLoad 26/10 Q Sepharose High Performance, Q Sepharose Fast Flow, HiLoad 16/60 Superdex 200pg, and Superdex peptide 10/300 GL were obtained from GE Healthcare; endonucleases were obtained from Takara Bio; and DNA-modifying enzymes were obtained from Toyobo. A. aculeatus RhgB was a kind gift from Novozymes A/S.
Microorganisms and culture conditions.
To investigate growth on different carbon sources, strain 168 cells were aerobically cultured at 30°C for 48 h in a modified Spizizen's minimal medium (4) containing 5.0 mg/ml each carbon source (glucose, pectin, polygalacturonan, or RG-I), 1.0 mg/ml of sodium citrate, 6.0 mg/ml of KH2PO4, 14 mg/ml of K2HPO4, 2.0 mg/ml of ammonium sulfate, 0.39 mg/ml of MgSO4·7H2O, 0.05 mg/ml of tryptophan, 55 ng/ml of CaCl2, 135 ng/ml of FeCl2, 10 ng/ml of MnCl2·4H2O, 17 ng/ml of ZnCl2, 4.3 ng/ml of CuCl2·2H2O, 6.0 ng/ml of CoCl2·6H2O, 6.0 ng/ml of Na2MoO4·2H2O, and 4.7 ng/ml of Na2SeO4. B. subtilis strain natto (NBRC16449) isolated from natto was purchased from the NITE Biological Resource Center (NBRC), Japan. Strain natto cells were also aerobically grown at 30°C in modified Spizizen's minimal medium including 0.2 g/ml of steamed soybean as a carbon source instead of saccharides.
As a host for plasmid amplification, cells of Escherichia coli strain DH5α (Toyobo) were routinely cultured at 37°C in Luria-Bertani (LB) medium (46) containing ampicillin (100 μg/ml). E. coli strains HMS174(DE3) and HMS174(DE3)(pLysS) (Novagen) were used as hosts for the overexpression of YesW and YesX, respectively. For the expression of YesW, E. coli cells were aerobically cultured at 30°C in LB medium supplemented with ampicillin (100 μg/ml). When turbidity at 600 nm reached 1.4, isopropyl-β-d-thiogalactopyranoside (IPTG) was added to the culture at a final concentration of 0.2 mM, and cells were further cultured at 16°C for 42 h. For the expression of YesX, E. coli cells were aerobically cultured at 30°C in LB medium supplemented with ampicillin (100 μg/ml), chloramphenicol (34 μg/ml), and sorbitol (1 M). When turbidity at 600 nm reached 0.6, IPTG was added to the culture at a final concentration of 0.4 mM and cells were further cultured at 16°C for 42 h.
Preparation of total RNA.
Total RNAs for DNA microarray were prepared as follows. Strain 168 cells were grown in synthetic minimal medium containing 10 mg/ml glucose or RG-I as a carbon source until the turbidity at 600 nm reached 1.0. Cells were collected and treated with RNAprotect bacterial reagent (QIAGEN) for stabilization of RNA. Based on the manufacturer's instructions, total RNAs were extracted with an RNeasy kit (QIAGEN) and treated with DNase (RNase-free DNase set; QIAGEN) supplied with the kit.
DNA microarray analysis.
The genome sequence and open reading frame (ORF) prediction for strain 168 were provided by GenBank. A strain 168 DNA microarray purchased from NimbleGen Systems includes 4,112 target genes fixed on the glass slide by fixing at least 20 unique probes consisting of 24-mer synthetic oligonucleotides for each gene. Biotin labeling, fragmentation, and hybridization were done by NimbleGen Systems. Arrays were scanned with an Axon Genepix 4000B scanner at 532 nm and a resolution of 5 μm and analyzed by quantile normalization and robust multiarray averaging (16). This normalized data was processed with NANDEMO Analysis 1.0.0 software (Genefrontier). Genes from cells in the presence of RG-I with an expression threefold higher than those in the presence of glucose were selected. Student's t test for analyzing the mean log ratios of two samples and subsequent Bonferroni adjustment for multiple testing (total of 4,112 ORFs on arrays) were applied as a rigorous criterion for significantly changed signal intensity. Changes with P < 0.05 were considered statistically significant.
RT-PCR.
For reverse transcription-PCR (RT-PCR), total RNAs were isolated from strain natto cells grown for 8 h at 30°C in 5 ml of soybean minimal medium until the turbidity at 600 nm reached 1.0. cDNAs were synthesized from 500 ng of total RNAs with a ReverTra-Plus kit (Toyobo) based on the manufacturer's protocol. Subsequent PCR was done as follows: preheating at 94°C for 2 min and 98°C for 10 s, 63°C for 20 s, and 68°C for 1 min, for a total of 30 cycles. Specific PCR primers were used for the YesW gene (forward, 5′-TCTCATGACGGGAAAGGCATGTACTC-3′; reverse, 5′-TCTGCTTTTGTGCCGTTGTTAGCC-3′); YesX gene (forward, 5′-TCCACGAAAAGGGCTTGAAGTGTTCC-3′; reverse, 5′-TCTTGGCCTGAAGAACCGGGTTT-3′), and 16S rRNA gene (forward, 5′-TCGCTGCAGCTAACGCATTAAGCAC-3′; reverse, 5′-TCTTGCAGACTGCGATCCGAACT-3′). PCR after treatment of total RNA without reverse transcriptase was also done as a negative control.
Construction of overexpression systems for YesW and YesX.
Overexpression systems for YesW and YesX were constructed in E. coli cells as follows. To clone the YesW and YesX genes, PCR was conducted in a reaction mixture (100 μl) consisting of 5 U of KOD polymerase (Toyobo), 0.25 μg of strain 168 genomic DNA, 40 pmol of forward and reverse primers, 20 nmol of deoxynucleoside triphosphates (dNTPs), 100 nmol of MgCl2, 5 μl of dimethyl sulfoxide, and the commercial reaction buffer supplied with KOD polymerase. The forward and reverse primers for the YesW gene were 5′-GGCATATGAGAAGGAGCTGTCTGATGATTA-3′ and 5′-CCCTCGAGAGGCGTATACATATTTGGTTTT-3′, and those for the YesX gene were 5′-GGCATATGAAACCAAAAAAGAGGCAAATGG-3′ and 5′-GGCTCGAGCAGCGGCGCCTCCGCTTTGCTT-3′, with NdeI and XhoI sites (underlined) added to each of their 5′ regions. PCR conditions were as follows: 94°C for 30 s, 60°C for 2 s, and 74°C for 40 s for a total of 30 cycles. PCR products were separated by 8-mg/ml agarose gel electrophoresis, and DNA fragments corresponding to YesW and YesX genes were isolated with a GENECLEAN II kit (Bio 101). Nucleotide sequences of their fragments were confirmed to completely match those of YesW and YesX genes by DNA sequencing with an Applied Biosystems model 377 sequencer as described elsewhere (47). Fragments were ligated with HincII-digested pUC119 (Takara Bio), and the resultant plasmids were digested with NdeI and XhoI to isolate YesW and YesX genes. NdeI-XhoI fragments containing YesW and YesX genes were ligated with NdeI- and XhoI-digested pET21b (Novagen). The resultant plasmids were designated as pET21b/YesW and pET21b/YesX. YesW and YesX genes cloned in plasmids lack their original stop codons and encode C-terminal histidine-tagged protein.
Assays for enzyme and protein.
