Abstract
By using the selective capture of transcribed sequences (SCOTS) approach, we identified 28 genes preferentially expressed by the major swine pathogen and zoonotic agent Streptococcus suis upon interaction with porcine brain microvascular endothelial cells. Several of these genes may be considered new S. suis candidate virulence factors. Results from this study demonstrate the suitability of SCOTS for the elucidation of gene expression in streptococcal species and may contribute to a better understanding of the pathogenesis of S. suis infections.
Streptococcus suis is a gram-positive bacterium responsible for, among other diseases, meningitis and septicemia in swine (14). S. suis is also a zoonotic agent. Many cases of human S. suis infection have been reported in different Asian and European countries, as well as in New Zealand, Australia, Argentina, and Canada (25). Very recently, the first case of human meningitis caused by S. suis was recorded in the United States (43). Indeed, S. suis is increasingly becoming a public health concern. For instance, during a recent outbreak in China more than 200 cases of human S. suis infection were reported, 39 of which resulted in death (33, 45). Despite increasing research in recent years, knowledge of the pathogenesis of S. suis infection remains limited (11, 14). Only the capsular polysaccharide and a recently described serum opacity-like factor have been shown to play a critical role in the pathogenesis of the infection (3, 14). Proposed putative virulence factors such as the suilysin, the extracellular protein factor, and the muramidase-released protein, although associated with virulence, have been found to be nonessential factors (6, 14). Other determinants, such as a fibronectin/fibrinogen-binding protein, were found to be partially involved in virulence (6, 14), while the actual roles of some other virulence candidates (e.g., the cell wall and several putative adhesins and proteases) in the pathogenesis of S. suis infection remain to be verified (11, 14).
S. suis needs to attain the central nervous system (CNS) in order to cause meningitis in swine. It has been suggested that this pathogen might reach the CNS by crossing the porcine blood-brain barrier (BBB) by transcytosis through porcine brain microvascular endothelial cells (PBMEC) and/or porcine choroid plexus epithelial cells, as well as by disruption of the barrier caused by toxic effects on BBB-forming cells (11, 36). Support for these mechanisms has been provided by recent studies showing that S. suis is able to affect the viability of porcine choroid plexus epithelial cells through necrotic and apoptotic mechanisms (37) and to adhere to and invade in vitro-cultured PBMEC (38). However, little is known about the molecular means by which S. suis accomplishes these processes.
Selective capture of transcribed sequences (SCOTS) is a PCR-based RNA analysis method that offers several advantages in comparison to other genomic approaches, such as in vivo expression technology (IVET) or signature-tagged mutagenesis (29). In fact, SCOTS directly identifies bacterial genes rather than promoter regions and is not confounded by polar effects when genes are arranged in polycistronic operons (29). The SCOTS approach has been used with success in many gram-negative bacteria, as well as in Mycobacterium tuberculosis and Listeria monocytogenes (5, 10, 13, 22). In this work, we used the SCOTS approach to identify genes preferentially expressed by S. suis during its interactions with cells of the BBB, a process that might be essential for the pathogenesis of the meningitis caused by this pathogen.
Experimental model and bacterial transcriptome recovery.
S. suis serotype 2 highly virulent strain 31533 (38) and the PBMEC line PBMEC/C1-2 (34) were used in this study. PBMEC were grown in Primaria six-well tissue culture plates (Becton Dickinson, Franklin Lakes, NJ) with IF culture medium (a 1:1 mixture of Iscove's modified Dulbecco's and Ham's F-12 media; Invitrogen, Burlington, Ontario, Canada) as previously described (38). S. suis was grown in Todd-Hewitt broth (Becton Dickinson, Sparks, MD) for 16 h at 37°C, harvested by centrifugation, washed twice in phosphate-buffered saline (pH 7.3), and resuspended in fresh IF culture medium at 106 CFU/ml. Confluent monolayers of PBMEC (at 3 × 106 cells per well) were inoculated with 3 ml of this bacterial suspension (multiplicity of infection = 1). Plates were centrifuged at 800 × g for 10 min and incubated for 4 h at 37°C with 5% CO2. After incubation, actual S. suis adhesion to and invasion of PBMEC were verified in selected wells and found to be in agreement with reported values (38; data not shown). For identification of the genes transcribed during interaction, total RNA from S. suis-infected PBMEC cells was prepared from 24 independent P6 wells with RNAwiz (Ambion, Austin, TX) according to the manufacturer's instructions. Total RNA from S. suis grown under the same conditions but without cells (mock infection) was prepared from five P6 wells. Samples were treated with TURBO DNase (Ambion), and absence of contaminating DNA was verified by PCR with primers AROA-F and BA9 (Table 1), which target the aroA gene. RNA was quantified by measurement of absorbance at 260 nm, and its integrity was verified by visualization on 1% denaturing agarose gels.
TABLE 1.
