Abstract
We found that species combinations such as Lactobacillus casei subsp. rhamnosus IFO3831 and Saccharomyces cerevisiae Kyokai-10 can form a mixed-species biofilm in coculture. Moreover, the Kyokai-10 yeast strain can form a biofilm in monoculture in the presence of conditioned medium (CM) from L. casei IFO3831. The active substance(s) in bacterial CM is heat sensitive and has a molecular mass of between 3 and 5 kDa. In biofilms from cocultures or CM monocultures, yeast cells had a distinct morphology, with many hill-like protrusions on the cell surface.
Biofilms are superficial microbial colonies found on solid surfaces at the solid-liquid interface. Studies of biofilm formation have primarily focused on biofilms formed by a single species of microorganism (5, 6, 8, 12). In natural environments, however, biofilms are thought to be composed of more than one species. Indeed, several investigators have done pioneering work on mixed-species biofilm formation (1, 3, 4, 7, 11, 16).
In the early period of traditional Japanese fermentation mashes to make rice wine and vinegar, experts recognized that lactic acid bacteria and yeasts coexisted in these mixtures, which contained steamed rice and rice koji (steamed rice grown with Aspergillus oryzae) (2, 10). Moreover, the steamed rice and rice koji in the mashes provide many solid surfaces. These observations prompted us to investigate the possibility that lactic acid bacteria and yeast can form a mixed-species biofilm.
Initially, we tested 10 strains of lactic acid bacteria and 13 strains of yeasts for their ability to form biofilms. Lactic acid bacterial strains B-1, B-56, L-14, L-54, and H-61 in Table 1 were isolated in our laboratory from yoghurt samples. Saccharomyces cerevisiae X2180-1A is a widely used laboratory yeast strain. Yeast strains designated Kyokai-[number] were kindly supplied by the Brewing Society of Japan. Other bacterial and yeast strains were obtained from stock culture centers.
TABLE 1.
Amount of biofilm formation in coculture between lactic acid bacteria and yeasts
| S. cerevisiae strain | Absorbance of crystal violet ina:
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Monoculture | L. mesenteroides IAM 1064 | P. acidilactici IAM 1233 | L. paracasei subsp. paracasei IFO3533 | L. sakei IFO3541 | L. casei subsp. rhamnosus IFO3831 | L. helveticus B-1 | L. bulgaricus B-56 | L. casei L-14 | L. acidophilus L-54 | S. cremoris H-61 | |
| None | 0.036 | 0.048 | 0.022 | 0.020 | 0.049 | 0.040 | 0.035 | 0.024 | 0.022 | 0.029 | |
| X2180-1A | 0.044 | 0.059 | 0.071 | 0.080 | 0.070 | 0.108 | 0.079 | 0.094 | 0.070 | 0.078 | 0.064 |
| Kyokai-6 | 0.028 | 0.071 | 0.077 | 0.065 | 0.053 | 0.071 | 0.051 | 0.092 | 0.066 | 0.086 | 0.068 |
| Kyokai-7 | 0.055 | 0.069 | 0.074 | 0.049 | 0.048 | 0.074 | 0.072 | 0.086 | 0.086 | 0.089 | 0.083 |
| Kyokai-9 | 0.039 | 0.044 | 0.047 | 0.039 | 0.063 | 0.048 | 0.072 | 0.070 | 0.051 | 0.069 | 0.077 |
| Kyokai-10 | 0.077 | 0.097 | 0.100 | 0.092 | 0.084 | 0.208 | 0.096 | 0.082 | 0.089 | 0.170 | 0.097 |
| Kyokai-11 | 0.033 | 0.107 | 0.149 | 0.091 | 0.113 | 0.134 | 0.085 | 0.062 | 0.090 | 0.127 | 0.092 |
| Kyokai-12 | 0.037 | 0.129 | 0.100 | 0.086 | 0.080 | 0.198 | 0.095 | 0.070 | 0.067 | 0.157 | 0.094 |
| Kyokai-13 | 0.048 | 0.124 | 0.151 | 0.074 | 0.092 | 0.210 | 0.095 | 0.115 | 0.093 | 0.092 | 0.100 |
| IAM 4125 | 0.040 | 0.035 | 0.051 | 0.057 | 0.047 | 0.038 | 0.040 | 0.043 | 0.044 | 0.042 | 0.035 |
