Abstract
Little is known about the size and kinetics of treponemal burdens in blood and tissues during acquired or experimental syphilitic infection. We used real-time quantitative PCR to measure Treponema pallidum DNA levels in rabbits infected intratesticularly with the prototype Nichols strain. At the outset, we performed a series of in vitro blood spiking experiments to determine the effect of blood processing procedures on the distribution of treponemes in various blood components. T. pallidum DNA levels in plasma and whole blood were approximately 10-fold higher than those in serum and more than 200-fold greater than those in peripheral blood mononuclear cells (PBMCs). Ten rabbits were inoculated intratesticularly with doses of treponemes ranging from 4 × 107 to 2 × 108 organisms. In five rabbits, T. pallidum DNA levels were measured sequentially in serum, plasma, whole blood, and PBMCs until sacrifice at peak orchitis, at which time brain, kidney, liver, spleen, and testicles were harvested; blood and organs were also harvested at orchitis from the other five rabbits. T. pallidum DNA was detected in plasma within 24 h postinfection. Treponeme levels in whole blood and blood components increased significantly with the development of peak orchitis. Overall, levels in serum and PBMCs were lower than those in plasma and whole blood; this disparity was particularly marked at early time points. Significantly greater numbers of spirochetes were found in the spleen than in liver, kidney, or brain tissue at the time of sacrifice. Our findings highlight the remarkable capacity of T. pallidum to disseminate from the site of infection to blood and tissues, and they identify the spleen as a prime target for treponemal invasion.
Syphilis, a sexually transmitted disease of humans, is caused by the noncultivatable spirochetal pathogen Treponema pallidum subsp. pallidum (10, 23). The disease often presents clinically as distinct primary, secondary, and tertiary stages (8, 10). Early and widespread dissemination of spirochetes appears to be the rule during acquired human syphilis, as evidenced by the many organ systems, including the central nervous system, T. pallidum can invade (23). Rabbit infectivity testing has been the traditional method for isolating viable treponemes from clinical and research specimens (17, 34). Because this method is time-consuming and expensive and can be performed only in specialized research laboratories, our understanding of the microbiological events that transpire during early syphilitic infection in humans has been limited.
Much of our understanding of syphilis pathogenesis has evolved from studies in the rabbit model (1, 5, 12, 35). Infection in rabbits is typically induced by intratesticular, intradermal, or intravascular inoculation with the virulent Nichols strain of T. pallidum (23). Following intratesticular challenge with a high dose of treponemes (typically approximately 1 × 108 organisms), orchitis develops within 7 to 11 days and then resolves as the spirochetes are cleared by host effector mechanisms, principally activated macrophages and opsonic antibodies (11, 13, 29). Although replication and clearance of T. pallidum within infected testes have been studied extensively (1, 32, 36, 37), little is known about the kinetics of spirochetal dissemination during these events. While rabbit infectivity testing has been used to detect the presence of treponemes in various organs (23), this method is qualitative and does not provide information about spirochetal burdens within infected tissues.
The advent of real-time quantitative PCR (qPCR) makes it feasible to directly quantify bacterial loads within infected body fluids and tissues (4, 18). Champion et al. (4) used qPCR to measure treponeme numbers within the skin of rabbits intradermally inoculated with T. pallidum following the administration of a partially protective monoclonal antibody, but treponeme burdens in blood or extracutaneous sites were not assessed. Investigators also have used molecular amplification techniques to detect T. pallidum DNA in blood and tissues from patients with untreated syphilis, both as an adjunct diagnostic method (3, 9, 16, 19, 24, 28, 38) and for molecular typing (16, 20-22, 31). In this study, we used a sensitive qPCR assay to monitor T. pallidum DNA levels in whole blood and blood fractions (serum, plasma, and peripheral blood mononuclear cells [PBMCs]) of rabbits from shortly after intratesticular inoculation to the time of peak orchitis. In addition, we determined spirochetal burdens in selected organs at the time of peak orchitis. Our findings highlight the remarkable capacity of T. pallidum to disseminate to blood and tissues, and they identify the spleen as a prime target for treponemal invasion.
MATERIALS AND METHODS
Experimental infection of rabbits.
