Abstract
Molecular analysis of the virulence mechanisms of the emerging pathogen Campylobacter fetus has been hampered by the lack of genetic tools. We report the development and functional analysis of Escherichia coli-Campylobacter shuttle vectors that are appropriate for C. fetus. Some vectors were constructed based on the known Campylobacter coli plasmid pIP1455 replicon, which confers a wide host range in Campylobacter spp. Versatility in directing gene expression was achieved by introducing a strong C. fetus promoter. The constructions carry features necessary and sufficient to detect the expression of phenotypic markers, including molecular reporter genes in both subspecies of C. fetus, while retaining function in C. jejuni. The capacity to express several gene products from different vectors in a single host can be advantageous but requires distinct plasmid replicons. To this end, replication features derived from a cryptic plasmid of C. fetus subsp. venerealis strain 4111/108, designated pCFV108, were adapted for a compatible series of constructions. The substitution of the C. coli replication elements reduced vector size while apparently limiting the host range to C. fetus. The complementation of a ciprofloxacin-resistant mutant phenotype via vector-driven gyrA expression was verified. Cocultivation demonstrated that shuttle vectors based on the pCFV108 replicon were compatible with pIP1455 replication functions, and the stable maintenance of two plasmids in a C. fetus subsp. venerealis host over several months was observed. The application of both vector types will facilitate the investigation of the genetics and cellular interactions of the emerging pathogen C. fetus.
Campylobacter species are important human and animal pathogens highly adapted to mucosal surfaces (23). The species Campylobacter fetus has received attention in the past primarily because it causes substantial veterinary problems associated with ruminant infertility (44). There are two subspecies, Campylobacter fetus subsp. fetus and Campylobacter fetus subsp. venerealis, both of which are considered primary pathogens. Differentiation of the taxa relies on phenotypical assays, including tolerance to glycine and hydrogen sulfide production, PCR analysis, and other molecular methods, including genome sizing, amplified fragment length polymorphism fingerprinting, and multilocus sequence typing (24, 39, 52, 53, 55). C. fetus shows a clonal population structure (25, 52, 61), but otherwise the subspecies display striking differences in host specificity. C. fetus subsp. venerealis is an important bovine pathogen colonizing the genital tract. The resulting induction of epidemic abortion is economically significant for the cattle industry. Human infections with this subspecies are rare (44). In contrast, C. fetus subsp. fetus colonizes the intestinal tract of humans and animals. It is an important agent in ovine abortion worldwide (40) and is the predominant Campylobacter species isolated from human blood (6). Human infections with C. fetus subsp. fetus can cause serious systemic disease and even death. This subspecies is considered an emerging pathogen, placing infants and immunocompromised and debilitated persons at risk (6, 41). Yet our understanding of C. fetus pathogenesis remains quite limited. To date, most progress has been made in describing the C. fetus surface layer (S-layer), a paracrystalline array of specialized proteins on the outermost surface of the cell (44). The C. fetus S-layer renders bacteria serum resistant (7, 8) and contributes to the evasion of host immunity (19, 20, 47, 48). Recognition of the significance of C. fetus as a human and animal pathogen initiated a complete genome-sequencing effort for C. fetus subsp. fetus 82-40 (GenBank accession number NC_008599). The availability of the genome sequence will be invaluable to future research activities involving C. fetus, yet the lack of molecular genetics tools to manipulate the organism remains a severely limiting factor. The range of techniques currently available for efficient DNA delivery, gene inactivation, and complementation, including established shuttle and suicide vectors, is limited for C. fetus subsp. fetus and completely lacking for C. fetus subsp. venerealis.
The aims of this study were to expand our understanding of C. fetus genetics and to develop tools that would generally facilitate research of Campylobacter. To these ends, we surveyed the majority of existing chimeric plasmids, originally developed as Escherichia coli-C. jejuni shuttle vectors, for their application in C. fetus. We found that the extension of these tools to work with C. fetus requires the exclusive use of C. fetus promoter elements for the expression of phenotypic selection markers. Modifications to enable gene expression in C. fetus did not restrict function in Campylobacter jejuni. Thus, the new series of generally applicable E. coli-Campylobacter shuttle vectors exhibit an expanded host range that extends to C. fetus. Moreover, constructions based on a C. fetus-specific replicon, derived from a cryptic plasmid endogenous to C. fetus subsp. venerealis strain 4111/108, are compatible with plasmid pIP1455-based vectors and can be simultaneously maintained in C. fetus subsp. venerealis over months. pCFV108-based constructions are substantially reduced in size; thus, independent application of this series and the additional option of combining vectors of both lineages in a common host offer increased capacity and flexibility for genetic complementation and reporter gene expression in C. fetus-specific applications.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and identification of bacterial isolates.
Campylobacter strains were grown on Columbia blood agar (CBA) plates containing 5% sheep blood (bioMérieux, Marcy l′Etoile, France) at 37°C in a microaerobic atmosphere (GENbag/GENbox MicroAir; bioMérieux). E. coli strains were grown on Luria-Bertani (LB) plates or in LB broth at 37°C. Plasmid-containing Campylobacter and E. coli cells were grown with antibiotic selection (100 μg ml−1 ampicillin or tetracycline, kanamycin, or chloramphenicol at 30 μg ml−1). The ciprofloxacin MIC was determined by the E-test (AB Biodisk, Solna, Sweden) according to the manufacturer's instructions. Bacterial strains used throughout this study are listed in Table 1. A total of 105 C. fetus strains were taken from our culture collection and analyzed for the presence of repE homologues (see below) (see Table S1 in the supplemental material). Subspecies identification was performed biochemically according to growth in the presence of 1% (wt/vol) glycine and the reduction of 0.1% sodium selenite in liquid culture (54). Additionally, a subspecies-specific PCR assay was applied to all isolates (24), and amplified fragment length polymorphism analyses (55) and pulsed-field gel electrophoresis (38) were performed where needed.
TABLE 1.
Bacterial strains used in this study
| Strain | Descriptiona | Source or reference |
|---|---|---|
| C. fetus subsp. fetus ATCC 27374 | Type strain, Nalr | ATCC |
| C. fetus subsp. fetus BT 10/98 | Sheep isolate, Nalr | J. Wagenaarb |
| C. fetus subsp. fetus BT 34/99 | Bovine isolate, aborted fetus, Nalr | J. Wagenaar |
| C. fetus subsp. fetus F12 | Human isolate, septicemia, Austria, Cipr Nalr | 26 |
| C. fetus subsp. venerealis ATCC 19438 | Type strain, Nalr | ATCC |
| C. fetus subsp. venerealis v311 | Animal isolate, Nalr | J. Wagenaar |
| C. fetus subsp. venerealis 4111/108 | Bovine isolate, infected bull, Australia, Nalr | 24 |
| C. jejuni H02/52 | Human isolate, diarrhea, no plasmid, Nalr | G. Feierlc |
| C. jejuni B02/55 | Human isolate, diarrhea, no plasmid, Nalr Kmr | G. Feierl |
| E. coli DH5α | endA1 recA1 gyrA96 thi-l hsdR17 supE44 λ−relA1 deoR Δ(lacZYA-argF)-U169 φ80dlacZΔ(M15) | 62 |
| E. coli S17-1 λpir | Tpr SmrrecA thi pro hsdR−M+ RP4:2-Tc:Mu:Km Tn7 λ pir | 12 |
Nalr, nalidixic acid resistance phenotype; Cipr, ciprofloxacin resistance phenotype; Smr, streptomycin resistance phenotype; Tpr, trimethoprim resistance phenotype.
Department of Infectious Disease and Immunology, Utrecht University.
Institute of Hygiene, Medical University Graz.
DNA preparation and PCR amplification.
Plasmids were purified routinely from E. coli by alkaline lysis (3); for large-scale preparations, the NucleoBond PC 2000 kit (Macherey-Nagel, Düren, Germany) or the PureYield Plasmid Midiprep system (Promega, Mannheim, Germany) was employed. Plasmid DNA from Campylobacter cells was isolated with the QIAprep Spin Miniprep kit (QIAGEN, Hilden, Germany). Chromosomal Campylobacter DNA was isolated using a lysozyme lysis method as described previously (18). Restriction endonucleases and DNA modifying enzymes, obtained from New England Biolabs, Inc. (Beverly, MA) and Fermentas GmbH (St. Leon-Rot, Germany), were used as recommended by the suppliers. The Wizard SV gel and PCR cleanup kit (Promega) was used to purify PCR products from agarose gels. DNA fragments for cloning were amplified using Phusion high-fidelity DNA polymerase (Finnzymes Oy, Espoo, Finland). All other PCR amplifications utilized DyNAzyme II DNA polymerase (Finnzymes Oy) according to the manufacturer's specifications.
Bacterial transformation.
Electrotransformation was as previously described (21). In brief, Campylobacter cells were grown on CBA plates for 48 h, harvested with a sterile loop, and resuspended in 5 ml of ice-cold 15% (vol/vol) glycerol-9% (wt/vol) sucrose to obtain a suspension with approximately 2 × 1011 viable cells. An optical density at 600 nanometers (d = 1 cm) of 0.1 corresponds to approximately 5 × 108 CFU/ml C. fetus, as determined by plating serial dilutions. Cells were collected by centrifugation (5,000 rpm × 10 min at 4°C) in a tabletop centrifuge. The cell pellet was resuspended in 2 ml 15% glycerol-9% sucrose solution and centrifuged again. The final bacterial pellet was resuspended in 1 ml 15% glycerol-9% sucrose solution to obtain a suspension of approximately 109 to 1010 cells/50 μl. Aliquots were transferred to fresh tubes, frozen in liquid nitrogen, and stored at −70°C. For plasmid uptake, a maximum of 1 μg DNA was mixed with a 50-μl portion of Campylobacter cells, which had been thawed on ice. The mixtures were transferred to a chilled 0.1-cm cuvette and subjected to electroporation over a range of 1.8 to 2.5 kV and 4.5 to 5.5 ms. Two-hundred microliters of prewarmed (37°C) LB or Brucella broth (Laboratories Conda, Madrid, Spain) was added immediately, and the suspension was spotted onto a prewarmed CBA plate and incubated at 37°C for 3 to 5 h under microaerophilic conditions. Cells were washed from the plate with 700 μl prewarmed (37°C) LB or brucella broth, and aliquots were plated on CBA plates containing the appropriate antibiotics and incubated for up to 12 days at 37°C under microaerophilic conditions. When heat pretreatment of C. fetus was tested, cells were harvested in ice-cold phosphate-buffered saline (PBS) and, after determination of the optical density at 600 nm, incubated at 50°C for 0, 5, 10, and 15 min. Electrotransformation as described above followed immediately. The effect of heat treatment on viability after various times of incubation was determined by serial plating.
