Abstract
In response to osmotic stress, proline is accumulated in many bacterial and plant cells as an osmoprotectant. The yeast Saccharomyces cerevisiae induces trehalose or glycerol synthesis but does not increase intracellular proline levels during various stresses. Using a proline-accumulating mutant, we previously found that proline protects yeast cells from damage by freezing, oxidative, or ethanol stress. This mutant was recently shown to carry an allele of PRO1 which encodes the Asp154Asn mutant γ-glutamyl kinase (GK), the first enzyme of the proline biosynthetic pathway. Here, enzymatic analysis of recombinant proteins revealed that the GK activity of S. cerevisiae is subject to feedback inhibition by proline. The Asp154Asn mutant was less sensitive to feedback inhibition than wild-type GK, leading to proline accumulation. To improve the enzymatic properties of GK, PCR random mutagenesis in PRO1 was employed. The mutagenized plasmid library was introduced into an S. cerevisiae non-proline-utilizing strain, and proline-overproducing mutants were selected on minimal medium containing the toxic proline analogue azetidine-2-carboxylic acid. We successfully isolated several mutant GKs that, due to extreme desensitization to inhibition, enhanced the ability to synthesize proline better than the Asp154Asn mutant. The amino acid changes were localized at the region between positions 142 and 154, probably on the molecular surface, suggesting that this region is involved in allosteric regulation. Furthermore, we found that yeast cells expressing Ile150Thr and Asn142Asp/Ile166Val mutant GKs were more tolerant to freezing stress than cells expressing the Asp154Asn mutant.
In response to environmental osmotic stresses, a variety of bacteria and plants accumulate proline as an osmoprotectant (12, 14). In the yeast Saccharomyces cerevisiae, however, the intracellular proline content does not increase during high osmotic stress. In agreement with this observation, recent genome-wide expression analyses of S. cerevisiae showed that a set of genes involved in proline metabolism did not respond to various environmental changes, including changes in temperature, freezing, ethanol content, oxidation, pH, and osmolarity (1, 9, 28, 30, 38, 43). As it is in other organisms, the synthesis of trehalose (5, 22, 55), glycerol (3, 6, 31), or glycogen (43) is induced by many stress conditions in S. cerevisiae. These results suggest that during the evolutionary process yeast cells may choose trehalose or glycerol, not proline, as a stress protectant. In addition to being an osmoprotectant, proline has been shown to have multiple functions; it enhances the stability of proteins and membranes during freezing, dehydration, or high temperature (40), it lowers the melting temperature of DNA due to destabilization of the double helix during salinity stress tests (35), it inhibits aggregation during protein refolding (42), and it reduces the levels of reactive oxygen species under oxidative stress conditions (10, 20), although the in vivo protective mechanism is poorly understood. We previously found that proline protects yeast cells from damage by freezing, desiccation, oxidative, or ethanol stress (25-27, 48-52).
S. cerevisiae synthesizes proline from glutamate via the same pathway found in bacteria, which consists of three enzymes: γ-glutamyl kinase (GK) (the PRO1 gene product, EC 2.7.2.11), γ-glutamyl phosphate reductase (GPR) (the PRO2 gene product, EC 1.2.1.41), and Δ1-pyrroline-5-carboxylate (P5C) reductase (the PRO3 gene product, EC 1.5.1.2) (8, 53). Proline is converted to glutamate within mitochondria in two steps by the enzymes proline oxidase (the PUT1 gene product, EC 1.4.3.2) and P5C dehydrogenase (the PUT2 gene product, EC 1.5.1.12) (7, 56). Previous studies indicated that the PRO1 and PRO3 genes are not responsive to either proline or glutamate in the medium, but PRO1 is regulated by the general amino acid control system (8, 24). In bacteria and plants, proline synthesis is regulated by end product inhibition of GK (29, 46) and the GK domain of P5C synthetase (47, 58), respectively. Mutants that overproduced proline due to diminished sensitivity to the feedback inhibition of GK clearly showed enhanced osmotolerance (13, 20, 30, 39). These observations suggest that allosteric control of GK plays a key role in proline synthesis in bacteria and plants. However, the characteristics of S. cerevisiae GK have not been reported, probably because yeast cells do not accumulate proline in response to environmental stresses. To examine the stress-protective effect of proline, we previously isolated a mutant, derived from mutants of S. cerevisiae resistant to the proline analogue azetidine-2-carboxylic acid (AZC), which exhibited both proline accumulation and freeze tolerance (48). This mutant was recently found to carry an allele of PRO1 encoding GK and to have a single amino acid replacement at position 154 (Asp replaced by Asn) (26, 52).
Our objectives in this study were (i) to characterize the wild-type GK of S. cerevisiae, (ii) to elucidate the mechanism of proline accumulation by the D154N mutant GK, and (iii) to isolate and analyze mutant GKs which increase intracellular proline levels. We found that S. cerevisiae GK is subject to feedback inhibition by proline and that the D154N mutant is less sensitive than the wild-type enzyme, leading to proline accumulation. Using error-prone PCR random mutagenesis, we then successfully isolated several mutant GKs that, due to desensitization to feedback inhibition, enhance the ability to synthesize proline. In addition, we examined whether the increased intracellular proline levels correlate with a higher tolerance to freezing and other stresses.
MATERIALS AND METHODS
Strains and plasmids.
The S. cerevisiae strain with an S288C background used in this study was INVDput1pro1 (MATα his3-Δ1 leu2 trp1-289 ura3-52 put1::URA3 pro1::CgHIS3), which is a PUT1 and PRO1 disruptant (27). The 2μm-based high-copy-number plasmids pAD-WTPRO1 and pAD-D154NPRO1, which contain the selection marker LEU2, were used for expressing wild-type and Asp154Asn mutant GKs, respectively, in S. cerevisiae (52). Plasmid pTV-PRO2, which contains the selection marker TRP1, was used for expressing wild-type GPR in S. cerevisiae (52).
Escherichia coli strain DH5α [F− λ− φ80dlacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17(rK− mK+) supE44 thi-1 gyrA96] and strain JM109 [endA recA1 gyrA96 thi hsdR17(rK− mK+) relA1 supE44 λ− Δ(lac-proAB) (F′ traD36 proAB lacIqZΔM15)] were used for construction of the plasmids. The isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible vector TAGZyme pQE2 (QIAGEN, Valencia, CA) with a sequence encoding six consecutive His residues at the 5′ end of the cloning sites was used for expression of PRO1 and PRO2 in E. coli strain M15(pREP4) (lac ara gal mtl recA+ uvr+ [lacI Kanr]). E. coli proline auxotrophic strain KC1325 [BL21(DE3)(pLysS), which carries the proB1658::Tn10 insertion] (56) (supplied by L. N. Csonka) was used for PRO1 expression.
Culture media.