YesW or YesX was incubated at 30°C for 5 min in a reaction mixture (1 ml) consisting of 0.5 mg/ml of RG-I, 50 mM of Tris-HCl (pH 7.5), and 2 mM of CaCl2. Activity was determined by monitoring the increase in absorbance at 235 nm arising from the double bond formed in reaction products. One unit of enzyme activity was defined as the amount of enzyme required to produce an increase of 1.0 in absorbance at 235 nm per min by using a cuvette with a light path 1 cm long. With azo-galactan as the substrate, the enzyme was assayed based on the manual supplied by Megazyme. Protein content was determined by the method of Bradford (2), with bovine serum albumin as the standard.
Purification of recombinant YesW and YesX from E. coli.
Recombinant YesW was purified from E. coli cells. Unless otherwise specified, all procedures were done at 0 to 4°C. CaCl2 was included in buffers to activate the enzyme, and NaCl was included to repress its aggregation. E. coli cells harboring pET21b/YesW were grown in 40.5 liters of LB medium (1.5 liter/flask), collected by centrifugation at 6,000 × g for 5 min, washed with 20 mM Tris-HCl (pH 7.5) containing 2 mM CaCl2, and resuspended in the same buffer. Cells were ultrasonically disrupted at 9 kHz for 20 min, and the clear solution obtained on centrifugation at 20,000 × g for 20 min was used as the cell extract. After overnight dialysis against 20 mM of Tris-HCl (pH 7.5) containing 2 mM of CaCl2, 0.2 M of NaCl, and 20 mM of imidazole, cell extract was applied to an Ni2+-chelating Sepharose Fast Flow column (2.5 by 5 cm) equilibrated beforehand with 20 mM of Tris-HCl (pH 7.5) containing 2 mM of CaCl2, 0.2 M of NaCl, and 20 mM of imidazole. YesW containing a histidine tag at the C terminus was eluted with a linear gradient of imidazole (20 to 500 mM) in 20 mM of Tris-HCl (pH 7.5) containing 2 mM of CaCl2 and 0.2 M of NaCl (200 ml), and 5-ml fractions were collected every 5 min. Fractions containing the enzyme, which was eluted with between 100 and 200 mM of imidazole, were combined and dialyzed overnight against 20 mM of Tris-HCl (pH 7.5) containing 2 mM of CaCl2 and 0.2 M of NaCl. After dialysis, the sample was divided into five fractions of 5 ml each, and each fraction was applied to a HiLoad 16/60 Superdex 200pg column (1.6 by 60 cm) equilibrated beforehand with 20 mM of Tris-HCl (pH 7.5) containing 2 mM of CaCl2 and 0.2 M of NaCl. YesW was eluted with the same buffer (120 ml), and 2-ml fractions were collected every 2 min. Fractions containing YesW were combined and used as the purified enzyme source.
E. coli cells harboring pET21b/YesX were grown in 13.5 liters of LB medium (1.5 liters/flask) as described above. The purification procedures for YesX are the same as those for YesW.
Size-exclusion chromatography.
To clarify the mode of action of YesW and YesX, the degradation profile of RG-I through the YesW or YesX reaction was analyzed by size-exclusion chromatography. Substrate RG chain suitable for this analysis was prepared from RG-I as described elsewhere (37). Briefly, 1 g of de-esterified RG-I was hydrolyzed in 100 ml of 0.1 M HCl at 80°C for 72 h. The hydrolysate was neutralized with NaOH and centrifuged at 20,000 × g for 10 min. The clear solution was applied to an ion-exchange column (Q Sepharose Fast Flow) equilibrated beforehand with 5 mM of (NH4)HCO3, and the RG chain was eluted with a linear gradient of (NH4)HCO3 (5 to 500 mM) and freeze-dried. The resultant RG chain was a mixture consisting of RG backbones with different degrees of polymerization. Appropriate amounts of YesW, YesX, or Aspergillus aculeatus RhgB were incubated at 30°C for 60 min in a reaction mixture (1 ml) consisting of 0.5 mg/ml of the RG chain, 50 mM of Tris-HCl (pH 7.5), and 2 mM of CaCl2. Saccharides derived from the RG chain through the enzyme reaction were regarded as final reaction products, because they were not degraded further by the addition of a fresh enzyme. Products were analyzed by Superdex peptide 10/300 GL with an AKTA purifier (Amersham Biosciences). Saccharides were eluted at a flow of 0.5 ml/min with 10 mM of potassium phosphate (pH 7.0) and detected using a UV detector at 235 nm.
TLC analysis.
Products derived from the RG chain through the YesW or YesX reaction were separated by TLC using a solvent system consisting of 1-butanol-acetic acid-water (3/2/2 [vol/vol]). Products were visualized by heating the TLC plate at 130°C for 5 min after spraying it with 10% (vol/vol) sulfuric acid in ethanol.
Mass spectrometry.
The smallest reaction product through the YesX reaction was purified by gel filtration of a Bio-Gel P-2 column (Bio-Rad), and analyzed by electrospray ionization-mass spectrometry (ESI-MS) using the Micromass ZMD liquid chromatography/MS system (Waters). The mass spectrometer was operated in the negative mode. The needle voltage was −2.5 kV, and the cone voltage was −10 V. During analysis, the mass spectrum was scanned from 10 to 1,000 m/z.
RESULTS
Strain 168 growth.
The growth of strain 168 cells was investigated in minimal media containing different carbon sources (Fig. 1B). Although all minimal media contained sodium citrate in addition to saccharides, strain 168 cells did not show any growth on minimal medium containing no added saccharides. As indicated previously (39), strain 168 cells grew well on glucose, pectin, and polygalacturonan. Although strain 168 cells did not show rapid growth in the RG-I minimal medium, it was apparent that cells can assimilate RG-I as a carbon source and that the genes responsible for RG-I degradation are present in the bacterial genome.
Gene clusters responsible for degrading RG-I.
To clarify genes responsible for RG-I degradation, DNA microarray analysis was done and mRNA expression in strain 168 cells grown on RG-I was compared to that of cells grown on glucose. The expression of 183 genes out of a total of 4,112 on the designed DNA microarray was up-regulated over threefold in RG-I-grown cells. Three gene clusters (yesOPQRSTUVWXYZ, ytePQRST, and ybcMOPST-ybdABDE) were inducibly expressed in the presence of RG-I (Table 1). Although some genes in the yes cluster appear, based on homology analysis, to be involved in RG degradation, the function of yte and ybc-ybd clusters remains to be clarified. Since we have recently identified function-unknown YteR and YesR as novel unsaturated galacturonyl hydrolases responsible for degradation of the unsaturated RG disaccharide (17), the yte cluster probably plays an important role in the transport and degradation of RG. Although P after Bonferroni adjustment of genes such as yesR and yesT (RG acetylesterase homolog gene) in the yes cluster was higher than the cutoff, other genes in the cluster, such as yesW and yesX, were significantly up-regulated in the presence of RG-I (P < 0.05) (Table 1). Together with our previous results from the molecular identification of YesR (17), this transcriptome analysis indicates experimentally that the yes cluster is crucial for degrading RG-I (Fig. 2C).
TABLE 1.