Oligonucleotide primers used in this study
| Primer | Sequence (5′-3′) |
|---|---|
| AROA-F | AACGTGACCTACCTCCGTTG |
| AROA-R | CGGTCATCGTAGAATTCGAGT |
| CELL-RNA | ACACTCTCGAGACATCACCGGTACCNNNNNNNNN |
| MOCK-RNA | CTTAGCCACTACGTGCGGATCCAGACNNNNNNNNN |
| RDNA-F | GGCTCAGGACGAACGCTG |
| RDNA-R | GCTAAGCGACTACCGTATCT |
| MOCK-PCR | CTTAGCCACTACGTGCGGATCCAGAC |
| CELL-PCR | GACACTCTCGAGACATCACCGGTACC |
| RPOD-F | TCTTTCAAATACATGCGGACTG |
| RPOD-R | ATTCCATTTACGCTTGATGCTG |
| SSU0424-F | AATCAAAGATTGGACGAGCC |
| SSU0424-R | CAATCCATCCCAATTCAGACAG |
| SSU0870-F | GGTATCATGAATACGGACGAAG |
| SSU0870-R | GAATGGATGGGCAATGAGAG |
| SSU0067-F | ATCAATCATCAAGGGATGCG |
| SSU0067-R | GATAGCCACCTCTTTTTCCAC |
| SSU1448-FQ | TTCTCTCTGTACTTGCTCCC |
| SSU1448-RQ | GGTCGCTCTAACCTTTGATG |
| SSU0457-F | ACCCAGATAGCCACTATTCC |
| SSU0457-R | CTGATCATAAGTGAAGTCGCC |
| SSU0597-F | TGCGTCTGGTTAAGACTTTG |
| SSU0597-R | GTTCTTGCCCAGCTTTTTTTC |
Selective capture of transcribed sequences.
Five micrograms of total RNA prepared from both infected and mock-infected samples was reverse transcribed by random priming with SuperScript II (Invitrogen). Primer CELL-RNA or MOCK-RNA, with a defined terminal sequence at the 5′ end and a random nonamer at the 3′ end, was used (Table 1). Thereafter, cDNA sequences corresponding to bacterial mRNAs were selectively captured from the mixture of total PBMEC-S. suis RNAs or total S. suis RNAs by performing three rounds of SCOTS as previously described (5). Briefly, samples were normalized by self-hybridization and then hybridized overnight at 68°C to biotinylated genomic S. suis 31533 DNA that had previously been blocked with PCR-generated DNA representing 16S and 23S S. suis rRNA sequences (primers RDNA-F and RDNA-R, Table 1). Bacterial cDNAs were then separated with streptavidin-coupled magnetic beads. After elution, cDNAs were PCR reamplified with primer CELL-PCR or MOCK PCR (Table 1), which corresponds to the defined sequence added during reverse transcription and is specific to each condition. Sequences preferentially transcribed by S. suis upon interaction with PBMEC were obtained after three rounds of enrichment carried out as previously described (5). Briefly, cDNAs obtained during PBMEC interaction were subjected to the procedure outlined above, but this time the biotinylated genomic S. suis DNA used for capture had previously been prehybridized with DNA sequences corresponding to 16S and 23S S. suis rRNAs and cDNAs from the mock infection. The resulting interaction-specific cDNAs were cloned into vector pCR4 (TOPO TA cloning kit; Invitrogen) and sequenced. DNA sequences were determined at the DNA Sequencing Facility of the University of Maine (Orono) on a 373A DNA Sequencing System (Applied Biosystems, Foster City, CA).
Identification of preferentially expressed genes.
The BLAST software package was used to determine sequence homologies in the GenBank databases (http://www.ncbi.nlm.nih.gov/BLAST/). Sequence comparison was also performed against sequence data produced by the S. suis Sequencing Group at the Sanger Institute (http://www.sanger.ac.uk/Projects/S_suis) for European strain P1/7 and at the Joint Genome Institute Microbial Genomics website (http://genome.jgi-psf.org/cgi-bin/runAlignment?db=strsu&advanced=1) for North American strain 89-1591. We report here the identification, by SCOTS, of 28 genes as being preferentially expressed by S. suis upon interaction with PBMEC. These genes can be divided into the following eight functional groups: metabolism/housekeeping, cell envelope, secreted proteases, cell division/replication, regulatory, protein sorting- and transport/binding-related genes, and genes with unknown function. To the best of our knowledge, none of the identified genes has ever before been associated with the pathogenesis of S. suis infection. Similar to other studies of host-pathogen interaction (2, 5, 31), most of the genes identified by SCOTS in this study are putatively involved in metabolic/housekeeping functions and do not encode “genuine” virulence factors. However, identification of these genes may be of importance, since new information about the metabolism of S. suis is rendered that may eventually prove useful for vaccine development. On the other hand, some genes identified by SCOTS in this study are known to be important for the virulence of other gram-positive bacteria (including at least three different streptococcal species). The relevance of these genes will be discussed below. For all of the other genes identified in this study, putative functions and references to publications describing their in vivo expression and/or involvement in virulence in other organisms are listed in Table 2.
TABLE 2.
