Abstract
While plasmids are very commonly associated with the majority of the lactic acid bacteria, they are only very rarely associated with Lactobacillus delbrueckii, with only four characterized to date. In this study, the complete sequence of a native plasmid, pDOJ1, from a strain of Lactobacillus delbrueckii subsp. bulgaricus was determined. It consisted of a circular DNA molecule of 6,220 bp with a G+C content of 44.6% and a characteristic ori and encoded six open reading frames (ORFs), of which functions could be predicted for three—a mobilization (Mob) protein, a transposase, and a fused primase-helicase replication protein. Comparative analysis of pDOJ1 and the other available L. delbrueckii plasmids (pLBB1, pJBL2, pN42, and pLL1212) revealed a very similar organization and amino acid identities between 85 and 98% for the putative proteins of all six predicted ORFs from pDOJ1, reflecting a common origin for L. delbrueckii plasmids. Analysis of the fused primase-helicase replication gene found a similar fused organization only in the theta replicating group B plasmids from Streptococcus thermophilus. This observation and the ability of the replicon to function in S. thermophilus support the idea that the origin of plasmids in L. delbrueckii was likely from S. thermophilus. This may reflect the close association of these two species in dairy fermentations, particularly yogurt production. As no vector based on plasmid replicons from L. delbrueckii has previously been constructed, an Escherichia coli-L. delbrueckii shuttle cloning vector, pDOJ4, was constructed from pDOJ1, the p15A ori, the chloramphenicol resistance gene of pCI372, and the lacZ polylinker from pUC18. This cloning vector was successfully introduced into E. coli, L. delbrueckii subsp. bulgaricus, S. thermophilus, and Lactococcus lactis. This shuttle cloning vector provides a new tool for molecular analysis of Lactobacillus delbrueckii and other lactic acid bacteria.
Bacteria of the genus Lactobacillus are gram-positive rods and constitute a major genus of the lactic acid bacteria, a group of economically important microorganisms that produce lactic acid as a major metabolite and are used in many fermentation processes as well as for probiotic purposes (20, 25, 32). Lactobacillus delbrueckii subsp. bulgaricus is used extensively in food fermentations, but especially in the production of yogurt. The organism was the original probiotic culture proposed by Metchnikoff (22), but its limited acid tolerance subsequently decreased its probiotic relevance. While the complete genome sequence of this bacterium has recently been published (18), molecular studies of this important organism have been few due to very limited tools for its genetic manipulation. Cloning vectors have been constructed for several different Lactobacillus species, such as L. acidophilus (17), L. curvatus (14), Lactobacillus delbrueckii subsp. lactis (42), L. plantarum (7), L. pentosus (29), and L. reuteri (16). Electroporation procedures have been developed for a number of Lactobacillus species, including L. acidophilus (41), L. casei (10, 29), L. delbrueckii subsp. lactis (42), and L. helveticus (5, 11).
Unlike the majority of Lactobacillus species, plasmids are extremely rare for L. delbrueckii (21). In a study of 48 strains of L. delbrueckii subsp. bulgaricus, a small cryptic plasmid was observed for only one strain, suggesting a frequency of <2% for these bacteria (3). To date, only one L. delbrueckii subsp. bulgaricus plasmid, pLBB1, has been isolated and sequenced (3). The plasmids pJBL2 and pN42 (6) and pLL1212 (GenBank accession number AF109691) have also been isolated and sequenced from the closely related L. delbrueckii subsp. lactis. While the origin of replication from pLBB1 has been cloned (3), cloning vectors based on replicons from L. delbrueckii subsp. bulgaricus or subsp. lactis plasmids have not been developed. The objective of this study was to conduct a comparative analysis of a native plasmid (pDOJ1) from L. delbrueckii subsp. bulgaricus B36 and the available four published plasmids from L. delbrueckii and to develop pDOJ1 into a functional shuttle cloning vector for this industrially relevant organism.
MATERIALS AND METHODS
Bacteria and growth conditions.
The bacterial strains used in this study are listed in Table 1. Lactobacillus delbrueckii subsp. bulgaricus strains and Propionibacterium freudenreichii subsp. shermanii were cultured in lactobacillus MRS medium (Difco, Detroit, MI) under anaerobic conditions at 37°C. Escherichia coli was grown in Luria-Bertani (LB) medium (30) at 37°C. Lactococcus lactis and Streptococcus thermophilus were cultured in M17 medium (Difco) supplemented with 0.5% d-glucose at 30°C and 42°C, respectively. Agar plates were made by adding 1.5% agar (Difco) to broth medium. The antibiotic chloramphenicol was added at a concentration of 20 μg/ml for E. coli; 3.0 μg/ml for L. delbrueckii subsp. bulgaricus, P. freudenreichii subsp. shermanii, and S. thermophilus; and 5.0 μg/ml for Lactococcus lactis. Isopropylthio-β-d-galactoside (IPTG; Sigma, St. Louis, MO) and 5-bromo-4-chloro-3-indolyl-β-d-galactoside (X-Gal; Sigma) were added to LB medium plates for blue/white color selection of colonies at final concentrations of 0.1 mM and 40 μg/ml, respectively.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s)a | Source or referenceb |
|---|---|---|
| Bacterial strains | ||
| Lactobacillus delbrueckii subsp. bulgaricus | ||
| B36 | Wild-type dairy isolate, host of pDOJ1 | Dairy culture collection, UMN |
| 449 | Electroporation host | Dairy culture collection, UMN |
| ATCC 11842 | Electroporation host | ATCC |
| E. coli HB101 | Cloning host | Promega |
| Lactococcus lactis LM0230 | Electroporation host | Dairy culture collection, UMN |
| Streptococcus thermophilus ST403 | Electroporation host | Dairy culture collection, UMN |
| Propionibacterium freudenreichii subsp. shermanii ATCC 9614 | Electroporation host | ATCC |
| Plasmids | ||
| pDOJ1 | 6.2-kb wild-type plasmid from L. delbrueckii B36 | This study |
| pDOJ2 | E. coli-L. delbrueckii shuttle cloning vector; Tcr | This study |
| pDOJ3 | E. coli-L. delbrueckii shuttle cloning vector; Cmr | This study |
| pDOJ4 | E. coli-L. delbrueckii shuttle cloning vector; Cmr; lacZ; MCS | This study |
| pACYC184 | E. coli cloning vector, source of p15A ori | 9 |
| pCI372 | E. coli-Lactococcus lactis shuttle cloning vector; Cmr | 12 |
| pUC18 | E. coli cloning vector, source of lacZ and MCS | 23 |
Tcr, tetracycline resistance; Cmr, chloramphenicol resistance; ori, origin of replication; MCS, multiple cloning site.
