Abstract
DNA-based microbial source tracking (MST) methods were developed and used to specifically and sensitively track the unintended aerosolization of land-applied, anaerobically digested sewage sludge (biosolids) during high-wind events. Culture and phylogenetic analyses of bulk biosolids provided a basis for the development of three different MST methods. They included (i) culture- and 16S rRNA gene-based identification of Clostridium bifermentans, (ii) direct PCR amplification and sequencing of the 16S rRNA gene for an uncultured bacterium of the class Chloroflexi that is commonly present in anaerobically digested biosolids, and (iii) direct PCR amplification of a 16S rRNA gene of the phylum Euryarchaeota coupled with terminal restriction fragment length polymorphism to distinguish terminal fragments that are unique to biosolid-specific microorganisms. Each method was first validated with a broad group of bulk biosolids and soil samples to confirm the target's exclusive presence in biosolids and absence in soils. Positive responses were observed in 100% of bulk biosolid samples and in less than 11% of the bulk soils tested. Next, a sampling campaign was conducted in which all three methods were applied to aerosol samples taken upwind and downwind of fields that had recently been land applied with biosolids. When average wind speeds were greater than 5 m/s, source tracking results confirmed the presence of biosolids in 56% of the downwind samples versus 3% of the upwind samples. During these high-wind events, the biosolid concentration in downwind aerosols was between 0.1 and 2 μg/m3. The application of DNA-based source tracking to aerosol samples has confirmed that wind is a possible mechanism for the aerosolization and off-site transport of land-applied biosolids.
The predominant method for reusing stabilized sewage sludge (biosolids) in the United States is land application to agricultural and forest land (3, 13). While regulations restrict access to application sites based on the recognized pathogen, respiratory irritant, and metal content in class B biosolids (42), the potential off-site exposure due to wind-aerosolized biosolids has not been considered (27). In agricultural fields, wind aerosolization events may occur several times in a year. Climate, soil moisture, soil texture, and the level of soil disturbance control the frequency and severity of these events (16, 17). Wind velocities as low as 4 to 10 m/s can aerosolize material from soils with sandy textures and low moisture content (15).
Direct confirmation of biosolid wind aerosolization may not be obtainable by common wastewater bioaerosol analysis, the focus of which has been coliform detection (14, 25). The large reduction of coliforms during sewage sludge stabilization, coupled with the inability of many gram-negative microorganisms to survive environmental and collection-associated stresses (8, 43), makes coliform bacteria a poor choice for tracking biosolid-derived aerosols. Indeed, the release of biosolid bioaerosols has only been documented at the source of overt aerosol-producing activities such as biosolid spreading, loading, or disk incorporation into soils (4, 31, 38). In these cases, the large aerosol mass concentrations of biosolids (ca. 500 to 1,000 μg/m3) were documented more readily by culturing spores of thermotolerant Clostridia and bacteriophages than by coliform detection (11). The potentially lower aerosol mass loading produced during wind aerosolization events suggests the need for a sensitive, culture-independent method to track these aerosols. Culture-independent analysis circumvents sampling stress and bioaerosol die-off and allows for the inclusion of highly enriched, uncultured microorganisms as indicators. Additionally, DNA-based sequence comparison provides more specific and definitive information from which source tracking can be conducted.
The central hypothesis of the following research was that DNA-based analysis can be used to specifically and sensitively confirm the unintended aerosolization of land-applied biosolids during high-wind events. Three biosolid source tracking methods based on the 16S rRNA gene sequences of Clostridia, Chloroflexi, and Euryarchaeota (“Clostridia, Chloroflexi, and Euryarchaeota methods” hereafter) were tested and then applied to downwind and upwind control aerosol samples collected during high- and low-wind events at class B biosolid land application sites.
MATERIALS AND METHODS
Field site description and sampler locations.
Biosolid, soil, and aerosol sampling was performed in agricultural fields southwest of Buckeye, AZ. Dewatered class B biosolids that were stabilized by anaerobic mesophilic digestion were land applied at coverage rates between 0.8 × 104 kg/hectare and 1.6 × 104 kg/hectare. The applied areas considered were typically 1.5 ha and in the shape of a rectangle with sides 100 m perpendicular and 150 m parallel to the wind direction. Two sets of wind aerosolization conditions were considered. In the first, termed “no disking,” aerosol samples were collected upwind and downwind of fields on which biosolids had been applied within the previous 36 h. The biosolids in these fields had not yet been disk incorporated into soils. In the second scenario, termed “disking,” aerosol samples were taken upwind and downwind of fields where previously applied biosolids were disk incorporated within 36 h of sampling. Disking refers to tilling soil to a depth of approximately 10 to 15 cm to incorporate biosolids previously applied on the surface. Disking is mandated through federal or state management guidelines to reduce odors and vector attraction of surface-applied biosolids. For both aerosolization conditions, high-wind experiments were conducted at times when the average wind speed exceeded 5 m/s. During the no-disking experiments, aerosol samples were also taken under low-wind conditions (average wind speed less than 2 m/s). All three scenarios were tested in at least three independent experimental runs held on different days in different fields. Experiments were conducted during the spring season, when high-wind events are prevalent in central Arizona (March through May 2005).
Three aerosol measurement stands were located at the downwind edge of the land application area and were spaced 20 m apart. Each sample stand contained two liquid impinger biosamplers (SKC, Inc., Eighty Four, PA) and one aerosol filter sampler. All samplers were suspended 1.5 m above ground and pointed directly into the wind. For upwind control, two sampling stands, each containing two liquid impingers and one filter sampler, were located approximately 150 m upwind of the biosolid application area. One real-time monitor for particulate matter smaller than 10 μm in aerodynamic diameter (PM10) (DustTrak aerosol monitor; TSI, Inc., St. Paul, MN) was located on a downwind sample stand, while another was located with the upwind samplers. Meteorological conditions were monitored every minute with a Weather monitor II (Davis Instrument Corporation, Hayward, CA); these conditions included temperature, relative humidity, wind speed, and wind direction.
Aerosol, biosolid, and soil sample collection.
Liquid impinger biosamplers were filled with 20 ml of sterile phosphate-buffered saline (PBS; 30 mM phosphate buffer, 125 mM NaCl [pH 7.2]) and collected aerosols at 12.5 liters/min. Impingers were refilled with sterile PBS during experimentation (120-min duration) to maintain volume. Impingers were also shielded from sunlight with aluminum foil. Impinger contents were used in both culture analysis and PCR-based analyses. Microbial aerosol sampling for PCR-based analyses was also carried out using filter samplers that contained 1.0-μm-pore-size polycarbonate track-etched (PCTE) filters (GE Osmonics, Minnetonka, MN) mounted on open face holders and operated at a flow rate of 33 liters/min. For long-term sample storage at −80°C, impinger contents were transferred into sterile glass tubes or filters were transferred into a sterile 5-cm-diameter petri dish and sealed with Teflon tape. Flow rates for all sampling equipment were calibrated prior to each field experiment.