Genes identified by SCOTS that are differentially expressed by S. suis upon interaction with PBMEC
| Function and clone | Genea | Putative function (organism) | GenBank identification | Reference(s) |
|---|---|---|---|---|
| Metabolism/housekeeping | ||||
| D3C3 | ssu0707 | Putative exonuclease RexB (S. suis 89/1591) | ZP_00874950 | |
| 2H6 | ssu0767 | 1-Phosphofructokinase (S. suis 89/1591) | ZP_00875124 | 24 |
| 2G7 | ssu1411 | Aminotransferase, classes I and II (S. suis 89/1591) | ZP_00875661 | |
| D3G2 | ssu1527 | Aminodeoxychorismate lyase-like protein (S. suis 89/1591) | ZP_00876086 | |
| 1B7 | ssu1444 | Uridine kinase (S. thermophilus CNRZ1066) | AAV_62804 | |
| D1B7 | ssu0844 | Haloacid dehalogenase-like hydrolase (S. suis 89/1591) | ZP_00874652 | |
| 1G10 | ssu1044 | Ribonucleoside diphosphate reductase (S. suis 89/1591) | ZP_00875241 | |
| D1H10 | ssu1159 | Ribosome recycling factor (S. suis 89/1591) | ZP_00875081 | |
| 1C7 | ssu0870 | Nucleotidyltransferase (S. suis 89/1591) | ZP_00874394 | 12 |
| 2F10 | ssu0871 | Glucose-1-phosphate adenylyltransferase (S. suis 89/1591) | ZP_00874395 | |
| 1A11 | ssu0764 | tRNA (guanine-N1-)-methyltransferase (S. suis 89/1591) | ZP_00875121 | |
| Cell envelope | ||||
| 2A11 | ssu0597 | Membrane-bound O-acyl transferase, DltB (S. suis 89/1591) | ZP_00875261 | 12 |
| 2A8 | ssu1184 | d-Alanine-d-alanine ligase (S. suis 89/1591) | ZP_00875052 | 18 |
| 1H9 | ssu1448 | Peptidoglycan polysaccharide deacetylase PgdA (S. suis 89/1591) | ZP_00876135 | 40, 41 |
| 1E9 | ssu1487 | VanZ-like protein (S. suis 89/1591) | ZP_00875572 | 12 |
| D1G11 | ssu1114 | Glycosyltransferase, group 1 (S. suis 89/1591) | ZP_00875224 | |
| Regulatory | ||||
| D2D4 | ssu0869 | Putative transcriptional regulator, LysR family (S. pneumoniae TIGR4) | AAK74821 | |
| Protein sorting | ||||
| 1C11 | ssu0424 | Signal peptidases S24, S26A, and S26B (S. suis 89/1591) | ZP_00875273 | |
| 1B8 | ssu0453 | Sortase-like protein SrtE (S. suis) | BAB83972 | 29 |
| Secreted protease | ||||
| D1E9 | ssu0457 | Collagenase-peptidase U32 (S. suis) | BAB83975 | |
| Cell division/replication | ||||
| 1G5 | ssu0007 | DNA polymerase III, β chain (S. suis 89/1591) | ZP_00875475 | |
| 2F4 | ssu0432 | Cell division protein FtsQ/DivIB (S. suis 89/1591) | ZP_00876117 | 12 |
| Transport/binding | ||||
| D1H3 | ssu1787 | Multidrug ABC transporter, ATP-binding protein (B. cereus E33L) | AAU18528 | |
| D3G1 | ssu1023 | Putative permease (S. suis 89/1591) | ZP_00874974 | |
| Unknown function | ||||
| D1G1 | ssu0067 | Protein of unknown function DUF925 (S. suis 89/1591) | ZP_00876271 | |
| 2F6 | ssu1424 | Hypothetical protein (S. suis 89/1591) | ZP_00875489 | |
| D1H2 | ssu0858 | Protein of unknown function UPF0153 (S. suis 89/1591) | ZP_00875788 | |
| D1A3 | ssu1792 | Conserved hypothetical protein (S. suis 89/1591) | ZP_00876058 |
Genes are named in accordance with the S. suis strain P1/7 sequencing project nomenclature.
Validation of SCOTS results by q-PCR.
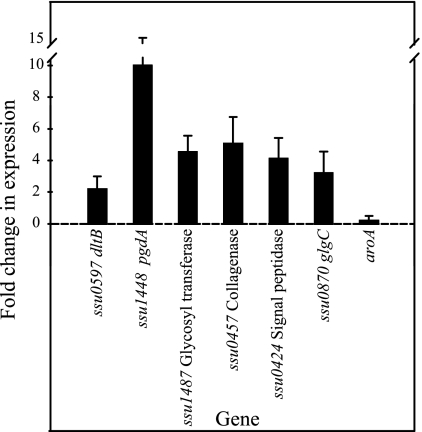
The SCOTS approach, as used in this study, should result in the identification of genes that are upregulated by S. suis upon interaction with PBMEC (5). Therefore, to validate our SCOTS results, we used quantitative PCR (q-PCR) to measure the level of expression of random selected genes on a new series of infection replicates. Infection of PBMEC, mock infections, and RNA extraction from both samples were performed as described above. cDNAs were synthesized in triplicate by using SuperScript II with random hexamers (Roche, Laval, Quebec, Canada). q-PCR was performed by using the QuantiTect SybrGreen PCR kit (QIAGEN, Mississauga, Ontario, Canada) according to the manufacturer's instructions. For each sample, a no-reverse transcription reaction was run as a control. The primers used are described in Table 1. For each q-PCR run, the calculated threshold cycle (CT) was normalized to the CT of the internal control rpoD gene amplified from the corresponding sample, and the n-fold change was calculated by the 2−ΔΔCT method as previously described (23). Results of q-PCR analysis for these selected genes showed that they were indeed upregulated by S. suis upon interaction with PBMEC (Fig. 1), with changes ranging from 2.18- to 10-fold. The gene aroA, which is known to be expressed in equal amounts under both conditions (our unpublished results), was also used.