High-expression genes in strain 168 cells grown on RG-I compared to glucose
| ORF name | Gene | Change (fold)a | Pb | Functional class |
|---|---|---|---|---|
| BSU06970 | yesO | 9.0 | 0.0086 | Probable multiple-sugar transport system substrate-binding protein |
| BSU06980c | yesP | 4.0 | 0.67 | Probable multiple-sugar transport system permease protein |
| BSU06990 | yesQ | 6.2 | 0.0067 | Probable multiple-sugar transport system permease protein |
| BSU07000c | yesR | 4.0 | 0.30 | Unknown |
| BSU07010 | yesS | 6.6 | 0.00019 | Probable transcriptional regulator (AraC/XylS family) |
| BSU07020c | yesT | 3.1 | 0.56 | Similar to RG acetylesterase |
| BSU07030c | yesU | 3.6 | 1 | Unknown |
| BSU07040c | yesV | 3.9 | 1 | Unknown |
| BSU07050 | yesW | 3.1 | 0.030 | RG lyase |
| BSU07060 | yesX | 5.2 | 0.0031 | RG lyase |
| BSU07070 | yesY | 14.6 | 9.72E−06 | Similar to RG acetylesterase |
| BSU07080 | yesZ | 9.7 | 0.0087 | Probable β-galactosidase |
| BSU30100 | yteT | 7.5 | 0.023 | Putative oxidoreductase |
| BSU30110 | yteS | 11.5 | 0.00015 | Unknown |
| BSU30120 | yteR | 7.5 | 0.024 | Unknown |
| BSU30130 | yteQ | 10.1 | 0.00017 | Putative multiple-sugar transport system permease protein |
| BSU30140 | yteP | 15.3 | 3.17E−06 | Putative multiple-sugar transport system permease protein |
| BSU01900 | ybcM | 32.0 | 1.35E−08 | Similar to glucosamine-fructose-6-phosphate aminotransferase |
| BSU01910 | ybcO | 9.7 | 4.06E−06 | Probable cell killing factor precursor |
| BSU01920 | ybcP | 28.9 | 1.74E−08 | Cell killing factor production, similar to coenzyme PPQ synthesis protein |
| BSU01930 | ybcS | 16.9 | 0.00085 | Unknown |
| BSU01940 | ybcT | 25.4 | 1.46E−08 | Cell killing factor production, probable membrane-bound metalloprotease |
| BSU01950 | ybdA | 10.7 | 0.0016 | Exporter of cell killing factor, ABC transport system ATP-binding protein |
| BSU01960 | ybdB | 10.4 | 0.00044 | Exporter of cell killing factor, ABC transport system permease protein |
| BSU01970c | ybdD | 6.0 | 0.44 | Unknown |
| BSU01980 | ybdE | 8.7 | 0.00026 | Unknown |
Changes (fold) are defined as the expression of growth conditions in the presence or absence of RG-I.
P after Student's t test and with Bonferroni adjustment.
Gene for which the P value was higher than the cutoff (0.05).
FIG. 2.
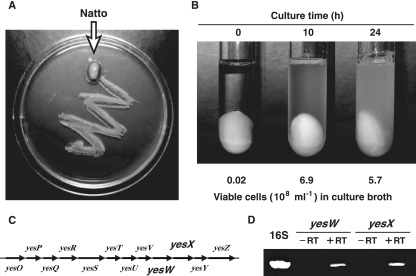
B. subtilis strain natto. (A) The soybean surface was covered with strain natto cells. The LB plate surface was streaked with natto and incubated at 37°C. (B) Growth of strain natto cells on boiled soybean minimal medium. Viable cells in culture broth were determined by measuring colony counts after spreading on LB medium. (C) Genetic organization of the yes cluster. (D) Expression of 16S rRNA, YesW, and YesX genes in strain natto cells grown on soybeans. Each gene was amplified by using total RNAs after treatment with (+RT) and without (−RT) reverse transcriptase.
Since RG lyase is thought to be the first enzyme for RG-I degradation, we focused on RG lyase homologs YesW and YesX. YesW and YesX significantly resemble each other in primary structure (i.e., 68.7% identity in a 597-amino-acid overlap). YesW and YesX are similar, in primary structure, to RG lyases such as Rgl11A from Cellvibrio japonicus (29) and Rgl11Y from Clostridium cellulolyticum (43): YesW versus Rgl11A, 59.0% identity in 600-amino-acid overlap; YesX versus Rgl11A, 53.0% identity in 610-amino-acid overlap; YesW versus Rgl11Y, 61.7% identity in a 614-amino-acid overlap; and YesX versus Rgl11Y, 59.2% identity in a 598-amino-acid overlap. This homology analysis was done on 25 December 2006, using the BLAST program on the GenomeNet server (http://BLAST.genome.jp). In the Carbohydrate-Active enZymes (CAZy) database (http://afmb.cnrs-mrs.fr), YesW, YesX, Rgl11A, and Rgl11Y are classified into polysaccharide lyase (PL) family 11 based on primary sequence similarities. YesW and YesX show no homology with RG lyase RhgB from A. aculeatus strain KSM 510 (36), classified into PL family 4.
Expression of the yes cluster in strain natto.
Natto is a traditional Asian food produced from boiled soybeans using B. subtilis. Strain natto cells industrially used for natto production showed sufficient growth (6.9 × 108 cells/ml culture broth) on a soybean as the sole carbon source at 10 h after inoculation of seed culture (0.02 × 108 cells/ml culture broth) (Fig. 2A and B), although cells attached to the soybean were not counted. Since RG and arabinogalactan are easily extracted as water-soluble polysaccharides from soybeans treated with boiling water (38), the expression of the yes cluster in strain natto (Fig. 2C), especially YesW and YesX genes, was examined by RT-PCR. Both YesW and YesX genes were amplified from reverse transcriptase-treated total RNAs isolated from strain natto cells grown on soybeans, although no genes were amplified from total RNAs without treatment by reverse transcriptase (Fig. 2D). This indicates that YesW and YesX genes are expressed in strain natto cells grown on soybeans and are probably crucial for soybean cell wall degradation.
Purification and characterization of recombinant YesW and YesX expressed in E. coli cells.
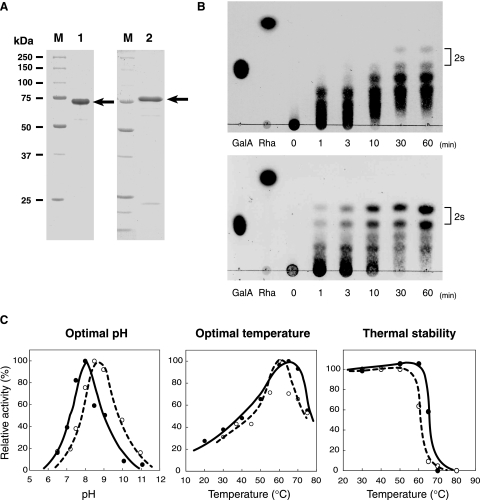
To clarify the significance of two copies of RG lyase in strain 168, YesW and YesX were overexpressed in E. coli cells, purified, and characterized. YesW and YesX were purified 73.5- and 18.8-fold from E. coli cells, with recovery rates of 19.8 and 8.94%, respectively (Table 2). Purified enzymes were confirmed to be almost homogeneous by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (Fig. 3A). TLC analysis of reaction products from the RG chain through the YesW or YesX reaction indicates that both enzymes directly interact with the RG backbone and release oligosaccharides with different degrees of polymerization (Fig. 3B). Absorbance at 235 nm in the reaction mixture containing RG-I incubated with purified YesW or YesX increased dependent on time and enzyme concentration (data not shown), indicating that reaction products contain unsaturated saccharides and that both YesW and YesX degrade RG-I through a β-elimination reaction. YesW and YesX were thus identified as an RG lyase.
TABLE 2.
Purification of YesW and YesX from E. coli cells
| Stepa | Total protein (mg) | Total activity (U) | Sp act (U/mg) | Yield (%) | Purification (fold) |
|---|---|---|---|---|---|
| YesW | |||||
| Cell extract | 4,819 | 13,616 | 2.83 | 100 | 1.0 |
| Ni2+-chelating Sepharose Fast Flow | 60.0 | 6,376 | 106 | 46.8 | 37.5 |
| Superdex 200pg | 13.0 | 2,700 | 208 | 19.8 | 73.5 |
| YesX | |||||
| Cell extract | 1,397 | 161 | 0.12 | 100 | 1.0 |
| Ni2+-chelating Sepharose Fast Flow | 11.0 | 24.0 | 2.18 | 14.9 | 18.2 |
| Superdex 200pg | 6.40 | 14.4 | 2.25 | 8.94 | 18.8 |
Purification procedures are detailed in Materials and Methods.