The Nichols strain of T. pallidum was passaged by intratesticular inoculation of New Zealand White rabbits (average weight of about 3.5 kg). Rabbits used in this study were prescreened for venereal disease using the rapid plasma reagin (RPR) test (Wampole Laboratories, Princeton, NJ) following the manufacturer's instructions. All rabbit experiments were conducted at Biocon, Inc. (Rockville, MD), under an Institutional Animal Care and Use Committee-approved protocol (Biocon A0570-04 and USAMRMC Animal Care and Use Review Office 03006003). Rabbits were housed individually in a biosafety level 2 containment room at 15.5 ± 1.6°C. Rabbits were fed antibiotic-free food and water for at least 3 weeks prior to inoculation. Ten non-RPR-reactive rabbits were infected by intratesticular inoculation (0.5 ml per testis) of freshly harvested T. pallidum at 4 × 107 to 2 × 108 treponemes/ml in BSK-H complete medium (Sigma-Aldrich Inc., St. Louis, MO). Orchitis was determined by daily observations of the testes for enlargement and by palpation for firmness. Infected rabbits were euthanized at orchitis. The testes were removed and processed within 1 h to obtain a harvest of T. pallidum essentially as described previously (6). Several lengthwise cuts were made in the testes, and the organs were incubated for 1 h at 30°C with shaking in 10 ml of prewarmed BSK-H complete medium for harvesting of viable treponemes. The medium was removed from the testes, placed in a sterile 15-ml conical tube, and centrifuged at 400 × g for 15 min. The supernatant containing eluted treponemes was removed and placed in a fresh tube for counting, aliquoting, and freezing. In order to determine the concentration of T. pallidum in the suspension, a 10-μl aliquot of a 1/500 dilution was placed into a Petroff-Hausser chamber, and the spirochetes were counted under a dark-field microscope. In addition to removal of the testes, blood was collected at the time of sacrifice (as described below), as were small sections (∼20 mg) of spleen, liver, kidney, and brain. Scalpels, scissors, and forceps were decontaminated with alcohol before each use to decrease the potential for cross-contamination between tissues. All tissues were snap-frozen during harvest. Blood specimens and tissues were stored at −80°C.
Ex vivo blood spiking experiment.
To examine the distribution of T. pallidum in blood fractions ex vivo, blood was collected from three non-RPR-reactive rabbits into one 3-ml serum collection tube and two 3-ml EDTA tubes (Becton Dickinson, San Jose, CA). Ten microliters of freshly harvested treponemes was added to each tube immediately after the draw (final concentrations of 1 × 106 and 2.7 × 106 organisms per ml by dark-field count and qPCR, respectively). All blood tubes were inverted 10 times after the addition of T. pallidum and placed at room temperature (RT) for 1 h to allow equilibration and clotting in the serum collection tubes. Portions (0.5 ml) of whole blood collected in EDTA tubes were frozen in a dry ice-ethanol bath and stored at −80°C. Plasma and PBMCs were also obtained from whole blood drawn in EDTA collection tubes. Plasma and cells were separated by centrifugation of the EDTA-treated blood, while PBMCs were purified by Ficoll banding as described below. Serum was obtained by centrifugation of clotted blood.
Collection and processing of blood.
Blood was drawn from the ear veins of rabbits following T. pallidum infection daily except for weekends. A 3-ml sample of blood was collected in a serum collection tube from each rabbit for qPCR testing and determination of RPR titers. Blood was allowed to clot for 30 to 60 min and serum was obtained by centrifugation of the clotted blood at 1,200 × g for 10 min. Five animals were monitored sequentially for determination of T. pallidum levels in whole blood, plasma, PBMCs, and serum. An additional 5 ml of blood was drawn from these animals in EDTA collection tubes at designated time points. Two 0.5-ml portions of EDTA-treated blood were transferred into cryovials, snap-frozen, and stored at −80°C. Plasma was separated from the remaining EDTA treated blood (4 ml) by centrifugation at 400 × g for 15 min at RT. Plasma was removed, aliquoted into two cryovials, and frozen. A volume of RPMI 1640 medium (Sigma-Aldrich, St. Louis, MO) equivalent to the plasma volume removed from the EDTA tube was added to the remaining blood. The blood and medium were mixed by pipetting and transferred to a 15-ml conical tube. The blood was diluted twofold with RPMI 1640 medium, and 3 ml of Ficoll-PaquePlus (Amersham Biosciences, Piscataway, NJ) was layered underneath the diluted blood. After centrifugation at 400 × g for 30 min at RT, the white cellular band (PBMCs) was removed, washed in phosphate-buffered saline (PBS) containing no magnesium or calcium, and counted (the average cell count with a Beckman Coulter [model Z1] was 8 × 105/ml blood). The PBMCs were split between two cryovials such that each cryovial contained cells equivalent to 2 ml of whole blood. PBMCs were pelleted by centrifugation at 5,000 × g for 5 min. The PBS was removed, and cell pellets were frozen in a dry ice-ethanol bath and stored at −80°C.