Plasmid mobilization from E. coli to Campylobacter.
Effective vector mobilization from E. coli to Campylobacter has been reported previously (9, 29). The process is conjugative and utilizes IncP plasmid transfer functions expressed from the E. coli S17-1 λpir chromosome (12). Briefly, in preparation for bacterial mating, Campylobacter cells were freshly streaked to CBA plates for 24 h of growth under microaerophilic conditions, harvested with a sterile loop, and resuspended in prewarmed (37°C) PBS. E. coli S17-1 λpir was transformed with the plasmids of interest. Donor cells were grown overnight in LB broth with the appropriate antibiotics under gentle shaking at 37°C. Three hours before conjugation, 4 ml fresh LB broth supplemented with antibiotics was inoculated with 1 ml of the E. coli overnight culture and incubated with gentle agitation at 37°C. Donor and recipient cells were mixed in a ratio of 1:100 (107 and 109 cells, respectively, in suspension). This cell suspension was centrifuged briefly in a tabletop centrifuge (13,000 rpm for 30 s), and the bacterial pellet was resuspended in 20 μl PBS (37°C). The suspension was spotted onto a nitrocellulose filter (25 mm, 0.22 μm pore size; Millipore), which was placed on a CBA plate and incubated under microaerophilic conditions at 37°C for 3 h in the absence of antibiotics. Cells were removed from the filter by vortexing in 1.5 ml PBS (37°C) and then collected by centrifugation (8,000 rpm for 2 min). The bacterial pellet was resuspended in 800 μl prewarmed PBS (37°C) and spread over eight CBA plates. Selection for transconjugant Campylobacter combined nalidixic acid (75 μg ml−1) with selection for the newly acquired plasmid. Agar plates were incubated under microaerophilic conditions at 37°C for 3 to 5 days in the case of C. jejuni or 8 to 14 days for C. fetus recipients. Viable cell counts were determined for donor and recipient strains by serial dilutions and subsequent plating. The conjugation frequency was expressed as transconjugants per donor cells. Verification of the Campylobacter recipients was performed throughout the study based on various techniques, including PCR analyses specific for C. jejuni (32) and for C. fetus (24), oxidase expression assessed with the BBL DrySlide oxidase test (Becton, Dickinson, and Co., Sparks, Maryland), and microscopic confirmation of typical Campylobacter spiral morphology.
Plasmid detection and rescue.
Putative Campylobacter transconjugants were restreaked onto fresh CBA plates containing the appropriate antibiotics and grown for at least 2 days. For PCR verification of plasmid uptake, a single bacterial colony was resuspended in 15 μl sterile water and cells were lysed at 95°C for 5 min. Debris were cleared by centrifugation, and 4 μl of the supernatant was transferred to the PCR mixture. The primers used are indicated in Table 2. For plasmid visualization on agarose gels, the putative transconjugants were subcultured for up to 6 days and then harvested and resuspended in ice-cold PBS prior to plasmid DNA isolation. If plasmids were poorly detectable on agarose gels, the isolated plasmid DNA was amplified intracellularly via transformation of E. coli DH5α cells. A maximum of 40% of the QIAprep (QIAGEN) yield was used to transform highly competent (108 to 109 cells/μg DNA) E. coli DH5α cells by electroporation. Single colonies of E. coli transformants were grown under selection in LB broth overnight, and plasmids were isolated in small scale by alkaline lysis.
TABLE 2.
Oligonucleotides used in this study
| Oligonucleotidea | Sequenceb (5′→ 3′) | Descriptionc | Accession number or reference |
|---|---|---|---|
| T7 | GTAATACGACTCACTATAGGG | pBluescript IIKS(−) | |
| T3 | AATTAACCCTCACTAAAG | pBluescript IIKS(−) | |
| aphA-3_f* | GAGGATCCGCTAAAATGAGAATATCACCG | aphA-3 from codon 2 (nt 607 to 626) | M26832d |
| aphA-3_r* | GAGGATCCCTTTTTAGACATCTAAATCTAGG | aphA-3 (nt 1423 to 1401) | M26832d |
| cat_BamHI_f* | TTGGATCCCAATTCACAAAGATTGATATA | cat from codon 2 (nt 312 to 332) | M35190d |
| cat_BamHI_r* | GGGATCCTATTTATTCAGCAAGTCTTG | cat (nt 931 to 912) | M35190d |
| gfp_BamHI_f | ACGGATCCAGTAAAGGAGAAGAACTTTTCACT | gfp from codon 2 (nt 894 to 917) | AF292556d |
| gfp_EcoRI_r | AGAATTCTTTCGACTGAGCCTTTCGTTTT | gfp (nt 1651 to 1630) | AF292556d |
| Km_PstI_f | GATCTGCAGTGTAGAAAAGAGGAAGGAA | aphA-3 (nt 578 to 596) | M26832d |
| Km_EcoRI_r | GTCGAATTCTCTAGGTACTAAAACAATTC | aphA-3 (nt 1406 to 1387) | M26832d |
| tetO-BamHI-f* | ATGGATCCAAAATAATTAACTTAGGCATTC | tetO from codon 2 (nt 210 to 321) | M18896d |
| tetO-BamHI-r* | GCGGATCCTTAAGCTAACTTGTGGAAC | tetO (nt 2108 to 2126) | M18896d |
| SKTetO_Bam_f | TTGGATCCAACAAAACAGATGTTTCAATGC | pATJ200, tetO from nt 190 | M18896d |
| TetO_SpeI_r | TTACTAGTTTAAGCTAACTTGTGGAAC | tetO (nt 2108 to 2126) | M18896d |
| sapA-2F-XbaI-f | TATCTAGAACTGAAAAAGTGCCAGAC | sapA promoter (nt 472 to 489) | 49 |
| sapA-BamHI-r | TTGGATCCCATTTAAGGACTCCTTAATA | sapA promoter (nt 724 to 741) | 49 |
| GyrA_BamHI_f | CTTGGATCCGAAGAAAATATTTTCAGTTCAAATC | gyrA from codon 2 (nt 314 to 338) | U25640d |
| GyrA_BamHI_r | CTTGGATCCTTCTAAGCAAAACTTATTCCATAT | gyrA (nt 2889 to 2912) | U25640d |
| VirD4_BamHI_f | GCGGATCCAAAAGGTCTCAGGTTGTTT | virD4 from codon 2, C. fetus subsp. venerealis ATCC 19438 | —e |
| VirD4_PstI_r | GCACTGCAGCCAATCTTTACAAATGGTAT | nt 161 to 141, distal to virD4 STOP, C. fetus subsp. venerealis ATCC 19438 | —e |
| BamHI_B9C2 | GCGGATCCATAAAAAATAATAAAAAATTATTTAATATTGC | virB9 from codon 2, C. fetus subsp. venerealis ATCC 19438 | —e |
| PstI_D4term | CACTGCAGCCAATCTTTACAAATGGTATAGGTATG | nt 161 to 134, distal to virD4 STOP, C. fetus subsp. venerealis ATCC 19438 | —e |
| p108-rep-1f | AAGAGTTTATAATAGACAAGC | pCFV108 (nt 2199 to 2216) | EF050075d |
| p108-rep-1r | TCTCGTTGGCTCTGATCTC | pCFV108 (nt 1450 to 1468) | |
| p108-rep-2f | AAGAACCGCTATTACTTGGG | pCFV108 (nt 2143 to 2162) | |
| p108-rep-2r | AATTTACCTACTCATTCCCC | pCFV108 (nt 1549 to 1568) | |
| p108-rep-5f | TTTGCTATGAAATGTCATAGCC | pCFV108 (nt 1759 to 1780) | |
| p108-rep-6r | CTGCTAAATTCAGCAAGTTC | pCFV108 (nt 2303 to 2322) | |
| p108-rep-7f | CCCATTACATCGAAGCTAG | pCFV108 (nt 3018 to 3036) | |
| p108_8r | ACCATCACTTAACCTTTTAAG | pCFV108 (nt 83 to 103) | |
| p108_9r | AATCTCCATTCTAGATCCTG | pCFV108 (nt 991 to 1010) | |
| p108_10f | GTGGGTAGAGCATACAGAC | pCFV108 (nt 371 to 389) | |
| p108_11r | TCTTTAGCTTCTATGCTGTC | pCFV108 (nt 579 to 598) | |
| p108_13f | GCTGACACAAGAAGCTGAC | pCFV108 (nt 2488 to 2506) | |
| p108_14r | CTAGCTTCGATGTAATGGG | pCFV108 (nt 3018 to 3036) |
Asterisks indicate primers applied in PCR verification of conjugative DNA uptake.
Relevant restriction sites are shown in bold.
nt, nucleotides.
GenBank accession number.
—, G. Gorkiewicz and E. Zechner, unpublished data.
Construction of shuttle vectors.
Oligonucleotide primers are listed in Table 2. Plasmids and cloning intermediates are described in Table 3. To construct vectors that would allow the expression of resistance markers in C. fetus, a 260-bp fragment homologous to the previously characterized sapA promoter of strain 23D (49) was amplified by PCR from strain C. fetus subsp. venerealis 4111/108 using primers sapA-2F-XbaI-f and sapA-BamHI-r. The reverse primer encoded the ribosomal binding site and a 5′ BamHI site immediately following in frame with an ATG as start codon. After amplification, the PCR product was digested with XbaI and BamHI, gel purified, and ligated to XbaI/BamHI-digested pRY111 to create pRYGG1. This vector carries the sapA promoter, followed by a multicloning site (MCS) and cat (about 3 kb downstream) with the endogenous Campylobacter coli promoter. Similar to the results of a previous study (60), pRYGG1 was designed to enable constitutive expression of various genes by inserting into the unique BamHI site the desired coding sequence from codon 2 to the stop codon. The recombinant gene product begins with an N-terminal methionine, glycine, and serine (due to the in-frame BamHI site), followed by the native protein sequence starting at amino acid 2. The BamHI site in pRYGG1 was used to insert a 1,917-bp tetO fragment, amplified from pATJ200 with primers tetO-BamHI-f and tetO-BamHI-r (creating pRYGG2) and an 833-bp aphA-3 coding sequence amplified from pILL600 with the primers aphA-3_f and aphA-3_r (for pRYSS1). To express more than one coding sequence in tandem, the 833-bp aphA-3 marker was amplified with the primers Km_PstI_f and Km_EcoRI_r and inserted into the pRYGG1 MCS directly downstream of the BamHI site to generate pRYVL1. A 2.2-kb virD4 PCR product, amplified from C. fetus subsp. venerealis chromosomal DNA with the primer pair VirD4_BamHI_f and VirD4_PstI_r was then inserted into BamHI/PstI-cut pRYVL1 to generate pRYVL2. For pRYMJ1, a 5.47-kb fragment harboring four coding sequences expressed as an operon from the C. fetus subsp. venerealis chromosome (G. Gorkiewicz and E. Zechner, unpublished data) was amplified using primers BamB9C2 and PstD4term and inserted into BamHI/PstI-cut pRYGG1. To construct the gfp-harboring vector pRYEL1, a 765-bp PCR product was amplified from pWM1007 with gfp_BamHI_f and gfp_EcoRI_r, digested with BamHI and EcoRI, and ligated to BamHI- and EcoRI-cut pRYGG1.