FIG. 3.
Properties of RG lyases. (A) SDS-PAGE of purified YesW and YesX followed by protein staining with Coomassie brilliant blue. Lane M contained molecular mass standards (from top to bottom)—synthetic polypeptides with molecular masses of 250, 150, 100, 75, 50, 37, and 25 kDa. Lane 1, purified recombinant YesW (5 μg protein); lane 2, purified recombinant YesX (5 μg protein). The arrow indicates the position of each enzyme. (B) Enzymatic degradation of RG-I. Degradation of the RG chain by YesW (upper) and YesX (lower). The reaction mixture consisted of 5 mg/ml of the RG chain, 50 mM of Tris-HCl (pH 7.5), 2 mM of CaCl2, and the purified enzyme. Reaction products were periodically sampled and analyzed on TLC plates, followed by staining with sulfuric acid. Reaction times (min) are indicated in the figure. The spots indicated by “2s” are unsaturated RG disaccharides with and without an addition of ammonium ion, which was used during preparation of the RG chain. Galacturonic acid (GalA) and rhamnose (Rha) were used for marker control. (C) Effects of pH and temperature on the activity and stability of YesW and YesX. The relative activity of YesW is indicated by a closed circle and solid line and that of YesX by an open circle and broken line. For optimal pH (left panel), activity was assayed with HEPES-NaOH (pH 6.5 and 7.2), Tris-HCl (pH 7.5, 8.0, and 8.5), and glycine-NaOH (pH 9.0, 10.0, and 11.0). Activity at pH 8.0 in YesW and pH 8.5 in YesX is taken as 100%. For optimal temperature (center panel), activity at 65°C in YesW and 60°C in YesX is taken as 100%. For thermal stability (right panel), purified enzymes were preincubated for 5 min at the temperatures indicated and residual enzyme activity was measured. The activity of enzymes preincubated at 4°C for 5 min is taken as 100%.
(i) Molecular mass and N-terminal amino acid sequence.
Molecular masses of YesW and YesX were determined to be 68 kDa and 77 kDa, respectively, through SDS-PAGE analysis (Fig. 3A). The N-terminal amino acid sequence of YesW was determined to be NH2-AARQMEALN, which corresponds to residues 38 to 46 of the N terminus of the YesW amino acid sequence published in DNA/protein databases (620 amino acid residues; accession number for GenPept, CAB12524), indicating that the N-terminal 37 residues of YesW are probably cleaved as a signal peptide in E. coli cells. The molecular mass of the mature form of YesW including the C-terminal histidine tag (8 amino acid residues; LEHHHHHH) was calculated as 64,444 Da (591 amino acid residues), which was in close agreement with SDS-PAGE results.
The N-terminal amino acid sequence of YesX was determined to be NH2-MKPKKR, which matched that of the YesX amino acid sequence published in the DNA/protein databases (612 amino acid residues; accession number for GenPept, CAB12525). The molecular mass of YesX including the C-terminal histidine tag (8 amino acid residues; LEHHHHHH) was calculated as 68,754 Da (620 amino acid residues), closely corresponding to that determined by SDS-PAGE.
In permeation chromatography on Superdex 200 10/300 GL, YesW and YesX purified from E. coli cells were eluted as proteins with molecular masses of about 65 kDa and 70 kDa, respectively (data not shown), indicating that both enzymes are active in the monomeric form.
(ii) pH and temperature.
Optimal pH and temperature and thermal stability for enzyme activity were investigated. Both YesW and YesX were most active at pH 8.0 to 8.5 and 60 to 65°C and stable below 50 to 55°C (Fig. 3C).
(iii) Chemicals.
Effects of different chemicals on enzyme activity were examined. After treatment of YesW and YesX with EDTA or EGTA at 10 mM, both lost enzyme activity (Table 3), indicating that YesW and YesX require bivalent metal ions for enzyme activity. Addition of bivalent metal ions such as Ca2+, Mn2+, Co2+, and Zn2+ to chelator-treated enzymes restored their enzyme activity (Table 3). Significant activation of RG lyases especially for YesX by Mn2+, Co2+, and Zn2+ was observed, suggesting that these ions rather than calcium ions are suitable for their enzyme activity. Thiol reagents at 1 mM (dithiothreitol, reduced glutathione, and 2-mercaptoethanol) and sugars at 5 mM (l-fucose, d-galactose, d-glucose, d-glucuronic acid, d-mannose, l-rhamnose, d-xylose, d-sucrose, 2-deoxyglucose, and d-glucosamine) had no significant effects on YesW and YesX activity. In contrast, half of the YesX activity was lost in the presence of 1 mM iodoacetic acid. d-Galacturonic acid reduced the activity of both enzymes to about their half-level (Table 4). Since d-galacturonic acid is a component of the substrate for YesW and YesX, the monosaccharide can be probably accommodated at the active site of both enzymes.
TABLE 3.
Effects of metal ions on YesW and YesX activity
| Pretreatmenta | Metal(s) added (1 mM concn) | Activity (%)b
|
|
|---|---|---|---|
| YesW | YesX | ||
| CaCl2 (1 mM) | CaCl2 | 100 | 100 |
| None | 0 | 7 | |
| EGTA (10 mM) | None | 0 | 0 |
| EDTA (10 mM) | None | 0 | 0 |
| AlCl3 | 0 | 16 | |
| MgCl2 | 4 | 0 | |
| MnCl2 | 134 | 661 | |
| CaCl2 | 54 | 89 | |
| CoCl2 | 54 | 111 | |
| ZnCl2 | 21 | 579 | |
YesW and YesX were treated with 10 mM EDTA or EGTA and then dialyzed against 20 mM of Tris-HCl (pH 7.5) containing 0.2 M NaCl. The activity of these enzymes pretreated with EDTA was determined in the presence of different metal ions at the indicated concentration.
The activity of chelator-treated enzymes in the presence of CaCl2 is taken as 100%.
TABLE 4.
Effects of different compounds on YesW and YesX activity
| Compound | Activity (%)a
|
|
|---|---|---|
| YesW | YesX | |
| None | 100 | 100 |
| Thiol reagents (1 mM) | ||
| Dithiothreitol | 115 | 107 |
| Glutathione | 92 | 98 |
| 2-Mercaptoethanol | 96 | 102 |
| Iodoacetic acid | 92 | 55 |
| Sugars (5 mM) | ||
| l-Fucose | 108 | 92 |
| d-Galactose | 89 | 104 |
| d-Glucose | 86 | 120 |
| d-Glucuronic acid | 117 | 125 |
| d-Mannose | 117 | 116 |
| l-Rhamnose | 108 | 124 |
| d-Xylose | 106 | 137 |
| d-Sucrose | 77 | 120 |
| d-Galacturonic acid | 43 | 59 |
| 2-Deoxyglucose | 73 | 102 |
| d-Glucosamine | 90 | 108 |
The enzyme assay was done in the presence of the compounds listed. Activity in the absence of a compound is taken as 100%.
(iv) Substrate specificity and kinetics.
To determine substrate specificity and the kinetic parameters of YesW and YesX, we assayed the enzyme using different substrates derived from pectin. Although RG-I appears to be the optimal substrate for YesW and YesX, both were active on pectin, pectic acid, and polygalacturonan, but inert on RG-I side-chain galactan (Table 5). This suggests that both YesW and YesX mainly act on RG-I backbone, although they show a slight activity on polygalacturonan. Kinetic parameters Km and Vmax of these enzymes toward RG-I were 0.35 mg/ml and 511 U/mg for YesW and 1.9 mg/ml and 11.3 U/mg for YesX.