FIG. 1.
n-Fold changes in the expression of selected S. suis genes identified by SCOTS as measured by q-PCR upon interaction of the bacterium with PBMEC. See the text for details.
Genes involved in cell envelope modification.
As stated above, some genes identified by SCOTS might be considered, on the basis of their functions in other organisms, potential S. suis candidate virulence factors. For instance, the ssu0597 gene (dltB) belongs to an operon comprising four genes, dltA, dltB, dltC, and dltD, which is present in all of the genomes of low-G+C bacteria determined so far (28). In all of the species where this operon has been studied, all four of the genes are required to catalyze the incorporation of d-alanine residues into the lipoteichoic acids (LTAs). d-Alanylation of LTAs allows gram-positive bacteria to modulate their surface charge, to regulate ligand binding, and to control the electromechanical properties of the cell wall (28). In addition, formation of d-alanyl-LTAs is required to resist the action of antimicrobial peptides in L. monocytogenes, Staphylococcus aureus, Streptococcus pneumoniae, Streptococcus pyogenes (group A Streptococcus), and Streptococcus agalactiae (group B Streptococcus [GBS]) (1, 20, 21, 30, 42). Besides, the virulence of mutants deficient in d-alanylation of LTAs of GBS, L. monocytogenes, or S. aureus was severely impaired in the murine or rabbit model of infection (1, 30, 42). The d-alanylation of S. suis LTAs has not been documented. However, it is known that wild-type S. suis LTAs are important for adhesion of this bacterium to PBMEC. Indeed, inhibition of the adhesion of S. suis to this cell type can be obtained by preincubation of PBMEC with purified LTA (39). From our SCOTS results, it might be hypothesized that S. suis might be able to modulate the degree of d-alanylation of its LTAs by upregulation of its dlt operon upon interaction with PBMEC. Further studies focusing on this operon of S. suis are under way to evaluate this hypothesis. However, it is interesting that in some gram-positive pathogens it has been shown that d-alanyl-LTAs contribute to adhesion to and invasion of various cell lines and that these steps may depend on a high ratio of d-alanine to glycerol/ribitol phosphate in their LTAs (1, 21, 30, 42).
The main clinical feature of S. suis is meningitis, and this bacterium is often isolated from the cerebrospinal fluid of animals or patients with meningitis (14). On the other hand, it has been reported that patients suffering from meningitis present increased titers of lysozyme in their cerebrospinal fluid (19). As shown in this study, S. suis differentially expresses a gene (ssu1448) highly homologous to S. pneumoniae pgdA, which encodes a peptidoglycan N-acetylglucosamine deacetylase. Peptidoglycan is an essential component of the bacterial cell wall and an important target for the innate immune system. Peptidoglycan modification by deacetylation seems to be important for gram-positive pathogens. Indeed, pneumococci in which pgdA was inactivated became hypersensitive to the action of lysozyme (41) and showed reduced virulence in a murine model of infection (40). In addition, it has very recently been reported that a pgdA mutant strain of L. monocytogenes was impaired in the ability to induce disease in the murine model of infection and that the pgdA gene was required by this species to resist the host innate immune response mediated by lysozyme (4). In this regard, it may be of interest to further evaluate the hypothesis that S. suis, through the action of the pgdA gene product, has the ability to modify its peptidoglycan by deacetylation and therefore resist a host innate response mediated by this enzyme. On the other hand, it is intriguing that in our in vitro model, where the immune response of the host would not be as relevant as in the in vivo situation, the pgdA gene was found to be highly upregulated. However, it has been proposed that, in vivo, S. suis might gain access to the CNS by transcytosis across PBMEC (38). It might therefore be plausible that during its interaction with PBMEC, in addition to genes required for adhesion to and invasion of these cells, S. suis also upregulates genes required for the steps immediately following the BBB crossing. Further studies are required to evaluate this hypothesis.
Identification of a putative pilus island in S. suis.