For bulk biosolid and soil analysis, approximately 50 g (wet weight) of material was collected at the time and location of aerosol sampling. Each sample was a composite, containing 10 g to 20 g of bulk material from at least three different field locations. Soil samples were taken in fields adjacent to where biosolids had been applied. For long-term storage, bulk biosolids and soil samples were archived at −80°C. Moisture content of all bulk samples was determined gravimetrically by oven drying 10 g of samples at 105°C for 24 h. Soil particle size distribution was determined via sieve analysis for the largest particles and hydrometer analysis for particles smaller than 75 μm, in accordance with previously described methods (2).
MST methods.
Microbial source tracking (MST) targets that were unique to and highly enriched in biosolids were first identified by constructing phylogenetic clone libraries for bulk biosolid and aerosol samples taken directly downwind of a biosolid spreading operation, and ambient aerosol samples taken upwind from biosolid spreading operations. (Further details on these aerosol samples are available in reference 28.) Based on the results of these libraries and a review of the literature concerning anaerobic digester microbial ecology, three MST methods were chosen. These methods include (i) the culture of sulfite-reducing Clostridia followed by PCR amplification of colonies to obtain 16S rRNA gene sequences; (ii) DNA extraction, PCR amplification, and sequencing of the 16S rRNA gene for a bacterium of the class Chloroflexi (hereafter “Chloroflexi sp.”) commonly found in bulk biosolids; and, (iii) DNA extraction, nested PCR, and terminal restriction fragment length polymorphism (tRFLP) analysis of 16S rRNA gene sequences of Euryarchaeota specific to biosolids. These methods were first applied to a variety of bulk biosolid and bulk soil samples to determine their average rate of correct classification and then applied to aerosol samples taken at land application sites during high-wind events.
Culture analysis.
The enumeration of sulfite-reducing Clostridia was performed using a modified membrane filtration technique (35), where cells filtered onto 0.22-μm Durapore membranes were anaerobically incubated at 37°C for 48 h on an antibiotic-supplemented egg yolk-free tryptose-sulfite-cycloserine (TSC) agar. For bulk biosolid analysis, microorganisms were extracted from 10 g (wet weight) of biosolids by being mixed with 100 ml of 0.25× Ringer's solution (38 mM NaCl, 1.4 mM KCl, 1.1 mM CaCl2, 0.6 mM NaHCO3) according to previously described methods (26). For aerosol samples, up to 35 ml of aerosol impinger collection fluid (pooled liquid impinger contents from the same sampler stand) was filtered to provide a detection level of approximately 1 CFU/m3.
DNA isolation.
DNA was extracted from frozen biosolid, soil, and aerosol samples. For aerosol filter samples, PCTE filters were eluted in the 5-ml petri dish used for storage by adding 3 ml sterile PBS with 0.05% Tween and a Teflon bar (4 cm long, 3-mm radius). This mixture was agitated on an orbital shaker at 36 rpm for 12 h. The eluant was concentrated by filtering through a 1-cm2 section of a 0.22-μm-pore-size PCTE filter. For aerosol liquid impinger samples, 5 to 10 ml of sample was also concentrated by filtering through a 1-cm2 section of a PCTE filter. After aerosol sample concentration, the 1-cm2 PCTE filters were placed in a 2-ml bead-beating tube and cells were extracted from the filters by vortexing at top speed for 2 min in 200 μl of extraction buffer I (150 mM Na2EDTA, 225 mM NaCl, 0.05% Tween 20 [pH 8.5]). DNA was then isolated by bead beating to lyse cells, phenol-chloroform extraction to separate DNA, and column filtration for DNA cleanup and concentration. This method followed a previously described method (30), with the exception that no lysozyme was added and the bead-beating rate was set at 2,500 rpm. The method described above was also used to isolate DNA from bulk soil and biosolid samples. In these cases, 200 mg of material was placed in bead-beating tubes with 200 μl of extraction buffer I.
PCR amplification.
16S rRNA genes were PCR amplified from genomic DNA extracted from bulk biosolid, bulk soil, and aerosol samples. All primers used in this study are listed in Table 1. PCR for the bulk biosolid and aerosol universal clone libraries was carried out with both bacterial (27F and 907R) and universal (515F and 1392R) primer sets. In addition to the two primer sets, both bulk biosolid and aerosol libraries were constructed from two sets of biosolid and aerosol samples collected on different days. PCR amplification was carried out in a 50-μl-volume reaction mixture containing 5 μl of 10× buffer (50 mM KCl, 10 mM Tris-HCl), 10 μl of 25 mM MgCl2, 1 μl of deoxynucleoside triphosphate (dNTP) mix (0.2 mM each dATP, dCTP, dGTP, and dTTP), 5 μl of 10-mg/ml bovine serum albumin (BSA), 4 μl of 5 μM each of the bacterial primers 27F and 907R or the universal primers 515F and 1392R, 2 μl of extracted DNA template, and 5 U of Taq DNA polymerase (Eppendorf, Westbury, NY). The following thermocycler (Eppendorf Mastercycler; Perkin-Elmer, Inc., Boston, MA) temperature profile was used (for both bacterial and universal primers): hot start for 12 min at 94°C followed by 20 cycles of 92°C for 30 s, 65°C minus 1°C/cycle for 30 s, and 72°C for 90 s. This initial amplification was followed by 20 additional cycles at 92°C for 30 s, 45°C for 90 s, and 72°C for 90 s and a final extension at 72°C for 20 min. PCR for colonies of sulfite-reducing Clostridia was performed using universal primers 515F and 1392R in accordance with reagents and the temperature profile described above. DNA template was supplied by picking single black colonies of Clostridia from plates and suspending cells in the 50-μl PCR mix. For the Chloroflexi sp., 16S rRNA gene amplification was performed with forward primer CH608 and reverse primer CH1124. PCR was carried out with 5 μl of extracted aerosol, bulk soil, or bulk biosolid DNA template in a 50-μl PCR mixture containing 5 μl of 10× PCR buffer (50 mM KCl, 10 mM Tris-HCl), 3 μl of 1.5 mM MgCl2, 1 μl dNTP (0.2 mM each dATP, dCTP, dGTP, and dTTP), 5 μl of 10-mg/ml BSA, 4 μl of 5 μM each primer, and 2.5 U Taq DNA polymerase (Eppendorf). The following temperature program was used: hot start of 5 min at 80°C, after which the Taq DNA polymerase was added; followed by 5 min at 94°C; followed by 34 cycles of 60 s at 94°C, 90 s at 56°C, and 90 s at 72°C. A final extension at 72°C for 6 min concluded the amplification. Finally, amplification of 16S rRNA genes Euryarchaeota were performed using nested PCR. The primary primer set was primer A21F specific for the domain Archaea and the universal primer 1392R. The secondary primer set included MB301F (6-carboxyfluorescein labeled at the 5′ end) which circumscribes the genera Methanobacterium, Methanobrevibacter, and Methanosphaera (41) and primer AR912R specific for the domain Archaea. The primary amplification PCR mixture contained 5 μl of DNA template in 50 μl PCR mix containing 10 μl of 5× PCR buffer (Phusion HF buffer), 1 μl dNTP (0.2 mM each dATP, dCTP, dGTP, and dTTP), 1.5 μl of dimethyl sulfoxide, 2.5 μl of 5 μM each primer, and 1 U Phusion DNA polymerase (Finnzymes, Espoo, Finland). PCR for the primary amplification was performed using the following temperature program: 30 s at 98°C, followed by 34 cycles of 10 s at 98°C, 30 s at 59°C, and 30 s at 72°C. A final extension at 72°C for 7 min completed the program. The secondary PCR was carried out with 2 μl of the primary product in 50 μl PCR mix. PCR was performed as described for the primary PCR, with the exception of an annealing temperature of 66°C. In all PCRs, amplicon lengths shown in Table 1 were confirmed by gel electrophoresis. Sterile filtered water-negative controls were included in all PCRs, and no amplification was observed in these controls.