TABLE 5.
Substrate specificity of YesW and YesX
| Substrate | Sp act in U/mg (%)a
|
|
|---|---|---|
| YesW | YesX | |
| RG-I | 133 (100) | 0.93 (100) |
| Pectin | 18 (14) | 0.03 (3) |
| Pectic acid | 21 (16) | 0.17 (18) |
| Polygalacturonan | 22 (17) | 0.21 (23) |
| Azo-galactan | ND | ND |
Activity toward RG-I is taken as 100%. ND, not detected.
(v) Mode of action.
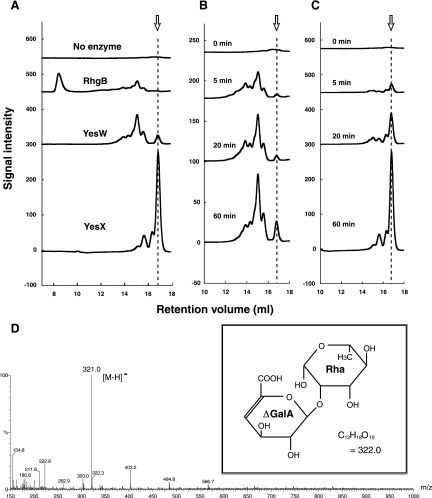
Since we found no reports on the mode of action of PL family 11 enzymes, we studied that of YesW and YesX using the RG chain—a mixture of different RG backbone sizes—as the substrate. The degradation profile of the RG chain by the reaction of PL family 4 RhgB or PL family 11 YesW or YesX is shown in Fig. 4A. The molecular mass of final products through the YesW or YesX reaction appears to be smaller than that of those through the RhgB reaction. RhgB acts on dodecasaccharide as a smallest substrate and releases tetrasaccharide as the smallest product (8, 37), so both YesW and YesX probably produce the disaccharide as a smallest product (arrow in Fig. 4A). To identify products as disaccharides, the product was purified and subjected to mass spectrometry. In the ESI-MS spectrum (Fig. 4D), the major peak of the deprotonated ion [M-H]− in negative mode indicates m/z 321.0, demonstrating that the molecular weight of the product is 322. Since the unsaturated RG disaccharide (4-deoxy-β-l-threo-hex-4-enopyranuronosyl-(1->2)-α-l-rhamnopyranoside; C12H18O10) has a molecular weight of 322 (Fig. 4D, inset), the smallest product released from the substrate through the YesW or YesX reaction was identified as the unsaturated RG disaccharide.
FIG. 4.
Degradation profiles of RG chain with RG lyases. (A) Analysis of final products from the RG chain through the RhgB, YesW, or YesX reaction by size-exclusion chromatography. Each product was analyzed after a 60-min reaction by Superdex peptide 10/300 GL. Product from the RG chain without enzyme added was used as the negative control. All profiles are overlaid, and the smallest product through the YesW or YesX reaction is indicated by an arrow. Degradation profiles of the RG chain with YesW (B) and YesX (C) were periodically analyzed. Reaction times were 0, 5, 20, and 60 min. The smallest product is indicated by an arrow. (D) ESI-MS spectrum of the smallest product released from the RG chain through the YesX reaction. The main peak in the negative mode coincided with the molecular mass of the deprotonated ion from the unsaturated RG disaccharide indicated in the box.
The profiles of the final reaction products differ for YesW and YesX (Fig. 4A), suggesting that these enzymes show different modes of action, so we studied the reaction products. As shown in Fig. 4B and 4C, YesW released both disaccharide and larger saccharides, while YesX released disaccharide as a major product during the reaction time tested. This indicates that YesW acts on the substrate endolytically and YesX exolytically. TLC analysis of YesW (Fig. 3B, upper panel) shows good agreement with the reaction profile of YesW from size-exclusion chromatography (Fig. 4B). In contrast to the YesW reaction, disaccharides are released from the RG chain even in the initial stage of the YesX reaction (Fig. 3B, lower panel, and Fig. 4C). Although some oligosaccharides other than disaccharides were observed on the TLC plate in the YesX reaction products (Fig. 3B, lower panel), YesX released unsaturated disaccharides as a major product, as shown in Fig. 4C. Therefore, the oligosaccharides on the TLC plate are thought to be saturated saccharides like other products after release of unsaturated disaccharides from substrates.
DISCUSSION
In our analysis of the B. subtilis system for plant cell wall polysaccharide (RG-I) degradation, we found that although gene functions in the three clusters (yes, yte, and ybc-ybd) inducibly expressed in strain 168 cells grown on RG-I have not yet been identified, yes and yte clusters played a crucial role in RG-I degradation because some of them (YesR, YesW, YesX, and YteR) were experimentally identified as RG-degrading enzymes. Since all three clusters contain genes coding for the ATP-binding cassette (ABC) transporter, gene products probably function as an importer for RG, especially for the unsaturated RG disaccharide.
In addition to the three clusters, up-regulated genes were found to encode putative sporulation-related proteins and flagellin. The growth experiment indicates that, compared to growth on glucose, pectin, and polygalacturonan, RG-I is not an excellent carbon source for B. subtilis (Fig. 1B). This may be why the bacterium tends to form endospores in RG-I minimal medium. B. subtilis shows high motility before the formation of endospores (32). Due to this motility, the gene coding for flagellin, a component of helical filament in flagellum, may be expressed in response to endospore formation.
In the CAZy database, RG lyases are grouped into PL families 4 and 11. Distinct from fully characterized PL family 4 RG lyase RhgB, YesW and YesX were identified as novel RG lyases in that both release the unsaturated RG disaccharide from the substrate. Products from RG through the reaction of PL family 11 RG lyases Rgl11A and Rgl11Y have yet to be determined (29, 43). Four PL family 11 RG lyases analyzed thus far (Rgl11A, Rgl11Y, YesW, and YesX) are similar in optimal pH, thermal stability, and calcium ion requirement but not in molecular mass or substrate specificity. The molecular masses of Rgl11A (93 kDa) and Rgl11Y (84 kDa) are greater than those of YesW (68 kDa) and YesX (78 kDa). Rgl11A contains two additional C-terminal domains (fibronectin type 3 module and cellulose-binding domain) (29), and Rgl11Y appears to be glycosylated due to a cellulosome component (43). Although both Rgl11A and Rgl11Y act on galactan (29, 43), the polymer was not degraded by either YesW or YesX. Since we recently determined the crystal structure of YesW (40; unpublished data), the difference in substrate specificity among PL family 11 RG lyases may be structurally explained in the near future.