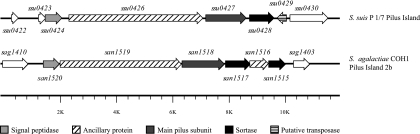
Pili in several gram-positive bacteria have recently been described, and it has been proposed that they may play an important role in virulence (35). For instance, in GBS, pili participate in adhesion to human epithelial cells (7) and their role in adhesion to extracellular matrix (ECM) proteins has been suggested (27). In this work, we identified a gene (ssu0424) putatively encoding a signal peptidase homologous to the LepB-type signal peptidases of gram-negative bacteria. A homologous LepB-type signal peptidase is the first gene in GBS pilus island 2b (PI-2b), one of the three identified pilus islands in this species (35). GBS PI-2b contains five other downstream genes, encoding two LPXTG proteins (suggested to be an ancillary protein and the main pilus subunit), a class C sortase, a third LPXTG protein (ancillary protein), and a second class C sortase (35) (Fig. 2). The presence of thin, pilus-like structures on the surface of S. suis has been revealed by electron microscopy (15). Interestingly, analysis of data from the two S. suis sequencing projects strongly suggests that S. suis possesses a truncated version of this pilus island. In fact, in sequenced S. suis strains P1/7 and 89-1591, two genes encoding LPXTG proteins (highly homologous to the ancillary and main pilus subunits of GBS, respectively) and a gene encoding an undescribed putative class C sortase-like protein are found downstream of the LepB signal peptidase that was identified by SCOTS (Fig. 2). Although the S. suis putative pilus island lacks the last two genes in comparison to that of GBS, the similarity in genetic organization, the strong homology showed by the LPXTG proteins to the main and ancillary pilus proteins of the latter species, and the current proposed mechanism for pilus formation in gram-positive bacteria (7, 27) suggest that a pilus might be formed by the gene products of this island. In addition, we speculate that this pilus might participate in S. suis adhesion to or invasion of PBMEC. In fact, pili have been very recently shown to be important for GBS adhesion to and invasion of human BMEC (26). Although functional analysis of this S. suis putative pilus island is needed to fully evaluate this hypothesis, it is interesting that the LepB signal peptidase was found to be highly upregulated by q-PCR (fourfold change) upon interaction of S. suis with PBMEC (Fig. 1).
FIG. 2.
Putative pilus island in S. suis (top) and its counterpart, PI-2b, found in S. agalactiae strain COH1 (bottom). In S. suis, the first gene of the locus is a LepB-type signal peptidase (identified by SCOTS and q-PCR as upregulated upon interaction with PBMEC), which is followed downstream by the genes for a putative ancillary protein and a main pilus subunit. A previously undescribed class C sortase is encoded by the last gene of the island. S. agalactiae PI-2b, which is organized in a similar way, comprises an additional ancillary pilus subunit and a second class C sortase (GenBank entry NZ_AAJR01000022).
Additionally, in this study we identified the ssu0453 gene, which was previously named srtE and encodes one of the four class C sortases already described in S. suis (8, 29). In GBS and group A Streptococcus models of pilus assembly, class C sortases have been proposed to catalyze the covalent polymerization of the pilin subunits encoded by genes within the pilus island bearing the class C sortases (7, 27). However, previous work with S. suis (29), as well as analysis of sequenced strains P1/7 and 89-1591, indicates that the S. suis srtE gene is not flanked by genes encoding LPXTG proteins and thus does not seem to be part of a pilus island. Therefore, the putative participation of S. suis ssu0453/srtE in pilus formation following the proposed model is unlikely. However, it might be interesting to evaluate whether this sortase is required for, or contributes to, the assembly of pilin subunits encoded by the island described in this work or by other, as yet unidentified, islands.
Interaction of S. suis with ECM proteins.
It has been shown that S. suis is able to interact with ECM proteins (9). S. suis also has the ability to degrade ECM proteins through the upregulation of metalloproteinase 9 production by human macrophages (16), which may result in tissue destruction and BBB disruption. However, the ability of S. suis to degrade ECM proteins directly has not been demonstrated. Interestingly, one of the genes identified by SCOTS (ssu0457) encodes a putative collagenase which, in sequenced strains P1/7 and 89-1591, is located upstream of a gene that putatively encodes a second collagenase, in an operon-like organization. It has been suggested that the impairment of BBB function during infection with different S. suis strains may depend on proteases produced by this pathogen (17). It is thus tempting to speculate, even if we lack evidence regarding its exact function, that upregulating the expression of the collagenase identified in this work upon interaction with PBMEC might, in vivo, be useful to increase the permeability of the BBB and therefore contribute to the migration of S. suis to the CNS.
Suitability of the SCOTS approach for elucidation of gene expression in S. suis.