TABLE 1.
PCR primers used in this study
| Primer name | Primer target group | Primer sequence (5′→3′) | Amplicon size (bp) | Reference |
|---|---|---|---|---|
| 27F | Domain Bacteria | AGAGTTTGATCCTGGCTCAG | 900 | 22 |
| 907R | Universal | CCGTCAATTCCTTTRAGTTT | 22 | |
| 515F | Universal | GTGCCAGCMGCCGCGG | 899 | 22 |
| 1392R | Universal | ACGGGCGGTGTGT[A:G]C | 22 | |
| A21F | Domain Archaea | TTCCGGTTGATCC[C:T]GCCGGA | 1,351 | 9 |
| 1392R | Universal | ACGGGCGGTGTGT[A:G]C | 22 | |
| MB301F | Methanobacteriales | CTTGTCTCAGGTTCCATCTCCG | 608 | 41 |
| AR912R | Domain Archaea | CTCCCCCGCCAATTCCTTTA | 24 | |
| CH608 | Chloroflexi sp. | CCAAGCTTGAGGATGGTAGA | 516 | This study |
| CH1124 | Chloroflexi sp. | GACTTAATATCGGCAGTACCGTG | This study |
Cloning and sequencing.
Universal clone libraries were created for both biosolid and aerosol samples to guide MST microorganism selection. Libraries of Euryarchaeota were constructed for bulk biosolid and soil samples to guide tRFLP enzyme selection. For the construction of all libraries, PCR amplicons were purified with a QIAquick PCR purification kit (QIAGEN, Inc., Valencia, CA), ligated into the PCR 4-TOPO vector (TOPO TA cloning kit; Invitrogen, Carlsbad, CA), and transformed into chemically competent Escherichia coli cells (One Shot E. coli; Invitrogen) following the manufacturer's protocol for optimal plasmid generation. Transformed cells were plated onto Luria-Bertani agar medium augmented with 50 μg of ampicillin per ml and grown overnight at 37°C. To confirm that colonies contained plasmids with the correctly sized insert, single colonies were suspended in 50 μl of PCR mix and amplified using primers T3 and T7, which circumscribe the vector insert site. PCR amplicons with the correct insert were used for further sequencing. For sequencing, excess primers and nucleotides were first removed using ExoSAP-IT (U.S. Biochemicals Corporation, Cleveland, OH) according to the manufacturer's protocol. Samples combined with 20 ng of T3 primer were sequenced with a capillary column sequencer (no. 3730; Applied Biosystems, Foster City, CA) at the Arizona State University DNA laboratory. In addition to sequencing clones, PCR amplicons from colonies of sulfate-reducing Clostridia and the the Chloroflexi sp. amplicons were prepared for sequencing on a capillary column sequencer by first removing nucleotides (ExoSAP-IT) and then combining the amplicons with 20 ng of primer (515F for Clostridia or CH608 for the Chloroflexi sp.).
All retrieved sequences were compared with available 16S rRNA gene sequences by using the Basic Local Alignment Search Tool (BLAST) and the National Center of Biotechnology Information (NCBI) database (1). Sequences were aligned using CLUSTALX (40), and construction of phylogenetic trees was performed by the neighbor-joining method using MEGA2 software (21).
tRFLP analysis.
A 2.5-μl master mix containing 0.3 μl restriction enzyme solution (5,000 U/ml SfcI; New England Bolas, Ipswich, MA), 2 μl of 10× NE buffer (20 mM Tris-acetate, 50 mM potassium acetate, 10 mM magnesium acetate, 1 mM dithiothreitol), and 0.2 μl BSA (100 μg/ml) was added to 17.5 μl of sterile water containing 0.3 μg PCR of product. Samples were incubated at 25°C for 16 h and then inactivated by heating to 65°C for 20 min. Digested PCR product was purified using the QIAquick nucleotide removal kit (QIAGEN, Inc., Valencia, CA) according to the manufacturer's protocol and eluted in 30 μl of 10 mM Tris buffer. A 3-μl volume containing 5 parts loading buffer (50 mg/ml blue dextran, 25 mM EDTA, GeneScan-1000 [ROX] red; Applied Biosystems, Foster City, CA) and 1 part standard (GeneScan-1000 [ROX] red; Applied Biosystems, Foster City, CA) was mixed with 2 μl of the cleaned digested product and loaded into a gel sequencer (no. 377; Applied Biosystems, Foster City, CA) to separate terminal fragments. Fragment data were viewed using Genescan and Genotyper software (Applied Biosystems).
Nucleotide sequence accession numbers.
Sequences obtained from the bulk biosolid, bulk soil, and aerosol samples have been deposited in the NCBI GenBank database under accession no. EF029242 to EF029507 (biosolid and aerosol clone libraries) and EF029508 to EF029712 (MST method development and application).