The media used for growth of S. cerevisiae were a synthetic minimal SD medium (2% glucose, 0.67% Bacto yeast nitrogen base without amino acids [Difco Laboratories, Detroit, MI]) and a nutrient YPD medium (2% glucose, 1% yeast extract, 2% peptone). The SD medium contains 10 mM ammonium sulfate as the nitrogen source. When appropriate, required amino acids were added to the media for auxotrophic strains. The E. coli recombinant cells were grown in Luria-Bertani (LB) medium (41) containing ampicillin (50 μg/ml) or in M9 medium (41) plus 2% Casamino Acids (M9CA) containing ampicillin (50 μg/ml) and kanamycin (25 μg/ml). If necessary, 2% agar was added to solidify the medium.
PCR random mutagenesis.
The full-length PRO1 gene was amplified from pAD-WTPRO1 (52) by error-prone PCR with 5′-TAG ACG TCA AGC TTG CGA ACT AAT GCT TTC TCG-3′ and 5′-CTG GAC TAG GGA GCT CGT TCG TTT CAA CGA GGT GGG A-3′ (the underlining indicates the positions of HindIII and SacI sites, respectively). Ten nanograms of pAD-WTPRO1 DNA was added as a template to a solution containing 10 mM Tris-HCl (pH 9.0), 50 mM KCl, 1.5 mM MgCl2, 0.1% Triton X-100, 0.5 mM dATP, 0.5 mM dTTP, 0.2 mM dCTP, 0.2 mM dGTP, 1.0 μM each of the two primers, 0.25 μl of Taq DNA polymerase (5 U/μl), and enough distilled water to bring the total volume to 50 μl. Twenty-five cycles (94°C for 1 min, 55°C for 1 min, and 72°C for 1 min) of PCR were carried out. The unique 1.4-kb amplified band was digested with HindIII and SacI and ligated to the HindIII and SacI sites of pAD4. The ligated DNA was used to transform E. coli DH5α on LB solid medium containing ampicillin, and plasmid DNAs prepared from the ampicillin-resistant colonies (approximately 12,000) were used to isolate mutant GKs as the mutagenized plasmid library.
Intracellular proline contents and freeze tolerance test.
Yeast cells were grown to the stationary phase in 5 ml of SD medium at 30°C for 48 h with shaking. For determination of the intracellular proline content, 4 ml of the cell suspension (approximately 2 × 108 cells) was removed, and the cells were washed twice and suspended in 0.4 ml of distilled water. The cells were transferred to a boiling water bath and incubated for 10 min, and intracellular amino acids were extracted. After centrifugation (5 min at 15,000 × g), each supernatant was subsequently quantified with an amino acid analyzer (model L-8500A; Hitachi, Tokyo, Japan). The proline content was expressed as a percentage of the dry weight.
For the freeze tolerance test, 1 ml of cell suspension was removed, and the cells were washed twice and suspended in 1 ml of distilled water. Aliquots (0.1 ml) of the cell suspension (approximately 5 × 106 cells) were stored at −20°C. After 0, 1, 2, 5, and 11 days, samples of the frozen cells were thawed at 30°C for 15 min, serial dilutions in distilled water were prepared, and aliquots (0.1 ml) were plated on YPD medium plates. After incubation at 30°C for 2 days, the numbers of colonies were counted and the survival cell rates were expressed as percentages, which were calculated as follows: [(number of colonies after freezing at −20°C)/(number of colonies before freezing)] × 100.
Construction of plasmids for expression of PRO1 and PRO2 in E. coli.
A DNA fragment of PRO1 was prepared by PCR with pAD-WTPRO1 and oligonucleotide primers 5′-GGC CGA GCT CAG ATG AAG GAT GCT AAT GAG AG-3′ and 5′-CGG GGT ACC GTT TCA ACG AGG TGG GAA TG-3′, and a DNA fragment of PRO2 was amplified from pTV-PRO2 with oligonucleotide primers 5′-GGC CGA GCT CAG ATG TCC AGT TCA CAA CAA ATA GCC-3′ and 5′-CGG GGT ACC ACG TCC TTC ATA CAT GG-3′ (the underlining indicates the positions of SacI and KpnI sites, respectively). The unique 1.3- and 1.4-kb amplified bands were digested with SacI and KpnI and then subcloned into TAGZyme pQE2 to construct pQE2-PRO1 and pQE2-PRO2, respectively. The nucleotide sequences were confirmed by DNA sequencing. The plasmids harboring the mutant PRO1 genes were constructed by the method described above.
Expression and purification of the recombinant GK and GPR in E. coli.
E. coli M15(pREP4) was transformed with pQE2-PRO1 and pQE2-PRO2, and then the transformed cells were grown at 37°C in 200 ml of M9CA containing ampicillin and kanamycin. When the absorbance at 600 nm reached 0.5, IPTG was added to the culture medium to a final concentration 1 mM to induce gene expression. After cultivation for 4 h at 37°C, the cells were harvested and suspended in 20 ml of ice-cold solution buffer [50 mM NaH2PO4, 50 mM Na2HPO4 (pH 7.5), 500 mM (NH4)2SO4, 5 mM β-mercaptoethanol]. Glass beads (1 g) were added, and cells were broken with a multibead shocker (model MB501; Yasui Kikai, Osaka, Japan) with cooling. After centrifugation (30 min at 10,000 × g), the soluble fraction of the supernatant was purified using Ni-nitrilotriacetic acid-agarose (QIAGEN) according to the procedure recommended by the supplier. The His-tagged fusion proteins were applied to an Ni-nitrilotriacetic acid-agarose column equilibrated with the same buffer containing 10 mM imidazole. The column was washed with 50 mM imidazole in the same buffer, and proteins were eluted with 300 mM imidazole in the same buffer. Fractions including proteins were pooled, and then imidazole was removed in 50 mM Tris-HCl buffer (pH 7.5) with a PD-10 column (GE Healthcare, Piscataway, NJ). Protein concentrations were determined using a Bio-Rad protein assay kit (Hercules, CA) with bovine serum albumin as the standard protein. Mutant GKs were purified by the method described above.
Enzyme assay.
The GK activity was assayed by the procedure of Smith et al. (46), with some modifications. The reaction mixture (final volume, 0.25 ml) contained the following: 50 mM Tris base containing 100 mM hydroxylamine-HCl (pH 7.0), 50 mM glutamate, 10 mM ATP, 20 mM MgCl2, and the purified recombinant enzymes (the molar ratio of GPR to GK was 5:1) plus water. The reaction was carried out at 30°C for 15 to 45 min and then terminated by addition of 1 ml of stop solution (55 g of FeCl3·6H2O, 20 g of trichloroacetic acid, and 21 ml of 12 N HCl per liter). Precipitated proteins were removed by centrifugation, and the absorbance at 535 nm was recorded against a blank identical to the one mentioned above but lacking ATP. The amount of γ-glutamyl hydroxamate was determined from the absorbance at 535 nm by comparison with a standard curve for γ-glutamyl hydroxamate (Sigma Chemical, St. Louis, MO). One unit of activity was defined as the amount of enzyme required to produce 1 μmol of γ-glutamyl hydroxamate per min.