UMN, University of Minnesota; ATCC, American Type Culture Collection.
General DNA techniques.
Plasmid DNA was isolated from E. coli by alkaline lysis and purified using affinity columns from QIAGEN (Valencia, CA). Mini-prep isolation of Lactobacillus, Lactococcus, and S. thermophilus plasmids was performed using the method of O'Sullivan and Klaenhammer (27). Restriction endonuclease digestions were conducted according to the supplier's instructions (Promega, Madison, WI). Agarose gel electrophoresis was performed using standard methods (30). DNA ligation was performed using the Fast-Link DNA Ligation Kit (Epicenter, Madison, WI) according to the manufacturer's instructions.
Plasmid isolation from Lactobacillus delbrueckii subsp. bulgaricus.
A native L. delbrueckii subsp. bulgaricus plasmid designated pDOJ1 (6.2 kb) was isolated using the following procedure. One hundred milliliters of an 18-h culture was pelleted by centrifugation at 10,000 rpm in a Beckman J2-21 centrifuge using a JA-14 rotor (15,344 × g) for 15 min. The pellet was washed with TES buffer (50 mM Tris, pH 7.0, 20 mM EDTA, 0.2 mM sucrose) and resuspended in 2 ml of a solution containing 20% (wt/vol) sucrose and 30 mg/ml lysozyme. After 1 h incubation at 37°C with periodic mixing, it was quickly frozen in a −70°C freezer for 10 min and then thawed for 10 min at 37°C. A 4-ml aliquot of an alkaline sodium dodecyl sulfate solution (3% sodium dodecyl sulfate, 0.2 N NaOH) was added and quickly mixed, and the mixture was incubated at room temperature for 7 min. Three milliliters of a 3.0 M sodium acetate solution (pH 4.8) was added, mixed, and centrifuged at 10,000 rpm in a Beckman J2-21 centrifuge using a JA-14 rotor for 25 min. The supernatant was transferred to a new tube, and an equal volume of isopropanol (room temperature) was added, mixed, and centrifuged at 10,000 rpm for 25 min. The pellet was dried and resuspended in 9.4 ml of molecular water. Six milliliters of a 7.5 M ammonium acetate solution was added, prior to extraction with an equal volume of phenol-chloroform-isoamyl alcohol (25:24:1) solution. The mixture was centrifuged at 10,000 rpm for 20 min, and the top layer was transferred into a new tube with 2 volumes of ice-cold absolute ethanol and centrifuged at 10,000 rpm for 25 min. The pellet was rinsed with ice-cold 70% ethanol and centrifuged at 10,000 rpm for 5 min. The DNA pellet was dried and resuspended in 250 μl of TE buffer with 0.1 mg/ml RNase A and incubated at 37°C for 1 h.
DNA sequencing and bioinformatic analysis.
To conduct a random shotgun sequencing of pDOJ1, its plasmid DNA was randomly sheared using a HydroShear DNA shearing machine (Genomic Solutions, Ann Arbor, MI) and the sheared fragments were randomly cloned into pSTBlue-1 (Invitrogen, Carlsbad, CA) to construct a library of random pDOJ1 fragments. Sequencing of library clones was performed at the Biomedical Genomics Center (BMGC), University of Minnesota, using an ABI Prism 3100 automatic sequencer. To assemble contigs, DNASTAR (DNASTAR, Inc., Madison, WI) was utilized and primer walking was used to fill in gaps in the plasmid sequence. Primers were designed based on the ends of the shotgun sequences and used to sequence directly from the purified plasmid. The DNA and amino acid sequences were analyzed using both DNASTAR and OMIGA (Accelrys, San Diego, CA) software programs. Prediction of open reading frames (ORFs) was conducted with GeneMark (4) and the FgenesB (Softberry, Inc., Mount Kisco, NY) programs. Sequence annotation and database searches for similar sequences were performed using the BLAST suite of programs at the National Center for Biotechnology Information (1). The functional protein domain analysis of predicted ORFs was performed by the InterProScan program at the European Bioinformatics Institute (http://www.ebi.ac.uk/InterProScan/) and the NCBI Conserved Protein Domain Database program (19). Multiple sequence alignments were performed using the BLAST2SEQ (33) and CLUSTAL X (34) programs.
Plasmid transformation and electroporation.