Extraction of DNA from blood and tissues.
DNA was extracted from 0.4 ml of whole blood, serum, and plasma using a QIAamp DNA blood minikit (QIAGEN Inc., Valencia, CA) following procedures recommended by the manufacturer; except that extra washes of the QIAGEN column were performed for DNA extractions from whole blood. The QIAamp blood kit also was used to extract DNA from diluted testis harvests and from frozen PBMC pellets. Cell pellets were resuspended in 0.4 ml of PBS just prior to extraction. DNA from tissues (15 to 25 mg) was extracted using the QIAamp DNA minikit. DNA was eluted from the QIAGEN columns in 100 μl of elution buffer at 70°C and stored at −80°C. The concentration of DNA obtained from tissues was determined spectrophotometrically by absorbance at 260/280 nm. The quality and integrity of the DNA extracted from tissues were determined by electrophoretic fractionation of 5 μl of extracted DNA through 1.2% agarose gels (E-gels: Invitrogen Corp., Carlsbad, CA) at 70 V for 30 min. DNA extractions were performed by two technicians and analyzed in separate qPCR assay runs.
Real-time PCR testing for T. pallidum flaA.
The sequences of the primers and probe used in this study were generously provided by David L. Cox (Centers for Disease Control and Prevention, Atlanta, GA). The sense (5′-GCGGTTGCACAGTGGGAG-3′) and antisense (5′-CAGCATGGGCGACAGGAT-3′) primers are positioned at bases 316 to 333 and 376 to 359 of the T. pallidum flaA gene (M63142.1), respectively (T. pallidum genome nucleotide positions 261665 to 261682 and 261708 to 261725). The probe, 5′-FAM (fluorescein phosphoramidite)-TTGTGCTGAATTCTTCCGCGCG-3′-BHQ1, is located at flaA base positions 335 to 356 (T. pallidum genome nucleotide positions 261684 to 261705). The primers were synthesized by Sigma-Genosys (Woodlands, TX); the probe was synthesized by Biosource International (Camarillo, CA). Real-time PCR quantifications were performed in 50-μl reaction volumes containing 10 μl of extracted DNA, 1× QuantiTech DNA PCR master mix (QIAGEN Inc., Valencia, CA), 0.6 μM sense and antisense primers, and 0.2 μM flaA probe. Molecular-grade deionized water was used as a no-template control. BSK-H complete medium and PBS were used as negative extraction controls. Real-time PCR was performed on an ABI Prism 7700 sequence detector system (Applied Biosystems, Foster City, CA). The amplification parameters consisted of enzyme activation at 95°C for 15 min followed by 40 cycles of 94°C for 15 s and 60°C for 1 min. All PCRs were run in duplicate. Separate amplification reactions where reaction tubes containing extracted DNAs were spiked with 2,000 flaA DNA copies were performed. No inhibition was observed in any of the PCRs. Amplification data were analyzed using Applied Biosystems SDS software, version 1.9.1. All amplification profiles were reviewed to exclude signals due to false fluorescence. A threshold cycle (CT) value greater than 38 was considered an amplification failure or “not detected.” Control reactions without template (buffer and negative extraction controls) were included in each assay: all negative controls had CT values of 40.
flaA DNA quantification standard.
The 61-bp flaA PCR product generated from amplification of a T. pallidum DNA stock was cloned into the pCRII 2.1 TOPO cloning vector (Invitrogen Inc., Carlsbad, CA). The gene copy numbers were calculated from the concentration of the plasmid DNA as determined by absorbance at 260 nm. Tenfold serial log dilutions of the flaA plasmid DNA were used to generate a standard curve ranging from 101 to 105 flaA DNA input copies. Five and two input copies of the flaA DNA standard were included in each assay run as a monitor of the lower limit of detection. The flaA DNA copy number for each sample was determined by plotting of the CT value versus the log of the copy numbers for the flaA standard. The raw data obtained from the amplifications were adjusted for quantity tested to generate the flaA DNA concentration, expressed as copies/ml or copies/μg of extracted cellular DNA from tissues. The flaA DNA concentrations in PBMCs were calculated per ml of blood from which the cells were isolated. The efficiencies of the PCRs were determined from the slopes of the plots of DNA concentrations versus CT values, where a slope of −3.327 indicated an efficiency of 100%.