TABLE 3.
Plasmids used in this study
| Plasmid (size in kb) | Relevant features | Reference or source |
|---|---|---|
| pRYGG1 (7.32) | sapA108 promoter (PsapA) in the pRY111 MCS | This study |
| pRYGG2 (9.24) | Fusion of PsapA to 1.9-kb tetO coding sequence in pRYGG1 | This study |
| pRYSS1 (8.15) | Φ(PsapA-aphA-3), 0.8-kb aphA-3 inserted in pRYGG1 | This study |
| pRYVL1 (8.15) | Φ(PsapA-aphA-3) in pRYGG1, PstI and EcoRI linker inserted between PsapA and aphA-3 | This study |
| pRYVL2 (10.49) | Φ(PsapA-virD4-aphA-3), 2.2-kb gene inserted between PsapA and aphA-3 of pRYVL1 | This study |
| pRYMJ1 (12.77) | Φ(PsapA-C. fetus subsp. venerealis operon), 5.47-kb coding sequence inserted in pRYGG1 | This study |
| pRYEL1 (8.06) | Φ(PsapA-gfp) in pRYGG1 | This study |
| pCFV108 (3.72) | Cryptic plasmid isolated from C. fetus subsp. venerealis 4111/108 | This study |
| pSK108-1 (6.17) | 3,207 bp of pCFV108 including mobA, repE and iterons in pBluescriptKSII(−) | This study |
| pRYSK1 (8.10) | C. coli oriV of pRYSS1 replaced by repE and iterons of pCFV108 | This study |
| pRYSK2 (6.58) | repE deletion of pRYSK1, pCFV108 iterons alone | This study |
| pRYSK3 (5.61) | pCFV108 iterons, Φ(PsapA-aphA-3) in pBlue-oriT | This study |
| pRYSK4 (6.72) | Φ(PsapA-tetO), aphA-3 of pRYSK3 replaced by tetO | This study |
| pRYSK5 (9.33) | Φ(PsapA-gyrA-tetO), 2.6-kb gyrA inserted between PsapA and tetO of pRYSK4 | This study |
| pBlue-oriT (3.72) | mobIncP of pILL570 in the pBluescriptKSII(−) MCS | This study |
| pBluescriptIIKS(−) | bla lacZ | Stratagene |
| pATJ200 | Φ(PCff-sapA-tetO) PCc-aphA-3 mobIncPoriVpIP1455oriVpBR322, source of tetO | 14 |
| pRY107 | PCc-aphA-3 mobIncPoriVpIP1455oriVpBR322 | 65 |
| pRY111 | PCc-cat mobIncPoriVpIP1455oriVpBR322, source of cat | 65 |
| pILL570 | Source of mobIncP | 30 |
| pILL600 | Source of aphA-3 | 28 |
| pWM1007 | Source of gfp | 36 |
Total plasmid DNA from C. fetus subsp. venerealis 4111/108 was isolated with the QIAprep Spin Miniprep kit (QIAGEN). After gel electrophoresis, a single plasmid band (designated pCFV108) was purified from the gel slice with the Wizard SV gel and PCR cleanup kit (Promega). The isolated DNA was partially digested with the restriction enzyme SwaI and ligated to EcoRV-cut pBluescriptIIKS(−) (Stratagene, La Jolla, CA) to generate pSK108-1. The 1,681-bp NdeI/XmnI fragment containing Campylobacter replication features was removed from pRYSS1 and replaced with a 1,630-bp NdeI/HincII fragment of pSK108-1. This construction, pRYSK1, was cut with NdeI and AflII, degraded to blunt ends with T4 DNA polymerase and religated to eliminate 1,519 bp of pCFV108 DNA and generate pRYSK2. For the construction of pRYSK3, the 1,544-bp AflII/SmaI fragment of pRYSK1 containing the iterons and the PsapAaphA-3 cassette was made blunt and ligated to pBlue-oriT linearized with SmaI. pBlue-oriT was generated by inserting a 790-bp SalI/PstI fragment of pILL570 (containing the IncP nick site) into SalI-/PstI-cut pBluescriptIIKS(−). For pRYSK4, the aphA-3 gene of pRYSK3 was excised with BamHI/SpeI and replaced by a tetO-containing amplicon of pATJ200 (primers SKTetO_Bam_f and TetO_SpeI_r). The gyrA gene of C. fetus subsp. fetus strain ATCC 27374 was amplified (gyrA primers) (Table 2), digested with BamHI, and inserted in the corresponding site between the sapA promoter and the tetO gene of pRYSK4 to create pRYSK5.
Sequence analysis of pCFV108.
Nucleotide sequence data were generated by means of primer walking using oligonucleotides listed in Table 2 and pSK108-1 or subclones generated by PCR amplification from MCS-flanking T7 and T3 primers and subsequent digestion with EcoRI or with HindIII. Cycle sequencing reactions with the BigDye termination cycle sequencing ready reaction kit, version 1.1 (Applied Biosystems, Foster City, CA), were performed on amplified pCFV108 DNA (30 to 80 ng), and the products were resolved on an Applied Biosystems ABI 310 genetic analyzer. Sequences were assembled using the SeqMan module from Lasergene (DNAStar, Inc., Madison, WI). The plasmid sequence was annotated, starting with the adenine following a SwaI recognition site. Open reading frames (ORFs) were predicted, with a cutoff value of 50 bp, using the NCBI ORF finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html) and Artemis software (http://www.sanger.ac.uk/Software/Artemis). Additional criteria to annotate the ORFs were the presence of a suitable initiation codon, allowing for alternative bacterial initiation codons as well as appropriate spacing to a putative ribosome binding site (63). Annotation of the identified ORFs was based on similarity searches using BLASTX (http://www.ncbi.nlm.nih.gov/BLAST/) against the nonredundant database on the protein level and against the NCBI conserved domain database (http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml). An analysis of secondary structural properties and the detection of specific domains or motifs in the predicted proteins were carried out with SMART (http://smart.embl-heidelberg.de). A final BLAST search was performed in April 2007. Direct repeats were predicted with WinSeq (ftp://ftp.pasteur.fr/pub/GenSoft/MS-Windows/sequence_tools/). All programs were used with default settings. The plasmid map was created with the software Vector NTI (Invitrogen, Carlsbad, CA).
DNA dot blots.
Chromosomal DNA from 68 C. fetus subsp. venerealis strains and 37 C. fetus subsp. fetus strains and plasmid DNA from 18 C. fetus subsp. fetus strains (see Table S1 in the supplemental material), taken from our laboratory collection of human and animal isolates as well as referenced isolates from culture collections (the American Type Culture Collection [ATCC], the Belgian Coordinated Collection of Micro-Organisms [BCCM/LMG], and the Culture Collection, University of Göteborg [CCUG]) were fixed to a nylon membrane (Hybond-XL; Amersham Bioscience Corp., Piscataway, NJ) using a 96-well dot blotting vacuum manifold assembled according to the manufacturer's instructions (BioDot Apparatus, Bio-Rad, Hercules, CA). DNA samples (300 ng of plasmid or 500 ng chromosomal DNA) were boiled for 10 min in the presence of 0.4 M NaOH and 0.01 M EDTA (500-μl total volume) and then centrifuged briefly and placed on ice. After vacuum application to the membrane, wells were washed with 500 μl of 0.4 M NaOH. The membrane was removed, rinsed in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), air dried, and cross-linked by UV irradiation (120 mJ/cm2).
Southern blotting.
Five hundred nanograms of HaeIII-cut chromosomal C. fetus DNA and a 40% total yield from extrachromosomal DNA were subjected to agarose gel electrophoresis. Subsequently, the gel was denaturated (1.5 M NaCl, 0.5 M NaOH) for 30 min and then neutralized (1.5 M NaCl, 0.5 M Tris HCl, pH 7.0) for 30 min. The DNA was transferred to a nylon membrane by capillary blotting using standard procedures (3) and cross-linked with UV irradiation (120 mJ/cm2).
Radiolabeling of DNA fragments and hybridization procedure.
PCR products amplified from pSK108-1 with different primer pairs (p108_13f and p108_14r or p108_rep_5f and p108_rep_6r) were used to generate repE-specific probes. The DNA was radiolabeled internally with [α32P]dCTP (3,000 Ci/mmol) and the Klenow fragment of DNA polymerase I by using the random priming kit (Promega). Blots were prehybridized at 42°C for 6 h in a 30-ml solution (50% formamide, 5× SSC, 5× Denhardt's reagent, and 0.1% sodium dodecyl sulfate) supplemented with 100 μg/ml of sheared and denatured herring sperm. DNA probes were heat denatured for 5 min and thoroughly chilled on ice, and 15 ng was added to the solution for hybridization overnight at 42°C. Blots were washed and exposed to a phosphor storage screen for 5 h to 3 days. Radioactive signal was measured with a PhosphorImager Storm 860 (Amersham Bioscience Corp.).
Confocal microscopy.
Microscopy was performed using a Leica SP2 AOBS confocal laser scanning microscope (Leica Microsystems, Mannheim, Germany). Single optical sections were acquired using a Leica 100× oil immersion lens (HCX PL APO OIL; numerical aperture, 1.4). Green fluorescent protein (GFP) fluorescence was excited at 488 nm and detected in the range of 500 to 550 nm. Transmission images were recorded using differential interference contrast optics. Image contrast was adjusted using Photoshop 7.0 (Adobe Systems, Inc., San Jose, CA).
Nucleotide sequence accession number.
The nucleotide sequence of the plasmid pCFV108 has been submitted to the GenBank nucleotide database under accession number EF050075.