Plasmids were introduced into E. coli HB101 using standard heat shock transformation (30), and electroporation was used for plasmid transfer into Lactococcus lactis (26), Streptococcus thermophilus (28), Propionibacterium freudenreichii subsp. shermanii (13), and Lactobacillus delbrueckii subsp. bulgaricus. Electrocompetent cells of L. delbrueckii subsp. bulgaricus were prepared based on modifications to a previous procedure (41) as follows. An overnight culture in MRS broth was used to inoculate 500 ml of MRS broth containing 3.0% glycine. The culture was grown to an optical density at 600 nm of ∼0.30. The cells were collected by centrifugation and washed four times with an equal volume of ice-cold electroporation buffer (0.1 mM HEPES, 0.5 M sucrose) and resuspended in the minimum amount of buffer possible, generally 1 ml or less. For electroporation, 50 μl of the competent cells and 0.5 μg of plasmid DNA were electroporated in a 2-mm ice-cold cuvette at 2.45 kV per cm using the default settings of an Eppendorf Electroporator 2510 (Eppendorf, Madison, WI). The recovery medium (MRS broth containing 0.5 M sucrose and 10 mM CaCl2) was added to a 1-ml volume and incubated at 37°C for 3 h under anaerobic conditions. Transformants were selected using MRS agar containing 10 mM CaCl2 and 3.0 μg/ml chloramphenicol for 2 days at 37°C.
Plasmid segregational stability.
The segregational stability of pDOJ4 in L. delbrueckii subsp. bulgaricus ATCC 11842 without antibiotic selection was monitored as described previously (15).
Nucleotide sequence accession numbers.
Sequence data from this article have been deposited with the GenBank Data Library under the accession numbers EF196093 (pDOJ1) and EF196094 (pDOJ4).
RESULTS
Sequence analysis of pDOJ1.
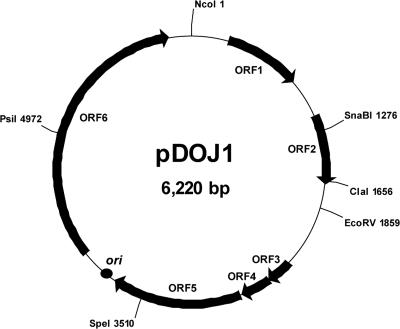
One plasmid was detected in Lactobacillus delbrueckii subsp. bulgaricus B36 using agarose gel electrophoresis. The identity of this isolate was confirmed by sequencing of the 16S gene, and its subspecies classification was confirmed by the size of its proline iminopeptidase gene as indicated by Torriani et al. (35). The linearization of the plasmid by NcoI digestion revealed a single DNA band of ∼6.2 kb, and the plasmid was designated pDOJ1 (data not shown). The complete DNA sequence of pDOJ1 was obtained using a shotgun sequencing strategy and completed using primer walking. This resulted in sequence coverage of ∼9.0-fold, and the sequence was checked manually to correct for any errors. The complete plasmid sequence of pDOJ1 consists of 6,220 bp with a G+C content of 44.6%, which is lower than that of Lactobacillus delbrueckii chromosomal DNA (49.7%) (39) (Fig. 1). Six putative ORFs containing one mobilization gene, one transposase gene, and one primase-helicase fusion gene, likely involved in its replication, were predicted by gene prediction programs (Table 2).
FIG. 1.
ORF and restriction enzyme map of pDOJ1 from Lactobacillus delbrueckii subsp. bulgaricus B36. ori, origin of replication. NcoI was chosen arbitrarily as the sequence start point.
TABLE 2.
ORF analysis of pDOJ1 from Lactobacillus delbrueckii subsp. bulgaricus
| ORF | Function | Position (sizea) | MWb | pIc | % Identityd | Best BLAST match | GenBank accession no. |
|---|---|---|---|---|---|---|---|
| ORF1 | Mobilization | 278-856 (192) | 21,833 | 9.25 | 97 over 192 aa | Putative mobilization protein (ORFB) of pLBB2, L. delbrueckii | NC_002191 |
| ORF2 | Transposase | 1,157-1,678 (173) | 19,475 | 6.62 | 98 over 171 aa | Hypothetical protein (ORF5) of pN42, L. delbrueckii | NC_004850 |
| ORF3 | Unknown | 2,333-2,527 (64) | 7,059 | 9.53 | 94 over 39 aa | Hypothetical protein (ORFA) of pLL1212, L. delbrueckii | NC_004937 |
| ORF4 | Unknown | 2,527-2,754 (75) | 8,708 | 5.00 | 94 over 75 aa | Hypothetical protein (ORFB) of pLL1212, L. delbrueckii | NC_004937 |
| ORF5 | Unknown | 2,769-3,740 (323) | 36,326 | 9.51 | 85 over 323 aa | Hypothetical protein (ORF3) of pN42, L. delbrueckii | NC_004850 |
| ORF6 | Primase-helicase | 4,035-6,101 (688) | 78,068 | 6.31 | 90 over 687 aa | Primase-helicase (ORFA) of pLBB2, L. delbrueckii | NC_002191 |
Number of amino acids.
MW, molecular weight.
pI, predicted isoelectric point.
Amino acid (aa) sequence identity.