Statistical analyses.
Assay reproducibility based upon CT values was determined using standard spreadsheet software (Excel; Microsoft Inc., Redmond, WA). GraphPad Prism 4.0 (GraphPad Software, San Diego, CA) was used for all other statistical analyses. The level of agreement between treponeme enumerations by dark-field microscopy and by qPCR was assessed by Pearson's correlation coefficient. The Wilcoxon matched-pairs test and the Mann-Whitney U test were used to determine (i) differences between the median T. pallidum DNA levels (copies/ml) in the blood compartments studied (whole blood, plasma, serum, and PBMCs) following in vitro spiking experiments and (ii) differences in median T. pallidum levels (copies/ml) between paired blood fractions studied at 3, 7, and 10 days after intratesticular inoculation of five rabbits. A two-tailed Student t test was used to compare the mean T. pallidum burdens (flaA DNA copies/μg of host DNA) in spleen with those in other tissues (i.e., brain, liver, and kidney) at the time of sacrifice (10 to 14 days after inoculation). All tests were two-tailed, and significance was defined as a P value of <0.05.
RESULTS
Performance characteristics of the flaA qPCR assay.
The flaA real-time qPCR assay consistently detected as few as five DNA copies based upon replicate testing of serially diluted cloned flaA DNA. Two flaA input copies of DNA were not always detected by qPCR. Among replicates, a standard deviation of 0.43 CT was observed, representing a two- to threefold variation in calculated flaA copy numbers for a given sample. Treponeme concentrations calculated by dark-field microscopy for six harvests were found to be in close agreement with values obtained by qPCR (Pearson correlation coefficient = 0.837: P = 0.04).
Analysis of rabbit blood and blood fractions spiked ex vivo with T. pallidum.
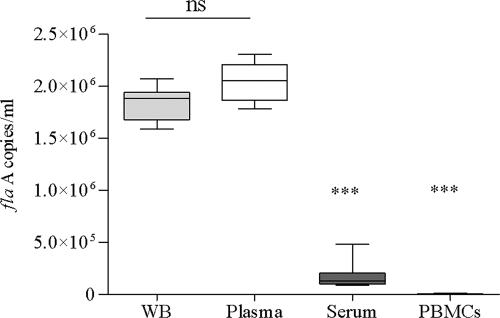
Treponemal DNA concentrations (flaA copies/ml) were measured by qPCR in whole blood, plasma, serum, and PBMCs. The distribution of T. pallidum in the various fractions isolated from the spiked blood is shown in Fig. 1. The median concentrations of T. pallidum DNA determined in plasma and whole blood were very similar. In contrast, the median T. pallidum DNA concentration in either plasma or whole blood was 10 times higher than that in serum and 200 times higher than that in PBMCs.
FIG. 1.
Analysis of rabbit blood and blood fractions spiked ex vivo with T. pallidum. Rabbit blood obtained from three animals was spiked with a similar amount (2.7 × 106 organisms/ml) of virulent T. pallidum, Nichols strain. The box plot shows the median T. pallidum DNA levels obtained 1 h after spiking in whole blood, plasma, serum, and PBMCs. Triple asterisks indicate values in serum and PBMCs that were significantly lower than those in plasma or whole blood (P < 0.0005). Median concentrations were not statistically different between whole blood and plasma (ns, not significant).
Quantitation of T. pallidum DNA in blood and blood fractions following intratesticular infection of rabbits.