RESULTS
To develop basic tools for the genetic manipulation of C. fetus, we focused initially on experimental systems currently in use with C. jejuni and C. coli. The E. coli-Campylobacter shuttle vector series, pILL* (29), pUOA* (57, 58), and pRY* (65), relies on replication functions derived from the C. coli plasmid pIP1455 (31). To our knowledge, the only shuttle vector constructed specifically for C. fetus subsp. fetus is the vector pATJ200 (14). The fact that this construction is a pILL550 (29) derivative indicates that the C. coli replicon is functional in C. fetus subsp. fetus and might be additionally replication proficient in C. fetus subsp. venerealis. A conjugative plasmid transfer system from an E. coli host to Campylobacter can be used to facilitate gene transfer as the vectors of these series harbor a mob sequence recognized by the conjugative machinery of a broad host range IncP plasmid. For selection of plasmid maintenance, the pILL* series carries the 3′-amino-glycoside phosphotransferase (aphA-3) gene derived from the C. coli plasmid pIP1433 (45), including the endogenous promoter, which is also expressed in E. coli and several other Campylobacter species (29). The pRY* series (e.g., pRY111) carries a chloramphenicol acetyltransferase (cat) gene and its endogenous promoter from the C. coli plasmid pNR9589 (57). Chloramphenicol and kanamycin resistance phenotypes (Cmr and Kmr) are expressed in both E. coli and Campylobacter species, including C. fetus subsp. fetus (9, 14). pATJ200 expresses the tetO gene of the C. jejuni plasmid pUA466 (33) from a C. fetus-specific promoter (the S-layer protein [sapA] gene promoter, PsapA). Whether the host range of this set of functional modules for conjugative interspecies transfer, plasmid replication, gene expression, and phenotypic selection would extend equally well to both C. fetus subspecies was unknown.
Conjugative transfer but not transformation is appropriate for plasmid transmission to C. fetus.
Systematic analysis of the applicability of these genetic elements required first an optimization of gene transfer techniques for C. fetus. A widely effective means to deliver recombinant DNA to gram-negative bacteria utilizes the broad host range conjugative functions of the IncP plasmid RP4. Plasmid DNA equipped with the RP4 origin of transfer (oriT) can be mobilized readily to recipient strains from donor strains expressing RP4 conjugative functions, such as E. coli S17-1 λpir (12). A conjugative transfer from E. coli to C. fetus subsp. fetus has been demonstrated previously (9, 14, 29), whereas reports of successful electrotransformation have not been published. We compared the efficiencies of introducing plasmid DNA into C. fetus with both approaches.
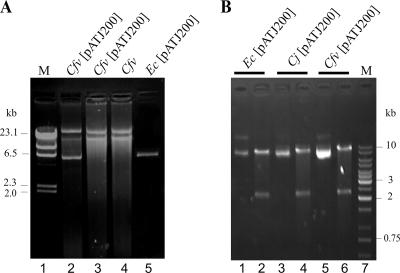
The uptake and phenotypic expression of resistance markers of plasmid pRY111 in C. fetus were compared to those of pATJ200, as the latter is known to be proficient at both, at least in C. fetus subsp. fetus. An interspecies transfer from E. coli to C. jejuni B02/55 and H02/52 was performed in parallel and served as a positive control. In each experiment, DNA transmission to the recipient host was first inferred from the acquisition of an antibiotic-resistant phenotype carried by the test plasmid. The acquisition of plasmid markers, tetO or cat, by putative transconjugants was then verified by PCR (data not shown). In a third step, plasmid DNA was isolated from the recipient cells. Visualization of these products on agarose gels did not always yield unambiguous confirmation of plasmid carriage, however, as illustrated for pATJ200 and C. fetus subsp. venerealis (Fig. 1A).
FIG. 1.
Recovery of mobilized vector DNA from C. fetus subsp. venerealis ATCC 19438 (Cfv) and C. jejuni H02/52 (Cj) transconjugants is demonstrated by direct isolation or following transformation of a surrogate E. coli (Ec) host. (A) Electrophoretic resolution in 0.8% agarose of a typical yield of plasmid DNA isolated directly from C. fetus subsp. venerealis. The pattern of bands obtained from the C. fetus subsp. venerealis type strain prior to conjugation (lane 4) or from two independent DNA preparations from a single transconjugant clone of C. fetus subsp. venerealis (lanes 2 and 3) is compared to plasmid isolated from pATJ200-carrying E. coli S17-1 λpir donor cells (lane 5), and 500 ng λHindIII-marker (M; lane 1). (B) DNA isolated from putative C. jejuni and C. fetus subsp. venerealis transconjugants was used to transform E. coli DH5α. Standard pATJ200 preparations from transformed E. coli and from the plasmid donor, E. coli S17-1 λpir, were analyzed by BamHI digestion, and pairs of untreated and treated DNA products, in that order, were resolved electrophoretically with a 1-kb DNA ladder (lane 7). Lanes 1 to 6 contain pATJ200 isolated from donor E. coli (1 and 2) or rescued from either C. jejuni (3 and 4) or C. fetus subsp. venerealis recipients (5 and 6).
In some cases, direct isolation of plasmid DNA from a transconjugant clone (Fig. 1A, lane 2) revealed the same pATJ200 band evident in DNA from the E. coli S17-1 λpir donor cells (lane 5). Yet, other preparations from the same transconjugant clone lacked an obvious plasmid band, and the bands revealed were indistinguishable from those of the recipient host prior to conjugation (Fig. 1A, compare lanes 3 and 4). Therefore, a plasmid rescue strategy was routinely used involving the transformation of E. coli hosts with DNA isolated from putative Campylobacter transconjugants. Confirmation of the identity of plasmids reisolated from the surrogate E. coli hosts enabled us to infer stable maintenance of those plasmids as extrachromosomal DNA in C. jejuni and C. fetus strains. Rescued plasmids were evaluated based on restriction enzyme fragment lengths (Fig. 1). The expected restriction patterns for pRY111 (data not shown) and for pATJ200 cut with BamHI, 8.92 kb and 2.35 kb, were obtained (Fig. 1B). As predicted, even those samples of isolated plasmid DNA from C. fetus, where the test vector was not visibly detectable (Fig. 1A, lane 3), produced unequivocal results when used to transform E. coli DH5α (Fig. 1B, lanes 5 and 6).
Based on these data, we conclude that interspecies transfer from E. coli to C. jejuni B02/55 and H02/52 occurred at frequencies of ∼10−4 transconjugants per donor with both plasmids as expected (29). Transconjugants of C. fetus subsp. fetus ATCC 27374 (type strain) were obtained at similar frequencies (∼10−4 transconjugants per donor) for each vector. By contrast, while uptake of pATJ200 by C. fetus subsp. venerealis ATCC 19438 (type strain) was observed at a comparable frequency, no Cmr transconjugants of this subspecies were obtained with pRY111.
In the case of C. jejuni, an effective alternative to conjugative transfer for DNA delivery is electrotransformation (29, 35, 58, 59). pRY111 and pATJ200 were again used to evaluate the capacity of C. fetus to take up DNA via electroporation. The transformation of C. jejuni H02/52 with these plasmids led to detectable antibiotic-resistant growth of C. jejuni after 3 to 4 days of incubation. Intact plasmid DNA was purified from the resistant bacteria and verified by agarose gel electrophoresis (data not shown). In contrast, no antibiotic-resistant growth was obtained after similarly treating the C. fetus subsp. venerealis type strain, even when the incubation period was extended to 14 days. Variation in the amount (1 μg maximum) or source (E. coli or C. jejuni H02/52) of DNA failed to produce transformants. To reduce the activity of putative intracellular restriction endonucleases, the C. fetus subsp. venerealis type strain was subjected to a short heat shock prior to transformation, as has been used successfully for E. coli and Salmonella enterica serovar Typhimurium (15). Growth on antibiotic selection remained negative in all experiments.
In summary, we conclude that IncP-mediated conjugative transmission of DNA, but not electrotransformation, is an efficient approach for C. fetus. The C. coli replication features originating from plasmid pIP1455 (45) are functional within both C. fetus subspecies, and the C. jejuni tetO gene present on pATJ200 is expressed in both C. fetus subspecies when under the control of the C. fetus sapA promoter. In addition, the selection of stable transconjugants carrying pRY111 was never observed with the C. fetus subsp. venerealis.
A C. fetus promoter permits phenotypic selection for plasmid markers in C. fetus transconjugants.
Given that pATJ200 and pRY111 share the same replicon, the failure of the E. coli S17-1 λpir (pRY111) donors to give rise to Cmr C. fetus subsp. venerealis recipients suggested that the expression of the pRY111 cat gene is reduced or absent in this host. By contrast, the tetO marker of pATJ200 is controlled by the promoter of the S-layer protein locus (PsapA) of C. fetus subsp. fetus strain 23D (14). It was thus reasonable to predict that pRY111 could be adapted for selection in C. fetus subsp. venerealis by the insertion of PsapA-driven resistance cassettes. Two pRY111 derivatives of this type were constructed to carry in the polylinker either a fusion of the PsapA upstream of tetO, pRYGG2, or a fusion of PsapA and the aphA-3 coding sequence, pRYSS1 (Table 3). Phenotypic selection of these markers in the C. fetus type strains was then evaluated following conjugative transfer of the new derivatives and compared to the parent, pRY111, and pATJ200 as a positive control.
Antibiotic-resistant colonies of both C. fetus subspecies were obtained for pATJ200, pRYGG2, and pRYSS1. Interestingly, the transfer of pRY111 gave rise to Cmr transconjugants of C. fetus subsp. fetus, but again, not C. fetus subsp. venerealis, during a maximum incubation of 14 days. The verification of plasmid uptake was performed as described previously, and the expected restriction fragment lengths were obtained (data not shown). These findings indicate that the antibiotic resistance markers originating from C. jejuni and C. coli plasmids confer a selective phenotype to C. fetus subsp. venerealis when under the control of a C. fetus promoter. Notably, the presence of PsapA on any of these constructions also correlated with the gain of Cmr phenotype expression in all C. fetus transconjugants (see below). We conclude, therefore, that optimal extension of the host range of E. coli-Campylobacter shuttle vectors to both C. fetus subspecies requires the presence of a C. fetus promoter for efficient gene expression.
Gene insertion between the sapA promoter and the resistance marker permits coexpression of a desired gene product.
