Abstract
In the aftermath of the anthrax letters of 2001, researchers have been exploring various analytical signatures for the purpose of characterizing the production environment of microorganisms. One such signature is stable isotope ratios, which in heterotrophs, are a function of nutrient and water sources. Here we discuss the use of stable isotope ratios in microbial forensics, using as a database the carbon, nitrogen, oxygen, and hydrogen stable isotope ratios of 247 separate cultures of Bacillus subtilis 6051 spores produced on a total of 32 different culture media. In the context of using stable isotope ratios as a signature for sample matching, we present an analysis of variations between individual samples, between cultures produced in tandem, and between cultures produced in the same medium but at different times. Additionally, we correlate the stable isotope ratios of carbon, nitrogen, oxygen, and hydrogen for growth medium nutrients or water with those of spores and show examples of how these relationships can be used to exclude nutrient or water samples as possible growth substrates for specific cultures.
The mailing of the anthrax letters in 2001 dramatically heightened concerns about the possibility of terrorist incidents involving microbiological agents. In the wake of the attacks, microbial forensics has emerged as a new focus area for research. Microbial forensics involves the characterization of microbial agents used as weapons for the purpose of identifying and eventually convicting those responsible (10, 11). Researchers in this nascent field have been working to develop methods that provide information useful in an investigation and evidence useful in a courtroom.
It became clear early on that genetic identity alone was not necessarily sufficient to lead investigators to a perpetrator. Analytical methods were needed that could differentiate genetically identical organisms produced under different conditions and also provide information about how a particular batch of organisms was produced. In response, scientists have been developing the use of a variety of analytical approaches to characterize changes in microbe signatures with culture conditions (9, 17, 59, 61, 63, 64). A major task faced in these studies is assessing how the profile of a given species varies in response to various culture conditions.
One analytical technique that has been explored as a microbial forensics tool is stable isotope ratio analysis (37, 43-46). Stable isotope ratios have proven to be of great forensic utility for natural products because of the direct link between the stable isotope ratios of organisms and their growth environments. In plants, stable isotope ratios of carbon (13C/12C), nitrogen (15N/14N), hydrogen (2H/1H), and oxygen (18O/16O), as well as soil elements such as strontium and lead, are controlled by growth environment and climate. Isotope ratios can link plant products to geographic growth regions, an application that has been referred to as geo-location. Geo-location through stable isotope ratios has been used to trace the origins of illicit drugs such as heroin and cocaine, as well as to determine the authenticity of plant-derived food products (1, 16, 25, 26, 33, 47, 65).
In animals, isotope ratios of tissues and molecules are a function of food and water sources (20, 21, 26a, 35, 48, 49). The isotope ratios of precipitation and therefore surface water are affected by geographically varying factors such as temperature and continentality (19, 40, 41), as are the O and H isotope ratios of plants that form the basis of the terrestrial food chain. Isotope ratios of animal tissues, particularly those of O and H, therefore, also show a geographic influence and can be used as natural tracers, for example, to follow migrations or to authenticate a region of origin (5, 14, 15, 30, 34).
A number of studies have clearly established that the stable isotope ratios of heterotrophic microorganisms, too, are a function of their nutrient and water sources (37, 43, 45, 46, 50, 56). Since the growth substrates of cultivated microorganisms are the water and nutrients mixed together in a container of culture medium, they are not necessarily related to the natural environment of the location in any way. The notion of geo-location through stable isotope ratios as used with plants and animals therefore does not apply to cultured microorganisms. Nevertheless, once isotopic relationships have been elucidated, stable isotope ratios can be used to limit the isotope ratios of media and water that could have been used to produce a batch of organisms, as well as to test whether samples of microorganisms could have been produced on specific batches of media or with specific sources of water.
Stable isotope ratios have also been suggested as a potential signature for sample comparisons. For this use of isotope ratios to be informative, there must exist the possibility for considerable variation in the stable isotope ratios of cultivated microorganisms. A survey of microbiological media sampled from laboratories around the United States has shown that there are substantial variations in C, N, O, and H isotope ratios of common growth media (44), and water isotope ratios are known to vary across the United States and across the world (see www.waterisotopes.org). There is thus sufficient potential isotopic variation in culture media to generate distinguishable isotope ratios among cultured organisms, provided within-sample and sample-to-sample variations can be thoroughly characterized.
Stable isotope ratios can provide useful, and potentially unique, information for forensic investigations. Here we illustrate applications in microbial forensics, using as a sample data set the C, N, H, and O stable isotope ratios from 247 spore cultures of Bacillus subtilis ATCC 6051 cultured on a total of 32 different media. We present an analysis of within-sample and sample-to-sample isotope ratio variation and show correlations between isotope ratios of growth environment and spores. In addition, we briefly describe approaches to the measurement of stable isotope ratios and discuss prospects for making measurements with very small samples.
MATERIALS AND METHODS
Media and culturing.
Spores of B. subtilis ATCC strain 6051 were cultured in liquid and on solid media as previously described (43, 45, 46). The nutrient media on which spores were cultured are listed in Table 1, along with the number of isotopically varying waters used to make the media. Previous experiments showed that autoclaving did not affect the isotope ratios of the medium water (46). Briefly, agar plates were streaked from a frozen stock culture, and isolated colonies from these plates were used to inoculate small overnight cultures. For experiments, liquid cultures were inoculated with a 1:200 dilution of the overnight culture, while agar plates were inoculated by spreading them with 500 μl of the overnight culture. All cultures were incubated at 37°C. Liquid cultures were shaken at 225 rpm, and their volume each occupied no more than 15% of the flask volume. We grew a minimum of three cultures in each nutrient medium-water combination. Broth cultures were incubated for about 48 to 72 h prior to harvest, while agar cultures were incubated for 2 to 14 days, but usually 2 to 4 days. Following harvest, spores were washed in the water used to make their growth medium a minimum of seven times over a minimum of 1 week to remove vegetative cells (31). Previous experiments showed that washing spores even in water that was isotopically very different from their growth water did not cause a statistically significant change in their O or H isotope ratios (46). Spores were lyophilized and stored in a vacuum desiccator prior to analysis. Each spore preparation was analyzed in duplicate for C, N, O, and H stable isotope ratios. The sample set consisted of 247 separate spore cultures produced on a total of 32 different nutrient media.
TABLE 1.
Media used for production of B. subtilis spores
| Nutrient medium | No. of isotopically distinct batches of water with which medium was prepared |
|---|---|
| Schaeffer's sporulation medium broth (31) made | |
| with three different nutrient broth powders | 4 |
| Schaeffer's sporulation medium agar made with | |
| two different nutrient broth powders | 4 |
| Schaeffer's sporulation medium broth with 0.2% | |
| glucose, made with two different nutrient | |
| broth powders | 4 |
| Nutrient broth | 4 |
| Tryptic soy broth | 4 |
| Tryptic soy agar | 4 |
| Luria broth | 4 |
| Columbia agar | 4 |
| 2 g beef extract, 6 g peptone per liter | 1 |
| 4 g beef extract, 4 g peptone per liter | 1 |
| 6 g beef extract, 2 g peptone per liter | 1 |
| 2 g yeast extract, 6 g peptone per liter | 1 |
| 4 g yeast extract, 4 g peptone per liter | 1 |
| 6 g yeast extract, 2 g peptone per liter | 1 |
| 2 g beef extract, 6 g tryptone per liter | 1 |
| 4 g beef extract, 4 g tryptone per liter | 1 |
| 6 g beef extract, 2 g tryptone per liter | 1 |
| 2 g yeast extract, 6 g tryptone per liter | 1 |
| 4 g yeast extract, 4 g tryptone per liter | 1 |
| 6 g yeast extract, 2 g tryptone per liter | 1 |
| 2 g beef extract, 6 g peptone, 2 g glucose per liter | 1 |
| 4 g beef extract, 4 g peptone, 2 g glucose per liter | 1 |
| 6 g beef extract, 2 g peptone, 2 g glucose per liter | 1 |
| 2 g yeast extract, 6 g peptone, 2 g glucose per liter | 1 |
| 4 g yeast extract, 4 g peptone, 2 g glucose per liter | 1 |
| 6 g yeast extract, 2 g peptone, 2 g glucose per liter | 1 |
| 2 g beef extract, 6 g tryptone, 2 g glucose per liter | 1 |
| 4 g beef extract, 4 g tryptone, 2 g glucose per liter | 1 |
| 6 g beef extract, 2 g tryptone, 2 g glucose per liter | 1 |
| 2 g yeast extract, 6 g tryptone, 2 g glucose per liter | 1 |
| 4 g yeast extract, 4 g tryptone, 2 g glucose per liter | 1 |
| 6 g yeast extract, 2 g tryptone, 2 g glucose per liter | 1 |
Stable isotope ratio measurements.