RESULTS
Clone libraries.
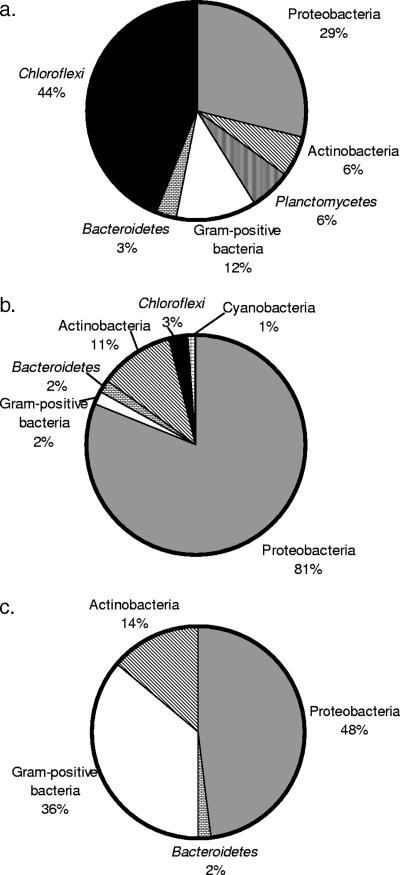
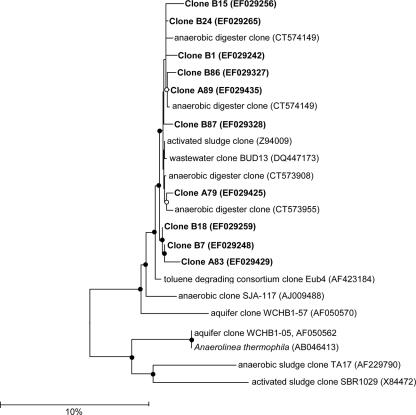
Universal clone libraries were constructed to identify 16S rRNA gene sequences that were highly enriched and unique to biosolids and to determine if these sequences could be detected in aerosols. In order to sample an aerosol that was know to contain biosolids, aerosol samples for these universal libraries were taken directly downwind of a biosolid spreading operation where PM10 monitoring revealed a 1,000-μg/m3 concentration of biosolid-derived aerosols (29). Ambient aerosol samples were also taken upwind from biosolid spreading operations for control. The resulting libraries for bulk biosolids, downwind aerosols, and ambient upwind aerosols contained 121, 138, and 64 clones, respectively. Figure 1a, b, and c show the phylogenetic distribution of the bulk biosolids, downwind aerosol clone sequences, and upwind aerosol clone sequences, respectively. Approximately 10% of downwind aerosol clone sequences were >98% similar to clone sequences derived from bulk biosolids, and 56% of the downwind aerosol sequences were most closely matched to database sequences derived from either sewage or wastewater treatment environments (7, 10, 37). More than 99% of all clones were bacteria, with the phyla Chloroflexi and Proteobacteria being the most abundant in bulk biosolids. In particular, most clones of the phylum Chloroflexi and 25% of all bulk biosolid clones closely matched a distinct group of uncultured microorganisms, listed here as “Chloroflexi sp.,” that were previously isolated from anaerobic municipal activated sludge (37) and anaerobic digesters (7) (Fig. 1a and 2). Downwind aerosol clone libraries also produced these Chloroflexi sp. sequences (listed under Chloroflexi in Fig. 1b), although at a lower enrichment. Ambient upwind aerosol libraries contained no Chloroflexi sp. sequences nor any other sequences similar to the clones described from the bulk biosolids (Fig. 1c).
FIG. 1.
Phylogenetic distribution of universal and bacterial clones derived from (a) bulk biosolids, (b) aerosols downwind of biosolid spreading operations, and (c) ambient control aerosols.
FIG. 2.
16S rRNA gene-based phylogenetic tree for selected isolates of Chloroflexi. The tree includes clones sequenced from biosolids (B) and soil (S), downwind aerosols (D), and selected type strains isolated from wastewater environments. The tree is based on the neighbor-joining method; the bar indicates 10% estimated sequence divergence. Closed circles represent >50% bootstrap values, and open circles represent <50% bootstrap values. The tree is rooted using E. coli (AM184252) as the out-group (not shown).
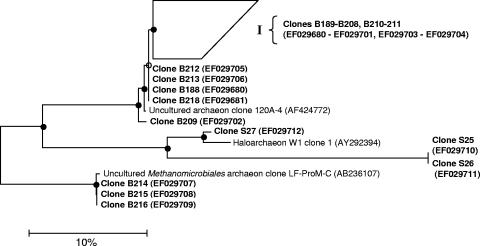
In a second set of libraries, clones of Euryarchaeota from bulk biosolids and soil provided sequence information from which to base tRFLP restriction enzyme choice upon. These libraries resulted in 30 clones from biosolids and three clones from soil (Fig. 3). The sequences of 87% of the biosolid clones were 99% similar to a clone of Archaea previously isolated from municipal wastewater anaerobic digesters, and 13% of clones had sequences that were 99% similar to species of Methanomicrobiales. Weak amplification of soil DNA using primers specific for Euryarchaeota resulted in the production of only three clones. Classified as Crenarchaeota, these soil clones were phylogenetically distinct from the biosolid clones.
FIG. 3.
16S rRNA gene-based phylogenetic tree of clones of Euryarchaeota sequenced from biosolids (B) and soil (S) and selected type strains. Branch I contains 22 sequences from biosolids. The tree is based on the neighbor-joining method. Closed circles represent >50% bootstrap values, and open circles represent <50% bootstrap values. The tree is rooted using Geoglobus ahangari (AF220165) as the out-group.
tRFLP.
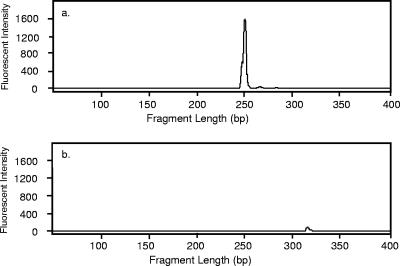
Biosolid and soil sequences from the libraries of Euryarchaeota were aligned and screened with the online software VectorDesigner (Invitrogen Corporation, Carlsbad, CA) to predict the terminal fragment lengths of a variety of different restriction enzymes. The enzyme SfcI (New England Biolabs, Ipswich, MA) provided the most promising result, cutting 27 of the 30 biosolid clones to produce a single 251-bp fragment, while not cutting soil clone sequences. To experimentally confirm the predicted fragment lengths, 16S rRNA gene sequences from the biosolid and soil libraries of Euryarchaeota were digested with SfcI and analyzed to measure the terminal fragment lengths. As predicted, SfcI cut 27 of the biosolid sequences of Euryarchaeota at 251 bp. None of the soil sequences was cut at 251 bp.