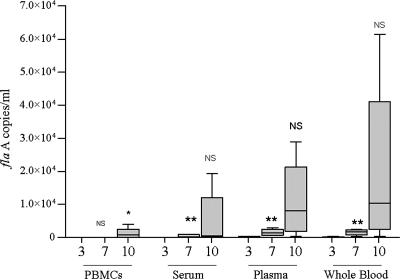
As described in Materials and Methods, T. pallidum DNA was assayed in whole blood and blood compartments from five rabbits at various time points following intratesticular injection (1, 3, 7, and 10 days). As early as 24 h postinjection, T. pallidum DNA was detected in plasma (17 to 668 flaA copies/ml) and/or whole blood (0 to 600 flaA copies/ml). As shown in Fig. 2, by day 3, T. pallidum DNA was detected at low concentrations in plasma and whole blood and was either very low or absent in serum and PBMC samples. By day 7, T. pallidum DNA concentrations steadily increased in plasma (range, 560 to 2,954 flaA copies/ml) and whole blood (410 to 2,380 flaA copies/ml) but remained low in serum (34 to 912 flaA copies/ml) and PBMCs (<50 flaA copies/ml). In contrast, by day 10 the median concentration increased in each of the four compartments. Two of five rabbits had detectable T. pallidum DNA in PBMCs at 3 days postinfection (<1 copy/106 PBMCs), whereas by day 10 all rabbits had detectable T. pallidum DNA in the PBMC compartment (26 to 599 flaA copies/106 PBMCs). These increases coincided clinically with the development of orchitis in the animals and serologically with RPR titers, which had doubled or tripled in comparison to titers on day 7, when serologic reactivity was first detected (data not shown). Consistent with the ex vivo spiking experiment results, median T. pallidum DNA concentrations were generally higher in plasma and whole blood than in serum and PBMCs at all time points.
FIG. 2.
Quantitation of T. pallidum DNA levels in blood and blood fractions following intratesticular infection of rabbits. Median T. pallidum DNA levels in PBMCs, serum, plasma, and whole blood obtained from five rabbits are shown in the box plot. Values for each blood compartment are shown for days 3, 7, and 10 following intratesticular inoculation. The single asterisk indicates that the median flaA DNA level on day 10 was significantly higher (P < 0.05) than that on day for each of the four blood compartments. Likewise, the double asterisk indicates that the median flaA DNA level was higher on day 7 than on day 3 for each blood compartment. Although the actual T. pallidum flaA levels were individually higher in most samples by day 10 in plasma and whole blood, the median values shown here for these two compartments were not statistically higher than the median values measured in serum or PBMCs (P > 0.05) by qPCR. NS, not significant.
Quantitation of T. pallidum levels in tissues.
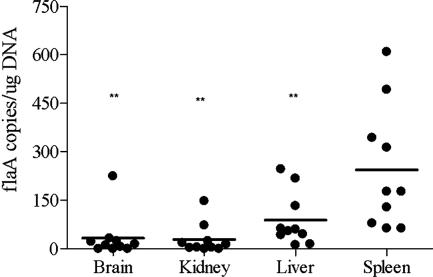
To further examine the extent of bacterial dissemination during early infection, we quantified T. pallidum DNA concentrations (flaA copies/μg of total host tissue DNA) in various organs from 10 animals 10 to 14 days postinoculation. As shown in Fig. 3, T. pallidum DNA was either undetected or present at less than 10 copies in 4/10 brain tissue samples and in 5/10 kidney specimens tested. On the other hand, 10 or more copies of T. pallidum DNA were detected in most livers and spleens. The mean concentration of T. pallidum DNA in the spleen was significantly higher than the corresponding values in all of the other organs studied (P < 0.01).
FIG. 3.
Quantitation of T. pallidum concentrations in organs. The T. pallidum concentrations, expressed as flaA copies/μg cellular DNA, are shown in the scatter plot for each of 10 rabbits studied. The bars show the mean values for each organ. Double asterisks indicate that flaA concentrations in the spleen were significantly different than those in brain, kidney, or liver (P < 0.05).