Microbiology studies involving stable isotopes often use labeled compounds, where isotopic content is high and expressed as the atom percent. In contrast, isotopic content at or near natural abundance, as in these studies, is expressed as a ratio (R) of the amount of the rare isotope to the abundant isotope (e.g., R = 2H/1H). Because the natural abundance of the rare isotope is low, these ratios are small numbers. Stable isotope contents are therefore usually expressed in “delta” notation as δ values in parts per thousand (per mille, or ‰), where δ‰ = (RA/RStd − 1) · 1,000‰, and RA and RStd are the molar ratios of the rare isotope to the abundant isotope (e.g., 2H/1H) in the sample and an internationally recognized standard, respectively. The standard used for both oxygen and hydrogen is Vienna Standard Mean Ocean Water (VSMOW [18]). The standard for carbon is the Peedee Belemnite (VPDB [18]), a fossil limestone from South Carolina; and the standard for nitrogen is air.
Light-element stable isotope ratios can be measured in bulk organic material by isotope ratio mass spectrometry. To measure C and N isotope ratios, the material is combusted in the presence of oxygen so that the carbon is converted to CO2; then, the combustion gases are reduced so that nitrogen oxides are converted to N2. The CO2 and N2 are separated chromatographically and flow sequentially into an isotope ratio mass spectrometer where the C and N stable isotope ratios of the gases are determined, reflecting the bulk isotopic composition of the sample. Bulk O and H isotope ratios are measured in a similar manner, except that pyrolysis in an oxygen-free atmosphere rather than combustion in the presence of oxygen is used to convert the sample to a gas. Oxygen in the sample is converted to CO and hydrogen to H2, which are chromatographically separated and flow into the isotope ratio mass spectrometer.
Stable isotope ratio measurements were made with isotope ratio mass spectrometers at the Stable Isotope Ratio Facility for Environmental Research (SIRFER) at the University of Utah. Laboratory standards calibrated to international standards are included as internal standards in every run. For C and N stable isotope analysis, 2 mg ± 10% of organic samples (growth media or spores) were weighed and placed into tin capsules. Carbon and nitrogen isotope ratios of each sample were determined using a Finnigan-MAT Delta S model isotope ratio mass spectrometer (IRMS, Thermo Fisher Scientific) interfaced with an Elemental Analyzer (model 1108; Carlo Erba, Milan, Italy). Samples were analyzed in duplicate, and the results were averaged. Measurement precision, reported as the standard deviations of measurements of laboratory standards accumulated over years, is 0.2‰ or less for carbon and nitrogen isotope ratios.
For O and H stable isotope analysis, 150 μg ± 10% was weighed and placed into silver capsules, which had been heated to 500°C for at least 30 min to remove silver oxide. The isotopic composition of each sample was determined with a Finnigan-MAT Delta Plus XL IRMS (Thermo Fisher Scientific) equipped with a thermal chemical elemental analyzer (Thermo Fisher Scientific) and a zero blank auto sampler (Costech Analytical, Valencia, CA). Samples were analyzed in duplicate, and the results were averaged. Measurement precision, reported as the standard deviations of measurements of laboratory standards accumulated over years, is 2‰ for hydrogen and 0.2‰ for oxygen.
RESULTS
Microbial forensics has been defined as “a scientific discipline dedicated to analyzing evidence from a bioterrorism act, biocrime, or inadvertent microorganism/toxin release for attribution purposes” (12). In a crime or a terrorist incident involving microorganisms, the first fact to be established about the microbe is its genetic identity. Further information that could be sought includes the method of production, the type of medium used, and whether samples used in two separate incidents appeared to be aliquots from the same batch of organisms. If a suspect was identified, it might become important to determine whether the organisms could have been cultured on a specific medium to which the suspect had access.
There are two ways in which stable isotope ratios could be useful in such an investigation. One way is as a signature for sample matching and comparison. In this application, two or more samples would be compared to determine their degree of similarity. As we will show below, stable isotope ratios have the potential to distinguish between genetically identical organisms grown in different batches under otherwise identical conditions. Bulk stable isotope ratios of C, N, O, and H can be determined quickly and relatively inexpensively and therefore potentially constitute a good screening tool for comparison.
The second application of stable isotope ratios is to associate a sample with potential growth media. The C, N, and H isotope ratios of spores have been shown to be linear functions of the ratios in powdered growth media, and the O and H isotope ratios are linear functions of those in growth water (43). Growth medium also contributes to the oxygen isotope ratios of spores, but the oxygen isotope ratios of spores are not a linear function of those measured in powdered growth media. Presumably, this observation is a result of biochemical processing of medium oxygen atoms during growth. For example, many media are rich in amino acids, and each amino acid has an oxygen atom in its hydroxyl group that would contribute to the O isotope ratio of the powdered medium. Since medium components such as casein hydrolysates and meat hydrolysates are precisely that, hydrolysates, the oxygen atoms in these hydroxyl groups presumably originated from the water used to process the casein or meat and may be isotopically distinct from the oxygen atoms that were incorporated into the proteins during biosynthesis. These same oxygen atoms would be lost during protein synthesis and, thus, would not be found in the spore biomass to the extent that they were present in the medium. Although one would make the same prediction concerning the hydrogen atoms in the hydroxyl groups of amino acids, there are apparently enough additional hydrogen atoms in the amino acid molecules to dampen any signal from them, since our observations are that spore H isotope ratios do correlate with those of growth media.
Thus, the C, N, and H isotope ratios of spores can be used to constrain those of the growth medium used to produce them or to test whether a particular batch of culture medium or a particular water supply could have been used to produce a particular batch of organisms. We shall illustrate these two applications of stable isotope ratios using data obtained with B. subtilis spores. The same general principles should apply to other bacteria, whether or not they form spores. It is important to note that the signatures of interest in a forensic investigation will be lost if the original material is subcultured; in this case, the isotope ratios of the organisms will reflect those of the medium used for subculturing.
Stable isotope ratios as signatures for sample comparisons.
The stable isotope ratios of a particular batch of microorganisms are a function of the nutrient medium and water used for culturing (34, 43, 45, 46, 55, 56, 58), as well as culturing practices. For example, incubation time can affect the oxygen isotope ratio, if conditions are such that significant evaporation of water from the medium occurs while spores are forming (45). Postculturing procedures such as washing (in the case of spores) or the addition of further components could also affect bulk isotope ratios. The contributions from both growth medium (nutrients and water) and culturing practices imply that stable isotope ratios should have discriminatory power as a signature for sample matching and comparison.
We begin our analysis by comparing within-batch to between-batch variations in spore isotope ratios. We hypothesized that if two samples were taken from the same spore production batch, their isotope ratios should be the same, within measurement precision. The combination of isotope ratios should constitute an isotopic signature for that production batch. We further hypothesized that organisms cultured on the same medium but at different times should be isotopically very similar to one another but not necessarily as similar as organisms from the same production batch.
Our data allow us to compare variations in isotope signatures of genetically identical spore preparations in the following situations: different samplings of the same batch (referred to as replicate samples), multiple batches made on the same day with the same materials by the same individual (referred to as tandem batches), preparations done with the same materials but at different times and by different people (referred to as temporally different batches), preparations made with the same nutrients but with different waters, preparations made with the same waters but with different nutrients, and preparations that vary in both water and nutrients.
Table 2 shows the variability in isotope ratios observed at three different levels. Table 2, duplicate measures, shows the average standard deviations of C, N, H, and O isotope ratio measurements made with duplicate aliquots taken from the 247 spore preparations. In some cases one of the duplicate analyses failed, accounting for the discrepancies between n, the number of samples analyzed for each element, and 247. Table 2, triplicate cultures, shows the average standard deviations of isotope ratio measurements made with triplicate cultures produced in tandem on the same day and processed together. The values compared were the averages of the two duplicate measurements made with each sample. Table 2, cultures produced in the same medium but different production runs, shows the average standard deviations of isotope ratio measurements made on cultures produced on the same medium (both nutrients and water) but grown and processed at different times. The values compared, like those of triplicate cultures, were the averages of the two duplicate measurements made with each sample. The broth cultures in this section could have been incubated for 2 or 3 days, while the agar cultures were produced on plates stored for various lengths of time prior to inoculation and could have been incubated anywhere from 2 to 14 days but usually 2 to 4 days.