The GPR activity was assayed as described by Hayzer and Leisinger (18). The activity in the forward (biosynthetic) direction was not detectable due to the lability of γ-glutamyl phosphate. To distinguish this reaction from the P5C dehydrogenase reaction, which converts P5C to proline with the aid of NAD+, we measured the reverse reaction of GPR by phosphate-dependent reduction of NADP+ with glutamate-γ-semialdehyde (derived from equilibrium with P5C) as the substrate. The reaction mixture (final volume, 1 ml) contained the following: 2 mM P5C, 1 mM NADP+, 100 mM KH2PO4, 50 mM imidazole base (pH 7.0), and the purified recombinant GPR plus water. The increase in the absorbance at 340 nm was recorded at 30°C against a blank identical to the one described above but lacking KH2PO4. One unit of activity was defined as the amount of enzyme required to produce 1 μmol of NADPH per min.
Nucleotide sequence accession numbers.
The GenBank accession numbers for PRO1 and PRO2 are M85293 and U43565, respectively. The PDB ID code for Campylobacter jejuni GK is 2AKO.
RESULTS AND DISCUSSION
S. cerevisiae GK activity requires GPR and is sensitive to feedback inhibition by proline.
In E. coli, it has been suggested that GK and GPR, both of which catalyze the first two steps of proline synthesis, function as an enzyme complex to channel the unstable intermediate, γ-glutamyl phosphate, in vivo (44, 46). In plants, a bifunctional P5C synthetase consists of a GK domain at the N terminus and a GPR domain at the C terminus (47, 57, 58). Our previous results also suggested that the yeast GK and GPR together form a complex to function with each other in vivo (27, 52). When plasmid pQE2-PRO1 was introduced into E. coli strain KC1352 with proB deleted, which lacks the GK activity, the transformants showed the proline-prototrophic phenotype, indicating that S. cerevisiae GK is functionally expressed in E. coli (data not shown).
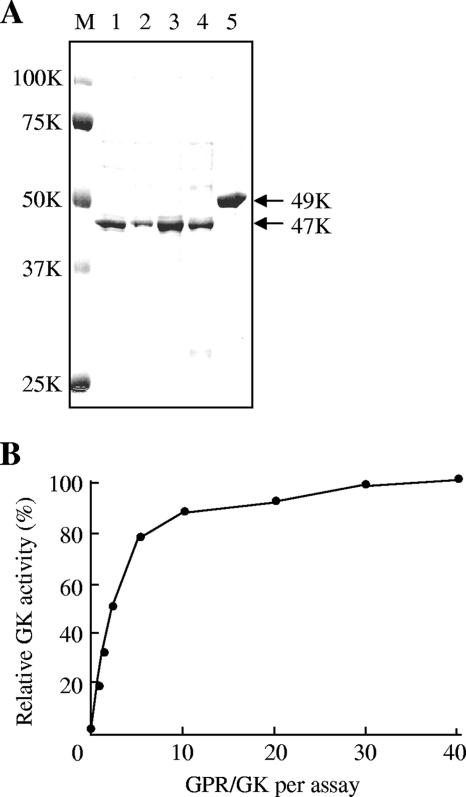
To understand the enzymatic properties of S. cerevisiae GK, we purified the recombinant GK and GPR from E. coli cells. The His-tagged fusion enzymes were purified so that they gave single bands, corresponding to relative molecular masses of approximately 47 and 49 kDa, respectively, on a sodium dodecyl sulfate-polyacrylamide gel (Fig. 1A). Interestingly, the purified GK showed absolutely no activity when the activity was measured by the commonly used hydroxamate assay (46), but the production of γ-glutamyl hydroxamate could be detected after the addition of purified GPR (Fig. 1B). The amount of GPR required to restore maximal activity far exceeded the amount of GK present in the reaction mixture, and a molar ratio of GPR to GK of 40:1 was necessary for maximal activity (18.4 U/mg). However, approximately half-maximal activity was observed when the ratio of GPR to GK was only 2:1. The heat-inactivated GPR could not promote the GK activity, showing that some specific property of the native GPR protein is required for full display of the GK activity.
FIG. 1.
GK and GPR of S. cerevisiae. (A) Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (12%) of recombinant GK (lanes 1 to 4) and GPR (lane 5). Lane 1, wild type; lane 2, D154N; lane 3, E149K; lane 4, I150T. The gel was stained with Coomassie brilliant blue R-250. The positions of molecular mass standards are shown on the left (lane M). The upper and lower arrows indicate the positions of the GPR (49 kDa) and GK (47 kDa) proteins, respectively. (B) Effect of GPR on the GK activity. The GK activity as determined by the hydroxamate assay was measured as a function of the amount of GPR added to each assay mixture. The assay mixture contained 0.15 μg of GK and 0 to 6.25 μg of GPR. The molar ratio of GPR to GK was calculated by using molecular weights of 47,000 for GK and 49,000 for GPR. The GK activity with a GPR/GK ratio of 40 was defined as 100% (18.4 U/mg). The values are the means of results from three independent experiments. The standard deviations for the values were <10% of the mean values.
It is also noteworthy that the purified GPR activity was virtually invariant in the absence (1.68 ± 0.11 U/mg) and in the presence (1.77 ± 0.09 U/mg) of the purified GK, suggesting that GK is not required for the reverse reaction of GPR. Previous studies and our results suggest that GK and GPR form a complex or that there is protein-protein interaction to directly transfer the intermediate from one enzyme to the other. Therefore, we have attempted to confirm that there is a physical interaction between the two enzymes using various methods, including a gel filtration assay, native polyacrylamide gel electrophoresis, a yeast two-hybrid assay, an S-protein-tagged pull-down assay, and an immunoprecipitation assay, but no evidence has been obtained so far (data not shown). This discrepancy cannot currently be resolved, and further study is thus needed. A possible mechanism is that GK and GPR are in close proximity (but are not physically linked) in such a manner that the γ-glutamyl phosphate formed on the kinase interacts with an active site moiety of the reductase to facilitate the catalytic reaction. The results also do not exclude the possibility that there are additional intermediate steps, such as steps involving acyl derivatives of the kinase or phosphorylation of the kinase (44).
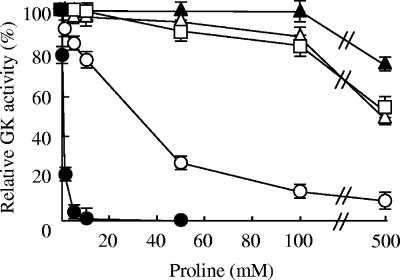
In many bacteria and plants, proline synthesis is strictly regulated through feedback inhibition of GK by proline (29, 46, 47, 58). We analyzed the GK activity of S. cerevisiae using the purified enzymes. The purified GK was assayed for activity with the purified GPR (the molar ratio of GPR to GK was 5:1, which resulted in approximately 80% of the maximal GK activity) (Table 1). The activity decreased significantly in the presence of proline (Fig. 2). As shown in Table 1, the concentration of proline needed to achieve 50% inhibition (IC50) was 0.5 mM, which was the same order of magnitude as the apparent Ki of E. coli (0.1 mM) (46), tomato (0.09 mM) (17), or mothbean (1.0 mM) (58). On the other hand, no inhibitory effect of proline (10 mM) on the GPR activity was detected (data not shown). These results indicate that the GK activity of S. cerevisiae is sensitive to feedback inhibition by proline; that is, GK is the rate-limiting enzyme in the proline biosynthesis of S. cerevisiae.