To facilitate the analysis of gene function in C. fetus, we sought to create expression vectors suitable for genetic complementation applications. Mutational analyses of genes of unknown function are complicated by the absence of direct evidence that the desired gene provided in trans is actually expressed in vivo. As an operon fused to a selectable marker is a useful alternative, we asked whether the short sapA promoter fragment we had used thus far was strong enough to regulate more than one downstream gene. Indeed, the presence of the PsapA-driven resistance cassettes on pRYGG2 and pRYSS1 enabled transconjugant C. fetus cells to express not only the immediately adjacent resistance marker but also the previously silent cat gene located 5.1 kb and 4 kb downstream, respectively, on the pRY111 backbone. Selection for the expression of the adjacent resistance marker (for cat expression) or simultaneous selection for both resistance phenotypes routinely yielded equivalent numbers of transconjugant cells.
A series of expression vectors was then constructed placing various lengths of coding sequence (up to 5.5 kb; each lacking termination signals) between the PsapA and an adjacent selectable marker, the aphA-3 gene, or between PsapA and the distally located cat gene (described in Table 3). Transconjugant C. fetus subsp. venerealis cells carrying pRYVL1 or pRYVL2 exhibited the Kmr phenotype; moreover, simultaneous selection for both Kmr and Cmr phenotypes was possible with these cells. Transconjugants carrying pRYGG1, pRYVL1, pRYVL2, pRYMJ1, and pRYEL1, which contain up to 8.6 kb between PsapA and cat, all expressed the Cmr phenotype. In each case, putative transconjugants were detected with a frequency of about 10−4 transconjugants per donor. Consistent with expectations, the transmission of these constructions to C. fetus subsp. fetus was uniformly observed following selection for the same resistance phenotypes. Plasmid uptake by C. fetus subsp. venerealis and C. fetus subsp. fetus was verified, and the expected restriction fragment lengths for the new constructions were obtained in every case (data not shown). For all of these vectors, the presence of the C. fetus sapA promoter clearly enhanced gene expression in C. fetus, while effective selection in C. jejuni host cells was maintained.
In summary, these constructions exhibit a host range that includes C. jejuni while extending to both subspecies of C. fetus for applications of plasmid delivery, extrachromosomal maintenance, and phenotypic selection via efficient expression of plasmid-borne markers. Moreover, the 260-bp C. fetus sap promoter appears strong enough to drive the expression of resistance markers distally located to over 8 kb of coding and intergenic sequences (e.g., pRYMJ1).
GFP is detectably expressed in C. fetus.
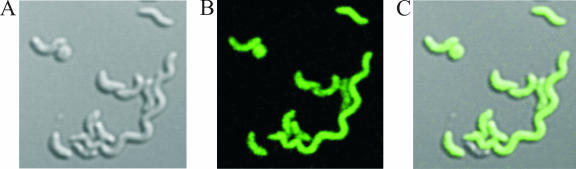
To directly demonstrate the coexpression of a gene of interest inserted between PsapA and the cat gene 3.1 kb downstream, a promoterless gfp allele was introduced to pRYGG1, generating pRYEL1 (Table 3). The expression of the green fluorophore by living C. fetus (pRYEL1) transconjugants was readily detectable with epifluorescence microscopy (data not shown) and confocal laser scanning microscopy (Fig. 2). The expression of the PsapA-gfp cassette in C. jejuni hosts carrying pRYEL1 was also readily detected by epifluorescence microscopy (data not shown). We conclude that PsapA-driven expression of multiple genes, including tandem insertions of coding sequences, is sufficient for genes of interest, including selectable antibiotic resistance and molecular reporter genes. Moreover, efficient expression of the PsapA-gfp cassette provides an excellent reporter sufficient for the visualization of single bacterial cells and particularly suited to the investigation of host-pathogen interactions.
FIG. 2.
Confocal microscopic detection of GFP expression in C. fetus subsp. venerealis ATCC 19438. Vector pRYEL1 was mobilized to C. fetus subsp. venerealis, and Cmr transconjugants were selected. Bacteria were scraped from a 48-h CBA plate, suspended in PBS, and observed by confocal microscopy for differential interference contrast (A) and green fluorescence (B). Merged images are shown in panel C.
Sequence and functional analysis of native plasmid pCFV108.
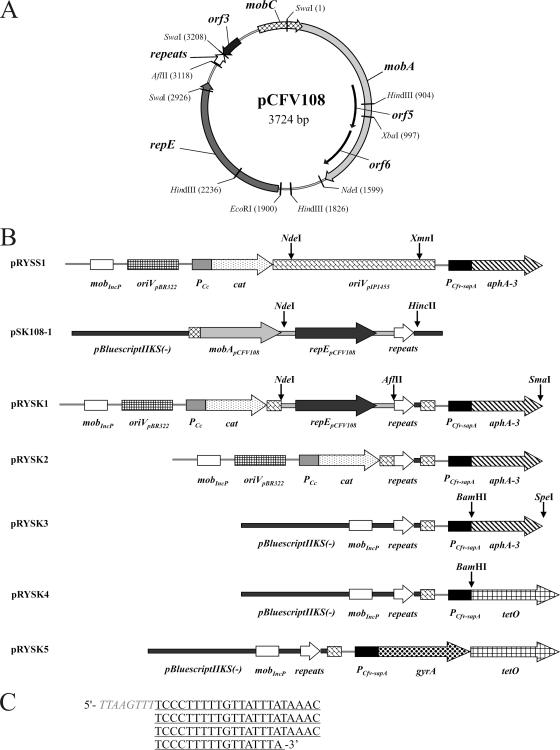
For many studies, the ability to shuttle more than one vector into a C. fetus host to achieve combinations of protein or molecular reporter expression is desirable but prevented by the presence of identical (pIP1455-derived) replication functions in pATJ200 and in the vectors we developed. To gain flexibility in gene expression from a compatible vector series, we sought to adapt replication functions from natural plasmids endogenous to C. fetus. To this end, a small cryptic plasmid carried by the C. fetus subsp. venerealis strain 4111/108 was characterized. The isolation of extrachromosomal DNA from this strain reveals three distinct plasmids. The intermediate sized 3.7-kb plasmid, named pCFV108, was analyzed in detail. The purified plasmid was partially digested with SwaI and ligated to pBluescriptIIKS(−) for propagation in E. coli. The determination of the pCFV108 sequence was accomplished using primer walking with the chimeric plasmid pSK108-1 and additional subclones. To address the possibility that a portion of original pCFV108 sequence may have been lost in generating pSK108-1 (due to restriction digestion), primers were designed for sequencing across the relevant SwaI site directly from C. fetus subsp. venerealis 4111/108 purified plasmid DNA. Indeed an additional fragment of pCFV108 sequence flanked by SwaI sites, not present in pSK108-1, was thus identified. The complete nucleotide sequence of pCFV108 (3,724 bp) was determined with a minimum of sevenfold coverage and deposited in GenBank.
Analyses of the sequence revealed an overall G+C content of 28%. A physical map is shown in Fig. 3A. A total of six putative ORFs were identified with ATG, GTG, and TTG as possible translational start codons. In each case, a conserved AAAGG sequence, suggestive of the ribosomal binding sites found in other Campylobacter species (64), was located 4 to 10 bp upstream of the predicted initiation codon. The deduced amino acid sequences were searched against the sequence database using the BLAST server. The best homologies of the putative ORFs imply two functional categories for pCFV108 genes, replication control and conjugative mobilization. One unknown function, ORF3, encodes a hypothetical protein (50 amino acids), including a signal peptide sequence and lacking significant matches in the database. The cluster of mobilization-like ORFs (Fig. 3A, positions 3522 to 1565) shares homology to conjugative mobilization (mob) regions of colicinogenic plasmids, including the prototypic plasmid ColE1 (17). Predicted protein MobA (492 amino acids) exhibits a maximum identity (36%) to a putative conjugative relaxase, the MobA-like protein of C. jejuni plasmid pCJ1170 (GenBank accession number YP618234). The vast majority of matches are other plasmid-encoded mobilization proteins from gram-negative bacteria. Predicted protein MobC (103 amino acids) of pCFV108 exhibits the highest homology within the same ColE1 superfamily of mobilizable plasmids. Maximum identity (35%) was to the Lactococcus lactis subsp. cremoris SK11 mobilization protein, MobC (GenBank accession number YP_796481.1). Also consistent with the genetic organization of the ColE1 superfamily is the presence of two smaller ORFs (ORF5, positions 778 to 1125; and ORF6, positions 1138 to 1506) entirely within the third MobA-like sequence, as found in pCFV108. The four mob homologues of the prototypic plasmid ColE1 are essential for conjugative mobilization (10).
FIG. 3.
Annotated replication features of pCFV108 were used to develop new shuttle vectors for C. fetus. (A) Physical map and genetic organization of the cryptic plasmid pCFV108. Positions for unique restriction sites AflII, EcoRI, NdeI, and XbaI are indicated as well as multiple sites for HindIII and SwaI. The ORFs of the putative mobilization region are predicted to encode a relaxase, MobA (light gray arrow), overlapped by the terminal five codons (−1 frameshift) of an auxiliary mobilization protein, MobC (hatched arrow), and two entirely overlapping ORFs, orf5 and orf6 (black, recessed arrows). The putative replication initiation protein, RepE (dark gray arrow), is followed by a region of direct repeats (open arrow). orf3 encodes a hypothetical protein of unknown function. (B) Linear representation of the pRYSK* series of shuttle vectors based on pCFV108 replication functions and vectors used for their construction. Important modules are identified beneath each feature. Restriction sites used for cloning steps (described in Materials and Methods) are indicated. The pBluescript extension (black line) downstream of the iteron region in pSK108-1 and subsequent constructions represents 21 bp of polylinker sequence from EcoRV to HincII. (C) The pCFV108 sequence conferring C. fetus replication activity to recombinant vectors is shown with the apparent 22-bp iterons in alignment (underlined).
Within the putative replication module (Fig. 3A, positions 1919 to 3208), one predicted product (351 amino acids) shares 38% identity to the putative replication initiation proteins of Enterococcus faecalis plasmids pS68 (GenBank accession number CAA11136) (34), pEF47 (GenBank accession number AAX44237) (42), and pAMα1 (GenBank accession number NP863355) (16). All of the closest matches shared homology to the first 220 N-terminal amino acids of several known or putative replication initiation proteins of gram-positive and gram-negative plasmids. In accordance with this putative functional assignment, the ORF was designated RepE (E to highlight the relatedness to enterococcal plasmids). The repE gene is followed by an A+T-rich sequence, including a region with three complete copies and one 3′ truncated copy of a 22-bp repeat (Fig. 3C; positions 3128 to 3208). The presence of direct reiterations of short DNA sequences (iterons) in prokaryotic plasmids is often indicative of an origin of replication (oriV), as a specific replication initiator protein controls replication frequency via interactions with these binding sites (13, 27). This mode of replication control is widespread among plasmids, including plasmids from Campylobacter (1).