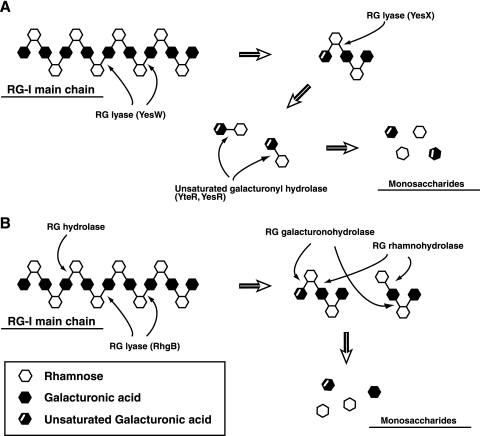
YesW and YesX are similar in optimal pH and temperature, thermal stability, metal ion requirement, behavior toward the different chemicals tested, and substrate specificity but not in kinetic parameters for RG-I (YesW, Km, 0.35 mg/ml, and Vmax, 511 U/mg; YesX, Km, 1.9 mg/ml, and Vmax, 11.3 U/mg) or mode of action (YesW, endotype; YesX, exotype). Kinetic analysis clearly indicates that YesW is more active than YesX on RG-I. Subcellular localization prediction using the PSORT program on the ExPASy proteomics server (http://www.psort.org) strongly suggests that both YesW and YesX are secreted into the extracellular fraction in B. subtilis. In fact, the N-terminal 37 residues of YesW probably function as a signal peptide in E. coli, although no N-terminal peptide of YesX is excised in E. coli. We also identified YesR and YteR as a novel intracellular enzyme, unsaturated galacturonyl hydrolase, which catalyzes the hydrolytic reaction and cleaves the glycosidic bond between unsaturated galacturonic acid and rhamnose of the unsaturated RG disaccharide (17). Based on previous studies and the present work, we show the enzymatic route for the complete degradation of the RG-I main chain in B. subtilis in Fig. 5A. B. subtilis secretes YesW, which catalyzes the initial cleavage of the glycosidic bond in the RG-I main chain, and the resultant oligosaccharides with different degrees of polymerization are converted to the unsaturated RG disaccharide through the extracellular exotype YesX reaction. The disaccharide is finally incorporated through the putative ABC transporter and degraded into its constituent monosaccharides through the reaction of intracellular unsaturated galacturonyl hydrolases YesR and YteR. During depolymerization of the RG-I main chain, RG acetylesterase-homologous proteins (YesT and YesY) and β-galactosidase-homologous YesZ encoded in the yes cluster possibly function as the enzymes responsible for the removal of the acetyl group from the RG-I main chain and for the degradation of the RG-I side chain, respectively. This enzymatic route for RG-I degradation in strain 168 differs significantly from that in A. aculeatus strain KSM 510. In strain KSM 510 (Fig. 5B), RG hydrolase cleaves the α-1,2 glycosidic bond between galactopyranouronic acid and rhamnose of the RG backbone, and RG lyase cleaves the α-1,4 glycosidic bond between rhamnose and galactopyranouronic acid. The resultant products (saturated and unsaturated tetrasaccharides as the smallest products) by RG hydrolase and RG lyase are degraded into their constituent monosaccharides through the reactions of RG rhamnohydrolase and RG galacturonohydrolase, which release rhamnose and galacturonic acid, respectively (33-35, 37, 50). The strain 168 genome, however, contains no genes coding for proteins similar to RG rhamnohydrolase and RG galacturonohydrolase.
FIG. 5.
Bacterial and fungus systems for degradation of the RG main chain. (A) B. subtilis. (B) A. aculeatus. Thin arrows indicate the cleavage site for RG-I-degrading enzymes, and thick arrows indicate the degradation pathway. Details are given in the text.
Strain 168 RG lyases, especially YesX, release the unsaturated RG disaccharide from substrates. Oligosaccharides called oligosaccharins show physiological functions such as a bifidus factor, an elicitor of plant growth, a growth enhancer of human endothelial cells and keratinocytes, and an inducer of cytokine production from mouse macrophage cells (1, 5, 18, 19, 21, 22, 27). The unsaturated RG disaccharide “lepidimoide” has been isolated from seeds of cress (Lepidium sativum L.) and shown to function as an allelopathic factor for plants (12, 53). Hirose et al. developed a convenient chemical method to synthesize the unsaturated RG disaccharide from RG (13), and Saranpuetti et al. found a way to release the disaccharide from okra polysaccharide in the culture fluid of the fungus Colletotrichum sp. (48). YesW and YesX may provide further alternatives for the production of the disaccharide.
Acknowledgments
We thank Novozymes A/S for kindly supplying A. aculeatus RG lyase RhgB.
This work was supported in part by grants-in-aid from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (no. 18031018 to K.M. and no. 18580075 to W.H.) and by COE for Microbial-Process Development Pioneering Future Production Systems from the Ministry of Education, Culture, Sports, Science, and Technology of Japan. Part of this work was also supported by Research Fellowships from the Japan Society for the Promotion of Science for Young Scientists (A.O.).
Footnotes
Published ahead of print on 20 April 2007.
REFERENCES
- 1.Akiyama, H., T. Endo, R. Nakakita, K. Murata, Y. Yonemoto, and K. Okayama. 1992. Effect of depolymerized alginates on the growth of bifidobacteria. Biosci. Biotechnol. Biochem. 56:355-356. [DOI] [PubMed] [Google Scholar]
- 2.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 3.Carpita, N. C., and D. M. Gibeaut. 1993. Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. Plant J. 3:1-30. [DOI] [PubMed] [Google Scholar]
- 4.Clements, L. D., U. N. Streips, and B. S. Miller. 2002. Differential proteomic analysis of Bacillus subtilis nitrate respiration and fermentation in defined medium. Proteomics 2:1724-1734. [DOI] [PubMed] [Google Scholar]
- 5.Darvill, A., C. Bergmann, F. Cervone, G. De Lorenzo, K. S. Ham, M. D. Spiro, W. S. York, and P. Albersheim. 1994. Oligosaccharins involved in plant growth and host-pathogen interactions. Biochem. Soc. Symp. 60:89-94. [PubMed] [Google Scholar]
- 6.Darvill, A. G., M. McNeil, and P. Albersheim. 1978. Structure of plant cell walls. VIII. A new pectic polysaccharide. Plant Physiol. 62:418-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davies, G., and B. Henrissat. 1995. Structures and mechanisms of glycosyl hydrolases. Structure 3:853-859. [DOI] [PubMed] [Google Scholar]
- 8.Deng, C., M. A. O'Neill, and W. S. York. 2006. Selective chemical depolymerization of rhamnogalacturonans. Carbohydr. Res. 341:474-484. [DOI] [PubMed] [Google Scholar]
- 9.de Vries, R. P., and J. Visser. 2001. Aspergillus enzymes involved in degradation of plant cell wall polysaccharides. Microbiol. Mol. Biol. Rev. 65:497-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dubnau, D. 1991. Genetic competence in Bacillus subtilis. Microbiol. Rev. 55:395-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gummadi, S. N., and D. S. Kumar. 2005. Microbial pectic transeliminases. Biotechnol. Lett. 27:451-458. [DOI] [PubMed] [Google Scholar]