TABLE 1.
Specific activities and IC50 values of the recombinant GKs
| Enzyme | Sp act (U/mg)a | IC50 (mM)b |
|---|---|---|
| Wild type | 14.7 ± 0.42 | 0.50 ± 0.02 |
| D154N | 26.3 ± 1.91 | 32 ± 1.4 |
| E149K | 4.2 ± 0.16 | 840 ± 61 |
| N142D/I166V | 3.2 ± 0.08 | 540 ± 13 |
| I150T | 29.0 ± 1.88 | 500 ± 55 |
The specific activity was determined as described in Materials and Methods.
The IC50 was estimated from Fig. 2. The data are the means ± standard deviations for three independent experiments.
FIG. 2.
Effect of proline on the GK activity. The activities of wild-type (•), D154N (○), E149K (▴), N142D/I166V (□), and I150T (▵) GKs were measured using the hydroxamate assay in the presence of the proline concentrations indicated. Each assay mixture contained 0.15 μg of GK and 0.78 μg of GPR (the molar ratio GPR to GK was 5:1). The enzyme activities of wild-type, D154N, N142D/I166V, E149K, and I150T GKs in the absence of proline were defined as 100% (Table 1). The values are the means and standard deviations for three independent experiments.
D154N mutant GK is less sensitive to feedback inhibition, leading to proline accumulation.
We previously isolated a mutant of S. cerevisiae that was resistant to the toxic proline analogue AZC, accumulated proline, and tolerated freezing (48). This mutant has a single mutation in PRO1 that results in a D154N amino acid substitution in GK (27). To analyze the mechanism of proline accumulation in non-proline-utilizing cells expressing the D154N mutant GK, we purified the mutant enzyme from E. coli cells. The D154N mutant GK showed a twofold increase in specific activity compared with the wild-type enzyme (Table 1). The level of activity was 85% even in the presence of 5 mM proline, while wild-type GK was markedly inhibited (Fig. 2). The D154N mutant GK was desensitized by a factor of 64 compared to the wild-type enzyme (Table 1). These results indicate that diminished sensitivity to proline feedback inhibition in GK causes proline accumulation. It is interesting that the D154N mutant GK is more thermostable than the wild-type enzyme (52). This mutation in microbial GKs is novel in that different mutations in other amino acid residues required for the feedback inhibition of GK by proline were identified in bacterial genes that have been studied (11, 13, 23, 30). A similar mutation in the tomato GK (D162N) corresponding to position 154 resulted in the removal of feedback inhibition but a drastic reduction in the specific activity (16). This is probably because the regional sequence similarity in the two GKs is very low. Residue 154 in S. cerevisiae GK may be responsible in part for the catalysis and stability, as well as the proline inhibition.
Mutant GKs that increase intracellular proline levels were isolated by PCR random mutagenesis.
To further analyze and improve the enzymatic properties of GK, PCR random mutagenesis of PRO1 was employed. Proline-accumulating cells could be isolated on minimal medium containing the toxic compound AZC (27, 48), which is incorporated into proteins competitively with proline (21, 37, 54). Error-prone PCR was carried out as described in Materials and Methods. The mutagenized plasmid library was introduced into S. cerevisiae INVDput1pro1(pTV-PRO2), and transformants were selected on SD agar medium containing AZC (15 μg/ml). Transformants harboring pAD-WTPRO1 and pAD-D154NPRO1 were prepared as negative and positive controls, respectively. As a result, we obtained 18 AZC-resistant colonies capable of growing faster than the colonies expressing the wild-type GK, suggesting that these clones accumulate intracellular proline due to desensitization to feedback inhibition by proline. Plasmids prepared from these colonies were then shuttled into E. coli DH5α and back into strain INVDput1pro1(pTV-PRO2) to retest AZC resistance. Of the 18 plasmids, 15 were proved to confer AZC resistance, confirming that this phenotype was caused by a mutation(s) on the plasmid. The PRO1 genes in the plasmids were sequenced, and 10 types of amino acid substitutions were identified (Table 2). Multiple amino acid substitutions due to hypermutagenic PCR were also found.
TABLE 2.
Summary of amino acid and DNA substitutions in the S. cerevisiae mutant GKs
| Mutant GKa | Amino acid substitution(s) (base substitution[s]) | Proline content (% of dry wt)b |
|---|---|---|
| pAD-WTPRO1c | 0.10 ± 0.08 | |
| pAD-D154NPRO1d | D154N (G460 → A) | 0.69 ± 0.13 |
| 1 | E149K (G445 → A) | 2.80 ± 0.33 |
| 2 | N142D (A424 → G), I166V (A496 → G) | 2.24 ± 0.48 |
| 3 | I150T (T449 → C) | 1.87 ± 0.13 |
| 4 | S146P (T436 → C) | 1.77 ± 0.39 |
| 5 | H306R (A917 → G) | 1.00 ± 0.27 |
| 6 | A105V (C314 → T) | 0.79 ± 0.06 |
| 7 | N309D (A925 → G), R428C (C1282 → T) | 0.72 ± 0.28 |
| 8 | R148G (A442 → G), Q351R (A1052 → G) | 0.69 ± 0.04 |
| 9 | R148G (A442 → G) | 0.66 ± 0.11 |
| 10 | T126A (A376 → G) | 0.51 ± 0.01 |
The mutant GKs obtained in this study (GKs 1 to 10) are arranged in order of proline content.
The intracellular proline content was determined after cultivation in SD liquid medium at 30°C for 2 days. The data are the means ± standard deviations for three independent experiments.
Plasmid harboring wild-type PRO1.
Plasmid harboring D154N mutant PRO1.
In general, greater resistance to AZC reflects a higher level of intracellular proline. To examine whether the amino acid substitutions in GK lead to proline accumulation in yeast cells, INVDput1pro1 cells expressing wild-type or mutant GK were cultivated in SD liquid medium, and the cellular proline levels were examined (Table 2). A small amount of proline was detected in the case of wild-type GK (0.10% of the dry weight), due to deficiency of the degradation pathway. It was found that there was a significant accumulation of proline with the D154N mutant GK (0.69%), as expected from previous studies (52). Interestingly, cells with 4 of 16 mutant GKs clearly accumulated larger amounts of proline than cells with the D154N mutant GK accumulated. Cells with the N142D/I166V, S146P, E149K, and I150T mutant GKs showed prominent 2.6- to 4.1-fold increases in the proline content (1.77 to 2.80%). It is also noteworthy that these mutations are located close to position 145.