Ninety base pairs of pCFV108 DNA are sufficient to support vector propagation in C. fetus.
To assess what regions of pCFV108 actually mediate replication, fragments carrying the annotated replication features (repE and iterons) were used to replace the pIP1455-derived replicon in pRYSS1 (Fig. 3B). In pRYSK1, the C. coli replication features were replaced with an NdeI/HincII fragment carrying the entire repE gene and distal sequences, including the 83-bp repeat region. Replication proficiency was tested by mobilization to C. jejuni H02/52 and the C. fetus type strains. Both Kmr and Cmr transconjugants of C. fetus subsp. fetus and C. fetus subsp. venerealis were obtained routinely at equivalent frequencies of 10−4 transconjugants per donor. Plasmid pRYSK1 was reisolated from these transconjugants, and its identity was verified by restriction enzyme analysis (data not shown). By contrast, E. coli S17-1 λpir (pRYSK1) donors did not give rise to transconjugants of C. jejuni in three independent mobilization experiments. This finding indicates that the 1.6-kb NdeI/SwaI fragment of pCFV108 is sufficient to mediate replication in C. fetus but not in C. jejuni. The absence of function in C. jejuni may be directly related to the replication process or possibly due to poor or disrupted expression of the repE gene but was not further investigated. The identification of the minimum sequence necessary to promote plasmid replication in C. fetus was pursued by deleting 1.5 kb of pCFV108 DNA, including the entire repE gene from pRYSK1. The remaining 90 bp of pCFV108 sequence present in pRYSK2 contain just the iteron region. Replication proficiency was again inferred from the ability to propagate C. fetus recipient cells under kanamycin or chloramphenicol selection for several days and subsequently recover pRYSK2 from the putative transconjugants. Both the C. fetus subsp. fetus and C. fetus subsp. venerealis type strains yielded pRYSK2 DNA in this experiment, but again, colony formation of C. jejuni under selection for pRYSK2 resistance markers was never observed. Recovered plasmid DNA was verified by restriction analysis.
Lack of a requirement for the repE gene in cis to the iteron region for replication in C. fetus caused us to investigate two possible explanations for this finding. Given that prior constructions have not all been sequenced, the first aim was to remove all nonessential sequences from the vector and eliminate the trivial possibility that a residual fragment harbored a replication-promoting function. The pCFV108 iterons and the selectable PsapA-aphA-3 cassette were excised from pRYSK1 and ligated to a mobIncP-harboring derivative of pBluescriptIIKS(−). The latter is replication deficient in C. fetus and, thus, can be maintained in that host by only gain of function through the additional pCFV108 DNA. The test plasmid pRYSK3 was mobilized to C. jejuni and the C. fetus type strains. Again, no colonies were detected for C. jejuni, but putative transconjugants were obtained for both C. fetus type strains at equivalent frequencies. Plasmid carriage was verified with the rescue strategy and restriction analyses (data not shown). All of the screened transconjugants carried pRYSK3, confirming that the 90-bp iteron-containing region of pCFV108 is sufficient to mediate replication in C. fetus.
Another possible explanation for the observed repE-independent replication would be that the chromosomes of the C. fetus type strains carry a repE gene or highly conserved homologues that would produce a functional replication initiator protein in trans. The implications for vector utility in that case would be important since host range would be limited to those strains carrying a related gene. To address this hypothesis, we performed a broad hybridization analysis of chromosomal DNA preparations from 105 C. fetus strains of various origins to detect the presence of repE homologues (see Table S1 of the supplemental material). The screen was performed in dot blot format using a repE-specific probe. Seven percent (7 of 105) exhibited homology to repE DNA (data not shown). Of those repE-positive strains, four were C. fetus subsp. fetus and three were C. fetus subsp. venerealis, including the pCFV108 parent strain 4111/108. Notably, neither the C. fetus subsp. fetus nor the C. fetus subsp. venerealis type strain contained repE homologous sequences. In good agreement with these results, hybridization of two nonoverlapping repE probes to a Southern blot containing chromosomal and plasmid DNA isolated from selected strains indicated no homology in the C. fetus type strains (data not shown). In conclusion, repE homologues are not widely distributed in C. fetus and additionally are not found in the C. fetus type strains shown to support the replication of pRYSK3. Thus, the identity of a specific replication initiator protein remains unknown. The proteins required to replicate this plasmid are apparently entirely of chromosomal origin. As a consequence, the amount of vector DNA dedicated to replication in the Campylobacter host is quite small. The versatility of vectors based on this replicon and their capacity to carry multiple genes or large insertion sequences are enhanced as a result.
Vectors of both lineages are stably maintained and compatible in a common C. fetus host.
We find that the pRYGG1-based constructions are stably maintained in C. fetus subsp. venerealis hosts when cultivated under selective pressure (over several months), and we also observe remarkable stability of these vectors during sequential passage on CBA plates without antibiotics (>2 months). Efficient plasmid maintenance was verified when DNA isolated from a random selection of 10 colonies each transformed an E. coli DH5α host and was subsequently reisolated (data not shown). To achieve greater flexibility in directing gene expression, the capacity of a single host to maintain a combination of vectors simultaneously was expanded. An additional derivative of pRYSK3 was generated that replaced the aphA-3 gene with tetO under the control of PsapA (pRYSK4) (Fig. 3B). Carriage of both pRYSK* plasmids and pRYGG1-based constructions by the same host is thus selectable with distinct pairs of antibiotics as desired. Stable coresidence of pRYSK3 and pRYEL1 was observed over months of cultivation in the C. fetus subsp. venerealis type strain on CBA medium containing chloramphenicol and kanamycin. The presence of both plasmids was again confirmed by transformation of E. coli DH5α cells with DNA isolated from a random selection of C. fetus colonies (data not shown). These data demonstrate that vectors derived from the distinct replicon lineages are compatible and stably maintained in C. fetus subsp. venerealis over extended periods of laboratory growth. It follows that vectors of both lineages are suitable for genetic complementation studies singularly and in combination.
Complementation of a mutant ciprofloxacin resistance phenotype to wild-type sensitivity is observed.
C. fetus strains are intrinsically resistant to the quinolone antibiotic nalidixic acid but are generally susceptible to ciprofloxacin (43). The verification of effective complementation activity from a pRYSK* vector took advantage of the known ciprofloxacin resistance phenotype (MIC, >32 μg/ml) of human blood isolate C. fetus subsp. fetus strain F12 (Table 1) (26). The resistant strain bears a C-to-T transition in the gyrA gene, corresponding to a substitution of Thr-86 to Ile in the A subunit of DNA gyrase. The mutation thus maps to a small region of gyrA called the quinolone resistance-determining region, which has been linked to ciprofloxacin resistance in clinical and induced laboratory mutant isolates of C. fetus subsp. fetus as well as C. jejuni strains (11, 43, 56). We reasoned that vector-driven expression of the wild-type gyrA allele in strain F12 would increase its sensitivity to ciprofloxacin. The complementation allele was placed under Psap control in pRYSK5 (Fig. 3B), and the expression construction was mobilized from E. coli S17-1 λpir to the mutant C. fetus subsp. fetus strain F12. A determination of the MIC of ciprofloxacin in transconjugant cells (0.38 μg/ml) revealed a substantial gain of sensitivity to this antibiotic compared to the sensitivity of the resistant isolate (>32 μg/ml). We conclude, therefore, that the Psap-driven expression cassettes harbored by both series of shuttle vectors established in this study are suitable for genetic complementation and gene expression applications in C. fetus hosts.
DISCUSSION
To meet the aims of this study, we identified features of existing E. coli-Campylobacter shuttle vectors that enable their range of application to extend to C. fetus while retaining utility in C. jejuni hosts. The analysis shows that constructions employing the C. fetus sapA promoter to drive transcription of genes of interest enable robust expression of a versatile range of selection markers, molecular reporter genes or tandem arrays of genes of interest and downstream genes allowing phenotypic selection. The sapA promoter is the only functionally characterized C. fetus promoter used in shuttle vector applications to date. The promoter sequence as well as the whole sap region appears to be very unique for C. fetus (48), underlining apparent genetic differences between C. jejuni, C. coli, and C. fetus (46). GFP expression has not been demonstrated previously for C. fetus, and its use has well-known advantages for monitoring pathogen-host interactions in vitro and in vivo (50, 51). Consistent with the observations of others (48), we obtained efficient delivery of vector DNAs via only conjugative mobilization from an E. coli host. Unlike C. jejuni, which exhibits natural and induced competence (58, 59), all strains of C. fetus subsp. fetus or C. fetus subsp. venerealis that we have ever tested have remained refractory to DNA uptake by transformation, despite a wide range of conditions applied to induce competence (48).
To expand the repertoire of replicons suitable for C. fetus vector development, we analyzed the replication features of a small cryptic plasmid from C. fetus subsp. venerealis. Plasmid pCFV108 is the first reported C. fetus plasmid sequenced thus far. Similar to cryptic plasmids from many gram-negative and gram-positive bacteria, pCFV108 carries the minimum of functional modules expected for its persistence as a mobile genetic element. Eighty percent of this genome is apparently dedicated to coding sequence. The MobA sequence shows strong amino acid sequence identity to the family of DNA transesterase or “relaxase” proteins expressed both by large, self-transmissible conjugative plasmids and small mobilizable plasmids. Relaxase proteins are essential for the initiation of plasmid DNA strand transfer in conjugative DNA transfer mechanisms (66). The application of a classification scheme for mobilization regions of bacterial plasmids proposed recently by Francia and colleagues (17) places the putative polypeptide in the ColE1 superfamily subgroup MOBHEN comprised of predominantly Proteobacteria. In accordance with this putative functional classification, the gene was designated mobA. This region of pCFV108 shares the same complex organization of mobilization genes conserved in the ColE1 superfamily. mobA sequences overlap with three other ORFs, including the upstream gene mobC, which terminates in mobA, and the two putative proteins encoded in tandem within a different reading frame of the mobA sequence. A mobilization region of similar organization was previously identified on the cryptic plasmid pHel4 of Helicobacter pylori strain P8 (22). At present, we have no information concerning the occurrence of conjugative plasmids in Campylobacter that would mediate the horizontal transfer of pCFV108 or other potentially mobilizable small plasmids. Thus, the capacity of this plasmid to be mobilized among C. fetus or other hosts was not addressed experimentally.