| IAM 4140 | 0.035 | 0.024 | 0.031 | 0.043 | 0.044 | 0.033 | 0.038 | 0.042 | 0.048 | 0.048 | 0.050 |
| IAM 4178 | 0.030 | 0.025 | 0.031 | 0.039 | 0.038 | 0.031 | 0.036 | 0.026 | 0.065 | 0.046 | 0.041 |
| IAM 4274 | 0.049 | 0.063 | 0.052 | 0.066 | 0.060 | 0.069 | 0.056 | 0.038 | 0.053 | 0.074 | 0.038 |
| IAM 4954 | 0.028 | 0.022 | 0.039 | 0.046 | 0.043 | 0.028 | 0.038 | 0.053 | 0.040 | 0.035 | 0.053 |
Numerals given in the table represent absorbance of crystal violet extracted from crystal violet-stained biofilms. Values in boldface represent combinations with distinct biofilm formation. Results are shown for the species Leuconostoc mesenteroides, Pediococcus acidilactici, Lactobacillus paracasei, Lactobacillus sakei, Lactobacillus casei, Lactobacillus helveticus, Lactobacillus bulgaricus, Lactobacillus acidophilus, and Streptococcus cremoris.
To prepare seed cultures, lactic acid bacteria were grown in DeMan, Rogosa, and Sharpe broth (MRS; Becton, Dickinson and Co., Sparks, MD) at 30°C for 24 h. Yeasts were grown in YPD broth (Becton Dickinson and Co.) at 30°C for 24 h. Cells were cultured to stationary phase.
Quantification of biofilm formation was done using the conventional titer plate method with minor modifications (14, 15). To assay biofilm formation in monoculture, stationary-phase lactic acid bacteria or yeast cells were diluted 1:100 in fresh YPD (total, 200 μl) and grown in microtiter plate wells. To assay biofilm formation in coculture, stationary-phase cultures of lactic acid bacteria and yeasts were diluted by 1:200 into fresh YPD broth (total, 200 μl), mixed, and cocultured in microtiter plate wells. After inoculation, both mono- and coculture samples were incubated at 30°C for 24 h.
Alternatively, biofilms were formed on glass slides. To do this, either a 500-μl monoculture of one cell type or 250-μl monoculture of each cell type was added to 50 ml YPD in a beaker containing a glass slide and the chamber was incubated at 30°C for 24 h. After cultivation, the slide was washed three times with distilled water and dried at room temperature for 30 min. Dried biofilms on slides were Gram stained using the FAVOR-G SET-F kit (Nissui Pharmaceutical Co., Ltd., Tokyo, Japan) and examined under a microscope.
To prepare conditioned medium (CM), a 15-ml culture of Lactobacillus casei subsp. rhamnosus was transferred into 1,500 ml of YPD and incubated at 30°C for 24 h. Cells were removed by centrifugation (5,600 × g for 20 min), and the supernatant was sterilized via passage through a 0.2-μm-pore-size sterile filter (Toyo Roshi Co. Ltd., Tokyo, Japan). The sterilized flowthrough was considered CM. Further fractionation of CM was performed by ultrafiltration with centrifugal filter devices with a 10-kDa normal molecular mass limit, 5-kDa normal molecular mass limit, and 3-kDa molecular mass cutoffs (all from Millipore Co., Bedford, MA).
Light microscopic observation was performed using an Olympus BX60 equipped with a UPlanFl oil immersion lens (Olympus Co., Tokyo, Japan) and a DP90 digital camera (Olympus Co.). Scanning electron microscopy (SEM) was used to visualize the cell surface of the biofilm-forming yeast strain Kyokai-10. SEM was performed as described by Guzel-Seydim et al. (9). Biofilm samples were prepared on a glass slide as described, shaved off with a rubber spatula, suspended in 0.1 M sodium phosphate buffer, and fixed with 2% glutaraldehyde. The samples were observed using a Keyence VE-8800 scanning electron microscope (Keyence Co., Osaka, Japan).