TABLE 2.
Variation in C, N, O, and H stable isotope ratios of B. subtilis spores
| Element | Avg SD (‰)
|
|||||
|---|---|---|---|---|---|---|
| Instrument precisiona | Duplicate measures of one spore culture (no. of cultures)b | Triplicate cultures produced in same medium and processed in tandem
|
Cultures produced in the same medium but in different production runs with potentially different incubation times
|
|||
| Broth cultures (n = 41) | Agar cultures (n = 20) | Broth cultures (n = 15 media, 49 total) | Agar cultures (n = 8 media, 53 total) | |||
| C | 0.20 | 0.08 (238) | 0.12 | 0.09 | 0.34 | 0.36 |
| N | 0.20 | 0.10 (239) | 0.16 | 0.29 | 0.39 | 0.36 |
| O | 0.20 | 0.20 (245) | 0.25 | 0.27 | 0.71 | 0.79 |
| H | 2.0 | 1.5 (246) | 1.8 | 2.4 | 4.4 | 3.1 |
Standard deviation of laboratory reference material measured over time.
Broth and agar cultures.
We note that the variability reported in Table 2 is inherently nested. In particular, the tandem batch measurements include variability arising from the tandem cultures as well as from measurement variability. Similarly, measurements from temporally different batches include measurement variability, tandem batch variability, and variability due to culture preparation at different times by different people. Taking advantage of this variability structure, we performed a nested analysis of variance (ANOVA) (36) to identify significant sources of variation.
The results indicate that the tandem-batch variation is significantly larger than the replicate sample variability for all isotope ratios (P < 0.001 for C, N, O, and H). Water is a significant source of variation in O and H (P < 0.001 for O and H). Different lots of the same medium preparation produced significant variation in C, N, and H isotope ratios (P < 0.001). Additionally, spores prepared in different lots of the same medium preparation showed as much variability in C, N, and, H as spores prepared in different media (i.e., P > 0.3 for C, N, and H when comparing between-medium variability to lot-to-lot variability).
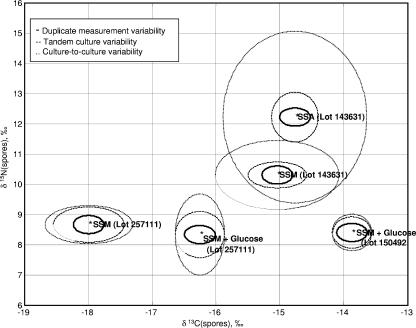
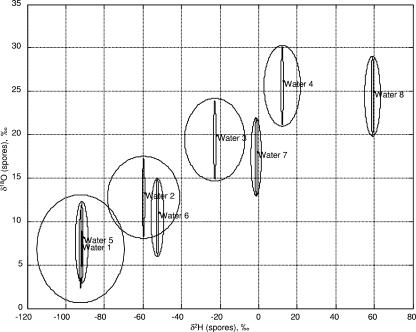
As part of nested ANOVA, the contribution of different sources of variability was computed. Figure 1 (and Fig. 2) shows plots of an uncertainty region around C and N (and O and H) isotope ratios for spores prepared in Schaeffer's sporulation medium (SSM [31]), Schaeffer's sporulation medium plus agar (SSA), and SSM plus glucose media. The uncertainty region is estimated to be three times the standard deviation of each variability measurement. Three times the standard deviation was chosen because for most statistical distributions, 99% or more of the population falls within three times the standard deviation of the mean. We note that a variability contribution of zero (as with the between-culture variability in 13C for spores prepared in SSM plus glucose) indicates insufficient replicates were present to compute a variability estimate, and does not imply a standard deviation of zero.
FIG. 1.
Computed uncertainty intervals (with three times the standard deviation) surrounding the means for 13C and 15N isotope ratios of B. subtilis 6051 spores produced in closely related media.
FIG. 2.
Computed uncertainty intervals (with three times the standard deviation) surrounding the means for 18O and 2H isotope ratios of B. subtilis 6051 spores produced on various media in isotopically varying water.
Figures 1 and 2 visually confirm the results of the nested ANOVA. In particular, the tandem batch variability is larger than the measurement variability but smaller than the variability in the temporally different batches. In the case of C and N, the spores prepared with different lots of the same medium appear to have as much variation as spores prepared with different media. For O and H, water appears to be the most significant source of variation.
The data were used to explore the potential of isotope ratios for signature matching. In particular, a simple statistical test was developed to address whether or not isotope ratios can be used to determine if two samples (i) originated from the same batch or if they (ii) were grown under identical conditions (but not necessarily the same batch). The test was constructed by placing a prediction interval around the average C, N, O, and H measurements for a given sample. (To address the first question, the replicate sample variability was used to construct the prediction interval. To address the second question, the temporally different batch variability was used.) The remaining measurements in the database were then treated as unknowns and were compared to the prediction interval for that sample. A set of measurements was determined to be consistent with the sample if all values fell within the prediction interval. Otherwise, it was determined to be inconsistent. This process was repeated for all measurements for a select subset of the data.
Table 3 shows the results of tests performed to determine whether or not two samples could have been prepared in the same batch. Three types of errors are reported: the number of same-batch comparisons erroneously resulting in “inconsistent,” the number of across-batch (tandem and temporally different) comparisons erroneously resulting in “consistent,” and the number of different water, medium, and/or lot comparisons erroneously resulting in “consistent.” Out of a total of 46 within-batch comparisons, four (9%) resulted in an erroneous “inconsistent” conclusion. This low overall error rate suggests that isotope ratios have the potential to reliably associate cocultured spores. On the other hand, the number of across-batch comparisons resulting in a “consistent” conclusion was 63 out of 220 (29%). This implies that if the isotopic signatures of an unknown sample fell within the prediction limits for a reference sample, it would not necessarily imply that the samples were cocultured. However, these false consistent results were obtained with spores that were cultured on the same nutrient medium made with the same water and, thus, were produced under very similar conditions. The number of different water, medium, and or lot comparisons resulting in a false “consistent” determination was 20 out of 10,740 (0.2%), supporting our hypothesis that isotope ratios provide a distinct signature for culture conditions.
TABLE 3.
Comparison results for testing whether or not two spore samples were cocultured
| Water sample no./medium/lot ID | No. of false inconsistent decisions (no. of comparisons) using:
|
||
|---|---|---|---|
| Cocultured spores | Same water/medium/lot spores | Different water/medium/lot spores | |
| 1/LB/819278-2 and 823252-3 | 0 (3) | 3 (7) | 3 (447) |
| 1/SSA/150492 | 2 (18) | 40 (135) | 3 (3,624) |
| 1/SSM/143631 | 0 (3) | 3 (12) | 11 (942) |
| 1/SSM/150492 | 0 (1) | 1 (27) | 0 (453) |
| 2/SSA/150492 | 0 (6) | 7 (9) | 1 (1,338) |
| 3/SSA/150492 | 0 (6) | 1 (9) | 0 (1,320) |
| 4/SSA/150492 | 1 (6) | 4 (9) | 0 (1,302) |
| 5/SSA/150492 | 1 (3) | 4 (12) | 2 (1,314) |
| Total | 4 (46) | 63 (220) | 20 (10,740) |
Table 4 shows the results of the test to determine whether or not two samples were cultured under identical conditions (not necessarily in the same batch). In this case, the number of same water, medium, and/or lot comparisons resulting in an erroneous “inconsistent” conclusion was 12 of 266 (4.5%). The number of different water, medium, and/or lot comparisons resulting in an erroneous “consistent” conclusion was 260 of 10,740 (2.4%). Both of these error rates are quite small, providing further evidence that isotope ratios have the potential to become a valuable tool for sample matching in microbial forensics.
TABLE 4.
Comparison results for testing whether or not two spore samples were cultured under the same conditions
| Water sample no./medium/lot ID | No. of false inconsistent decisions (no. of comparisons) using:
|
|
|---|---|---|
| Same water/medium/lot spores | Different water/medium/lot spores | |
| 1/LB/819278-2 & 823252-3 | 0 (10) | 7 (447) |
| 1/SSA/150492 | 7 (153) | 81 (3,624) |
| 1/SSM/143631 | 1 (15) | 92 (942) |
| 1/SSM/150492 | 3 (28) | 40 (453) |
| 2/SSA/150492 | 0 (15) | 6 (1,338) |
| 3/SSA/150492 | 1 (15) | 0 (1,320) |
| 4/SSA/150492 | 0 (15) | 3 (1,302) |
| 5/SSA/150492 | 0 (15) | 31 (1,314) |
| Total | 12 (266) | 260 (10,740) |
Stable isotopic correlations to growth environment.