Many amino acid biosynthetic genes in S. cerevisiae are regulated by a global system known as general amino acid control (19). In a gcn4 (general control nonderepressible) mutant, RNA and enzymatic activities cannot increase in response to amino acid starvation. The PRO1 and PRO2 genes are believed to be under general control, while PRO3 is not (8, 24). In this study, the PRO1 and PRO2 genes in each plasmid were constitutively expressed under the ADH1 promoter in SD medium. Therefore, one might expect that the mutant GKs lead to proline accumulation in a gcn4 mutant cell.
Newly derived mutant GKs were extremely insensitive to feedback inhibition by proline.
Three mutant GKs (N142D/I166V, E149K, and I150T) were purified and assayed for activity with the purified GPR. The specific activities of N142D/I166V, E149K, and I150T GK in the absence of proline were 3.2, 4.2, and 29.0 U/mg, respectively (Table 1). The activities of these mutant GKs were very insensitive to feedback inhibition at proline concentrations up to 50 mM, while the activity of the D154N mutant GK fell to 27% of that in the absence of proline (Fig. 2). When the IC50 values of proline were determined, the mutant GKs showed approximately 1,000-fold or more desensitization to proline feedback inhibition compared to the wild-type enzyme (Table 1). Despite their reduced specific activities, yeast cells expressing the N142D/I166V and E149K mutant GKs accumulated large amounts of proline. These results indicate that extreme desensitization to feedback regulation in mutant GKs results in proline accumulation and that the feedback inhibition is more important than the specific activity for the regulation of proline biosynthesis.
Structural and functional features of mutant GKs.
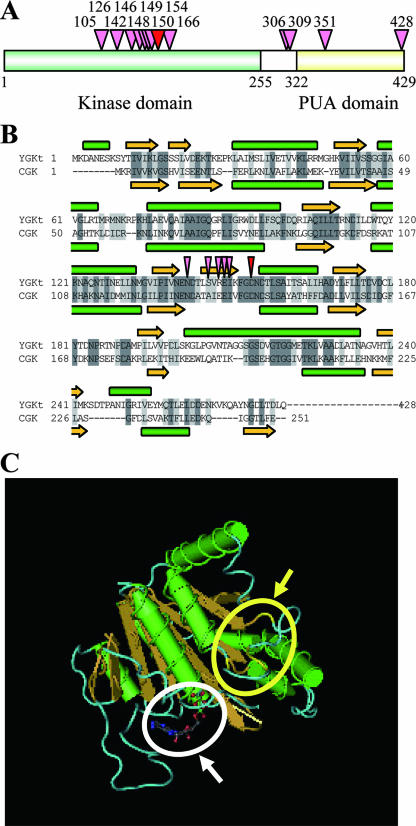
Figure 3A shows the positions in the primary structure of GK where the amino acids were replaced. It should be emphasized that most of the amino acid substitutions were in the N-terminal amino acid kinase domain found in the majority of microbial GKs (36). In E. coli, the C-terminal PUA domain (4) was proposed to modulate the function of the kinase domain (34). The PUA domain is not essential for either catalysis or proline inhibition, but deletion of this domain greatly reduces the concentration of Mg needed for activation of GK and increases the concentration of proline needed for inhibition of GK. Some mutations (H306R and N309D) that trigger proline accumulation were located at the junction between two domains (Fig. 3A). Although the function of this junction is unclear, each mutation might alter the conformation of GK to a conformation having a lower affinity for proline or might inhibit the binding of some group to proline. In the PUA domain, we found some mutations (Q351R and R428C). It is unlikely that these mutations increase the intracellular proline level, because the R148G mutant accumulated proline and residue R428 is located near the C terminus.
FIG. 3.
Features of mutant GKs. (A) Schematic representation of S. cerevisiae GK. The positions of 13 amino acids where mutations occur in the proline-accumulating mutants are indicated by red (D154) and pink arrowheads. The numbers are residue numbers. The kinase and PUA domains are green and yellow, respectively. (B) Comparison of the amino acid sequence of S. cerevisiae GK (YGKt) with that of C. jejuni GK (CGK). Identical and similar amino acids in the two proteins are indicated by dark and light shading, respectively. Yellow arrows and green cylinders above and below the sequences indicate β-sheets and α-helices, respectively. Dashes indicate the absence of corresponding amino acid residues at the positions. Residues N142, S146, R148, E149, I150, and D154, where mutations resulted in proline accumulation, are indicated by red (D154) and pink arrowheads. (C) Three-dimensional structure of C. jejuni GK. Yellow arrows and green cylinders depict β-sheets and α-helices, respectively. The yellow arrow and circle indicate a large loop region corresponding to residues 142 to 154 in S. cerevisiae GK, which lies on the molecular surface. The white arrow and circle indicate the ADP binding pocket.
It is intriguing to see which amino acid residues are involved in the allosteric regulation and catalytic activity of GK. Although there are still limited data on the three-dimensional structure (33), the tetrameric crystal structure of GK from the bacterium C. jejuni has been determined recently (and is available only from the Protein Data Bank). The amino acid sequence of C. jejuni GK with 251 residues lacking the PUA domain was homologous to the sequence of the N-terminal kinase domain of S. cerevisiae GK with 428 residues. In the overlapping region consisting of 280 amino acids, 36% of the amino acids were identical, and 34% were considered to be similar (Fig. 3B). Figure 3B also shows a comparison of the hypothetical secondary structures of the kinase domains of yeast GK and C. jejuni GK. The predicted structure was made up of eight α-helices and nine β-sheets in yeast GK, similar to the α-helices and β-sheets seen in C. jejuni GK.
In this study, seven amino acid substitutions at positions 142, 146, 148, 149, 150, and 154 in S. cerevisiae GK were shown to be important for increasing intracellular proline levels, probably due to elimination of allosteric regulation. These mutations were concentrated in a sequence highly homologous with that of C. jejuni GK corresponding to a large loop region between a β-sheet (I122 to E128) and an α-helix (D143 to F153), which is on the molecular surface (Fig. 3C). Based on the prediction of the secondary structure of S. cerevisiae GK, the sequence is situated in the region between the β-sheet (I136 to N140) and the α-helix (D156 to I166), which includes a β-sheet-like sequence (L145 to K151), of S. cerevisiae GK. Residues 142 to 154, where most of the mutations are concentrated, are mapped to the region that was identified as important for allosteric control of the tomato GK (16). Although the sequence similarity in the regions of the two GKs is low, the regions are suggested to constitute part of the proline-binding site. The amino acid substitutions that neutralize or reverse the charge on the protein surface (N142D, E149K, R148G, and D154N) may impair the electrical environment with the adjacent residues and thereby inhibit the proline binding. The Pro substitution at position 146 might break a β-sheet consisting of L145 to K151. Replacement of Ile150 by Thr may cause a structural alteration because of a decreased hydophobic interaction in the proline-binding site. It is very interesting that the two mutations at positions 150 and 154 increased the GK activity, in addition to removing feedback inhibition. It is likely that residues 150 and 154 are located at the border between the allosteric domain and the substrate-binding site in GK, and a conformational change in the region upon proline binding could affect the affinity for substrates.
Yeast cells expressing mutant GKs had enhanced freeze tolerance.