We set out to test for the ability to form a mixed-species biofilm for every combination of each of 10 species of lactic acid bacteria and 13 strains of the yeast Saccharomyces cerevisiae using the conventional titer plate method (14, 15). We observed distinct biofilm formation in seven combinations (Table 1). None of the lactic acid bacteria or yeasts formed biofilm in monoculture. However, some Kyokai yeast strains used in rice wine brewing did form biofilms when cocultured with suitable lactic acid bacterium partners. Culturing the cells in mixed populations had no significant effect on yeast or lactic acid bacterial cell numbers (data not shown).
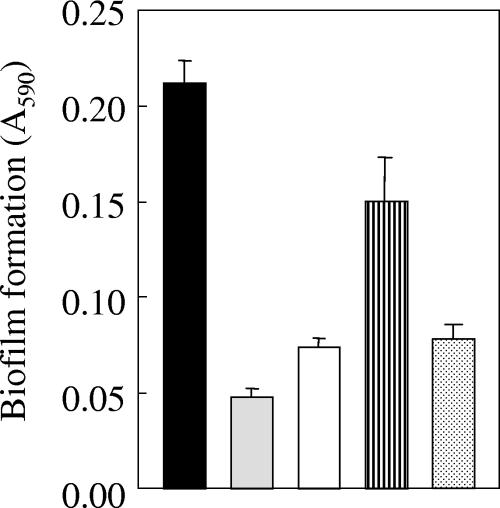
To investigate possible interactions between lactic acid bacteria and yeasts, we cultured the yeast strains that had proved capable of forming biofilms (henceforth, “respondent yeasts”) in monoculture in CM of partner lactic acid bacteria that had been grown in YPD medium. To our surprise, the respondent yeasts could form monospecies biofilms that reached biofilm mass levels similar to what was found for cocultures. The results obtained for the representative combination of Saccharomyces cerevisiae Kyokai-10 cells with CM from Lactobacillus casei subsp. rhamnosus IFO3831 (hereafter, L. casei IFO3831) are shown in Fig. 1.
FIG. 1.
Biofilm formation between Lactobacillus casei subsp. rhamnosus IFO3831and Saccharomyces cerevisiae Kyokai-10. The ordinate shows the absorption of crystal violet (CV) extracted from CV-stained biofilms, an indicator of biomass. Black bar, coculture; gray bar, lactic acid bacterium in monoculture; white bar, yeast in monoculture; striped bar, yeast cultured in the CM from the bacterium; dotted bar, yeast cultured in heat-treated CM (100°C, 10 min). Error bars indicate standard deviations.
We next determined the effects of the heat treatment at 100°C for 10 min on the ability of the CM to induce biofilm formation in respondent yeasts. We found that for the representative combination of Kyokai-10 yeast with CM from L. casei IFO3831, a biofilm did not form in the presence of heat-treated CM, suggesting that the putative biofilm-forming factor(s) in CM is heat sensitive (Fig. 1). Similar results were obtained for all seven biofilm-competent combinations (data not shown).
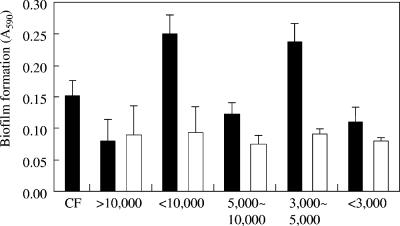
In attempt to isolate the factor(s) that induces biofilm formation, untreated CM was fractionated using filter-type molecular sieves. The fraction with a molecular mass of between 3 and 5 kDa could induce biofilm formation of Kyokai-10 yeast (Fig. 2). These data are consistent with the idea that CM contains a signal molecule(s) produced by L. casei IFO3831 that induces biofilm formation in Kyokai-10 yeast.
FIG. 2.
Fractionation of biofilm-forming factor(s) using molecular sieves. The ordinate is as described for Fig. 1. CF, S. cerevisiae Kyokai-10 monoculture in CM from L. casei IFO3831. Black bars, without heat treatment; white bars, with heat treatment (100°C, 10 min). Error bars indicate standard deviations.