The SCOTS approach has been used successfully with several bacterial species (5, 10, 13, 22). To the best of our knowledge, this is the first report of its use with a streptococcal species. Results presented here clearly demonstrate that SCOTS is also suitable for the elucidation of gene expression in streptococci and particularly in organisms like S. suis, for which very few molecular tools exist. Indeed, with the exception of the present study, only one genomic approach has been used to study this pathogen (32). In that work, an adapted IVET approach identified several S. suis iron-induced and/or in vivo (porcine infection model)-expressed genes (32). However, all of the genes identified in that study were also expressed in vitro under standard laboratory growth conditions. These results can be explained by the absence of promoter sequences exclusively expressed under the conditions tested (32). However, since a plasmid-based system was used instead of an integrative promoter trap system, the results obtained might also be explained by the inability of that system to detect in vitro silent genes because of gene dose effects (32). On the other hand, with SCOTS we clearly showed condition-specific differences in S. suis gene expression. In fact, SCOTS may be considered the only approach available for the direct study of global differential gene expression in S. suis. Despite the fact that IVET and SCOTS have identified the same genes in some cases (31), there were no overlapping genes in the IVET and SCOTS S. suis studies. This was not surprising, however, since only a small number of genes were identified in either study and, more importantly, the experimental conditions used were essentially different. Therefore, in this study, the use of the SCOTS approach resulted in original insights into the molecular mechanisms that this pathogen might use to cross the BBB. Indeed, the identification of the 28 genes preferentially expressed upon interaction of S. suis with PBMEC, several of which show great potential as virulence factors, may result in a better understanding of how this pathogen causes meningitis. In addition, extending the SCOTS analysis to identify transcriptional differences at different in vivo locations (i.e., brain, heart, tonsils), as well as at different stages of infection, may lead to comprehension of the mechanisms of disease progression and provide clues to prevention.
Addendum.
While this work was under revision, an article was published (44) describing the use of the signature-tagged mutagenesis approach to study genes important for the virulence of S. suis in a pig model of infection. The ssu0457 gene reported in the present article was also found in that study.
Acknowledgments
We are grateful to T. Sekizaki for critical reading of the manuscript. We thank P. Friedl for kindly providing the PBMEC/C1-2 cell line. We also thank D. Tremblay, S. Faucher, and M. Lamarche for helpful suggestions and M. Esgleas for assistance.
This work was supported by Natural Sciences and Engineering Research Council of Canada (NSERC) grant 0680154280 and by the Canadian Research Network on Swine Infectious Diseases. N.F. and G.V. are holders of NSERC postgraduate scholarships.
Footnotes
Published ahead of print on 4 May 2007.
REFERENCES
- 1.Abachin, E., C. Poyart, E. Pellegrini, E. Milohanic, F. Fiedler, P. Berche, and P. Trieu-Cuot. 2002. Formation of d-alanyl-lipoteichoic acid is required for adhesion and virulence of Listeria monocytogenes. Mol. Microbiol. 43:1-14. [DOI] [PubMed] [Google Scholar]
- 2.Autret, N., and A. Charbit. 2005. Lessons from signature-tagged mutagenesis on the infectious mechanisms of pathogenic bacteria. FEMS Microbiol. Rev. 29:703-717. [DOI] [PubMed] [Google Scholar]
- 3.Baums, C. G., U. Kaim, M. Fulde, G. Ramachandran, R. Goethe, and P. Valentin-Weigand. 2006. Identification of a novel virulence determinant with serum opacification activity in Streptococcus suis. Infect. Immun. 74:6154-6162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boneca, I. G., O. Dussurget, D. Cabanes, M.-A. Nahori, S. Sousa, M. Lecuit, E. Psylinakis, V. Bouriotis, J.-P. Hugot, M. Giovannini, A. Coyle, J. Bertin, A. Namane, J.-C. Rousselle, N. Cayet, M.-C. Prevost, V. Balloy, M. Chignard, D. J. Philpott, P. Cossart, and S. E. Girardin. 2007. A critical role for peptidoglycan N-deacetylation in Listeria evasion from the host innate immune system. Proc. Natl. Acad. Sci. USA 104:997-1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daigle, F., J. Y. Hou, and J. E. Clark-Curtiss. 2002. Microbial gene expression elucidated by selective capture of transcribed sequences (SCOTS). Methods Enzymol. 358:108-122. [DOI] [PubMed] [Google Scholar]
- 6.de Greeff, A., H. Buys, R. Verhaar, J. Dijkstra, L. van Alphen, and H. E. Smith. 2002. Contribution of fibronectin-binding protein to pathogenesis of Streptococcus suis serotype 2. Infect. Immun. 70:1319-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dramsi, S., E. Caliot, I. Bonne, S. Guadagnini, M. C. Prevost, M. Kojadinovic, L. Lalioui, C. Poyart, and P. Trieu-Cuot. 2006. Assembly and role of pili in group B streptococci. Mol. Microbiol. 60:1401-1413. [DOI] [PubMed] [Google Scholar]
- 8.Dramsi, S., P. Trieu-Cuot, and H. Bierne. 2005. Sorting sortases: a nomenclature proposal for the various sortases of gram-positive bacteria. Res. Microbiol. 156:289-297. [DOI] [PubMed] [Google Scholar]
- 9.Esgleas, M., S. Lacouture, and M. Gottschalk. 2005. Streptococcus suis serotype 2 binding to extracellular matrix proteins. FEMS Microbiol. Lett. 244:33-40. [DOI] [PubMed] [Google Scholar]
- 10.Faucher, S. P., S. Porwollik, C. M. Dozois, M. McClelland, and F. Daigle. 2006. Transcriptome of Salmonella enterica serovar Typhi within macrophages revealed through the selective capture of transcribed sequences. Proc. Natl. Acad. Sci. USA 103:1906-1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gottschalk, M., and M. Segura. 2000. The pathogenesis of the meningitis caused by Streptococcus suis: the unresolved questions. Vet. Microbiol. 76:259-272. [DOI] [PubMed] [Google Scholar]
- 12.Hava, D. L., and A. Camilli. 2002. Large-scale identification of serotype 4 Streptococcus pneumoniae virulence factors. Mol. Microbiol. 45:1389-1406. [PMC free article] [PubMed] [Google Scholar]
- 13.Haydel, S. E., and J. E. Clark-Curtiss. 2006. The Mycobacterium tuberculosis TrcR response regulator represses transcription of the intracellularly expressed Rv1057 gene, encoding a seven-bladed β-propeller. J. Bacteriol. 188:150-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Higgins, R., and M. Gottschalk. 2005. Streptococcal diseases, p. 769-783. In B. E. Straw, S. D'Allaire, W. L. Mengeling, and D. J. Taylor (ed.), Diseases of swine. Iowa State University Press, Ames.