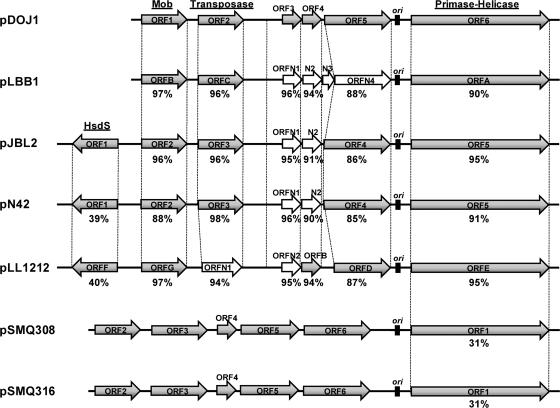
The predicted protein from ORF1 (putative mobilization protein) is very similar to ORFB of pLBB1 (3), ORF4 of pJBL2 (6), ORF4 of pN42 (6), and ORFG of pLL1212 (3) (Fig. 2). Interestingly, this putative mobilization protein has very low identity with other mobilization proteins predicted from the BLASTP database but is highly conserved in all the reported L. delbrueckii plasmids. Protein conserved domain analysis by InterProScan revealed that ORF1 has a conserved DNA binding domain which is consistent with a mobilization protein (Table 3). However, an oriT similar to those of other plasmids with mobilization genes was not detected.
FIG. 2.
Comparative sequence analysis of pDOJ1 and the other Lactobacillus delbrueckii plasmids and the group B plasmids from Streptococcus thermophilus. The amino acid identities with the predicted protein sequences from the six ORFs from pDOJ1 are indicated. The amino acid identities for the hsdS gene products are compared to pJBL2. The gene annotations are as originally published for each plasmid, except for the open arrows, which reflect ORFs that were annotated in this study. ori, origin of replication; HsdS, specificity subunit for a type I R-M system; Mob, mobilization protein.
TABLE 3.
Conserved domains of predicted proteins from pDOJ1
| ORF | Function | Position | E value | Conserved domain | Database accession no.c |
|---|---|---|---|---|---|
| ORF1 | Mobilization | 290-622 | 6.3e-09 | Putative DNA binding domain | SSF46955 |
| ORF2 | Transposase | 1160-1291 | 0.00017 | Homeodomain-like | SSF46689 |
| ORF3 | Unknown | 2336-2443 | 0.0089 | Lambda repressor-like DNA binding domain | SSF47413 |
| ORF6 | Primase-helicase | 4035-4382 | 8.4e-14 | Zinc-beta ribbon | SSF57783 |
| 4455-5042 | 1.2e-27 | DNA primase core | SSF56731 | ||
| 4500-4562 | 9.4e-05 | DNA primase catalytic core, N terminal | PF08275 | ||
| 4707-4940 | 2.4e-06 | TOPRIMa subdomain | SM00493 | ||
| 4707-4964 | 5.8e-07 | TOPRIM | PF01751 | ||
| 5046-5801 | 1.2e-08 | P loop containing NTPb hydrolase | SSF52540 | ||
| 5097-5681 | 3.6e-11 | DnaB-like helicase, C terminal | PS51199 |
TOPRIM, topoisomerase primase.
NTP, nucleoside triphosphate.
SSF, Superfamily database; PF, HMMPfam database; SM, HMMSmart database; PS, ProfileScan database.
The predicted protein from ORF2 (putative transposase) exhibits 94 to 98% identity with ORFC of pLBB1, ORF5 of pJBL2, and ORF5 of pN42. A protein domain analysis indicated that ORF2 has a homeodomain-like conserved DNA binding domain which is consistent with transposase functions (Table 3).
The predicted proteins of the short ORF3 and ORF4 have no significant database similarities other than those to the corresponding short ORFA and ORFB in pLL1212. Interestingly, reannotation of pLBB1, pJBL2, and pN42 revealed that they also have conserved homologs to both these ORFs at >90% identity at the amino acid level in all cases (Fig. 2). They are preceded by ribosome binding sites which substantiate their role as genes, and ORF3 has a lambda repressor-like DNA binding domain (Table 3).
The predicted protein of ORF5 shares similarity with ORF3 of pJBL2, ORF3 of pN42, and ORFD of pLL1212. Interestingly, there are no conserved domains or homolog sequences in the database, indicating that this is a unique, hypothetical protein. While the published annotation of pLBB1 does not include a homolog for ORF5, reannotation of this plasmid sequence shows that one is present with 88% amino acid sequence identity (Fig. 2).
The predicted protein from ORF6 is quite unique, as it is predicted to be a fusion of both a primase and a helicase protein. Its location, directly following the ori region, is consistent with a role in plasmid replication. This ORF is very highly conserved in all the reported Lactobacillus delbrueckii plasmids (Fig. 2) with E values of 0.0 using BLASTP. A protein conserved domain analysis by InterProScan showed that this gene has three different protein conserved domains: a zinc-beta ribbon domain believed to be involved in binding Zn2+, a DNA primase core, and the C-terminal end of a DnaB-like helicase. This latter helicase motif at the carboxy-terminal end has a P loop containing a purine nucleoside triphosphate DNA binding motif, which is frequently found in DNA and RNA helicases and is a common DNA binding motif (40). As the N-terminal part of the DnaB helicase is missing in ORF6, it suggests that other replication initiator proteins (possibly ORF3 to ORF5) may be involved, suggesting a novel replication mechanism of Lactobacillus delbrueckii plasmids compared to most other plasmids from gram-positive bacteria. This unusual fusion organization of the ORF6 predicted protein of pDOJ1 and its corresponding homologs in pLBB1, pJBL2, pN42, and pLL1212 has been observed elsewhere only in the group B plasmids from Streptococcus thermophilus (37).
Predicted origin of replication.