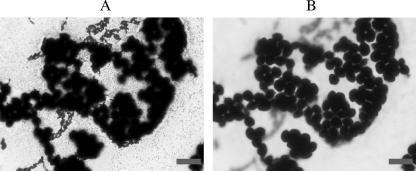
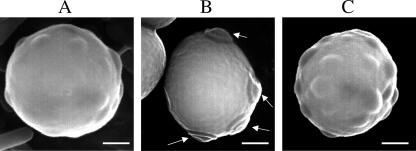
We next examined the morphology of cocultured biofilms. Figure 3 shows a representative biofilm formed on the surface of a glass slide in the presence of YPD medium. Examination under a light microscope after Gram staining reveals that the biofilm is composed of both Kyokai-10 yeast cells and L. casei IFO3831 cells. The lactic acid bacterial cells appear to form a basal layer, with the yeast cells above them. Moreover, Gram staining revealed that the yeast cells have small protrusions on their cell surfaces. To confirm this, we further examined biofilm-forming yeast cells via SEM after scraping the cells from the glass slide. After biofilm formation in the presence of L. casei IFO3831, Kyokai-10 yeast cells took on an ordinal spherical morphology with many small, hill-like protrusions visible on the cell surface (Fig. 4A). In contrast, planktonic Kyokai-10 yeast cells did not have this type of protrusion on the cell surface but had bud scars (Fig. 4B). In the case of the monoculture biofilm in the presence of CM from L. casei IFO3831, Kyokai-10 yeast cells again took on an ordinal spherical morphology with many protrusions visible on the cell surface (Fig. 4C). Bud scars were detectable using calcofluor, but the small protrusions were not stained with calcofluor, consistent with the idea that they are a distinct morphological structure (data not shown). The micrographs shown in Fig. 4 are representative of what we observed for most yeast cells under the corresponding culture conditions.
FIG. 3.
Gram-stained biofilm formed between L. casei IFO3831 and S. cerevisiae Kyokai-10 made on a slide glass. (A) Micrograph focusing on the lower layer of the biofilm, which is comprised primarily of bacteria. (B) Micrograph focusing on the yeast cells (upper layer). Bars, 10 μm.
FIG. 4.
Detection of protrusions on the cell surface of biofilm-forming yeasts by SEM. (A) Yeast and bacterial cells from a mixed-species biofilm (L. casei IFO3831 and S. cerevisiae Kyokai-10). (B) Planktonic Kyokai-10 cells. Arrows indicate bud scars. (C) Yeast cells in the monoculture biofilm formed by Kyokai-10 in the presence of CM from L. casei IFO3831. Bars, 1 μm.
To the best of our knowledge, ours is the first report that a mixed-species biofilm can be formed between lactic acid bacteria and yeast strains isolated from rice wine brewing cultures. This seems likely to have relevance to traditional Japanese fermentation of rice wine or vinegar, as in those settings, the rice provides plenty of solid surfaces on which biofilms could be formed. Traditionally, fermentation of rice wine and rice vinegar is done in the open air, and it has been established that under these conditions, various lactic acid bacteria and yeasts coexist in high density beginning at an early period of fermentation (2, 10).
We have also demonstrated that a yeast-biofilm-forming factor(s) of relatively low molecular weight was produced by lactic acid bacteria such as L. casei subsp. rhamnosus. Some lactic acid bacteria are known to produce lantibiotics, or peptide-derived bacteriocins. Lantibiotics are similar in molecular weight to what we found for the biofilm-forming factor(s). Unlike the factor characterized here, however, most bacteriocins are heat resistant (13). It seems likely that the biofilm-forming factor(s) would differ from lantibiotics.
Based on what is known about bacterial and yeast behaviors, we reason that one or more signaling ligands are produced by lactic acid bacteria and received via a receptor on the surface of respondent yeast cells. Intracellular transduction of the signal presumably brings about formation of protrusions on the cell surface of the respondent yeast. We propose that the protrusions we observed on respondent yeast are necessary to help the yeast stick to a solid surface, thus facilitating formation of a biofilm.
Acknowledgments
This work was supported by the 21st Century COE Program (T. Beppu Project) of the Ministry of Education, Culture, Sports, Science and Technology, Japan. T.K. was also supported by the COE program as a postdoctoral fellow.