The other application of microbe stable isotope ratios is as an indicator of the isotope ratios of the growth medium nutrients and water that were used to produce the sample. The O and H isotope ratios of organisms are a function of both medium nutrients and water, while C and N ratios are a function of medium nutrients alone.
Stable isotope ratios cannot identify the type of culture medium used to produce a given batch of spores, but they can reveal general information about the sources of carbon and nitrogen (for a discussion of sources of isotopic variation in culture media, see reference 44). For example, nitrogen derived from animal protein such as casein or meat digests has a different stable isotope ratio than nitrogen derived from yeast extracts (44). These differences impart corresponding differences to spores grown in animal- versus yeast-based medium (43). The ranges of observed isotope ratios of standard medium components such as glucose, tryptone, peptones, and yeast extracts can be used to predict the isotope ratios of complex media so that it is possible in some cases to use stable isotope ratios to rate certain media as highly unlikely potential growth substrates even in the absence of other data (K. H. Jarman and H. W. Kreuzer-Martin, unpublished data).
The unique usefulness of stable isotope ratios in correlating media with spores is that the isotope ratios of spores constrain the isotope ratios of their culture medium to specific ranges. This permits the testing of specific medium samples (such as traces of powder found at a suspect location or medium recovered from a suspect laboratory) and water samples to determine whether their isotope ratios are consistent with those of the spores. Such testing would be informative even if the identity of the culture medium used to produce the organisms could not be determined. If the identity of the culture medium had been determined, isotope ratios could eliminate specific lots of culture medium from consideration, since the isotope ratios of different lots of the same medium sold by the same manufacturer can vary (44).
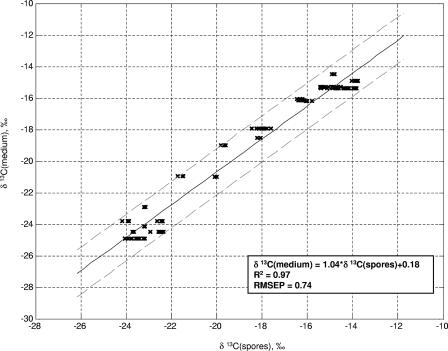
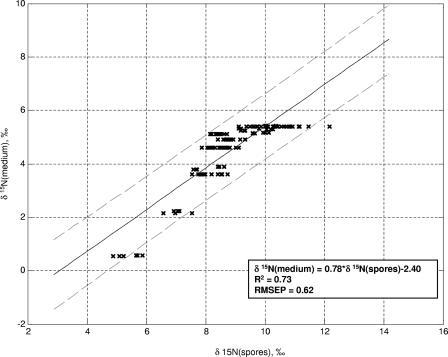
Figure 3 shows a regression of the carbon isotope ratio of liquid growth medium onto the carbon isotope ratio of spores, while Fig. 4 shows a corresponding regression for nitrogen isotope ratios. We present data only for spores produced in liquid media because most of the 32 different media used here were liquid media, and the presence of agar is often associated with evaporative enrichment of the water in the medium (evaporation increases the relative amounts of 2H and 18O in the residual water [40]) (45), thereby affecting the O and H isotope ratios of spores. We note that a method has been developed for detecting traces of agar on even well-washed spores (24); therefore, we will assume that it is possible to determine whether the medium used to produce a given batch of spores contained agar or not.
FIG. 3.
Relationship between C isotope ratios of spores and culture medium, measured in B. subtilis 6051. Spores were produced on the liquid media listed in Table 1. RMSEP, root-mean-squared error of prediction.
FIG. 4.
Relationship between N isotope ratios of spores and culture medium, measured in B. subtilis 6051. Spores were produced on the liquid media listed in Table 1. RMSEP, root-mean-squared error of prediction.
The stable isotope ratios of growth medium water can be of particular forensic interest. As stated above, the isotope ratios of surface water are influenced by geography and climate, and thus, vary geographically (23, 41). Isotope ratios of tap water vary geographically as well, in a manner similar to but not identical to that of surface water (7). Although the isotope ratios of precipitation water vary seasonally with temperature, sometimes by large amounts, the isotope ratios of surface water and tap water are far less variable (7, 23). Thus, if a particular location is suspected of being the site of production of microorganisms used in a criminal act, it is possible to determine whether the hydrogen and oxygen isotope ratios of the microorganisms are consistent with production in the local water or in water from a specific laboratory or other facility.
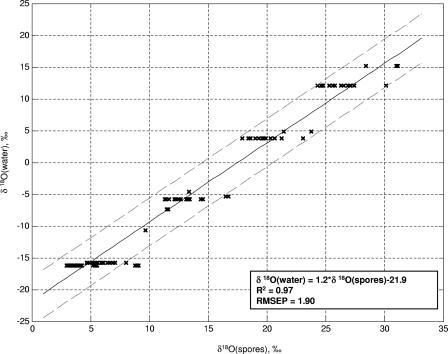
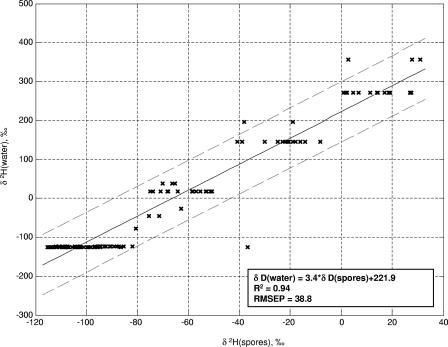
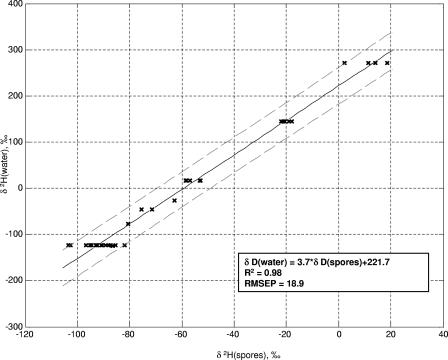
Figures 5 and 6 show, respectively, regressions of the oxygen and hydrogen isotope ratios of growth medium water onto those of spores produced in media made with those waters. The large, 95%, confidence interval of prediction is predominantly caused by the contribution of the medium to the spore isotope ratios. Approximately 70% of the oxygen atoms in B. subtilis spores originate in culture water, while only 30% of the hydrogen atoms do (43, 46). The curves in Fig. 5 and 6 use data from spores produced on many different media and thus should provide a good approximation of the variation that could be seen in spores produced in the same water using various growth substrates. Figure 7 shows a regression of water hydrogen isotope ratios onto spores that were all produced in the same nutrient medium; the error of prediction is dramatically reduced.
FIG. 5.
Relationship between O isotope ratios of spores and water from liquid culture media, measured in B. subtilis 6051. Spores were produced in the liquid media listed in Table 1 made with isotopically varying water. RMSEP, root-mean-squared error of prediction.
FIG. 6.
Relationship between H isotope ratios of spores and water from liquid culture media, measured in B. subtilis 6051. Spores were produced in the liquid media listed in Table 1 made with isotopically varying water. RMSEP, root-mean-squared error of prediction.
FIG. 7.
Relationship between O isotope ratios of spores and water from liquid SSM cultures, measured in B. subtilis 6051. SSM was made with the identical nutrients but isotopically varying water. RMSEP, root-mean-squared error of prediction.
Growth water stable isotope ratios are of greatest utility if a suspected production location has been identified. In this case, the O and H isotope ratios of the sample can be compared to those of the location water to determine whether it could have been used to produce the culture. If a microbiological sample has been recovered but no suspected production sites have been identified, the stable isotope range of potential growth water can be compared on maps showing the isotope ratios of surface and tap waters to identify locations that supply water within the target range. This information would probably be of very limited value, as water from large regions of the United States and the world has similar isotope ratios. Additionally, a perpetrator could have purchased water for culture production that was not isotopically linked to his or her geographic location, a possibility that must be considered even when a suspected production location has been identified. Finally, although deionization does not affect the hydrogen and oxygen isotope ratios of water, incomplete distillation does. If a perpetrator used incompletely distilled water, its isotope ratios could also be different from those of the local water from which it was derived. Thus, the situation in which predicted isotope ratios of growth water are most useful is when known samples are available for comparison.
A sample application.