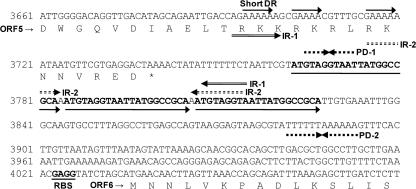
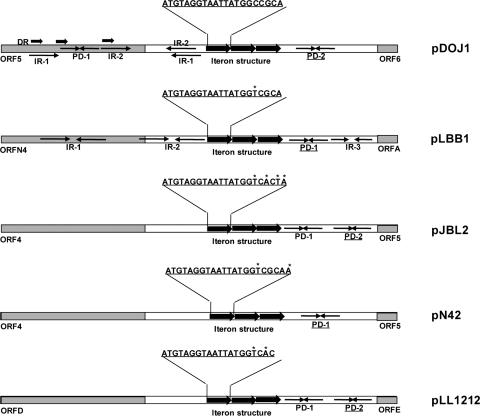
The predicted origin of replication (ori) sequence upstream from ORF6 consists of three contiguous 21-bp direct repeats, consistent with an iteron structure, and is preceded by three short direct repeats, two inverted repeats, and one palindrome structure and directly followed by a single palindrome structure (Fig. 3). A comparative analysis of other putative ori regions in L. delbrueckii plasmids as well as of the confirmed ori region from pLBB1 revealed highly conserved iteron structures and a palindrome structure directly following the iteron structure (Fig. 4). The first 16 bp of the iteron direct repeat is 100% conserved between the five plasmids, but the remaining sequence is less conserved, both in nucleotide identity and in length. This organization of the direct repeats in iteron structures is consistent with other characterized iteron regions and is believed to be involved in recognition and binding to replication proteins (15). Unlike ori regions of other plasmids that contain iteron structures, all the ori regions of L. delbrueckii plasmids lack AT-rich regions and DnaA box motifs preceding the iteron structure, further substantiating a likely novel mechanism of replication.
FIG. 3.
Structure of the predicted origin of replication in pDOJ1. It consists of contiguous conserved 21-bp direct repeats (iteron structure) surrounded by three short direct repeats (DR), two inverted repeats (IR), and two palindrome structures (PD). RBS, ribosome binding site.
FIG. 4.
Comparison of the predicted origins of replication in Lactobacillus delbrueckii plasmids. An iteron structure and a palindrome sequence (5′-TTTTTAAAAA-3′) are highly conserved in all L. delbrueckii plasmids. DR, direct repeat; IR, inverted repeat; PD, palindrome. A conserved palindrome sequence between all the plasmids is underlined. Stars indicate nonconserved nucleotides in the iteron repeat.
Construction of a shuttle cloning vector.
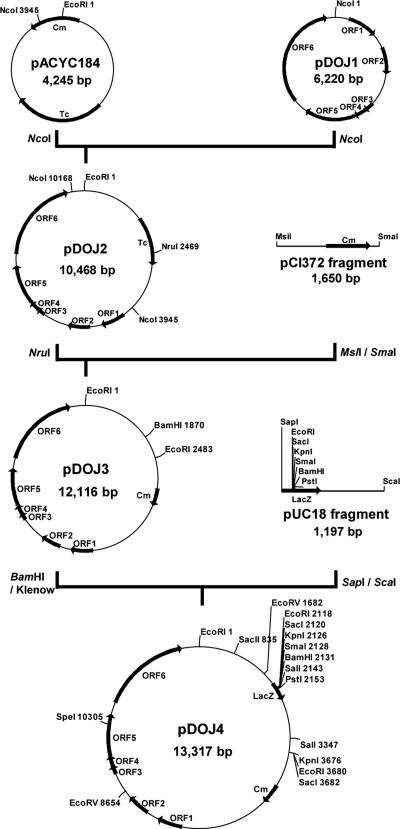
Plasmid pDOJ1 was linearized by NcoI digestion and cloned into the E. coli vector pACYC184, forming the recombinant plasmid pDOJ2 (Fig. 5). A chloramphenicol resistance gene from pCI372 was chosen as the selection marker as it has been previously used in the construction of various Lactobacillus, Lactococcus, and Bifidobacterium vectors (8, 12, 14, 15, 42). As this chloramphenicol-resistant vector, pDOJ3, contained only three unique restriction enzyme sites, a lacZ polylinker was cloned into a BamHI site to provide multiple cloning sites and blue/white colony screening capability in E. coli (Fig. 5).
FIG. 5.
Construction of the Lactobacillus delbrueckii subsp. bulgaricus shuttle cloning vector, pDOJ4.
Electroporation of pDOJ4 into Lactobacillus delbrueckii subsp. bulgaricus and other hosts.
An electroporation protocol was developed for L. delbrueckii subsp. bulgaricus, which was primarily based on a procedure developed for L. acidophilus (41), and was optimized to increase efficiency. The optimizations included a reduced optical density at 600 nm, an increase in glycine concentration, and different electroporator parameters. The pDOJ4 shuttle vector was introduced into L. delbrueckii subsp. bulgaricus 449 at a frequency of 1.4 × 103 CFU/μg. Plasmids pDOJ3 and pDOJ4 were successfully introduced into another strain of L. delbrueckii subsp. bulgaricus (ATCC 11842) at frequencies of 2.4 × 103 and 1.2 × 103 CFU/μg of DNA, respectively. Both plasmids were also transformed into Lactococcus lactis LM0230 at frequencies of 5.0 × 102 and 1.9 × 102 CFU/μg, respectively, demonstrating that the Lactobacillus delbrueckii subsp. bulgaricus native plasmid pDOJ1 replicon also functions in Lactococcus lactis and may also be used as a cloning vector for this host. As restriction and modification (R-M) systems can reduce transformation frequencies (2, 38), pDOJ3 was reisolated from an L. lactis LM0230 transformant and electroporated again into L. lactis LM0230. This resulted in a similar frequency, 4.6 × 102 CFU/μg, showing that the lower transformation frequency in L. lactis was not due to R-M systems. The pDOJ4 vector was also introduced into Streptococcus thermophilus at a frequency similar to that of L. lactis (6.6 × 102 CFU/μg) but could not be introduced into Propionibacterium freudenreichii subsp. shermanii.