We express our thanks to Makoto Tadenuma of the Brewing Society of Japan for providing rice wine brewing yeast strains and to Harushi Nakajima of Meiji University for providing the S. cerevisiae X2180-1A strain. We are also grateful to Hatsumi Shiratori of the Life Science Center, College of Bioresource Sciences, Nihon University, for teaching us fundamental techniques of SEM.
Footnotes
Published ahead of print on 25 May 2007.
REFERENCES
- 1.Adam, B., G. S. Baillie, and L. J. Douglas. 2002. Mixed species biofilms of Candida albicans and Staphylococcus epidermidis. J. Med. Microbiol. 51:344-349. [DOI] [PubMed] [Google Scholar]
- 2.Akiyama, Y. 1978. A microbial control of sake brewing from the standpoint of ecology of yeasts. Hakkokogaku 56:618-629. (In Japanese.) [Google Scholar]
- 3.An, D., T. Danhorn, C. Fuqua, and M. R. Parsek. 2006. Quorum sensing and motility mediate interactions between Pseudomonas aeruginosa and Agrobacterium tumefaciens in biofilm cocultures. Proc. Natl. Acad. Sci. USA 103:3828-3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burmølle, M., J. S. Webb, D. Rao, L. S. Hansen, S. J. Sørensen, and S. Kjelleberg. 2006. Enhanced biofilm formation and increased resistance to antimicrobial agents and bacterial invasion are caused by synergistic interactions in multispecies biofilms. Appl. Environ. Microbiol. 72:3916-3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 6.Davey, M. E., and G. A. O'Toole. 2000. Microbial biofilms: from ecology to molecular genetics. Microbiol. Mol. Biol. Rev. 64:847-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Egland, P. G., R. J. Palmer, and P. E. Kolenbrander. 2004. Interspecies communication in Streptococcus gordonii-Veillonella atypica biofilms: signaling in flow conditions requires juxtaposition. Proc. Natl. Acad. Sci. USA 101:16917-16922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Furukawa, S., S. L. Kuchma, and G. A. O'Toole. 2006. Keeping their options open: acute versus persistent infections. J. Bacteriol. 188:1211-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guzel-Seydim, Z., J. T. Wyffels, A. C. Seydim, and A. K. Greene. 2005. Turkish kefir and kefir grains: microbial enumeration and electron microscopic observation. Int. J. Dairy Technol. 58:25-29. [Google Scholar]
- 10.Haruta, S., S. Ueno, I. Egawa, K. Hashiguchi, A. Fujii, M. Nagano, M. Ishii, and Y. Igarashi. 2006. Succession of bacterial and fungal communities during a traditional pot fermentation of rice vinegar assessed by PCR-mediated denaturing gradient gel electrophoresis. Int. J. Food Microbiol. 109:79-87. [DOI] [PubMed] [Google Scholar]
- 11.Hogan, D. A., and R. Kolter. 2002. Pseudomonas-Candida interactions: an ecological role for virulence factors. Science 296:2229-2232. [DOI] [PubMed] [Google Scholar]
- 12.Kolter, R., and E. P. Greenberg. 2006. The superficial life of microbes. Nature 441:300-302. [DOI] [PubMed] [Google Scholar]
- 13.McAuliffe, O., R. P. Ross, and C. Hill. 2001. Lantibiotics: structure, biosynthesis and mode of action. FEMS Microbiol. Rev. 25:285-308. [DOI] [PubMed] [Google Scholar]
- 14.Narisawa, N., S. Furukawa, H. Ogihara, and M. Yamasaki. 2005. Estimation of the biofilm forming Escherichia coli K-12 by the cell number. J. Biosci. Bioeng. 99:78-80. [DOI] [PubMed] [Google Scholar]
- 15.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signaling pathways: a genetic analysis. Mol. Microbiol. 28:449-461. [DOI] [PubMed] [Google Scholar]
- 16.Rao, D., J. S. Webb, and S. Kjelleberg. 2005. Competitive interactions in mixed-species biofilms containing the marine bacterium Pseudoalteromonas tunicata. Appl. Environ. Microbiol. 71:1729-1736. [DOI] [PMC free article] [PubMed] [Google Scholar]