- 15.Jacques, M., M. Gottschalk, B. Foiry, and R. Higgins. 1990. Ultrastructural study of surface components of Streptococcus suis. J. Bacteriol. 172:2833-2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jobin, M.-C., M. Gottschalk, and D. Grenier. 2006. Upregulation of prostaglandin E2 and matrix metalloproteinase 9 production by human macrophage-like cells: synergistic effect of capsular material and cell wall from Streptococcus suis. Microb. Pathog. 40:29-34. [DOI] [PubMed] [Google Scholar]
- 17.Jobin, M. C., and D. Grenier. 2003. Identification and characterization of four proteases produced by Streptococcus suis. FEMS Microbiol. Lett. 220:113-119. [DOI] [PubMed] [Google Scholar]
- 18.Jones, A. L., K. M. Knoll, and C. E. Rubens. 2000. Identification of Streptococcus agalactiae virulence genes in the neonatal rat sepsis model using signature-tagged mutagenesis. Mol. Microbiol. 37:1444-1455. [DOI] [PubMed] [Google Scholar]
- 19.Klockars, M., S. Reitamo, T. Weber, and Y. Kerttula. 1978. Cerebrospinal fluid lysozyme in bacterial and viral meningitis. Acta Med. Scand. 203:71-74. [DOI] [PubMed] [Google Scholar]
- 20.Kovács, M., A. Halfmann, I. Fedtke, M. Heintz, A. Peschel, W. Vollmer, R. Hakenbeck, and R. Bruckner. 2006. A Functional dlt operon, encoding proteins required for incorporation of d-alanine in teichoic acids in gram-positive bacteria, confers resistance to cationic antimicrobial peptides in Streptococcus pneumoniae. J. Bacteriol. 188:5797-5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kristian, S. A., V. Datta, C. Weidenmaier, R. Kansal, I. Fedtke, A. Peschel, R. L. Gallo, and V. Nizet. 2005. d-Alanylation of teichoic acids promotes group A Streptococcus antimicrobial peptide resistance, neutrophil survival, and epithelial cell invasion. J. Bacteriol. 187:6719-6725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu, S., J. E. Graham, L. Bigelow, P. D. Morse II, and B. J. Wilkinson. 2002. Identification of Listeria monocytogenes genes expressed in response to growth at low temperature. Appl. Environ. Microbiol. 68:1697-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402. [DOI] [PubMed] [Google Scholar]
- 24.Loo, C. Y., K. Mitrakul, I. B. Voss, C. V. Hughes, and N. Ganeshkumar. 2003. Involvement of an inducible fructose phosphotransferase operon in Streptococcus gordonii biofilm formation. J. Bacteriol. 185:6241-6254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lun, Z. R., Q. P. Wang, X. G. Chen, A. X. Li, and X. Q. Zhu. 2007. Streptococcus suis: an emerging zoonotic pathogen. Lancet Infect. Dis. 7:201-209. [DOI] [PubMed] [Google Scholar]
- 26.Maisey, H. C., M. Hensler, V. Nizet, and K. S. Doran. 2007. Group B streptococcal pilus proteins contribute to adherence to and invasion of brain microvascular endothelial cells. J. Bacteriol. 189:1464-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mora, M., G. Bensi, S. Capo, F. Falugi, C. Zingaretti, A. G. Manetti, T. Maggi, A. R. Taddei, G. Grandi, and J. L. Telford. 2005. Group A Streptococcus produce pilus-like structures containing protective antigens and Lancefield T antigens. Proc. Natl. Acad. Sci. USA 102:15641-15646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neuhaus, F. C., and J. Baddiley. 2003. A continuum of anionic charge: structures and functions of d-alanyl-teichoic acids in gram-positive bacteria. Microbiol. Mol. Biol. Rev. 67:686-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Osaki, M., D. Takamatsu, Y. Shimoji, and T. Sekizaki. 2002. Characterization of Streptococcus suis genes encoding proteins homologous to sortase of gram-positive bacteria. J. Bacteriol. 184:971-982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poyart, C., E. Pellegrini, M. Marceau, M. Baptista, F. Jaubert, M.-C. Lamy, and P. Trieu-Cuot. 2003. Attenuated virulence of Streptococcus agalactiae deficient in d-alanyl-lipoteichoic acid is due to an increased susceptibility to defensins and phagocytic cells. Mol. Microbiol. 49:1615-1625. [DOI] [PubMed] [Google Scholar]