Segregational stability of pDOJ4 in Lactobacillus delbrueckii subsp. bulgaricus.
The stability of pDOJ4 in L. delbrueckii subsp. bulgaricus without antibiotic selection was monitored over 93 generations of growth in MRS broth medium. No loss of the plasmid was observed over this time, indicating a very high stability, consistent with other theta replicating plasmids.
DISCUSSION
Unlike most Lactobacillus species, and indeed the majority of lactic acid bacteria, plasmids are extremely rare for Lactobacillus delbrueckii subsp. bulgaricus (3). This is reflected in the low number of sequenced plasmids from L. delbrueckii that are deposited in GenBank, which is now five, including pDOJ1 from this study. A comparative analysis of the sequence of pDOJ1 with the four other sequenced plasmids from L. delbrueckii (pLBB1, pJBL2, pN42, and pLL1212) revealed a highly conserved organization. All the predicted proteins from the six ORFs of pDOJ1 had homologs in each of the other four plasmids with amino acid identities ranging from 85 to 98% (Fig. 2). This is strong evidence of a common ancestor for all five plasmids. It is noteworthy that the two plasmids isolated from L. delbrueckii subsp. bulgaricus (pDOJ1 and pLBB1) are cryptic plasmids (based on the available annotation) while the three plasmids isolated from L. delbrueckii subsp. lactis all carry genes for an HsdS protein, which is the specificity protein for type I R-M systems. The inclusion of hsdS genes on plasmids has previously been described for Lactococcus lactis (24) and Streptococcus thermophilus (31) and was proposed to be the result of selective pressure for greater R-M diversity due to the prevalence of bacteriophage in the cheese production environment. As Lactobacillus delbrueckii subsp. lactis is also predominantly from a cheese environment, this supports this hypothesis.
It is interesting that all five plasmids from L. delbrueckii carry a conserved mobilization gene, but no oriT is evident on either of the plasmids. As an oriT is needed for plasmid mobilization, it may suggest that this has been lost from the plasmid or alternatively that a different class of oriT may be present. Given the lower G+C content (<45%) of all five L. delbrueckii plasmids compared to the chromosomal G+C content for this species, 49.7%, it suggests that these plasmids were acquired via horizontal transfer in a recent evolutionary event. It is plausible that the mobilization gene had a direct role in this event. Further evidence for this hypothesis comes from the analysis of the fusion primase-helicase protein in all five plasmids. As this has previously been seen only in two plasmids from Streptococcus thermophilus, pSMQ308 and pSMQ316 (37), it may suggest a common ancestor. This is further substantiated by the ability of this L. delbrueckii subsp. bulgaricus replicon to function in S. thermophilus and by the finding that the amino acid identity between both of these fusion primase-helicase proteins encoded on S. thermophilus plasmids and the five L. delbrueckii plasmids is >30% in all cases. As S. thermophilus and L. delbrueckii subsp. bulgaricus are intimately connected by their role in yogurt manufacture, which likely dates to prehistoric times, it is intriguing to speculate that the emergence of plasmids in L. delbrueckii resulted from this association.
Recently, two complete genome sequences of two strains of L. delbrueckii subsp. bulgaricus were reported, and they are now available in GenBank (18). To functionally characterize the genomes of these commercially important bacteria, there is a need for different molecular tools that can be used in these bacteria. While tools developed for other lactic acid bacteria will likely be useful, vectors based on L. delbrueckii plasmid replicons will enhance this endeavor. As there are currently none developed, the cloning vector pDOJ4 was constructed based on the L. delbrueckii subsp. bulgaricus plasmid characterized in this study. The construction of pDOJ4 from pDOJ1, pACYC184, pCI372, and pUC18 represents the first E. coli-L. delbrueckii shuttle cloning vector. While the mode of replication of pDOJ4 has not been established, previous studies with the L. delbrueckii plasmids pLBB1, pJBL2, and pN42 and the S. thermophilus group B plasmids suggest that the most likely mode of replication is through theta replication (3, 6, 36). This would suggest that the vector pDOJ4 may be a useful and stable plasmid vector for L. delbrueckii and other lactic acid bacteria as it was successfully introduced into L. delbrueckii subsp. bulgaricus, E. coli, Lactococcus lactis, and Streptococcus thermophilus.
In summary, the comparative sequence analysis of pDOJ1 and the other four identified L. delbrueckii plasmids points to a common origin for plasmids from this genus, and the homology with the fusion primase-helicase replication protein from the theta replicating group B plasmids from S. thermophilus suggests that they may have been acquired via horizontal gene transfer. This hypothesis is substantiated by the close association of these two species during yogurt manufacture and the ability of the plasmid to replicate in S. thermophilus. The development of the shuttle cloning vector pDOJ4 will facilitate the functional analysis of the genome of this important bacterium for dairy food fermentations.
Acknowledgments
We acknowledge the financial support of the Minnesota Agricultural Experimental Station, the Midwest Dairy Association, and the USDA National Needs Fellowship Program.