The RepE ORF exhibits strong amino acid identity to several known or predicted replication initiator proteins of Enterococcus faecalis plasmids pS86 (34), pAMα1 (16), and pEF47 (42); Staphylococcus epidermidis plasmid pSK639 (2); Streptococcus bovis plasmid pSB01 (37); and Tetragenococcus halophila plasmid pUCL287 (5). The latter plasmid was shown to replicate using a theta mechanism (4, 5). Consistent with that observation, these plasmids also carry arrays of direct repeats upstream of their respective rep genes that suggest replication initiation is iteron controlled. Based on these conserved features, we predicted that the smallest fragment of pCFV108 DNA required to mediate autonomous replication of a plasmid vector in Campylobacter would contain both the entire repE gene and the downstream iterons. Instead, we found that repE coding sequences were dispensable for replication of the deletion derivatives pRYSK2 and pRYSK3 in both C. fetus subspecies. By contrast, transconjugants of C. jejuni with pCFV108-based plasmids were never observed with or without the repE gene. We concluded that the 90-bp repeat region of pCFV108 is sufficient to mediate the replication of recombinant plasmids in C. fetus and that the remarkably small size of this minimal replicon is well suited for vector adaptation. In the pIP1455-based series of E. coli-C. fetus shuttle vectors, this replicon is comparatively large (4.6 kb). Earlier attempts to reduce the vector backbone by partial HindIII digestion were shown to compromise vector stability (29). Thus, the very compact replication requirements for vectors based on pCFV108 provide the first opportunity to decrease their overall size. The distinct replicons of these series exhibit stable coexistence in C. fetus. Plasmid pairs have been maintained in a C. fetus host for several months of continuous cultivation. An important advantage to generating a second compatible lineage of vector molecules is to express desired combinations of genes in strains of interest more readily and with fewer cloning steps. The substitution of antibiotic resistance cassettes resulted in a more convenient range of options for dual or individual plasmid selection in E. coli and Campylobacter hosts.
In a final point, care was taken throughout this study to describe genetics tools suitable for experiments in both C. fetus subspecies. Although their requirements were generally similar, we observed more tolerance for heterologous promoters in C. fetus subsp. fetus than in the related C. fetus subsp. venerealis. C. fetus subsp. fetus is an important veterinary pathogen and emerging human pathogen. Human infection by C. fetus subsp. venerealis is rare, but infection in ruminants is not. Despite the substantial economic losses imposed on the cattle industry by C. fetus subsp. venerealis infection, this subspecies remains very poorly characterized. It is remarkable that C. fetus subspecies exhibit such distinct niche preferences, despite the observation that they are highly genetically related (52). Comparative analyses of these subspecies on the genome level, followed by functional analyses of conserved as well as subspecies-specific sets of candidate virulence genes, are expected to rapidly promote our understanding of C. fetus virulence mechanisms. It follows that an improved capacity to manipulate both subspecies genetically, as described in this report, should facilitate our ability to elucidate the mechanisms employed by this pathogen to infect human and animal hosts.
Supplementary Material
Acknowledgments
We thank P. Guerry, A. Labigne, W. G. Miller, and S. A. Thompson for generous gifts of plasmids and J. Wagenaar, A. Burnens, and S. Hum for providing C. fetus strains. We also thank G. Feierl, C. Schober, L. Opriessnig, V. Lippitz, and H. Wolinski for their contributions to the work.
This study was supported by the Austrian FWF (P18607-B12) and the Austrian Academy of Science's DOC scholarship, awarded to S.K. Print costs were financed by the Universität Graz.
Footnotes
Published ahead of print on 18 May 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Alfredson, D. A., and V. Korolik. 2003. Sequence analysis of a cryptic plasmid pCJ419 from Campylobacter jejuni and construction of an Escherichia coli-Campylobacter shuttle vector. Plasmid 50:152-160. [DOI] [PubMed] [Google Scholar]
- 2.Apisiridej, S., A. Leelaporn, C. D. Scaramuzzi, R. A. Skurray, and N. Firth. 1997. Molecular analysis of a mobilizable theta-mode trimethoprim resistance plasmid from coagulase-negative staphylococci. Plasmid 38:13-24. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. A. Smith, J. G. Seidman, and K. Struhl. 1987. Current protocols in molecular biology. John Wiley & Sons, New York, NY.
- 4.Benachour, A., J. Frere, P. Boutibonnes, and Y. Auffray. 1995. Characterization and replication mode determination of the minimal replicon of Tetragenococcus halophila ATCC 33315 plasmid pUCL287. Biochimie 77:868-874. [DOI] [PubMed] [Google Scholar]
- 5.Benachour, A., J. Frere, and G. Novel. 1995. pUCL287 plasmid from Tetragenococcus halophila (Pediococcus halophilus) ATCC 33315 represents a new theta-type replicon family of lactic acid bacteria. FEMS Microbiol. Lett. 128:167-175. [DOI] [PubMed] [Google Scholar]
- 6.Blaser, M. J. 1998. Campylobacter fetus—emerging infection and model system for bacterial pathogenesis at mucosal surfaces. Clin. Infect. Dis. 27:256-258. [DOI] [PubMed] [Google Scholar]
- 7.Blaser, M. J., P. F. Smith, and P. F. Kohler. 1985. Susceptibility of Campylobacter isolates to the bactericidal activity of human serum. J. Infect. Dis. 151:227-235. [DOI] [PubMed] [Google Scholar]
- 8.Blaser, M. J., P. F. Smith, J. E. Repine, and K. A. Joiner. 1988. Pathogenesis of Campylobacter fetus infections. Failure of encapsulated Campylobacter fetus to bind C3b explains serum and phagocytosis resistance. J. Clin. Investig. 81:1434-1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blaser, M. J., E. Wang, M. K. Tummuru, R. Washburn, S. Fujimoto, and A. Labigne. 1994. High-frequency S-layer protein variation in Campylobacter fetus revealed by sapA mutagenesis. Mol. Microbiol. 14:453-462. [DOI] [PubMed] [Google Scholar]
- 10.Boyd, A. C., J. A. Archer, and D. J. Sherratt. 1989. Characterization of the ColE1 mobilization region and its protein products. Mol. Gen. Genet. 217:488-498. [DOI] [PubMed] [Google Scholar]
- 11.Brown, J. C., and S. G. B. Amyes. 1998. Quinolone resistance, p. 617-639. In N. Woodford and A. P. Johnson (ed.), Molecular bacteriology: protocols and clinical applications, vol. 15. Humana Press, Totowa, NJ. [Google Scholar]
- 12.de Lorenzo, V., and K. N. Timmis. 1994. Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 235:386-405. [DOI] [PubMed] [Google Scholar]
- 13.del Solar, G., R. Giraldo, M. J. Ruiz-Echevarría, M. Espinosa, and R. Díaz-Orejas. 1998. Replication and control of circular bacterial plasmids. Microbiol. Mol. Biol. Rev. 62:434-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dworkin, J., O. L. Shedd, and M. J. Blaser. 1997. Nested DNA inversion of Campylobacter fetus S-layer genes is recA dependent. J. Bacteriol. 179:7523-7529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edwards, R. A., R. A. Helm, and S. R. Maloy. 1999. Increasing DNA transfer efficiency by temporary inactivation of host restriction. BioTechniques 26:892-900. [DOI] [PubMed] [Google Scholar]
- 16.Francia, M. V., and D. B. Clewell. 2002. Amplification of the tetracycline resistance determinant of pAMα1 in Enterococcus faecalis requires a site-specific recombination event involving relaxase. J. Bacteriol. 184:5187-5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Francia, M. V., A. Varsaki, M. P. Garcillán-Barcia, A. Latorre, C. Drainas, and F. de la Cruz. 2004. A classification scheme for mobilization regions of bacterial plasmids. FEMS Microbiol. Rev. 28:79-100. [DOI] [PubMed] [Google Scholar]
- 18.Giesendorf, B. A., H. Goossens, H. G. Niesters, A. Van Belkum, A. Koeken, H. P. Endtz, H. Stegeman, and W. G. Quint. 1994. Polymerase chain reaction-mediated DNA fingerprinting for epidemiological studies on Campylobacter spp. J. Med. Microbiol. 40:141-147. [DOI] [PubMed] [Google Scholar]
- 19.Grogono-Thomas, R., M. J. Blaser, M. Ahmadi, and D. G. Newell. 2003. Role of S-layer protein antigenic diversity in the immune responses of sheep experimentally challenged with Campylobacter fetus subsp. fetus. Infect. Immun. 71:147-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grogono-Thomas, R., J. Dworkin, M. J. Blaser, and D. G. Newell. 2000. Roles of the surface layer proteins of Campylobacter fetus subsp. fetus in ovine abortion. Infect. Immun. 68:1687-1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guerry, P., R. Yao, R. A. Alm, D. H. Burr, and T. J. Trust. 1994. Systems of experimental genetics for Campylobacter species. Methods Enzymol. 235:474-481. [DOI] [PubMed] [Google Scholar]
- 22.Hofreuter, D., and R. Haas. 2002. Characterization of two cryptic Helicobacter pylori plasmids: a putative source for horizontal gene transfer and gene shuffling. J. Bacteriol. 184:2755-2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu, L., and D. J. Kopecko. 2000. Interactions of Campylobacter with eukaryotic cells: gut luminal colonization and mucosal invasion mechanisms, p. 191-215. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter, 2nd ed. ASM Press, Washington, DC.
- 24.Hum, S., K. Quinn, J. Brunner, and S. L. On. 1997. Evaluation of a PCR assay for identification and differentiation of Campylobacter fetus subspecies. Aust. Vet. J. 75:827-831. [DOI] [PubMed] [Google Scholar]
- 25.Kalka-Moll, W. M., M. A. Van Bergen, G. Plum, M. Kronke, and J. A. Wagenaar. 2005. The need to differentiate Campylobacter fetus subspecies isolated from humans. Clin. Microbiol. Infect. 11:341-342. [DOI] [PubMed] [Google Scholar]
- 26.Krause, R., S. Ramschak-Schwarzer, G. Gorkiewicz, W. J. Schnedl, G. Feierl, C. Wenisch, and E. C. Reisinger. 2002. Recurrent septicemia due to Campylobacter fetus and Campylobacter lari in an immunocompetent patient. Infection 30:171-174. [DOI] [PubMed] [Google Scholar]
- 27.Krüger, R., S. A. Rakowski, and M. Filutowicz. 2004. Participating elements in the replication of iteron-containing plasmids, p. 25-45. In B. E. Funnell and G. J. Phillips (ed.), Plasmid biology. ASM Press, Washington, DC.