- 31.Rediers, H., P. B. Rainey, J. Vanderleyden, and R. De Mot. 2005. Unraveling the secret lives of bacteria: use of in vivo expression technology and differential fluorescence induction promoter traps as tools for exploring niche-specific gene expression. Microbiol. Mol. Biol. Rev. 69:217-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith, H. E., H. Buijs, R. de Vries, H. J. Wisselink, N. Stockhofe-Zurwieden, and M. A. Smits. 2001. Environmentally regulated genes of Streptococcus suis: identification by the use of iron-restricted conditions in vitro and by experimental infection of piglets. Microbiology 147:271-280. [DOI] [PubMed] [Google Scholar]
- 33.Sriskandan, S., and J. D. Slater. 2006. Invasive disease and toxic shock due to zoonotic Streptococcus suis: an emerging infection in the East? PLoS Med. 3:e187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Teifel, M., and P. Friedl. 1996. Establishment of the permanent microvascular endothelial cell line PBMEC/C1-2 from porcine brains. Exp. Cell Res. 228:50. [DOI] [PubMed] [Google Scholar]
- 35.Telford, J. L., M. l. A. Barocchi, I. Margarit, R. Rappuoli, and G. Grandi. 2006. Pili in gram-positive pathogens. Nat. Rev. Microbiol. 4:509-519. [DOI] [PubMed] [Google Scholar]
- 36.Tenenbaum, T., R. Adam, I. Eggelnpohler, D. Matalon, A. Seibt, K. N. GE, H. J. Galla, and H. Schroten. 2005. Strain-dependent disruption of blood-cerebrospinal fluid barrier by Streptococcus suis in vitro. FEMS Immunol. Med. Microbiol. 44:25-34. [DOI] [PubMed] [Google Scholar]
- 37.Tenenbaum, T., F. Essmann, R. Adam, A. Seibt, R. U. Janicke, G. E. Novotny, H. J. Galla, and H. Schroten. 2006. Cell death, caspase activation, and HMGB1 release of porcine choroid plexus epithelial cells during Streptococcus suis infection in vitro. Brain Res. 1100:1-12. [DOI] [PubMed] [Google Scholar]
- 38.Vanier, G., M. Segura, P. Friedl, S. Lacouture, and M. Gottschalk. 2004. Invasion of porcine brain microvascular endothelial cells by Streptococcus suis serotype 2. Infect. Immun. 72:1441-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vanier, G., M. Segura, and M. Gottschalk. 2007. Characterization of the invasion of porcine endothelial cells by Streptococcus suis serotype 2. Can. J. Vet. Res. 71:81-89. [PMC free article] [PubMed] [Google Scholar]
- 40.Vollmer, W., and A. Tomasz. 2002. Peptidoglycan N-acetylglucosamine deacetylase, a putative virulence factor in Streptococcus pneumoniae. Infect. Immun. 70:7176-7178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vollmer, W., and A. Tomasz. 2000. The pgdA gene encodes for a peptidoglycan N-acetylglucosamine deacetylase in Streptococcus pneumoniae. J. Biol. Chem. 275:20496-20501. [DOI] [PubMed] [Google Scholar]
- 42.Weidenmaier, C., A. Peschel, V. A. J. Kempf, N. Lucindo, M. R. Yeaman, and A. S. Bayer. 2005. DltABCD- and MprF-mediated cell envelope modifications of Staphylococcus aureus confer resistance to platelet microbicidal proteins and contribute to virulence in a rabbit endocarditis model. Infect. Immun. 73:8033-8038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Willenburg, K. S., D. E. Sentochnik, and R. N. Zadoks. 2006. Human Streptococcus suis meningitis in the United States. N. Engl. J. Med. 354:1325. [DOI] [PubMed] [Google Scholar]
- 44.Wilson, T. L., J. Jeffers, V. J. Rapp-Gabrielson, S. Martin, L. K. Klein, D. E. Lowery, and T. E. Fuller. 2007. A novel signature-tagged mutagenesis system for Streptococcus suis serotype 2. Vet. Microbiol. 122:135-145. [DOI] [PubMed] [Google Scholar]
- 45.Yu, H., H. Jing, Z. Chen, H. Zheng, X. Zhu, H. Wang, S. Wang, L. Liu, R. Zu, L. Luo, N. Xiang, H. Liu, X. Liu, Y. Shu, S. S. Lee, S. K. Chuang, Y. Wang, J. Xu, and W. Yang. 2006. Human Streptococcus suis outbreak, Sichuan, China. Emerg. Infect. Dis. 12:914-920. [DOI] [PMC free article] [PubMed] [Google Scholar]