Footnotes
Published ahead of print on 25 May 2007.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Auad, L., M. A. A. Peril, A. A. P. R. Holgado, and R. R. Raya. 1998. Evidence of a restriction/modification system in Lactobacillus delbrueckii subsp. lactis CNRZ 326. Curr. Microbiol. 36:271-273. [DOI] [PubMed] [Google Scholar]
- 3.Azcarate-Peril, M. A., and R. R. Raya. 2002. Sequence analysis of pLBB1, a cryptic plasmid from Lactobacillus delbrueckii subsp. bulgaricus. Can. J. Microbiol. 48:105-112. [DOI] [PubMed] [Google Scholar]
- 4.Besemer, J., and M. Borodovsky. 1999. Heuristic approach to deriving models for gene finding. Nucleic Acids Res. 27:3911-3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhowmik, T., and J. L. Steele. 1993. Development of an electroporation procedure for gene disruption in Lactobacillus helveticus CNRZ32. J. Gen. Microbiol. 139:1433-1439. [Google Scholar]
- 6.Bourniquel, A. A., M. G. Casey, B. Mollet, and R. D. Pridmore. 2002. DNA sequence and functional analysis of Lactobacillus delbrueckii subsp. lactis plasmids pN42 and pJBL2. Plasmid 47:153-157. [DOI] [PubMed] [Google Scholar]
- 7.Bringel, F., L. Frey, and J. C. Hubert. 1989. Characterization, cloning, curing, and distribution in lactic acid bacteria of pLP1, a plasmid from Lactobacillus plantarum CCM 1904 and its use in shuttle vector construction. Plasmid 22:193-202. [DOI] [PubMed] [Google Scholar]
- 8.Chagnaud, P., C. K. Chan Kwo Chion, R. Duran, P. Naouri, A. Arnaud, and P. Galzy. 1992. Construction of a new shuttle vector for Lactobacillus. Can. J. Microbiol. 38:69-74. [DOI] [PubMed] [Google Scholar]
- 9.Chang, A. C., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chassy, B. M., and J. L. Flickinger. 1987. Transformation of Lactobacillus casei by electroporation. FEMS Microbiol. Lett. 44:173-177. [Google Scholar]
- 11.Hashiba, H., R. Takiguchi, S. Ishii, and K. Aoyama. 1990. Transformation of Lactobacillus helveticus subsp. jugurti with plasmid pLHR by electroporation. Agric. Biol. Chem. 54:1537-1541. [PubMed] [Google Scholar]
- 12.Hayes, F., C. Daly, and G. F. Fitzgerald. 1990. Identification of the minimal replicon of Lactococcus lactis subsp. lactis UC317 plasmid pCI305. Appl. Environ. Microbiol. 56:202-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jore, J. P., N. van Luijk, R. G. Luiten, M. J. van der Werf, and P. H. Pouwels. 2001. Efficient transformation system for Propionibacterium freudenreichii based on a novel vector. Appl. Environ. Microbiol. 67:499-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klein, J. R., C. Ulrich, and R. Plapp. 1993. Characterization and sequence analysis of a small cryptic plasmid from Lactobacillus curvatus LTH683 and its use for construction of new Lactobacillus cloning vectors. Plasmid 30:14-29. [DOI] [PubMed] [Google Scholar]
- 15.Lee, J. H., and D. J. O'Sullivan. 2006. Sequence analysis of two cryptic plasmids from Bifidobacterium longum DJO10A and construction of a shuttle cloning vector. Appl. Environ. Microbiol. 72:527-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin, C. F., and T. C. Chung. 1999. Cloning of erythromycin-resistance determinants and replication origins from indigenous plasmids of Lactobacillus reuteri for potential use in construction of cloning vectors. Plasmid 42:31-41. [DOI] [PubMed] [Google Scholar]
- 17.Lin, M. Y., S. Harlander, and D. Savaiano. 1996. Construction of an integrative food-grade cloning vector for Lactobacillus acidophilus. Appl. Microbiol. Biotechnol. 45:484-489. [DOI] [PubMed] [Google Scholar]
- 18.Makarova, K., A. Slesarev, Y. Wolf, A. Sorokin, B. Mirkin, E. Koonin, A. Pavlov, N. Pavlova, V. Karamychev, N. Polouchine, V. Shakhova, I. Grigoriev, Y. Lou, D. Rohksar, S. Lucas, K. Huang, D. M. Goodstein, T. Hawkins, V. Plengvidhya, D. Welker, J. Hughes, Y. Goh, A. Benson, K. Baldwin, J. H. Lee, I. Diaz-Muniz, B. Dosti, V. Smeianov, W. Wechter, R. Barabote, G. Lorca, E. Altermann, R. Barrangou, B. Ganesan, Y. Xie, H. Rawsthorne, D. Tamir, C. Parker, F. Breidt, J. Broadbent, R. Hutkins, D. O'Sullivan, J. Steele, G. Unlu, M. Saier, T. Klaenhammer, P. Richardson, S. Kozyavkin, B. Weimer, and D. Mills. 2006. Comparative genomics of the lactic acid bacteria. Proc. Natl. Acad. Sci. USA 103:15611-15616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marchler-Bauer, A., J. B. Anderson, P. F. Cherukuri, C. DeWeese-Scott, L. Y. Geer, M. Gwadz, S. He, D. I. Hurwitz, J. D. Jackson, Z. Ke, C. J. Lanczycki, C. A. Liebert, C. Liu, F. Lu, G. H. Marchler, M. Mullokandov, B. A. Shoemaker, V. Simonyan, J. S. Song, P. A. Thiessen, R. A. Yamashita, J. J. Yin, D. Zhang, and S. H. Bryant. 2005. CDD: a conserved domain database for protein classification. Nucleic Acids Res. 33:192-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McKay, L. L., and K. A. Baldwin. 1990. Applications for biotechnology: present and future improvements in lactic acid bacteria. FEMS Microbiol. Rev. 7:3-14. [DOI] [PubMed] [Google Scholar]
- 21.Mercenier, A., P. H. Pouwels, and B. M. Chassy. 1994. Genetic engineering of Lactobacilli, Leuconostocs and Streptococcus thermophilus, p. 252-293. In M. J. Gasson and W. M. de Vos (ed.), Genetics and biotechnology of lactic acid bacteria. Blackie Academic & Professional, Glasgow, United Kingdom.