For an illustration of how stable isotope signatures might be useful in a forensic investigation, Table 5 shows stable isotope ratios of six samples of B. subtilis 6051. All of the samples were produced in Schaeffer's sporulation medium, which consists of nutrient broth powder (8 g/liter) with added salts. The samples were inoculated from agar plates or slants that were streaked from the same frozen stock culture, and thus, their genetic profiles should be within the degree of similarity expected of cultures produced in the same laboratory. (Other biochemical signatures indicative of medium type, such as peptide profiles [59, 63] or elemental content [17], could potentially distinguish genetically identical samples cultured on different growth media and might also be indistinguishable among these samples.)
TABLE 5.
C, N, O, and H stable isotope ratios of genetically identical spore cultures of B. subtilis produced in different batches of liquid Schaeffer's sporulation mediuma
| Sample | Stable isotope ratio (‰)
|
|||
|---|---|---|---|---|
| δ13C | δ15N | δ18O | δ2H | |
| 1 | −14.2 | 10.8 | 13.4 | −62.8 |
| 2 | −14.9 | 9.1 | 11.6 | −71.5 |
| 3 | −14.3 | 10.6 | 9.7 | −80.6 |
| 4 | −14.3 | 10.9 | 11.5 | −75.5 |
| 5 | −14.6 | 10.3 | 6.0 | −92.9 |
| 6 | −18.0 | 8.6 | 5.6 | −109.3 |
All of these cultures can be excluded from identity with one another by bracketing their stable isotope ratios with a ±3-standard deviation interval, using the standard deviations in Table 2, duplicate measures column.
A comparison of the stable isotope ratios of the samples shows that they are inconsistent with any of the samples having been cocultured, using three times the standard deviations listed in Table 2, replicate samples, as confidence intervals. The C and N isotope ratios of samples 1 to 5 are consistent with culturing on the same nutrient medium, using three times the standard deviations listed in Table 2, temporally different batches, or using the regressions in Fig. 3 and 4. In contrast, the C and N isotope ratios of sample 6 are inconsistent with culturing on the same medium as samples 1 to 5. The O and H isotope ratios of samples 1 to 5 are not consistent with all of them as temporally different batches grown under the same conditions, though some could be (samples 1, 2, and 4 or 3 and 5). The relationships in Fig. 3 to 6 can be used to constrain the isotope ratios of the culture media and water used to produce the samples.
Because the O and H stable isotope ratios of spores are a function of both nutrients and water, it is not possible to determine precisely the isotope ratios of water that was used to produce a given sample from its bulk isotope ratios alone. However, the bulk isotope ratios can be used to determine whether a given sample could have been produced using water of given isotope ratios. For example, assume two laboratories were under suspicion as production locations, in which laboratory A has water isotope ratios δ18O of −16.2‰ and δ2H of −122‰ and laboratory B has water isotope ratios δ18O of −4.6‰ and δ2H of −27.3‰. The oxygen isotope ratios of samples 5 and 6 are consistent with production in water from laboratory A, while those of samples 1 to 4 are not. At the same time, the oxygen isotope ratios of samples 1, 2, and 4 are consistent with having been cultured in water from laboratory B, while those of samples 3, 5, and 6 are not.
In actuality, samples 1 to 5 were produced in laboratories in different cities using media made with the same production lot of nutrient broth, the same added salt solutions, and the local laboratory water (46). Sample 6 was produced in the same laboratory with the same water as sample 5 was, but the medium was made using a different bottle of nutrient broth powder. Although the samples were cultured in the same medium using the same method, their stable isotope signatures were inconsistent with their derivation from different aliquots of one large culture (coculture). The stable isotope ratios clearly showed that sample 6 had been grown in a different nutrient medium (the different bottle of nutrient broth) and that the cultures could not all have been produced using water from the same source.
DISCUSSION
The growth environment of a microorganism can leave signatures in the form of external residue associated with or clinging to the organisms or as intrinsic signatures in the form of variations in the composition of the organism itself. Within the nascent field of microbial forensics, techniques are being explored to detect both external signatures such as traces of heme or agar on spores (24, 62) and intrinsic signatures such as elemental composition (17), peptide fingerprints (59, 63), and stable isotope ratios. It is likely that combining data from a variety of external and intrinsic signatures will yield the most information about the method and medium used to produce a given sample. Recent trials involving multiple cultures produced on various media support this assumption. In these experiments, the greatest accuracy in predicting the culture media used to produce various batches of spores was obtained when data from elemental analysis, peptide fingerprints, and stable isotope ratio analyses were integrated (38, 39; K. H. Jarman, unpublished data).
Sample homogeneity and batch-to-batch variation.
Sample homogeneity is a critical issue for any type of chemical analysis, since heterogeneity can lead to significantly different results when two aliquots of the same sample are analyzed. The data shown in Table 2, duplicate measures, demonstrate that our samples were homogeneous. Homogeneity is required for accurate sample matching by isotope ratios or any other means. Our samples were washed extensively in water; if spores were not separated from vegetative cells and the remnants of spent growth media by washing, they might present far less homogeneous material. This type of heterogeneity would present a problem for many types of analyses. It is possible that “dirty” cell or spore preparations could be cleaned in such a way as to preserve the contaminating material for analysis of external indicators of production method and the cells or spores for analysis of intrinsic signatures. The development of such procedures would be desirable.
We did observe more batch-to-batch variation in the isotope ratios of spores cultured on the same medium than we observed in duplicate analyses of the same batch of spores. This result could be explained by different washing efficiencies from one batch to the next, leaving different amounts of contaminating material in the spore preparations (which we did not assess). The variation in isotope ratios could also be a reflection of intrinsic variation in the different batches of spores.
One factor that might have contributed to batch-to-batch variation in spores produced on the same medium at different times could be storage of the salt solutions added to many of the media. Solutions of FeCl2 and MnCl2 are made separately and added to Schaeffer's sporulation medium to promote sporulation. We used this medium for many of our cultures, both broth and agar, and we stored the salt solutions at room temperature and used them over and over again over time. Other researchers have observed that storage of such solutions before they are added to growth media changes the response of organisms to the media, presumably because of oxidation of the metals (N. Valentine, personal communication). If sporulation is less efficient on media made with old salt solutions, the resulting spore preparations could be contaminated with more vegetative cells, changing the isotope ratios of the preparations. We have not tested the effect of storage of salt solutions or metal ion oxidation on isotope ratios of spore preparations.
Another issue that could have had minor effects on batch-to-batch variation in oxygen and hydrogen isotope ratios is the potential for environmental water to affect the measured isotope ratio. There are two ways in which this could occur: by adsorption of vapor-phase water molecules or by chemical exchange of hydroxyl and amino hydrogen atoms with ambient vapor (54). Measurements in our laboratory suggest that about 3% of H atoms in B. subtilis spores exchange with ambient water (44 and unpublished data). This value is lower than what has been observed for hair keratin and other large organic molecules, where exchange on the order of 11% has been observed (6, 54). Thus, two aliquots of the same material, analyzed in, for example, Salt Lake City and Houston, where ambient water hydrogen isotope ratios differ by about 100‰, would differ in δ2H values by about 3‰ due to hydrogen exchange.
Hydrogen exchange and vapor adsorption could have had minor effects on the results reported here. The samples analyzed in this study remained submerged in the water used to wash them (which was the same water used to make their growth media), until they were lyophilized and then immediately put into a vacuum desiccator for storage. They were removed from the desiccator for weighing, which required various amounts of time but usually about 4 h for a sample set. Trays containing weighed samples were returned to the desiccator for various amounts of time prior to analysis. During the weighing interval, both water adsorption and hydrogen exchange could have occurred.
No studies of the kinetics of hydrogen exchange or water adsorption of spores have been published, but available data from hair equilibration studies, in which complete hydrogen exchange was achieved after 2 to 4 days at room temperature (6), suggest that exchange would not have been extensive. Adsorbed water would be removed during subsequent desiccation, but the different lengths of storage prior to analysis could mean that some samples contained residual adsorbed water. The larger variation in hydrogen and oxygen isotope ratios in cultures produced in the same medium but at different times, as opposed to those produced and processed in tandem, could be due in part to these effects.
Potential issues arising from hydrogen exchange or water adsorption can be controlled by the development of standardized sample handling procedures, such as those recently proposed by Bowen et al. for isotopic analysis of hair (6). After careful measurements of the kinetics of both hydrogen exchange and water adsorption, Bowen et al. recommended that, prior to analysis, both samples and calibrated hair standards be equilibrated with laboratory air, thoroughly vacuum dried, and then transferred to the helium atmosphere of the IRMS with minimal exposure to laboratory air. This method would minimize the adsorbed vapor content of the sample and permit correction to the standard. A similar approach should be applicable to the analysis of microorganisms and other organic samples. Once the kinetics of adsorption and exchange in spores have been described, specific recommendations for processing of spore samples could be developed.