- 28.Labigne-Roussel, A., P. Courcoux, and L. Tompkins. 1988. Gene disruption and replacement as a feasible approach for mutagenesis of Campylobacter jejuni. J. Bacteriol. 170:1704-1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Labigne-Roussel, A., J. Harel, and L. Tompkins. 1987. Gene transfer from Escherichia coli to Campylobacter species: development of shuttle vectors for genetic analysis of Campylobacter jejuni. J. Bacteriol. 169:5320-5323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Labigne, A., V. Cussac, and P. Courcoux. 1991. Shuttle cloning and nucleotide sequences of Helicobacter pylori genes responsible for urease activity. J. Bacteriol. 173:1920-1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lambert, T., G. Gerbaud, P. Trieu-Cuot, and P. Courvalin. 1985. Structural relationship between the genes encoding 3′-aminoglycoside phosphotransferases in Campylobacter and in gram-positive cocci. Ann. Inst. Pasteur Microbiol. 136B:135-150. [DOI] [PubMed] [Google Scholar]
- 32.Linton, D., A. J. Lawson, R. J. Owen, and J. Stanley. 1997. PCR detection, identification to species level, and fingerprinting of Campylobacter jejuni and Campylobacter coli direct from diarrheic samples. J. Clin. Microbiol. 35:2568-2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manavathu, E. K., K. Hiratsuka, and D. E. Taylor. 1988. Nucleotide sequence analysis and expression of a tetracycline-resistance gene from Campylobacter jejuni. Gene 62:17-26. [DOI] [PubMed] [Google Scholar]
- 34.Martínez-Bueno, M., E. Valdivia, A. Galvez, and M. Maqueda. 2000. pS86, a new theta-replicating plasmid from Enterococcus faecalis. Curr. Microbiol. 41:257-261. [DOI] [PubMed] [Google Scholar]
- 35.Miller, J. F., W. J. Dower, and L. S. Tompkins. 1988. High-voltage electroporation of bacteria: genetic transformation of Campylobacter jejuni with plasmid DNA. Proc. Natl. Acad. Sci. USA 85:856-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller, W. G., A. H. Bates, S. T. Horn, M. T. Brandl, M. R. Wachtel, and R. E. Mandrell. 2000. Detection on surfaces and in Caco-2 cells of Campylobacter jejuni cells transformed with new gfp, yfp, and cfp marker plasmids. Appl. Environ. Microbiol. 66:5426-5436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakamura, M., K. Ogata, T. Nagamine, K. Tajima, H. Matsui, and Y. Benno. 2001. The replicon of the cryptic plasmid pSBO1 isolated from Streptococcus bovis JB1. Curr. Microbiol. 43:11-16. [DOI] [PubMed] [Google Scholar]
- 38.On, S. L., and C. S. Harrington. 2001. Evaluation of numerical analysis of PFGE-DNA profiles for differentiating Campylobacter fetus subspecies by comparison with phenotypic, PCR and 16S rDNA sequencing methods. J. Appl. Microbiol. 90:285-293. [DOI] [PubMed] [Google Scholar]
- 39.Salama, S. M., M. M. Garcia, and D. E. Taylor. 1992. Differentiation of the subspecies of Campylobacter fetus by genomic sizing. Int. J. Syst. Bacteriol. 42:446-450. [DOI] [PubMed] [Google Scholar]
- 40.Skirrow, M. B. 1994. Diseases due to Campylobacter, Helicobacter and related bacteria. J. Comp. Pathol. 111:113-149. [DOI] [PubMed] [Google Scholar]
- 41.Skirrow, M. B., and M. J. Blaser. 2000. Clinical aspects of Campylobacter infections, p. 69-88. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter, 2nd ed. ASM, Washington, DC.
- 42.Sprincova, A., V. Stovcik, P. Javorsky, and P. Pristas. 2005. Occurrence of pS86/pEF47-related plasmids in gram-positive cocci. Curr. Microbiol. 51:198-201. [DOI] [PubMed] [Google Scholar]
- 43.Taylor, D. E., and A. S. Chau. 1997. Cloning and nucleotide sequence of the gyrA gene from Campylobacter fetus subsp. fetus ATCC 27374 and characterization of ciprofloxacin-resistant laboratory and clinical isolates. Antimicrob. Agents Chemother. 41:665-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thompson, S. A., and M. J. Blaser. 2000. Pathogenesis of Campylobacter fetus infections, p. 321-347. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter, 2nd ed. ASM Press, Washington, DC.
- 45.Trieu-Cuot, P., G. Gerbaud, T. Lambert, and P. Courvalin. 1985. In vivo transfer of genetic information between gram-positive and gram-negative bacteria. EMBO J. 4:3583-3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tu, Z. C., F. E. Dewhirst, and M. J. Blaser. 2001. Evidence that the Campylobacter fetus sap locus is an ancient genomic constituent with origins before mammals and reptiles diverged. Infect. Immun. 69:2237-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tu, Z. C., C. Gaudreau, and M. J. Blaser. 2005. Mechanisms underlying Campylobacter fetus pathogenesis in humans: surface-layer protein variation in relapsing infections. J. Infect. Dis. 191:2082-2089. [DOI] [PubMed] [Google Scholar]
- 48.Tu, Z. C., T. M. Wassenaar, S. A. Thompson, and M. J. Blaser. 2003. Structure and genotypic plasticity of the Campylobacter fetus sap locus. Mol. Microbiol. 48:685-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tummuru, M. K., and M. J. Blaser. 1992. Characterization of the Campylobacter fetus sapA promoter: evidence that the sapA promoter is deleted in spontaneous mutant strains. J. Bacteriol. 174:5916-5922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Valdivia, R. H., and S. Falkow. 1997. Fluorescence-based isolation of bacterial genes expressed within host cells. Science 277:2007-2011. [DOI] [PubMed] [Google Scholar]
- 51.Valdivia, R. H., A. E. Hromockyj, D. Monack, L. Ramakrishnan, and S. Falkow. 1996. Applications for green fluorescent protein (GFP) in the study of host-pathogen interactions. Gene 173:47-52. [DOI] [PubMed] [Google Scholar]
- 52.van Bergen, M. A., K. E. Dingle, M. C. Maiden, D. G. Newell, L. van der Graaf-Van Bloois, J. P. van Putten, and J. A. Wagenaar. 2005. Clonal nature of Campylobacter fetus as defined by multilocus sequence typing. J. Clin. Microbiol. 43:5888-5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vandamme, P. 2000. Taxonomy of the family Campylobacteraceae, p. 3-26. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter, 2nd ed. ASM Press, Washington, DC.
- 54.Véron, M., and R. Chatelain. 1973. Taxonomic study of the genus Campylobacter Sebald and Véron and designation of the neotype strain for the type species. Campylobacter fetus (Smith and Taylor) Sebald and Véron. Int. J. Syst. Bacteriol. 23:122-134. [Google Scholar]
- 55.Wagenaar, J. A., M. A. van Bergen, D. G. Newell, R. Grogono-Thomas, and B. Duim. 2001. Comparative study using amplified fragment length polymorphism fingerprinting, PCR genotyping, and phenotyping to differentiate Campylobacter fetus strains isolated from animals. J. Clin. Microbiol. 39:2283-2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang, Y., W. M. Huang, and D. E. Taylor. 1993. Cloning and nucleotide sequence of the Campylobacter jejuni gyrA gene and characterization of quinolone resistance mutations. Antimicrob. Agents Chemother. 37:457-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang, Y., and D. E. Taylor. 1990. Chloramphenicol resistance in Campylobacter coli: nucleotide sequence, expression, and cloning vector construction. Gene 94:23-28. [DOI] [PubMed] [Google Scholar]
- 58.Wang, Y., and D. E. Taylor. 1990. Natural transformation in Campylobacter species. J. Bacteriol. 172:949-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wassenaar, T. M., B. N. Fry, and B. A. van der Zeijst. 1993. Genetic manipulation of Campylobacter: evaluation of natural transformation and electro-transformation. Gene 132:131-135. [DOI] [PubMed] [Google Scholar]
- 60.Wiesner, R. S., D. R. Hendrixson, and V. J. DiRita. 2003. Natural transformation of Campylobacter jejuni requires components of a type II secretion system. J. Bacteriol. 185:5408-5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Willoughby, K., P. F. Nettleton, M. Quirie, M. A. Maley, G. Foster, M. Toszeghy, and D. G. Newell. 2005. A multiplex polymerase chain reaction to detect and differentiate Campylobacter fetus subspecies fetus and Campylobacter fetus -species venerealis: use on UK isolates of C. fetus and other Campylobacter spp. J. Appl. Microbiol. 99:758-766. [DOI] [PubMed] [Google Scholar]
- 62.Woodcock, D. M., P. J. Crowther, J. Doherty, S. Jefferson, E. DeCruz, M. Noyer-Weidner, S. S. Smith, M. Z. Michael, and M. W. Graham. 1989. Quantitative evaluation of Escherichia coli host strains for tolerance to cytosine methylation in plasmid and phage recombinants. Nucleic Acids Res. 17:3469-3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wösten, M. M., M. Boeve, W. Gaastra, and B. A. van der Zeijst. 1998. Cloning and characterization of the gene encoding the primary sigma-factor of Campylobacter jejuni. FEMS Microbiol. Lett. 162:97-103. [DOI] [PubMed] [Google Scholar]
- 64.Wösten, M. M., M. Boeve, M. G. Koot, A. C. van Nuene, and B. A. van der Zeijst. 1998. Identification of Campylobacter jejuni promoter sequences. J. Bacteriol. 180:594-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yao, R., R. A. Alm, T. J. Trust, and P. Guerry. 1993. Construction of new Campylobacter cloning vectors and a new mutational cat cassette. Gene 130:127-130. [DOI] [PubMed] [Google Scholar]
- 66.Zechner, E. L., F. de la Cruz, R. Eisenbrandt, A. M. Grahn, G. Koraimann, E. Lanka, G. Muth, W. Pansegrau, C. M. Thomas, B. M. Wilkins, and M. Zatyka. 2000. Conjugative-DNA transfer processes, p. 87-174. In C. M. Thomas (ed.), the horizontal gene pool: bacterial plasmids and gene spread. Harwood Academic Publishers, Amsterdam, The Netherlands.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.