- 12.Hasegawa, K., J. Mizutani, S. Kosemura, and S. Yamamura. 1992. Isolation and identification of lepidimoide, a new allelopathic substance from mucilage of germinated cress seeds. Plant Physiol. 100:1059-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirose, K., K. Endo, K. Hasegawa, K. Yamada, H. Matsumoto, K. Ishizuka, K. Miyamoto, S. Kosemura, S. Yamamura, and J. Mizutani. 2004. A convenient synthesis of lepidimoide from okra mucilage and its growth-promoting activity in hypocotyls. Carbohydr. Res. 339:9-19. [DOI] [PubMed] [Google Scholar]
- 14.Hugouvieux-Cotte-Pattat, N. 2004. The RhaS activator controls the Erwinia chrysanthemi 3937 genes rhiN, rhiT and rhiE involved in rhamnogalacturonan catabolism. Mol. Microbiol. 51:1361-1374. [DOI] [PubMed] [Google Scholar]
- 15.Hugouvieux-Cotte-Pattat, N., G. Condemine, W. Nasser, and S. Reverchon. 1996. Regulation of pectinolysis in Erwinia chrysanthemi. Annu. Rev. Microbiol. 50:213-257. [DOI] [PubMed] [Google Scholar]
- 16.Irizarry, R. A., B. Hobbs, F. Collin, Y. D. Beazer-Barclay, K. J. Antonellis, U. Scherf, and T. P. Speed. 2003. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4:249-264. [DOI] [PubMed] [Google Scholar]
- 17.Itoh, T., A. Ochiai, B. Mikami, W. Hashimoto, and K. Murata. 2006. A novel glycoside hydrolase family 105: the structure of family 105 unsaturated rhamnogalacturonyl hydrolase complexed with a disaccharide in comparison with family 88 enzyme complexed with the disaccharide. J. Mol. Biol. 360:573-585. [DOI] [PubMed] [Google Scholar]
- 18.Iwamoto, M., M. Kurachi, T. Nakashima, D. Kim, K. Yamaguchi, T. Oda, Y. Iwamoto, and T. Muramatsu. 2005. Structure-activity relationship of alginate oligosaccharides in the induction of cytokine production from RAW264.7 cells. FEBS Lett. 579:4423-4429. [DOI] [PubMed] [Google Scholar]
- 19.Iwamoto, Y., X. Xu, T. Tamura, T. Oda, and T. Muramatsu. 2003. Enzymatically depolymerized alginate oligomers that cause cytotoxic cytokine production in human mononuclear cells. Biosci. Biotechnol. Biochem. 67:258-263. [DOI] [PubMed] [Google Scholar]
- 20.Kato, J., M. Kisumi, T. Takagi, and I. Chibata. 1977. Increase in arginine and citrulline production by 6-azauracil-resistant mutants of Bacillus subtilis. Appl. Environ. Microbiol. 34:689-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawada, A., N. Hiura, M. Shiraiwa, S. Tajima, M. Hiruma, K. Hara, A. Ishibashi, and H. Takahara. 1997. Stimulation of human keratinocyte growth by alginate oligosaccharides, a possible co-factor for epidermal growth factor in cell culture. FEBS Lett. 408:43-46. [DOI] [PubMed] [Google Scholar]
- 22.Kawada, A., N. Hiura, S. Tajima, and H. Takahara. 1999. Alginate oligosaccharides stimulate VEGF-mediated growth and migration of human endothelial cells. Arch. Dermatol. Res. 291:542-547. [DOI] [PubMed] [Google Scholar]
- 23.Kimura, K., L. S. Tran, and Y. Itoh. 2004. Roles and regulation of the glutamate racemase isogenes, racE and yrpC, in Bacillus subtilis. Microbiology 150:2911-2920. [DOI] [PubMed] [Google Scholar]
- 24.Kimura, K., L. S. Tran, I. Uchida, and Y. Itoh. 2004. Characterization of Bacillus subtilis γ-glutamyltransferase and its involvement in the degradation of capsule poly-γ-glutamate. Microbiology 150:4115-4123. [DOI] [PubMed] [Google Scholar]
- 25.Kobayashi, K., S. D. Ehrlich, A. Albertini, G. Amati, K. K. Andersen, M. Arnaud, K. Asai, S. Ashikaga, S. Aymerich, P. Bessieres, F. Boland, S. C. Brignell, S. Bron, K. Bunai, J. Chapuis, L. C. Christiansen, A. Danchin, M. Debarbouille, E. Dervyn, E. Deuerling, K. Devine, S. K. Devine, O. Dreesen, J. Errington, S. Fillinger, S. J. Foster, Y. Fujita, A. Galizzi, R. Gardan, C. Eschevins, T. Fukushima, K. Haga, C. R. Harwood, M. Hecker, D. Hosoya, M. F. Hullo, H. Kakeshita, D. Karamata, Y. Kasahara, F. Kawamura, K. Koga, P. Koski, R. Kuwana, D. Imamura, M. Ishimaru, S. Ishikawa, I. Ishio, D. Le Coq, A. Masson, C. Mauel, R. Meima, R. P. Mellado, A. Moir, S. Moriya, E. Nagakawa, H. Nanamiya, S. Nakai, P. Nygaard, M. Ogura, T. Ohanan, M. O'Reilly, M. O'Rourke, Z. Pragai, H. M. Pooley, G. Rapoport, J. P. Rawlins, L. A. Rivas, C. Rivolta, A. Sadaie, Y. Sadaie, M. Sarvas, T. Sato, H. H. Saxild, E. Scanlan, W. Schumann, J. F. Seegers, J. Sekiguchi, A. Sekowska, S. J. Seror, M. Simon, P. Stragier, R. Studer, H. Takamatsu, T. Tanaka, M. Takeuchi, H. B. Thomaides, V. Vagner, J. M. van Dijl, K. Watabe, A. Wipat, H. Yamamoto, M. Yamamoto, Y. Yamamoto, K. Yamane, K. Yata, K. Yoshida, H. Yoshikawa, U. Zuber, and N. Ogasawara. 2003. Essential Bacillus subtilis genes. Proc. Natl. Acad. Sci. USA 100:4678-4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessieres, A. Bolotin, S. Borchert, R. Borriss, L. Boursier, A. Brans, M. Braun, S. C. Brignell, S. Bron, S. Brouillet, C. V. Bruschi, B. Caldwell, V. Capuano, N. M. Carter, S.-K. Choi, J.-J. Codani, I. F. Connerton, N. J. Cummings, R. A. Daniel, F. Denizot, K. M. Devine, A. Dusterhoft, S. D. Ehrlich, P. T. Emmerson, K. D. Entian, J. Errington, C. Fabret, E. Ferrari, D. Foulger, C. Fritz, M. Fujita, Y. Fujita, S. Fuma, A. Galizzi, N. Galleron, S.-Y. Ghim, P. Glaser, A. Goffeau, E. J. Golightly, G. Grandi, G. Guiseppi, B. J. Guy, K. Haga, J. Haiech, C. R. Harwood, A. Henaut, H. Hilbert, S. Holsappel, S. Hosono, M.-F. Hullo, M. Itaya, L. Jones, B. Joris, D. Karamata, Y. Kasahara, M. Klaerr-Blanchard, C. Klein, Y. Kobayashi, P. Koetter, G. Koningstein, S. Krogh, M. Kumano, K. Kurita, A. Lapidus, S. Lardinois, J. Lauber, V. Lazarevic, S.-M. Lee, A. Levine, H. Liu, S. Masuda, C. Mauel, C. Medigue, N. Medina, R. P. Mellado, M. Mizuno, D. Moestl, S. Nakai, M. Noback, D. Noone, M. O'Reilly, K. Ogawa, A. Ogiwara, B. Oudega, S.-H. Park, V. Parro, T. M. Pohl, D. Portetelle, S. Porwollik, A. M. Prescott, E. Presecan, P. Pujic, B. Purnelle, G. Rapoport, M. Rey, S. Reynolds, M. Rieger, C. Rivolta, E. Rocha, B. Roche, M. Rose, Y. Sadaie, T. Sato, E. Scanlan, S. Schleich, R. Schroeter, F. Scoffone, J. Sekiguchi, A. Sekowska, S. J. Seror, P. Serror, B.-S. Shin, B. Soldo, A. Sorokin, E. Tacconi, T. Takagi, H. Takahashi, K. Takemaru, M. Takeuchi, A. Tamakoshi, T. Tanaka, P. Terpstra, A. Tognoni, V. Tosato, S. Uchiyama, M. Vandenbol, F. Vannier, A. Vassarotti, A. Viari, R. Wambutt, E. Wedler, H. Wedler, T. Weitzenegger, P. Winters, A. Wipat, H. Yamamoto, K. Yamane, K. Yasumoto, K. Yata, K. Yoshida, H.-F. Yoshikawa, E. Zumstein, H. Yoshikawa, and A. Danchin. 1997. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 27.Kurachi, M., T. Nakashima, C. Miyajima, Y. Iwamoto, T. Muramatsu, K. Yamaguchi, and T. Oda. 2005. Comparison of the activities of various alginates to induce TNF-α secretion in RAW264.7 cells. J. Infect. Chemother. 11:199-203. [DOI] [PubMed] [Google Scholar]