MST verification.
Each MST method was tested against a suite of field-collected bulk biosolid and soil samples to confirm both presence in biosolids and absence in surrounding soils. Soils were used for comparison, as they are the major source of non-biosolid aerosols in the agricultural areas considered.
Sulfite-reducing Clostridia were successfully cultured from all biosolid samples and in 43% of soil samples, although concentrations between the two bulk matrices were markedly different. The average concentrations (mean ± standard deviation) were 3.72 × 106 ± 6.65 × 106 CFU/dry g in biosolids and 1.54 ± 2.27 CFU/dry g for soil samples. The ecologies of culturable sulfite-reducing Clostridia in biosolids and soils were also dissimilar, as shown in Fig. 4. Sequences >99% similar to Clostridium bifermentans (92% of the 78 biosolid sequences) dominated biosolid populations, while sequences similar to Clostridium subterminale (56%) dominated in bulk soil samples. C. bifermentans was identified in only 1 of 15 soil samples (Table 2). Clostridium perfringens strains were commonly isolated from both biosolids and soils. Based on these results, the 16S rRNA gene sequence for C. bifermentans was chosen as a marker for biosolid source tracking. Similarly, the Chloroflexi sp. and Euryarchaeota MST methods were applied to the same group of bulk soil and biosolid samples. Using specific primers, sequences of the Chloroflexi sp. were found in all 14 biosolid samples tested and in 2 of the 14 bulk soil samples analyzed; these sequences were >99% similar to the sequences of Chloroflexi sp. retrieved from the universal biosolid and source aerosol clone libraries. Finally, all bulk biosolid samples analyzed by the tRFLP method produced 251-bp fragments, while only 1 of the 10 soil samples considered showed these fragments (Fig. 5a and b). The average rates of correct classification (ARCC) (defined here as 100 − the percentage of positive responses in soils) were 93.3%, 85.7%, and 90% for the Clostridia, Chloroflexi sp., and Euryarchaeota methods, respectively. No single soil sample was positive for all three MST methods, yielding an ARCC of 100% when all MST methods were used to test a sample. Table 2 summarizes the results for MST method testing of the bulk biosolid and soil samples.
FIG. 4.
16S rRNA gene-based phylogenetic tree sequences from cultured sulfite-reducing Clostridia isolated from biosolids (B) and soil (S) and selected type strains. Branch I contains C. bifermentans sequences from biosolids and from soils, and branch II contains C. subterminale sequences from soil. The tree is based on the neighbor-joining method. Closed circles represent >50% bootstrap values, and open circles represent <50% bootstrap values. The tree is rooted using Geoglobus ahangari (AF220165) as the out-group (not shown).
TABLE 2.
Results of MST testing in bulk soil and bulk biosolids
| MST method | % Positive (no. of samples)
|
ARCC (%) | |
|---|---|---|---|
| Biosolids | Soil | ||
| C. bifermentans | 100 (13) | 6.7 (15) | 93.3 |
| Chloroflexi sp. | 100 (14) | 14.3 (14) | 85.7 |
| Euryarchaeota | 100 (11) | 10 (10) | 90 |
FIG. 5.
Characteristic tRFLP electrophoretic output for the Euryarchaeota MST method. The 251-bp fragment length and fluorescence intensity of amplified 16S rRNA genes are shown for biosolids (a) and were not present in sequences amplified from soils (b).
Application of MST methods to aerosol samples.
The three MST methods were next applied to determine if high-wind events resulted in the aerosolization of land-applied biosolids. Measurements to test this hypothesis included PM10 concentrations, aerosol concentrations of culturable sulfite-reducing Clostridia, and application of the three MST methods to the collected aerosol samples. Samples were collected for both the no-disking and disking scenarios. Table 3 lists average wind speed, PM10 data, and relevant soil and biosolid characteristics for all experiments.
TABLE 3.
Conditions during field experiments
| Expt (no. of independent expts) | Location | Avg wind speed (m/s) | Avg PM10 (μg/m3) | % Moisture
|
Texture of soil or soil-biosolid mixture | |
|---|---|---|---|---|---|---|
| Biosolidsa | Soil or soil-biosolid mixtureb | |||||
| No disking | ||||||
| High wind (4) | Upwind | 5.04 | 13 | 47 | 2.5-4 | Sandy loam |
| Downwind | 10 | |||||
| Low wind (3) | Upwind | 1.84 | 28 | 58 | 2.5-4 | Sandy loam |
| Downwind | ||||||
| Disking | ||||||
| High wind (3) | Upwind | 6.04 | 33 | 61 | 8-12 | Loam and sandy loam |
| Downwind | 110 | |||||
Moisture content was 70% to 80% during application but decreased due to drying in the time after land application and before aerosol measurement.
Moistures reported for no disking are for soil only; moistures reported for disking are for the soil-biosolid mixture.
For the no-disking scenario, there was no observable increase in the PM10 concentration during high-wind events (greater than 5 m/s). Indeed, the average PM10 concentration was 2.6 times higher during low-wind events (less than 2 m/s) than during high-wind events. Lower PM10 concentrations during high wind speeds are common when the threshold wind velocity required to aerosolize large amounts of soils or biosolids is not exceeded. While PM10 increases were not observed, application of MST methods provided evidence that biosolids were aerosolized during the high-wind, no-disking scenario. All four high-wind experiments resulted in significant concentrations of sulfite-reducing Clostridia downwind of the land-applied fields, while no sulfite-reducing Clostridia were detected upwind (Fig. 6a). The average concentration of downwind sulfite-reducing Clostridia was 7.39 ± 3.15 CFU/m3. One of the four low-wind samples (<2 m/s) produced an aerosol concentration of sulfite-reducing Clostridia. In this case, only one colony was detected—yielding an aerosol concentration that was an order of magnitude lower than average downwind concentrations of sulfite-reducing Clostridia measured during high-wind events. When all aerosol-cultured, sulfite-reducing Clostridia were PCR amplified and sequenced, aerosolization of biosolids was confirmed in 50% of high-wind event experiments and in none of the low-wind event experiments. For the Chloroflexi sp. and Euryarchaeota tRFLP methods, aerosolized biosolids were detected in 25% and 75% of high-wind experiments, respectively. The Chloroflexi and Euryarchaeota methods also detected aerosolized biosolids in 50% of samples for low-wind events (Table 4). All nine upwind control samples were negative for the no-disking experiments.