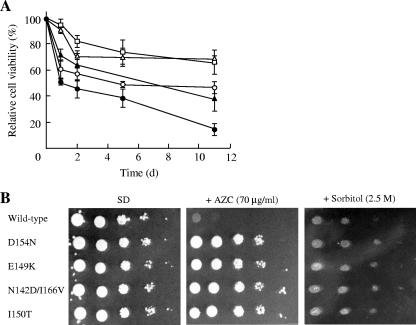
We previously found that yeast strains that accumulated proline were tolerant to freezing stress (25-27, 48, 50, 52). The natural protectants proline and trehalose were reported to preserve membrane structure and function during freezing (40). It has been suggested that proline can prevent ice nucleation and dehydration by forming strong hydrogen bonds with intracellular free water. We therefore examined the cell viabilities of proline-accumulating strains after freezing stress. As shown in Fig. 4A, in proportion to the cellular proline level, yeast cells expressing the N142D/I166V or I150T mutant GK showed an increased survival rate compared with that of cells expressing the D154N mutant GK when the cell suspensions were exposed to freezing at −20°C. However, the E149K mutant GK, which accumulated the highest level of proline, decreased the cell viability, and the level was close to that in cells with D154N mutant GK (Fig. 4A). In S. cerevisiae, free radicals are also generated during freezing and thawing (15), and cytoplasmic Cu/Zn-superoxide dismutase is required for freezing tolerance (32). Therefore, oxidative stress causes serious injury to yeast cells during the freeze-thaw process, in addition to physical damage caused by ice nucleation and dehydration. It has been proposed that proline may act as a free radical scavenger to protect plants from damage by oxidative stress caused during osmotic stress (20). We also found that overexpression of the D154N mutant GK could protect yeast cells from oxidative stress during exposure to H2O2 (52). In addition to its cryoprotective activity, proline has an important role in reducing the oxidative stress induced during freezing stress.
FIG. 4.
Effect of mutant GKs on yeast cells under stress conditions. (A) Freezing stress tolerance of S. cerevisiae strains expressing GK. The cell viabilities of each strain were measured after freezing at −20°C for the times indicated. The strains used were INVDput1pro1(pTV-PRO2) expressing wild-type (•), D154N (○), E149K (▴), N142D/I166V (□), and I150T (▵) GKs. The number of colonies before freezing was defined as 100%. The values are the means and standard deviations for three independent experiments. (B) Growth phenotypes on SD agar medium of S. cerevisiae strains expressing GK. After cultivation in SD liquid medium at 30°C for 48 h, approximately 106 cells of each strain and 10−1 to 10−4 serial dilutions (from left to right) were spotted and incubated on SD agar medium containing 70 μg/ml AZC or 2.5 M sorbitol at 30°C for 5 days.
Freeze-thaw injuries of yeast cells depend on numerous factors, including the physiological condition of the yeast cells, the genetic background of the yeast strains, and the freezing conditions. A decrease in freezing tolerance is reportedly strongly related to trehalose degradation, and the level of intracellular trehalose affects the freezing tolerance of baker's yeast (45). On the basis of DNA microarray analysis, Odani et al. (28) suggested that freeze-thaw stress causes damage to the structure of the cell wall and to cellular organelles. Ando et al. also recently performed genome-wide screening using the complete deletion strain collection of diploid S. cerevisiae (2). They pointed out the presence of two different mechanisms of freeze-thaw injury: oxidative stress generated during the freeze-thaw process and defects in cell wall assembly. Before freezing, yeast cells are exposed to a low temperature. Two transcription factors, Msn2 and Msn4, which are involved in stress-induced expression of many genes, were shown to mediate the stress response pattern observed during the late cold response, but the transcriptional response of the early cold response genes was Msn2 and Msn4 independent (43). Thus, despite extensive studies of freeze-thaw injuries in yeast, the molecular mechanisms behind the freezing tolerance remain unclear.
We also tested the growth phenotype of proline-accumulating strains with various stresses. Contrary to our expectations for the stress-protective effect of proline, there were no significant differences in growth between yeast cells expressing the D154N, N142D/I166V, E149K, or I150T mutant GK on SD agar medium with heat shock (50°C, 3 to 5 h), ethanol (16 to 20%), and sorbitol (2.0 to 2.5 M) stresses, although the proline-accumulating cells were more resistant to these stresses than cells expressing wild-type GK (Fig. 4B). We have recently reported that excess intracellular proline might be toxic to yeast cells only when it accumulates in the cytosol (25) or might delay yeast cell growth in the presence of ethanol (49). These results suggest that an appropriate proline level in yeast cells is important for its stress-protective effect.
This study is the first study to report the characteristics and engineering of S. cerevisiae GK. In summary, the feedback inhibition of GK by proline is a rate-limiting step in proline biosynthesis in S. cerevisiae. Desensitization of the allosteric regulation of GK results in proline accumulation and freezing tolerance in yeast cells. Since S. cerevisiae cells are exposed to various stresses, including freezing, desiccation, ethanol, oxidation, and osmolarity, during fermentation processes, stress tolerance is the key for application to industrial yeasts. Proline could be useful for breeding novel yeast strains that are resistant to various stresses, and improvements in proline accumulation are expected to involve an engineered GK which shows a higher level of feedback desensitization and has a higher specific activity.
Acknowledgments
We thank L. S. Csonka (Purdue University, West Lafayette, IN) for providing an E. coli strain. We also thank S. Nakamori and M. Takahashi for discussions concerning this work.
This work was supported by a grant to H.T. from the Program for Promotion of Basic Research Activities for Innovative Biosciences (PROBRAIN).
Footnotes
Published ahead of print on 20 April 2007.