Sample size considerations.
In this study we measured stable isotope ratios by isotope ratio mass spectrometry (IRMS), which offers the advantage of high throughput and high-precision measurements. For example, the measurement precision for carbon stable isotope ratios at the University of Utah facility, expressed as the standard deviation of repeated measurements of a laboratory standard over time, is 0.2‰. In this study the average standard deviation of repeated measures of spore samples was 0.08‰. The sample size required for the procedures we used was normally around 100 μg for an analysis of O and H isotope ratios and another 150 μg for an analysis of C and N, although we used larger samples for the C and N measurements in these studies.
The requirement for these sample sizes is not a function of the mass spectrometer itself but rather a consequence of the configuration for chromatography of the combustion gases prior to their introduction into the instrument. Isotope ratio mass spectrometers can be coupled to gas or liquid chromatographs so that precise stable isotope ratios of individual compounds in eluant streams can be measured. In these cases, only a few nanograms of material is required per peak (13, 32, 57). In principle, the sample introduction equipment for combusting or pyrolyzing solids prior to IRMS measurements could be modified so that far smaller samples could be analyzed.
For forensic applications, it would obviously be an advantage to be able to measure isotope ratios of very small samples. Secondary ion mass spectrometry (SIMS) (3), also called ion microprobe analysis, can provide isotopic content data for much smaller samples than is currently possible with IRMS (53). With SIMS, a primary ion beam erodes a sample surface, resulting in the release of secondary ions whose mass/charge ratios are measured by mass spectrometry. SIMS analyses require far smaller samples than do the IRMS procedures we used in this study and can be used to determine elemental or isotopic content. Cliff et al. used time-of-flight (TOF)-SIMS (60) for elemental analysis of spores and reported that their analyses could theoretically be carried out with 1 ng of material (about 1,000 spores) (17). The precision at which natural abundance stable isotope ratios could be measured in bacteria by TOF-SIMS has not been determined but may approach the precision available for magnetic sector instruments, albeit at the cost of longer analysis times (28).
High-spatial-resolution magnetic sector SIMS instruments have been used to make isotope ratio measurements from individual bacterial cells (51, 52). However, there are measurement precision tradeoffs for small samples (29). In preliminary studies, Weber found that he could measure C isotope ratios in a single spore with a 95% confidence interval of ∼6‰ (P. K. Weber, personal communication). With a sample size of 30 spores, the confidence interval was 2‰. Given that the potential C isotope ratio range for spores cultured in common growth media is probably about 15‰ (35), a confidence interval of 6‰ would make it impossible to distinguish most samples from one another or make useful inferences about growth media. A confidence interval of 2‰ would allow exclusion of some growth media as possible production media and would permit exclusion of the possibility of coculture for some samples as well. While increased precision is possible with the analysis of more spores, systematic studies to determine the precision with which high-resolution SIMS can measure C and other stable isotope ratios in bacteria are needed.
The prospect of obtaining isotope ratios from single spores or small numbers of spores is exciting, but it raises particular issues regarding sample homogeneity. For example, microheterogeneities in the growth environment of a single culture, such as whether a spore was formed next to the agar or on top of other spores or whether it formed earlier or later during culturing, could affect its isotope ratios. There is a small amount of published data suggesting that cell-to-cell isotopic variation could be significant. In reference experiments, Orphan et al. (51) measured C isotope ratios of individual cells from cultures of two different cyanobacteria. Although the average C isotope ratios of the combined measurements were comparable to bulk measurement, the authors observed standard deviations of 7.5‰ in the cell-to-cell measurements for one strain (n = 13) and 6.3‰ for the other (n = 23). They attributed these measurements to heterogeneity in the organisms. We are not aware of any published studies of cell-to-cell isotopic variation in cultured heterotrophic bacteria.
The effects of any culture microheterogeneities would likely be smoothed over in analyses that involved many cells or spores, such as the IRMS approaches described here and the SIMS analyses carried out with samples consisting of many spores. However, the data from the cyanobacterial studies highlight the need for careful study of spore-to-spore isotopic variation using spores produced under a variety of conditions before data acquired from single or very small numbers of spores can be interpreted and defended with confidence.
Potential contributions of stable isotope ratio analysis to microbial forensics.
The most powerful contribution that stable isotope ratio analysis can make to microbial forensics will probably be its ability to distinguish between otherwise chemically identical samples and to associate recovered microorganisms with culture media and water. Isotopic analyses will have to be performed with original material, since subculturing will cause the organisms to take on the signature of that environment. As described above with the data in Table 5, stable isotope ratios can distinguish between batches of genetically identical organisms produced on media identical in every aspect except for the isotope ratios of the water used to make them. Stable isotope ratios can also distinguish between batches of genetically identical organisms grown in nominally identical media made with different lots of reagents, if those reagents are isotopically distinct. There is no guarantee that different batches of any powdered growth medium will be isotopically distinct, but the likelihood of this increases as more elements are included in a signature. It is likely that there is also variation in other elements such as S and Sr, which could also be reflected in spores or cells. Though no studies of isotope ratios of these elements in bacteriological media have been published, data for their variations in plant and animal materials suggest that microbiological media should vary as well (2, 4, 5, 8, 27, 42). If that variation is translated into microbes, then incorporation of these elements into an isotopic signature would add to its discriminating power.
Stable isotope ratio signatures could also be measured as records of production batches of organisms at the time of production, at facilities that produce and store select agents or other critical reagents. This would enable a producer to compare a seized sample to their production records to determine whether or not that material might have come from their facility. The stable isotope ratios of organisms could also be deliberately manipulated through labeling to create a unique, nonnatural signature that could act as a unique identifier of a particular producer. Genetically identical organisms cultured under different conditions might possess the same genotype, but would not carry the same isotopic fingerprint.
Another potentially important application of stable isotope ratios might be to help determine whether a pathogen could be a natural product of the local environment, if genetic analysis could not provide an unambiguous answer. The stable isotope ratios of, for example, Escherichia coli O157 produced in local cattle should be predictable based on the diets of local animals and the isotope ratios of local water, although baseline studies to establish these isotopic relationships have not been carried out.
For an analytical signature of a microorganism to be useful in a forensic investigation, there must be a robust scientific foundation to support interpretation of the signature. Before any signature is useful, its inherent variability from one sample to the next must have been determined using samples produced under a variety of conditions and in whatever sample size is relevant to the analytical technique in question. If the signature is to be used for making inferences about the production environment of the agent, then that foundation must include an understanding of the basis for the variation in the signature and the relationship of the signature to the growth and/or production conditions.
The scientific basis for isotopic variation in microorganisms, as in other heterotrophs, is well understood. We have presented data that describe batch-to-batch variation in isotope ratios of B. subtilis 6051 as well as the isotopic relationships between B. subtilis 6051 spores and growth environment. Although these data demonstrate proof of principle, they need to be collected from B. anthracis spores to determine whether these same relationships or slightly modified ones hold true for that species.
Another variable that has not been explored in published literature to date is whether there is significant variation in the growth environment/microbe isotopic relationships among different strains of the same species. It would be useful to assess whether that variation is likely to be large or small. Studies that define the isotopic relationship between non-spore-forming organisms and their growth environment are needed, too. Although the same uses of stable isotope ratios as fingerprints and as recorders of growth environment should be true of non-spore-former organisms, there are additional variables such as the growth phase at harvest time that need to be addressed.
Finally, our studies assumed that no additional information about the spore growth environment was available other than whether or not it had contained agar. If additional techniques for characterizing other aspects of growth environment are developed and validated, it may well be possible to make more precise interpretations of isotopic evidence. Our measurements were also all made on bulk samples. By characterizing the isotope ratios of individual compounds purified from spores, it might also be possible to decrease the uncertainty of interpretations.
Acknowledgments
We thank John Cort, John Cliff, and Karen Wahl of Pacific Northwest National Laboratory, Peter K. Weber of Lawrence Livermore National Laboratory, and Christopher H. House and Jennifer F. Biddle of Pennsylvania State University for helpful discussions. We thank Lesley A. Chesson, Michael J. Lott, and James R. Ehleringer of the University of Utah for assistance with sample production and analysis.
Footnotes
Published ahead of print on 27 April 2007.