FIG. 6.
Concentrations of sulfite-reducing (red.) Clostridia in aerosols during high-wind events for (a) no-disking and (b) disking scenarios. During the no-disking scenario, two of the three dates for low wind showed no detection and all seven upwind controls showed no detection. For disking, all three upwind controls showed no detection.
TABLE 4.
Results of MST application to aerosol samples
| MST method | % Positive (no. of samples)
|
|||
|---|---|---|---|---|
| Downwind
|
Upwinda | |||
| No disking
|
Disking (high wind) | |||
| High wind | Low wind | |||
| C. bifermentans | 50 (4) | 0 (2) | 33 (3) | 0 (9) |
| Chloroflexi sp. | 25 (4) | 50 (2) | 75 (4) | 10 (10) |
| Euryarchaeota | 75 (4) | 50 (2) | 75 (4) | 0 (10) |
The positive upwind control was used for the disking scenario.
Additional wind aerosolization experiments were undertaken to determine if biosolids were more susceptible to aerosolization once they are incorporated into dry soils (disking scenario) and the soil crust is disturbed. In contrast to the no-disking scenario, high-wind experiments after disking produced elevated levels of PM10 downwind of the land application site (Table 3). PM10 values during high-wind events averaged 110 μg/m3 for experiments on 3 different days and were 3.3 times greater than levels upwind—where soils were undisturbed. Downwind PM10 averaged 200 μg/m3 on the highest-wind-speed day, with a peak value exceeding 1,500 μg/m3. Figure 7 demonstrates the relationship between PM10 generation and wind speed during this high-wind event.
FIG. 7.
Time-resolved wind speed and PM10 concentrations during wind aerosolization experiments. Measurements were taken downwind of a field in which biosolids had recently been disk incorporated.
Figure 6b presents aerosol concentrations of sulfite-reducing Clostridia during these high-wind events. Clostridia were detected downwind of fields in all three independent high-wind aerosol experiments; average concentrations of Clostridia were 7.1 ± 9.1 CFU/m3. PCR and sequencing of colonies of sulfite-reducing Clostridia confirmed biosolid aerosolization in 33% of downwind aerosols. MST results from both the Chloroflexi sp. and Euryarchaeota MST analyses verified biosolid aerosolization in 75% of downwind samples (Table 4). Ten of 11 upwind control samples taken during the disking scenario were negative for all MST analyses.
DISCUSSION
MST methods.
The three bacteria considered for MST development fulfill the criteria for a successful aerosol source tracking target. Chloroflexi sp. and Euryarchaeota are enriched in anaerobically digested biosolids, typically comprising greater than 1% of total cells (7, 23, 34), although it is noted that isolates of the Chloroflexi sp. have not been observed in all anaerobic digester microbial ecology studies. Culturable isolates of sulfite-reducing Clostridia are typically enriched to 105 to 107 CFU/dry g biosolids (0.01% or less of the total microorganism counts in biosolids) (31). The Chloroflexi sp. and Euryarchaeota methods do not require culturability and therefore circumvent the limitations associated with aerosolization and sampling stresses. Although the sulfite-reducing Clostridia method is culture dependent, Clostridia form spores, and the resistance of bacterial spores to aerosolization and collection stresses has been documented (20, 36). The MST validation experimental results revealed that each method provides a unique biosolid indicator. When each MST analysis was applied to bulk biosolids and soils, 100% of samples were positive for biosolids while less than 11% were positive for soils. ARCC for the individual methods ranged from 85% to 93%, and these values compare favorably with the wide variety of phenotypic and genotypic MST methods used to identify fecal contamination in water (65% to 99%) (5, 6, 19). Due to aerosol die-off and low enrichment and sampling limitations for coliphages based on size, the MST methods most commonly described in the literature that are based on culturable E. coli, coliforms, or coliphages are not relevant for aerosols, which require approaches based on environmentally resistant bacteria or culture-independent methods. The use of rRNA gene sequences to track aerosolized Clostridia back to bulk biosolid material has been previously established (12, 33). In these previous studies, Clostridia were obtained from aerosol samples taken downwind of biosolid loading operations and from grab samples of biosolid storage piles. The size of 16S to 23S interspacer region amplicons of isolates of Clostridia, as well as the patterns generated from HindIII digestion of these amplicons, provided links between some of the aerosol isolates and isolates from the biosolid storage piles. In the research described here, the sequence-based comparison of isolates of Clostridia supports the use of rRNA gene sequence information to track biosolid-derived Clostridia, estimates ARCC, and extends this MST method to quantitatively confirm wind aerosolization of land-applied biosolids.
No MST method produced correct classification rates of 100%. Several factors may have contributed. First, to be representative, soil samples were taken in fields adjacent to and approximately 100 m upwind of the biosolid land application location. Deposition of aerosols produced during nearby biosolid land application or other dairy or livestock activities also could not be unambiguously accounted for. While available records from biosolid land application contractors did not indicate previous biosolid application at these control sites, the possibility of prior, undocumented biosolid or animal compost application could not be definitively excluded. Although these uncertainties do not allow for a strictly controlled field experiment, they are inherent to both the agricultural setting and the biosolid land application process. For these reasons, a multiple-MST approach may be required to produce very high rates (greater than 95%) of correct classification.
Application of MST methods to aerosol samples.
The application of MST methods to aerosols demonstrated that biosolid-derived aerosols could be emitted from land-applied fields during high-wind events. For high-wind events in both the no-disking and disking scenarios, the majority of downwind aerosol samples contained 16S rRNA gene sequences originating from biosolids (13 of 23 downwind). When MST methods were applied to aerosol samples taken at upwind control locations, only 1 of 29 samples was positive. PM10 concentrations (only in disked fields) and culturable analysis of sulfite-reducing Clostridia in aerosol samples also support the conclusion of wind aerosolization (Fig. 6 and 7). The lower ARCC for field-measured aerosols compared to the ARCC for bulk soils may be due to the influence of detection levels: aerosols provide approximately 1 mg of material for analysis, while sample material in bulk soils is unlimited.