- 28.Linhardt, R. J., P. M. Galliher, and C. L. Cooney. 1986. Polysaccharide lyases. Appl. Biochem. Biotechnol. 12:135-176. [DOI] [PubMed] [Google Scholar]
- 29.McKie, V. A., J. P. Vincken, A. G. Voragen, L. A. van den Broek, E. Stimson, and H. J. Gilbert. 2001. A new family of rhamnogalacturonan lyases contains an enzyme that binds to cellulose. Biochem. J. 355:167-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McNeil, M., A. G. Darvill, and P. Albersheim. 1980. Structure of plant cell walls. X. Rhamnogalacturonan I, a structurally complex pectic polysaccharide in the walls of suspension-cultured sycamore cells. Plant Physiol. 66:1128-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McNeil, M., A. G. Darvill, S. C. Fry, and P. Albersheim. 1984. Structure and function of the primary cell walls of plants. Annu. Rev. Biochem. 53:625-663. [DOI] [PubMed] [Google Scholar]
- 32.Mirel, D. B., and M. J. Chamberlin. 1989. The Bacillus subtilis flagellin gene (hag) is transcribed by the σ28 form of RNA polymerase. J. Bacteriol. 171:3095-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mutter, M., G. Beldman, S. M. Pitson, H. A. Schols, A. G. Voragen, I. J. Colquhoun, and E. J. Bakx. 1998. Rhamnogalacturonan α-d-galactopyranosyluronohydrolase. An enzyme that specifically removes the terminal nonreducing galacturonosyl residue in rhamnogalacturonan regions of pectin. Plant Physiol. 117:153-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mutter, M., G. Beldman, H. A. Schols, and A. G. Voragen. 1994. Rhamnogalacturonan α-l-rhamnopyranohydrolase. A novel enzyme specific for the terminal nonreducing rhamnosyl unit in rhamnogalacturonan regions of pectin. Plant Physiol. 106:241-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mutter, M., I. J. Colquhoun, G. Beldman, H. A. Schols, E. J. Bakx, and A. G. Voragen. 1998. Characterization of recombinant rhamnogalacturonan α-l-rhamnopyranosyl-(1,4)-α-d-galactopyranosyluronide lyase from Aspergillus aculeatus. An enzyme that fragments rhamnogalacturonan I regions of pectin. Plant Physiol. 117:141-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mutter, M., I. J. Colquhoun, H. A. Schols, G. Beldman, and A. G. Voragen. 1996. Rhamnogalacturonase B from Aspergillus aculeatus is a rhamnogalacturonan α-l-rhamnopyranosyl-(1→4)-α-d-galactopyranosyluronide lyase. Plant Physiol. 110:73-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mutter, M., C. M. Renard, G. Beldman, H. A. Schols, and A. G. Voragen. 1998. Mode of action of RG-hydrolase and RG-lyase toward rhamnogalacturonan oligomers. Characterization of degradation products using RG-rhamnohydrolase and RG-galacturonohydrolase. Carbohydr. Res. 311:155-164. [DOI] [PubMed] [Google Scholar]
- 38.Nakamura, A., H. Furuta, H. Maeda, Y. Nagamatsu, and A. Yoshimoto. 2001. Analysis of structural components and molecular construction of soybean soluble polysaccharides by stepwise enzymatic degradation. Biosci. Biotechnol. Biochem. 65:2249-2258. [DOI] [PubMed] [Google Scholar]
- 39.Nasser, W., F. Chalet, and J. Robert-Baudouy. 1990. Purification and characterization of extracellular pectate lyase from Bacillus subtilis. Biochimie 72:689-695. [DOI] [PubMed] [Google Scholar]
- 40.Ochiai, A., M. Yamasaki, T. Itoh, B. Mikami, W. Hashimoto, and K. Murata. 2006. Crystallization and preliminary X-ray analysis of the rhamnogalacturonan lyase YesW from Bacillus subtilis strain 168, a member of polysaccharide lyase family 11. Acta Crystallogr. F Struct. Biol. Crystalliz. Comm. 62:438-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Olempska-Beer, Z. S., R. I. Merker, M. D. Ditto, and M. J. DiNovi. 2006. Food-processing enzymes from recombinant microorganisms—a review. Regul. Toxicol. Pharmacol. 45:144-158. [DOI] [PubMed] [Google Scholar]
- 42.O'Neill, M. A., D. Warrenfeltz, K. Kates, P. Pellerin, T. Doco, A. G. Darvill, and P. Albersheim. 1996. Rhamnogalacturonan-II, a pectic polysaccharide in the walls of growing plant cell, forms a dimer that is covalently cross-linked by a borate ester. In vitro conditions for the formation and hydrolysis of the dimer. J. Biol. Chem. 271:22923-22930. [DOI] [PubMed] [Google Scholar]
- 43.Pagès, S., O. Valette, L. Abdou, A. Bélaïch, and J. P. Bélaïch. 2003. A rhamnogalacturonan lyase in the Clostridium cellulolyticum cellulosome. J. Bacteriol. 185:4727-4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Piggot, P. J., and D. W. Hilbert. 2004. Sporulation of Bacillus subtilis. Curr. Opin. Microbiol. 7:579-586. [DOI] [PubMed] [Google Scholar]
- 45.Sakamoto, T., R. A. Hours, and T. Sakai. 1994. Purification, characterization, and production of two pectic transeliminases with protopectinase activity from Bacillus subtilis. Biosci. Biotechnol. Biochem. 58:353-358. [DOI] [PubMed] [Google Scholar]
- 46.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 47.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saranpuetti, C., M. Tanaka, T. Sone, K. Asano, and F. Tomita. 2006. Determination of enzymes from Colletotrichum sp. AHU9748 essential for lepidimoide production from okra polysaccharide. J. Biosci. Bioeng. 102:452-456. [DOI] [PubMed] [Google Scholar]
- 49.Schallmey, M., A. Singh, and O. P. Ward. 2004. Developments in the use of Bacillus species for industrial production. Can. J. Microbiol. 50:1-17. [DOI] [PubMed] [Google Scholar]
- 50.Schols, H. A., C. C. J. M. Geraeds, M. F. Searle-van Leeuwen, F. J. M. Kormelink, and A. G. J. Voragen. 1990. Rhamnogalacturonase: a novel enzyme that degrades the hairy regions of pectins. Carbohydr. Res. 206:105-115. [Google Scholar]
- 51.Stanley, N. R., and B. A. Lazazzera. 2005. Defining the genetic differences between wild and domestic strains of Bacillus subtilis that affect poly-γ-dl-glutamic acid production and biofilm formation. Mol. Microbiol. 57:1143-1158. [DOI] [PubMed] [Google Scholar]
- 52.Thakur, B. R., R. K. Singh, and A. K. Handa. 1997. Chemistry and uses of pectin—a review. Crit. Rev. Food Sci. Nutr. 37:47-73. [DOI] [PubMed] [Google Scholar]
- 53.Yamada, K., H. Matsumoto, K. Ishizuka, K. Miyamoto, S. Kosemura, S. Yamamura, K. Hasegawa, and J. Mizutani. 1998. Lepidimoide promotes light-induced chlorophyll accumulation in cotyledons of sunflower seedlings. J. Plant Growth Regul. 17:215-219. [DOI] [PubMed] [Google Scholar]