- 22.Metchnikoff, E. 1906. The prolongation of life: optimistic studies. G. P. Putnam's Sons, New York, NY.
- 23.Norrander, J., T. Kempe, and J. Messing. 1983. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene 26:101-106. [DOI] [PubMed] [Google Scholar]
- 24.O'Sullivan, D., D. P. Twomey, A. Coffey, C. Hill, G. F. Fitzgerald, and R. P. Ross. 2000. Novel type I restriction specificities through domain shuffling of HsdS subunits in Lactococcus lactis. Mol. Microbiol. 36:866-875. [DOI] [PubMed] [Google Scholar]
- 25.O'Sullivan, D. J. 2006. Primary sources of probiotic cultures, p. 91-107. In I. Goktepe, V. K. Juneja, and M. Ahmedna (ed.), Probiotics in food safety and human health. Taylor & Francis/CRC Press, Boca Raton, FL.
- 26.O'Sullivan, D. J., C. Hill, and T. R. Klaenhammer. 1993. Effect of increasing the copy number of bacteriophage origins of replication, in trans, on incoming-phage proliferation. Appl. Environ. Microbiol. 59:2449-2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O'Sullivan, D. J., and T. R. Klaenhammer. 1993. Rapid mini-prep isolation of high-quality plasmid DNA from Lactococcus and Lactobacillus spp. Appl. Environ. Microbiol. 59:2730-2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O'Sullivan, T. F., and G. F. Fitzgerald. 1999. Electrotransformation of industrial strains of Streptococcus thermophilus. J. Appl. Microbiol. 86:275-283. [DOI] [PubMed] [Google Scholar]
- 29.Posno, M., R. J. Leer, N. van Luijk, M. J. van Giezen, P. T. Heuvelmans, B. C. Lokman, and P. H. Pouwels. 1991. Incompatibility of Lactobacillus vectors with replicons derived from small cryptic Lactobacillus plasmids and segregational instability of the introduced vectors. Appl. Environ. Microbiol. 57:1822-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 31.Seegers, J. F., D. van Sinderen, and G. F. Fitzgerald. 2000. Molecular characterization of the lactococcal plasmid pCIS3: natural stacking of specificity subunits of a type I restriction/modification system in a single lactococcal strain. Microbiology 146:435-443. [DOI] [PubMed] [Google Scholar]
- 32.Sneath, P. H. A., N. S. Mair, M. E. Sharpe, and J. G. Holt. 1986. Bergey's manual of systematic bacteriology, vol. 2. Williams and Wilkins, Baltimore, MD.
- 33.Tatusova, T. A., and T. L. Madden. 1999. BLAST 2 Sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol. Lett. 174:247-250. [DOI] [PubMed] [Google Scholar]
- 34.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Torriani, S., G. Zapparoli, and F. Dellaglio. 1999. Use of PCR-based methods for rapid differentiation of Lactobacillus delbrueckii subsp. bulgaricus and L. delbrueckii subsp. lactis. Appl. Environ. Microbiol. 65:4351-4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Turgeon, N., M. Frenette, and S. Moineau. 2004. Characterization of a theta-replicating plasmid from Streptococcus thermophilus. Plasmid 51:24-36. [DOI] [PubMed] [Google Scholar]
- 37.Turgeon, N., and S. Moineau. 2001. Isolation and characterization of a Streptococcus thermophilus plasmid closely related to the pMV158 family. Plasmid 45:171-183. [DOI] [PubMed] [Google Scholar]
- 38.Twomey, D. P., L. L. McKay, and D. J. O'Sullivan. 1998. Molecular characterization of the Lactococcus lactis LlaKR2I restriction-modification system and effect of an IS982 element positioned between the restriction and modification genes. J. Bacteriol. 180:5844-5854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van de Guchte, M., S. Penaud, C. Grimaldi, V. Barbe, K. Bryson, P. Nicolas, C. Robert, S. Oztas, S. Mangenot, A. Couloux, V. Loux, R. Dervyn, R. Bossy, A. Bolotin, J. M. Batto, T. Walunas, J. F. Gibrat, P. Bessieres, J. Weissenbach, S. D. Ehrlich, and E. Maguin. 2006. The complete genome sequence of Lactobacillus bulgaricus reveals extensive and ongoing reductive evolution. Proc. Natl. Acad. Sci. USA 103:9274-9279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vetter, I. R., and A. Wittinghofer. 1999. Nucleoside triphosphate-binding proteins: different scaffolds to achieve phosphoryl transfer. Q. Rev. Biophys. 32:1-56. [DOI] [PubMed] [Google Scholar]
- 41.Walker, D. C., K. Aoyama, and T. R. Klaenhammer. 1996. Electrotransformation of Lactobacillus acidophilus group A1. FEMS Microbiol. Lett. 138:233-237. [DOI] [PubMed] [Google Scholar]
- 42.Zink, A., J. R. Klein, and R. Plapp. 1991. Transformation of Lactobacillus delbrueckii ssp. lactis by electroporation and cloning of origins of replication by use of a positive selection vector. FEMS Microbiol. Lett. 78:207-212. [Google Scholar]