REFERENCES
- 1.Anderson, K. A., and B. W. Smith. 2005. Use of chemical profiling to differentiate geographic growing origin of raw pistachios. J. Agric. Food Chem. 53:410-418. [DOI] [PubMed] [Google Scholar]
- 2.Beard, B. L., and C. M. Johnson. 2000. Strontium isotope composition of skeletal material can determine the birth place and geographic mobility of humans and animals. J. Forensic Sci. 45:1049-1061. [PubMed] [Google Scholar]
- 3.Benninghoven, A., F. Rudenauer, and W. Werner. 1987. Secondary ion mass spectrometry. Wiley, United Kingdom.
- 4.Bol, R., and C. Pflieger. 2002. Stable isotope (13C, 15N and 34S) analysis of the hair of modern humans and their domestic animals. Rapid Commun. Mass Spectrom. 16:2195-2200. [DOI] [PubMed] [Google Scholar]
- 5.Boner, M., and H. Forstel. 2004. Stable isotope variation as a tool to trace the authenticity of beef. Anal. Bioanal. Chem. 378:301-310. [DOI] [PubMed] [Google Scholar]
- 6.Bowen, G. J., L. Chesson, K. Nielson, T. E. Cerling, and J. R. Ehleringer. 2005. Treatment methods for the determination of delta H-2 and delta O-18 of hair keratin by continuous-flow isotope-ratio mass spectrometry. Rapid Commun. Mass Spectrom. 19:2371-2378. [DOI] [PubMed] [Google Scholar]
- 7.Bowen, G. J., J. R. Ehleringer, L. Chesson, E. Stange, and C. E. Cerling. 2007. Stable isotope ratios of tap water in the contiguous USA. Water Resour. Res. 43:W03419. [Google Scholar]
- 8.Braune, B. M., K. A. Hobson, and B. J. Malone. 2005. Regional differences in collagen stable isotope and tissue trace element profiles in populations of long-tailed duck breeding in the Canadian Arctic. Sci. Total Environ. 346:156-168. [DOI] [PubMed] [Google Scholar]
- 9.Breeze, R. G., B. Budowle, and S. E. Schutzer. 2005. Microbial forensics. Elsevier Academic Press, London, United Kingdom.
- 10.Budowle, B., M. D. Johnson, C. M. Fraser, T. J. Leighton, R. S. Murch, and R. Chakraborty. 2005. Genetic analysis and attribution of microbial forensics evidence. Crit. Rev. Microbiol. 31:233-254. [DOI] [PubMed] [Google Scholar]
- 11.Budowle, B., S. E. Schutzer, M. S. Ascher, R. M. Atlas, J. P. Burans, R. Chakraborty, J. J. Dunn, C. M. Fraser, D. R. Franz, T. J. Leighton, S. A. Morse, R. S. Murch, J. Ravel, D. L. Rock, T. R. Slezak, S. P. Velsko, A. C. Walsh, and R. A. Walters. 2005. Toward a system of microbial forensics: from sample collection to interpretation of evidence. Appl. Environ. Microbiol. 71:2209-2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Budowle, B., S. E. Schutzer, A. Einseln, L. C. Kelley, A. C. Walsh, J. A. L. Smith, B. L. Marrone, J. Robertson, and J. Campos. 2003. Building microbial forensics as a response to bioterrorism. Science 301:1852-1853. [DOI] [PubMed] [Google Scholar]
- 13.Burgoyne, T. W., and J. M. Hayes. 1998. Quantitative production of H2 by pyrolysis of gas chromatographic effluents. Anal. Chem. 70:5136-5141. [Google Scholar]
- 14.Cerling, T. E., G. Wittemyer, H. B. Rasmussen, F. Vollrath, C. E. Cerling, T. J. Robinson, and I. Douglas-Hamilton. 2006. Stable isotopes in elephant hair document migration patterns and diet changes. Proc. Natl. Acad. Sci. USA 103:371-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chamberlain, C., J. Blum, R. Holmes, X. Feng, T. Sherry, and G. Graves. 1997. The use of Iiotope tracers for identifying populations of migratory birds. Oecologia 109:132-141. [DOI] [PubMed] [Google Scholar]
- 16.Christoph, N., and A. Rossmann. 2005. Wine authentication using stable isotope ratio analysis: significance of geographic origin, climate, and viticultural parameters. Abstr. Pap. Am. Chem. Soc. 229:U40. [Google Scholar]
- 17.Cliff, J. B., K. H. Jarman, N. B. Valentine, S. L. Golledge, D. J. Gaspar, D. S. Wunschel, and K. L. Wahl. 2005. Differentiation of spores of Bacillus subtilis grown in different media by elemental characterization using time-of-flight secondary ion mass spectrometry. Appl. Environ. Microbiol. 71:6524-6530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coplen, T. B. 1996. New guidelines for reporting stable hydrogen, carbon and oxygen isotope-ratio data. Geochim. Cosmochim. Acta 60:3359-3360. [Google Scholar]
- 19.Craig, H. 1961. Isotopic variations in meteoric waters. Science 133:1702-1703. [DOI] [PubMed] [Google Scholar]
- 20.DeNiro, M. J., and S. Epstein. 1978. Influence of diet on the distribution of carbon isotopes in animals. Geochim. Cosmochim. Acta 42:495-506. [Google Scholar]
- 21.DeNiro, M. J., and S. Epstein. 1981. Influence of diet on the distribution of nitrogen isotopes in animals. Geochim. Cosmochim. Acta 45:341-351. [Google Scholar]
- 22.Reference deleted.
- 23.Dutton, A., B. H. Wilkinson, J. M. Welker, G. J. Bowen, and K. C. Lohmann. 2005. Spatial distribution and seasonal variation in O-18/O-16 of modern precipitation and river water across the conterminous USA. Hydrol. Process. 19:4121-4146. [Google Scholar]
- 24.Edberg, H. C., C. E. Petersen, N. B. Valentine, D. S. Wunschel, and K. L. Wahl. 2006. Detection of trace agar on Bacillus spores by agarase digestion ESI-MS, abstr. 403. Proc. 54th Am. Soc. Mass Spectrom. Conf. Mass Spectrom. Allied Topics, Seattle, WA, 28 May to 1 June 2006.
- 25.Ehleringer, J. R., J. F. Casale, M. J. Lott, and V. L. Ford. 2000. Tracing the geographical origin of cocaine. Nature 408:311-312. [DOI] [PubMed] [Google Scholar]
- 26.Ehleringer, J. R., D. A. Cooper, M. J. Lott, and C. S. Cook. 1999. Geo-location of heroin and cocaine by stable isotope ratios. Forensic Sci. Int. 106:27-35. [Google Scholar]
- 26a.Estep, M. F. 1981. Hydrogen isotope ratios of mouse tissues are influenced by a variety of factors other than diet. Science 214:1374-1376. [DOI] [PubMed] [Google Scholar]
- 27.Evans, J., N. Stoodley, and C. Chenery. 2006. A strontium and oxygen isotope assessment of a possible fourth century immigrant population in a Hampshire cemetery, southern England. J. Archaeol. Sci. 33:265-272. [Google Scholar]
- 28.Fahey, A. J., and S. Messenger. 2001. Isotopic ratio measurements by time-of-flight secondary ion mass spectrometry. Int. J. Mass Spectrom. 208:227-242. [Google Scholar]
- 29.Fitzsimons, I. C. W., B. Harte, and R. M. Clark. 2000. SIMS stable isotope measurement: counting statistics and analytical precision. Mineral. Mag. 64:59-83. [Google Scholar]
- 30.Franke, B. M., G. Gremaud, R. Hadorn, and M. Kreuzer. 2005. Geographic origin of meat: elements of an analytical approach to its authentication. Eur. Food Res. Technol. 221:493-503. [Google Scholar]
- 31.Harwood, C., and S. Cutting. 1990. Molecular biological methods for Bacillus. John Wiley & Sons, Ltd., Chichester, United Kingdom.
- 32.Hayes, J. M., K. H. Freeman, B. N. Popp, and C. H. Hoham. 1990. Compound-specific isotopic analyses: a novel tool for reconstruction of ancient biogeochemical processes. Org. Geochem. 16:1115-1128. [DOI] [PubMed] [Google Scholar]
- 33.Hays, P. A., G. S. Remaud, E. Jamin, and Y. L. Martin. 2000. Geographic origin determination of heroin and cocaine using site-specific isotopic ratio deuterium NMR. J. Forensic Sci. 45:552-562. [PubMed] [Google Scholar]
- 34.Hobson, K. A. 1999. Tracing origins and migration of wildlife using stable isotopes: a review. Oecologia 120:314-326. [DOI] [PubMed] [Google Scholar]
- 35.Hobson, K. A., L. Atwell, and L. I. Wassenaar. 1999. Influence of drinking water and diet on the stable-hydrogen isotope ratios of animal tissues. Proc. Natl. Acad. Sci. USA 96:8003-8006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hocking, R. R. 2003. Methods and applications of linear models: regression and the analysis of variance, 2nd ed. John Wiley & Sons, Hoboken, NJ.