Aerosol and bulk biosolid concentrations of sulfite-reducing Clostridia can also be used to estimate the biosolid content of aerosols. Using the average concentration of sulfite-reducing Clostridia in bulk biosolids sampled (3.72 × 106 ± 6.65 × 106 CFU/dry g), the 1- to 20-CFU/m3 range of aerosolized sulfite-reducing Clostridia corresponds to a 0.1- to 2-μg/m3 source aerosol concentration of pure biosolids and suggests limits of detection below this value. This aerosol concentration corresponds to less than 20% of total PM10 in the no-disking cases and less than 2% of the total PM10 in the disking scenario. These biosolid-derived PM10 values are markedly lower than the 500- to 1,000-μg/m3 source concentrations reported during biosolid spreading and disking of dewatered biosolids (29, 31). However, these low values should not be viewed as insignificant since the source emission area could be spread over several square kilometers and produce an equal or greater biosolid mass emission rate into the environment (mass biosolids emitted per time) than those produced by spreading and disking.
The low aerosol concentrations of released biosolids required sensitive MST methods. The direct PCR methods (Chloroflexi sp. and Euryarchaeota) appeared to be more sensitive than the Clostridia method as higher percentages of biosolid-positive results were produced in downwind aerosol samples using the Chloroflexi sp. and Euryarchaeota methods than the Clostridia method. Based on inhibition and inefficiencies in sample processing, realistic PCR detection levels of 100 to 200 cells have previously been established in environmental aerosol samples (32). In contrast, only one spore of sulfite-reducing Clostridia is necessary to form a detectable colony. These differences are negated when considering the enrichment in anaerobically digested biosolids of Chloroflexi sp. and Euryarchaeota (>1% total cells) compared to sulfite-reducing Clostridia (<0.01% total cells) (7, 23, 34). The two culture-independent methods also have greater potential for increased sensitivity. Higher volume filtration may be used for sampling without regard to collection inactivation. In addition, progress in filter extraction efficiency, PCR inhibition removal, and quantitative PCR will continue to improve the sensitivity and quantitativeness of culture-independent aerosol detection.
Finally, downwind concentrations of sulfite-reducing Clostridia and MST results were similar for the no-disking and disking experiments. However, PM10 aerosolization was markedly higher during disking experiments than with the no-disking scenario. These differences underscore the importance of understanding how the land application process affects ambient soil aerosolization. The threshold wind velocity required to aerosolize soil particles is depressed in soils with sandy textures, low soil moisture, and high levels of disturbance (18). Disking clearly disturbed soil structure, and thus larger concentrations of PM10 were released in the disking scenario compared to the no-disking scenario. A consequence of the similar aerosol concentrations of Clostridia for different PM10 levels is that the percentage of biosolid PM10 to total PM10 was approximately 10 times greater in the no-disking case. During disking, the incorporation of biosolids into the subsurface should render most of them unavailable for aerosolization and may partially explain the differences in these ratios. Before a comprehensive set of recommendations on how to reduce wind aerosolization can be made to practitioners, specific mechanistic wind aerosolization studies that include the MST methods developed here will be required to understand how climate, soil, biosolid characteristics, and land application influence wind aerosolization of biosolids. In addition, the three MST methods developed should be tested in new environments using different biosolids to ensure their applicability under all relevant conditions.
Three MST methods have been developed to track the aerosolization of anaerobically digested class B land-applied biosolids. Application of each MST method to field aerosol samples in conjunction with field PM10 measurements provides corroborating evidence that biosolids can become aerosolized during high-wind events. While the results presented here demonstrate the utility of the three biosolid source tracking methods, these methods and results may not be applicable to every land application location, given the diversity of wastewater sources, biosolid processing methods, and land application environments. However, the biosolid and field conditions of this study represent common land application practice. Anaerobic digestion of sewage sludge is the most common method for sludge stabilization (39), and land application is most prevalent in the western and midwestern United States, where arid soil conditions prevail (3). The application of culture-independent MST techniques to aerosols is a new field that has the broader potential for understanding the airborne exposure route and tracking the local, regional, and global transport of aerosols.
Acknowledgments
This work was supported by National Science Foundation grant BES 0348455, awarded to J. Peccia.
We thank Scott Bingham from Arizona State University for assistance with tRFLP and the City of Phoenix for assistance with land application site access.
Footnotes
Published ahead of print on 18 May 2007.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Meyers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Bardet, J. 1997. Experimental soil mechanics. Prentice Hall, Inc., Upper Saddle River, NJ.
- 3.Bastian, R. 1997. The biosolids (sludge) treatment, beneficial use, and disposal situation in the USA. Eur. Water Pollut. Control J. 7:62-72. [Google Scholar]
- 4.Brooks, J. R., B. D. Tanner, K. L. Josephson, C. Gerba, C. N. Haas, and I. Pepper. 2005. A national survey on the residential impact of biological aerosols from the land application of biosolids. J. Appl. Microbiol. 99:310-322. [DOI] [PubMed] [Google Scholar]
- 5.Carson, C. A., B. L. Shear, M. R. Ellersieck, and A. Asfaw. 2001. Identification of fecal Escherichia coli from humans and animals by ribotyping. Appl. Environ. Microbiol. 67:1503-1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choi, S., W. Chu, J. Brown, S. J. Becler, V. J. Harwood, and S. C. Jiang. 2003. Application of enterococci antibiotic resistance patterns for contamination source identification at Huntington Beach, California. Mar. Pollut. Bull. 46:748-755. [DOI] [PubMed] [Google Scholar]
- 7.Chouari, R., D. Le Paslier, P. Daegelen, J. Weissenbach, and A. Sghir. 2005. Novel predominant archaeal and bacterial groups revealed by molecular analysis of an anaerobic sludge digestor. Environ. Microbiol. 7:1104-1115. [DOI] [PubMed] [Google Scholar]
- 8.Cox, C. 1995. Stability of airborne microbes and allergens, p. 77-99. In C. Cox and C. Wathes (ed.), Bioaerosols handbook. Lewis Publishers, Washington, DC.
- 9.DeLong, E. F. 1992. Archaea in coastal marine environments. Proc. Natl. Acad. Sci. USA 89:5685-5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dennis, P. C., B. E. Sleep, R. R. Fulthorpe, and S. Liss. 2003. Phylogenetic analysis of bacterial populations in an anaerobic microbial consortium capable of degrading saturation concentrations of tetrachloroethylene. Can. J. Microbiol. 49:15-27. [DOI] [PubMed] [Google Scholar]
- 11.Dowd, S., K. Widmer, and S. Pillai. 1997. Thermotolerant Clostridia as an airborne pathogen indicator during land application of biosolids. J. Environ. Qual. 26:194-199. [Google Scholar]
- 12.Dowd, S. E., and S. D. Pillai. 1999. Identifying the sources of biosolid derived pathogen indicator organisms in aerosols by ribosomal DNA fingerprinting. J. Environ. Sci. Health Part A 34A:1061-1074. [Google Scholar]
- 13.Epstein, E. 2003. Land application of sewage sludge and biosolids. Lewis Publishers, Washington, DC.