REFERENCES
- 1.Alexandre, H., V. Ansanay-Galeote, S. Dequin, and B. Blondin. 2001. Global gene expression during short-term ethanol stress in Saccharomyces cerevisiae. FEBS Lett. 498:98-103. [DOI] [PubMed] [Google Scholar]
- 2.Ando, A., T. Nakamura, Y. Murata, H. Takagi, and J. Shima. 2007. Identification and classification of genes required for tolerance to freeze-thaw stress revealed by genome-wide screening of Saccharomyces cerevisiae deletion strains. FEMS Yeast Res. 7:244-253. [DOI] [PubMed] [Google Scholar]
- 3.Andre, L., A. Hemming, and L. Adler. 1991. Osmoregulation in Saccharomyces cerevisiae. Studies on the osmotic induction of glycerol production and glycerol-3-phosphate dehydrogenase (NAD+). FEBS Lett. 286:13-17. [DOI] [PubMed] [Google Scholar]
- 4.Aravind, L., and E. V. Kooin. 1999. Novel predicted RNA-binding domains associated with the translation machinery. J. Mol. Evol. 48:291-302. [DOI] [PubMed] [Google Scholar]
- 5.Attfield, P. V. 1987. Trehalose accumulates in Saccharomyces cerevisiae during exposure to agents that induce heat shock response. FEBS Lett. 225:259-263. [DOI] [PubMed] [Google Scholar]
- 6.Blomberg, A., and L. Adler. 1989. Roles of glycerol and glycerol-3-phosphate dehydrogenase (NAD+) in acquired osmotolerance of Saccharomyces cerevisiae. J. Bacteriol. 171:1087-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brandriss, M. C. 1983. Proline utilization in Saccharomyces cerevisiae: analysis of the cloned PUT2 gene. Mol. Cell. Biol. 3:1846-1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brandriss, M. C., and D. A. Falvey. 1992. Proline biosynthesis in Saccharomyces cerevisiae: analysis of the PRO3 gene, which encodes Δ1-pyrroline-5-carboxylate reductase. J. Bacteriol. 174:3782-3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Causton, H. C., B. Ren, S. S. Koh, C. T. Harbison, E. Kanin, E. G. Jennings, T. I. Lee, H. L. True, E. S. Lander, and R. A. Young. 2001. Remodeling of yeast genome expression in response to environmental changes. Mol. Biol. Cell. 12:323-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, C., and M. B. Dickman. 2005. Proline suppresses apoptosis in the fungal pathogen Colletotrichum trifolii. Proc. Natl. Acad. Sci. USA 102:3459-3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Csonka, L. N., S. B. Gelvin, B. W. Goodner, C. S. Orser, D. Siemieniak, and J. L. Slightom. 1988. Nucleotide sequence of a mutation in the proB gene of Escherichia coli that confers proline overproduction and enhanced tolerance to osmotic stress. Gene 64:199-205. [DOI] [PubMed] [Google Scholar]
- 12.Csonka, L. N., and A. D. Hanson. 1991. Prokaryotic osmoregulation: genetics and physiology. Annu. Rev. Microbiol. 45:569-606. [DOI] [PubMed] [Google Scholar]
- 13.Dandekar, A. M., and S. L. Uratsu. 1988. A single base pair change in proline biosynthesis genes causes osmotic stress tolerance. J. Bacteriol. 170:5943-5945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Delauney, A. J., and D. P. S. Verma. 1993. Proline biosynthesis and osmoregulation in plants. Plant J. 4:215-223. [Google Scholar]
- 15.Du, X., and H. Takagi. 2005. N-Acetyltransferase Mpr1 conferred freeze tolerance in Saccharomyces cerevisiae by reducing reactive oxygen species. J. Biochem. 138:391-397. [DOI] [PubMed] [Google Scholar]
- 16.Fujita, T., A. Maggio, M. García-Ríos, R. A. Bressan, and L. N. Csonka. 1998. Comparative analysis of the regulation of expression and structures of two evolutionarily divergent genes for Δ1-pyrroline-5-carboxylate synthetase from tomato. Plant Physiol. 118:661-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fujita, T., A. Maggio, M. García-Ríos, C. Stauffacher, R. A. Bressan, and L. N. Csonka. 2003. Identification of regions of the tomato γ-glutamyl kinase that are involved in allosteric regulation by proline. J. Biol. Chem. 278:14203-14210. [DOI] [PubMed] [Google Scholar]
- 18.Hayzer, D. J., and T. Leisinger. 1980. The gene-enzyme relationships of proline biosynthesis in Escherichia coli. J. Gen. Microbiol. 118:287-293. [DOI] [PubMed] [Google Scholar]
- 19.Hinnebusch, A. G. 1988. Mechanisms of gene regulation in the general control of amino acid biosynthesis in Saccharomyces cerevisiae. Microbiol. Rev. 52:248-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hong, Z., K. Lakkineni, Z. Zhang, and D. P. S. Verma. 2000. Removal of feedback inhibition of Δ1-pyrroline-5-carboxylate synthetase results in increased proline accumulation and protection of plants from osmotic stress. Plant Physiol. 122:1129-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoshikawa, C., M. Shichri, S. Nakamori, and H. Takagi. 2003. A nonconserved Ala401 in the yeast Rsp5 ubiquitin ligase is involved in degradation of Gap1 permease and stress-induced abnormal proteins. Proc. Natl. Acad. Sci. USA 100:111505-111510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kandror, O., N. Bretschneider, E. Kreydin, D. Cavalieri, and A. L. Goldberg. 2004. Yeast adapt to near-freezing temperatures by STRE/Msn2,4-dependent induction of trehalose synthesis and certain molecular chaperones. Mol. Cell 13:771-781. [DOI] [PubMed] [Google Scholar]
- 23.Kosuge, T., and T. Hoshino. 1998. Construction of a proline-producing mutant of the extremely thermophilic eubacterium Thermus thermophilus HB27. Appl. Environ. Microbiol. 64:4328-4332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li, W., and M. C. Brandriss. 1992. Proline biosynthesis in Saccharomyces cerevisiae: molecular analysis of the PRO1 gene, which encodes γ-glutamyl kinase. J. Bacteriol. 174:4148-4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matsuura, K., and H. Takagi. 2005. Vacuolar functions are involved in stress-protective effect of intracellular proline on Saccharomyces cerevisiae. J. Biosci. Bioeng. 100:538-544. [DOI] [PubMed] [Google Scholar]
- 26.Morita, Y., S. Nakamori, and H. Takagi. 2002. Effect of proline and arginine metabolism on freezing stress of Saccharomyces cerevisiae. J. Biosci. Bioeng. 94:390-394. [DOI] [PubMed] [Google Scholar]
- 27.Morita, Y., S. Nakamori, and H. Takagi. 2003. l-Proline accumulation and freeze tolerance in Saccharomyces cerevisiae are caused by a mutation in the PRO1 gene encoding γ-glutamyl kinase. Appl. Environ. Microbiol. 69:212-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Odani, M., Y. Komatsu, S. Oka, and H. Iwahashi. 2003. Screening of genes that respond to cryopreservation stress using yeast DNA microarray. Cryobiology 47:155-164. [DOI] [PubMed] [Google Scholar]
- 29.Omori, K., S. Suzuki, Y. Imai, and S. Komatsubara. 1991. Analysis of the Serratia marcescens proBA operon and feedback control of proline biosynthesis. J. Gen. Microbiol. 137:509-517. [DOI] [PubMed] [Google Scholar]
- 30.Omori, K., S. Suzuki, Y. Imai, and S. Komatsubara. 1992. Analysis of the mutant proBA operon from a proline-producing strain of Serratia marcescens. J. Gen. Microbiol. 138:693-699. [DOI] [PubMed] [Google Scholar]