- 37.Horita, J., and A. A. Vass. 2003. Stable-isotope fingerprints of biological agents as forensic tools. J. Forensic Sci. 48:122-126. [PubMed] [Google Scholar]
- 38.Jarman, K., N. Valentine, J. B. Cliff, H. W. Kreuzer-Martin, C. Petersen, H. C. Edberg, D. Wunschel, and K. Wahl. 2007. Bayesian integrated microbial forensics, abstr. 214(I), p. 63. Program and Abstracts of the ASM Biodefense and Emerging Diseases Research Meeting, Washington, DC, 27 February to 2 March 2007.
- 39.Jarman, K., K. Wahl, N. Valentine, J. B. Cliff, H. W. Kreuzer-Martin, C. Petersen, H. C. Edberg, and D. Wunschel. 2006. A statistical framework for integrating diverse mass spectral data, ref. MP240. Proc. 54th Am. Soc. Mass Spectrom. Conf. Mass Spectrom. Allied Topics, Seattle, WA, 28 May to 1 June 2006.
- 40.Kendall, C., and E. Caldwell. 1998. Fundamentals of isotope geochemistry, p. 51-68. In C. Kendall and J. J. McDonnell (ed.), Isotope tracers in catchment hydrology. Elsevier Science, New York, NY.
- 41.Kendall, C., and T. B. Coplen. 2001. Distribution of oxygen-18 and deuterium in river waters across the United States. Hydrol. Process. 15:1363-1393. [Google Scholar]
- 42.Kennedy, B. P., C. P. Chamberlain, J. D. Blum, K. H. Nislow, and C. L. Folt. 2005. Comparing naturally occurring stable isotopes of nitrogen, carbon, and strontium as markers for the rearing locations of Atlantic salmon (Salmo salar). Can. J. Fish. Aquat. Sci. 62:48-57. [Google Scholar]
- 43.Kreuzer-Martin, H. W., L. A. Chesson, M. J. Lott, J. V. Dorigan, and J. R. Ehleringer. 2004. Stable isotope ratios as a tool in microbial forensics. 1. Microbial isotopic composition as a function of growth medium. J. Forensic Sci. 49:954-960. [PubMed] [Google Scholar]
- 44.Kreuzer-Martin, H. W., L. A. Chesson, M. J. Lott, J. V. Dorigan, and J. R. Ehleringer. 2004. Stable isotope ratios as a tool in microbial forensics. 2. Isotopic variation among different growth media as a tool for sourcing origins of bacterial cells or spores. J. Forensic Sci. 49:961-967. [PubMed] [Google Scholar]
- 45.Kreuzer-Martin, H. W., L. A. Chesson, M. J. Lott, and J. R. Ehleringer. 2005. Stable isotope ratios as a tool in microbial forensics. 3. Effect of culturing on agar-containing growth media. J. Forensic Sci. 50:1372-1379. [PubMed] [Google Scholar]
- 46.Kreuzer-Martin, H. W., M. J. Lott, J. Dorigan, and J. R. Ehleringer. 2003. Microbe forensics: oxygen and hydrogen stable isotope ratios in Bacillus subtilis cells and spores. Proc. Natl. Acad. Sci. USA 100:815-819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lamoureux, C. 2003. Isotopic analyses in a French Customs laboratory. Actual. Chim. 267:31-33. [Google Scholar]
- 48.Longinelli, A. 1984. Oxygen isotopes in mammal bone phosphate: a new tool for paleohydrological and paleoclimatological research? Geochim. Cosmochim. Acta 48:385-390. [Google Scholar]
- 49.Luz, B., A. B. Cormie, and H. P. Schwarcz. 1990. Oxygen isotope variations in phosphate of deer bones. Geochim. Cosmochim. Acta 54:1723-1728. [Google Scholar]
- 50.MacGregor, B. J., V. Bruchert, S. Fleischer, and R. Amann. 2002. Isolation of small-subunit rRNA for stable isotopic characterization. Environ. Microbiol. 4:451-464. [DOI] [PubMed] [Google Scholar]
- 51.Orphan, V. J., C. H. House, K. U. Hinrichs, K. D. McKeegan, and E. F. DeLong. 2001. Methane-consuming archaea revealed by directly coupled isotopic and phylogenetic analysis. Science 293:484-487. [DOI] [PubMed] [Google Scholar]
- 52.Orphan, V. J., C. H. House, K. U. Hinrichs, K. D. McKeegan, and E. F. DeLong. 2002. Multiple archaeal groups mediate methane oxidation in anoxic cold seep sediments. Proc. Natl. Acad. Sci. USA 99:7663-7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Peteranderl, R., and C. Lechene. 2004. Measure of carbon and nitrogen stable isotope ratios in cultured cells. J. Am. Soc. Mass Spectrom. 15:478-485. [DOI] [PubMed] [Google Scholar]
- 54.Schimmelmann, A. 1991. Determination of the concentration and stable isotopic composition of nonexchangeable hydrogen in organic matter. Anal. Chem. 63:2456-2459. [Google Scholar]
- 55.Sessions, A. L., T. W. Burgoyne, A. Schimmelmann, and J. M. Hayes. 1999. Fractionation of hydrogen isotopes in lipid biosynthesis. Org. Geochem. 30:1193-1200. [Google Scholar]
- 56.Sessions, A. L., L. L. Jahnke, A. Schimmelmann, and J. M. Hayes. 2002. Hydrogen isotope fractionation in lipids of the methane-oxidizing bacterium Methylococcus capsulatus. Geochim. Cosmochim. Acta 66:3955-3969. [Google Scholar]
- 57.Sessions, A. L., S. P. Sylva, and J. M. Hayes. 2005. Moving-wire device for carbon isotopic analyses of nanogram quantities of nonvolatile organic carbon. Anal. Chem. 77:6519-6527. [DOI] [PubMed] [Google Scholar]
- 58.Valentine, D. L., A. Chidthaisong, A. Rice, W. S. Reeburgh, and S. C. Tyler. 2004. Carbon and hydrogen isotope fractionation by moderately thermophilic methanogens. Geochim. Cosmochim. Acta 68:1571-1590. [Google Scholar]
- 59.Valentine, N., S. Wunschel, D. Wunschel, C. Petersen, and K. Wahl. 2005. Effect of culture conditions on microorganism identification by matrix-assisted laser desorption ionization mass spectrometry. Appl. Environ. Microbiol. 71:58-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vickerman, J., and D. Briggs. 2001. ToF-SIMS: surface analysis by mass spectrometry. IM Publications, West Sussex, United Kingdom.
- 61.Wahl, K. L., N. B. Valentine, S. C. Wunschel, D. S. Wunschel, K. H. Jarman, and C. E. Petersen. 2003. Microorganism analysis and identification by MALDI-TOF-MS. Abstr. Pap. Am. Chem. Soc. 226:U121. [Google Scholar]
- 62.Whiteaker, J. R., C. C. Fenselau, D. Fetterolf, D. Steele, and D. Wilson. 2004. Quantitative determination of heme for forensic characterization of Bacillus spores using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Anal. Chem. 76:2836-2841. [DOI] [PubMed] [Google Scholar]
- 63.Wunschel, D. S., E. A. Hill, J. S. McLean, K. Jarman, Y. A. Gorby, N. Valentine, and K. Wahl. 2005. Effects of varied pH, growth rate and temperature using controlled fermentation and batch culture on matrix assisted laser desorption/ionization whole cell protein fingerprints. J. Microbiol. Methods 62:259-271. [DOI] [PubMed] [Google Scholar]
- 64.Wunschel, S. C., K. H. Jarman, C. E. Petersen, N. B. Valentine, K. L. Wahl, D. Schauki, J. Jackman, C. P. Nelson, and E. White. 2005. Bacterial analysis by MALDI-TOF mass spectrometry: an inter-laboratory comparison. J. Am. Soc. Mass Spectrom. 16:456-462. [DOI] [PubMed] [Google Scholar]
- 65.Zhang, D., W. Sun, Z. P. Yuan, H. X. Ju, X. J. Shi, and C. H. Wang. 2005. Origin differentiation of a heroin sample and its acetylating agent with C-13 isotope ratio mass spectrometry. Eur. J. Mass Spectrom. 11:277-285. [DOI] [PubMed] [Google Scholar]