- 14.Fannin, K. F., S. C. Vana, and W. Jakubowski. 1985. Effect of an activated sludge wastewater treatment plant on ambient air densities of aerosols containing bacteria and viruses. Appl. Environ. Microbiol. 49:1191-1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fécan, F., B. Marticorena, and G. Bergametti. 1999. Parameterization of the increase of the Aeolian erosion threshold wind friction velocity due to soil moisture for arid and semi-arid areas. Ann. Geophys. 17:149-157. [Google Scholar]
- 16.Fryrear, D. W. 1981. Southern Great Plains. Trans. ASAE 24:991-994. [Google Scholar]
- 17.Fryrear, D. W. 1990. Wind erosion: mechanics, prediction, and control. Adv. Soil Sci. 13:187-199. [Google Scholar]
- 18.Gillette, D. A. 1988. Threshold friction velocities for dust production for agricultural soils. J. Geophys. Res. 93:12645-12662. [Google Scholar]
- 19.Haznedaroglu, B. Z. 2005. Fatty acid methyl ester profiling of indicator organisms for microbial source tracking. Villanova University, Philadelphia, PA.
- 20.Jensen, P. A., W. F. Todd, G. N. Davis, and P. V. Scarpino. 1992. Evaluation of eight bioaerosol samplers challenged with aerosols of free bacteria. Am. Ind. Hyg. Assoc. J. 53:660-667. [DOI] [PubMed] [Google Scholar]
- 21.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 22.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 11-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. Wiley, Chichester, United Kingdom.
- 23.Leclerc, M., J. P. Delgenes, and J. J. Godon. 2004. Diversity of the archaeal community in 44 anaerobic digesters as determined by single strand conformation polymorphism analysis and 16S rDNA sequencing. Environ. Microbiol. 6:809-819. [DOI] [PubMed] [Google Scholar]
- 24.Lueders, T., and M. Friedrich. 2000. Archaeal population dynamics during sequential reduction processes in rice field soil. Appl. Environ. Microbiol. 66:2732-2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Melbostad, E., W. Eduard, A. Skogstad, P. Sandven, J. Lassen, P. Sostrand, and K. Heldal. 1994. Exposure to bacterial aerosols and work-related symptoms in sewage workers. Am. J. Ind. Med. 25:59-63. [DOI] [PubMed] [Google Scholar]
- 26.Mocé-Llivina, L., M. Muniesa, H. Pimenta-Vale, F. Lucena, and J. Jofre. 2003. Survival of bacterial indicator species and bacteriophages after thermal treatment of sludge and sewage. Appl. Environ. Microbiol. 69:1452-1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.NRC. 2002. Biosolids applied to land: advancing standards and practices. National Research Council of the National Academies, Washington, DC.
- 28.Paez-Rubio, T. 2006. Quantification of biological aerosols emitted during the land application of class B biosolids. Arizona State University, Tempe.
- 29.Paez-Rubio, T., A. Ramarui, J. Somer, H. Xin, J. Anderson, and J. Peccia. 2007. Aerosol emission rates of bacteria, endotoxins, metals, and PM10 produced during the land application of dewatered class B biosolids. Environ. Sci. Technol. 41:3537-3544. [DOI] [PubMed] [Google Scholar]
- 30.Paez-Rubio, T., E. Viau, S. Romero-Hernandez, and J. Peccia. 2005. Source bioaerosol concentration and rRNA gene-based identification of microorganisms aerosolized at a flood irrigation wastewater reuse site. Appl. Environ. Microbiol. 71:804-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paez-Rubio, T., H. Xin, J. Anderson, and J. Peccia. 2006. Particulate matter composition and emission rates from the disk incorporation of class B biosolids into soil. Atmos. Environ. 40:7034-7045. [Google Scholar]
- 32.Peccia, J., and M. Hernandez. 2006. Incorporating polymerase chain reaction-based identification, population characterization, and quantification of microorganisms into aerosol science: a review. Atmos. Environ. 40:3941-3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pillai, S., K. Widmer, S. E. Dowd, and S. C. Ricke. 1996. Occurrence of airborne bacteria and pathogen indicators during land application of sewage sludge. Appl. Environ. Microbiol. 62:296-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raskin, L., L. K. Poulsen, D. R. Noguera, B. E. Rittmann, and D. A. Stahl. 1994. Quantification of methanogenic groups in anaerobic biological reactors by oligonucleotide probe hybridization. Appl. Environ. Microbiol. 60:1241-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sartory, D. P., D. P. Prichard, and A. M. Holmes. 1993. Enumeration of sulfite-reducing Clostridia from potable water supplies. Water Sci. Technol. 27:279-282. [Google Scholar]
- 36.Setlow, P. 2006. Spores of Bacillus subtilis: their resistance to and killing by radiation, heat and chemicals. J. Appl. Microbiol. 101:514-525. [DOI] [PubMed] [Google Scholar]
- 37.Snaidr, J., R. Amann, I. Huber, W. Ludwig, and K.-H. Schleifer. 1997. Phylogenetic analysis and in situ identification of bacteria in activated sludge. Appl. Environ. Microbiol. 63:2884-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tanner, B. D., J. P. Brooks, C. N. Haas, C. P. Gerba, and I. L. Pepper. 2005. Bioaerosol emission rate and plume characteristics during land application of liquid class B biosolids. Environ. Sci. Technol. 39:1584-1590. [DOI] [PubMed] [Google Scholar]
- 39.Tchobanoglous, G., F. L. Burton, and H. D. Stensel (ed.). 2003. Wastewater engineering: treatment and reuse, 4th ed. Metcalf and Eddy, Inc.-McGraw-Hill, New York, NY.
- 40.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tokura, M., I. Chagan, K. Ushida, and Y. Kojima. 1999. Phylogenetic study of methanogens associated with rumen ciliates. Curr. Microbiol. 39:123-128. [DOI] [PubMed] [Google Scholar]
- 42.U.S. EPA. 1995. A guide to the biosolids risk assessment for the EPA. Part 503, rule EPA 832-B-93-005. Office of Wastewater Management, U.S. EPA, Washington, DC.
- 43.Willeke, K., X. J. Lin, and S. A. Grinshpun. 1998. Improved aerosol collection by combined impaction and centrifugal motion. Aerosol Sci. Technol. 28:439-456. [Google Scholar]