- 31.Panadero, J., C. Pallotti, S. Rodriguez-Vargas, F. Randez-Gil, and J. A. Prieto. 2006. A downshift in temperature activates the high osmolarity glycerol (HOG) pathway, which determines freeze tolerance in Saccharomyces cerevisiae. J. Biol. Chem. 281:4638-4645. [DOI] [PubMed] [Google Scholar]
- 32.Park, J. I., C. M. Grant, M. J. Davies, and I. W. Dawes. 1998. The cytoplasmic Cu, Zn superoxide dismutase of Saccharomyces cerevisiae is required for resistance to freeze-thaw stress. J. Biol. Chem. 273:22921-22928. [DOI] [PubMed] [Google Scholar]
- 33.Perez-Arellano, I., F. Gil-Ortiz, J. Cervera, and V. Rubio. 2004. Glutamate-5-kinase from Escherichia coli: gene cloning, overexpression, purification and crystallization of the recombinant enzyme and preliminary X-ray studies. Acta Crystallogr. Sect. D 60:2091-2094. [DOI] [PubMed] [Google Scholar]
- 34.Perez-Arellano, I., V. Rubio, and J. Cervera. 2005. Dissection of Escherichia coli glutamate 5-kinase: functional impact of the deletion of the PUA domain. FEBS Lett. 579:6903-6908. [DOI] [PubMed] [Google Scholar]
- 35.Rajendrakumar, C. S., T. Suryanarayana, and A. R. Reddy. 1997. DNA helix destabilization by proline and betaine: possible role in the salinity tolerance process. FEBS Lett. 410:201-205. [DOI] [PubMed] [Google Scholar]
- 36.Ramon-Maiques, S., A. Marina, F. Gil-Ortiz, I. Fita, and V. Rubio. 2002. Structure of acetylglutamate kinase, a key enzyme for arginine biosynthesis and a prototype for the amino acid kinase enzyme family, during catalysis. Structure 10:329-342. [DOI] [PubMed] [Google Scholar]
- 37.Reese, L. M., K. O. Cutler, and C. E. Deutch. 1996. Sensitivity of Escherichia coli to proline analogues during osmotic stress and anaerobiosis. Lett. Appl. Microbiol. 22:202-205. [DOI] [PubMed] [Google Scholar]
- 38.Rodriguez-Vargas, S., F. Estruch, and F. Randez-Gil. 2002. Gene expression analysis of cold and freeze stress in baker's yeast. Appl. Environ. Microbiol. 68:3024-3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roosens, N. H., R. Willem, Y. Li, I. I. Verbruggen, M. Biesemans, and M. Jacobs. 1999. Proline metabolism in the wild-type and in a salt-tolerant mutant of Nicotiana plumbaginifolia studied by (13)C-nuclear magnetic resonance imaging. Plant Physiol. 121:1281-1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rudolph, A. S., and J. H. Crowe. 1985. Membrane stabilization during freezing: the role of two natural cryoprotectants, trehalose and proline. Cryobiology 22:367-377. [DOI] [PubMed] [Google Scholar]
- 41.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 42.Samuel, D., T. K. S. Kumar, G. Ganesh, G. Jayaraman, P.-W. Yang, M.-M. Chang, V. D. Trivedi, S.-L. Wang, K.-C. Hwang, D.-K. Chang, and C. Yu. 2000. Proline inhibits aggregation during protein refolding. Protein Sci. 9:344-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schade, B., G. Jansen, M. Whiteway, K. D. Entian, and D. Y. Thomas. 2004. Cold adaptation in budding yeast. Mol. Biol. Cell 15:5492-5502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seddon, A. P., K. Y. Zhao, and A. Meister. 1989. Activation of glutamate by γ-glutamate kinase: formation of γ-cis-cycloglutamyl phosphate, an analog of γ-glutamyl phosphate. J. Biol. Chem. 264:11326-11335. [PubMed] [Google Scholar]
- 45.Shima, J., A. Hino, C. Yamada-Iyo, Y. Suzuki, R. Nakajima, H. Watanabe, K. Mori, and H. Takano. 1999. Stress tolerance in doughs of Saccharomyces cerevisiae trehalase mutants derived from commercial baker's yeast. Appl. Environ. Microbiol. 65:2841-2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith, C. J., A. H. Deutch, and K. E. Rushlow. 1984. Purification and characteristics of a γ-glutamyl kinase involved in Escherichia coli proline biosynthesis. J. Bacteriol. 157:545-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stines, A. P., D. J. Naylor, P. B. Hoj, and R. van Heeswijck. 1999. Proline accumulation in developing grapevine fruit occurs independently of changes in the levels of Δ1-pyrroline-5-carboxylate synthetase mRNA or protein. Plant Physiol. 120:923-931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takagi, H., F. Iwamoto, and S. Nakamori. 1997. Isolation of freeze-tolerant laboratory strain of Saccharomyces cerevisiae from proline-analogue-resistant mutants. Appl. Microbiol. Biotechnol. 47:405-411. [DOI] [PubMed] [Google Scholar]
- 49.Takagi, H., F. Matsui, A. Kawaguchi, H. Wu, H. Shimoi, and Y. Kubo. 2007. Construction and analysis of self-cloning sake yeasts that accumulate proline. J. Biosci. Bioeng. 103:377-380. [DOI] [PubMed] [Google Scholar]
- 50.Takagi, H., K. Sakai, K. Morida, and S. Nakamori. 2000. Proline accumulation by mutation or disruption of the proline oxidase gene improves resistance to freezing and desiccation stresses in Saccharomyces cerevisiae. FEMS Microbiol. Lett. 184:103-108. [DOI] [PubMed] [Google Scholar]
- 51.Takagi, H., M. Takaoka, A. Kawaguchi, and Y. Kubo. 2005. Effect of l-proline on sake brewing and ethanol stress in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 71:8656-8662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Terao, Y., S. Nakamori, and H. Takagi. 2003. Gene dosage effect of l-proline biosynthetic enzymes on l-proline accumulation and freeze tolerance in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 69:6527-6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tomenchok, D. M., and M. C. Brandriss. 1987. Gene-enzyme relationship in the proline biosynthetic pathway of Saccharomyces cerevisiae. J. Bacteriol. 169:5364-5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Trotter, E. W., L. Berenfeld, S. A. Krause, G. A. Petsko, and J. V. Gray. 2001. Protein misfolding and temperature up-shift cause G1 arrest via a common mechanism dependent on heat shock factor in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 98:7313-7318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van Dijck, P., D. Colavizza, P. Smet, and J. M. Thevelein. 1995. Differential importance of trehalose in stress resistance in fermenting and nonfermenting Saccharomyces cerevisiae cells. Appl. Environ. Microbiol. 61:109-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang, S.-S., and M. C. Brandriss. 1986. Proline utilization in Saccharomyces cerevisiae: analysis of the cloned PUT1 gene. Mol. Cell. Biol. 6:2638-2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yoshiba, Y., T. Kiyosue, T. Katagiri, H. Ueda, T. Mizoguchi, K. Yamaguchi-Shinozaki, K. Wada, Y. Harada, and K. Shinozaki. 1995. Correlation between the induction of a gene for Δ1-pyrroline-5-carboxylate synthetase and the accumulation of proline in Arabidopsis thaliana under osmotic stress. Plant J. 7:751-760. [DOI] [PubMed] [Google Scholar]
- 58.Zhang, C.-S., Q. Liu, and D. P. S. Verma. 1995. Removal of feedback inhibition of Δ1-pyrroline-5-carboxylate synthetase, a bifunctional enzyme catalyzing the first two steps of proline biosynthesis in plants. J. Biol. Chem. 270:20491-20496. [DOI] [PubMed] [Google Scholar]