DISCUSSION
For more than 80 years, the rabbit has been the animal model of choice for studying syphilis pathogenesis (1, 5, 11, 12, 15, 23, 29, 30, 35). Rabbits are highly susceptible to T. pallidum (14, 15, 35), and infected tissues histologically resemble those seen in human syphilis (35). Although PCR techniques have been used to study experimentally infected rabbits (4, 7), until now relatively little has been known about the size and kinetics of treponemal burdens in blood or tissues. In our study, T. pallidum flaA DNA was first identified in blood within 24 h of intratesticular injection, confirming the ability of the spirochete to rapidly disseminate from sites of initial infection (23, 25). The observation that treponemal loads in blood and blood compartments were substantial at day 7 postinoculation and continued to increase by day 10 suggests that the bacterium simultaneously replicates locally at the site of inoculation and rapidly disseminates to blood and tissues. Rapid bacterial dissemination, as shown here, is thought to be due to the spirochete's unusual molecular architecture, which lacks lipopolysaccharide and has a paucity of outer membrane proteins, which have yet to be fully identified (23). It is known that the major membrane immunogens are hydrophilic polypeptides tethered by covalently bound N-terminal lipids to the periplasmic leaflet of the cytoplasmic membrane (26), a topology which renders them inaccessible to the host's cellular innate immune responses.
Interestingly, the parallel increases in treponemal DNA levels in PBMC samples coincided with the development of a serologic response in the animal. Although not quantified in this study, previous studies have confirmed that by this time rabbits have T. pallidum opsonizing antibodies (11, 13, 29). It is also known from previous in vivo and ex vivo studies that these antibodies are required for efficient phagocytosis of the organism (27). Thus, the increase in spirochetal burdens seen in the PBMC compartment could be the result of enhanced uptake by circulating phagocytic cells. Additional experiments will be needed to examine this supposition.
The presence of T. pallidum DNA in spleen, liver, brain, and kidney is also consistent with the bacterium's capability to readily disseminate and penetrate many organs and tissues (6, 35). The higher concentration of T. pallidum DNA in spleen than in liver, kidney, or brain of the rabbits suggests that the spleen is a prime target for early infection. The absence of T. pallidum DNA in 60% of the brains at this time point insinuates that the Nichols strain is not as neuroinvasive as previously thought. This finding is in agreement with a previous study by Tantalo et al. (33) showing that the Nichols strain was present in the brain of only 2/8 rabbits studied and monitored over a 3-month period and, when present, was rapidly cleared. The lower DNA copy numbers in the organs studied, in contrast to the very high numbers in whole blood or plasma, suggest that replication in these tissues begins at a later point and perhaps in lower numbers following bacteremic spread from the infected testes.
Finally, our observation of an uneven distribution of bacteria in the various blood components studied also implies that serum samples are a less reliable source for detection of circulating spirochetes than whole blood or plasma. The lower concentrations in serum may be a consequence of spirochetal attachment to cellular structures, fibrin, or fibrin degradation products contained within the clot. This would not be surprising given the propensity of T. pallidum to attach to a variety of cell types and matrices (2, 8).
In summary, our findings highlight the remarkable propensity of T. pallidum for hematogenous dissemination and identify the spleen as a prime target organ for treponemal invasion during early infection. Our findings also indicate that whole blood and plasma are the ideal samples for future animal and human studies requiring accurate quantitation of treponemal loads. Further studies need to be done to determine the kinetics of treponemal spread to blood and tissues beyond the initial 10 to 14 days of infection in the rabbit model of experimental syphilis. Our data also suggest that the level of systemic dissemination is but a reflection of ongoing treponemal replication at the site of inoculation, similar to what probably occurs in humans at the time of development of the primary chancre and during the appearance of active secondary syphilis lesions in skin and other tissues.
Acknowledgments
This work was supported by the Tri-Service AIDS Clinical Consortium (L.L.J., N.L.M., and A.R.), a component of the U.S. Military HIV Research Program (cooperative agreement W81XWH-04-02-005). USMHRP is a DoD triservice program executed through the MRMC, WRAIR, and HJF, in direct partnership with DAIDS/NIAID/HHS, and is jointly planned and coordinated with these agencies. This work was also supported by Public Health Service grants 1K23 AI62439-01A1 (J.C.S.), AI-26756 (J.D.R.) and AI-38894 (J.D.R.).
We thank Jerlil Myrick, Dior Richards, and John Cooley for technical support and Frank Klotz at Biocon Inc. for management of the rabbits.
The opinions expressed herein are those of the authors and do not represent the official position of the U.S. Army or Department of Defense.
Editor: D. L. Burns
Footnotes
Published ahead of print on 16 April 2007.
REFERENCES
- 1.Baker-Zander, S., and S. Sell. 1980. A histopathologic and immunologic study of the course of syphilis in the experimentally infected rabbit. Demonstration of long-lasting cellular immunity. Am. J. Pathol. 101:387-413. [PMC free article] [PubMed] [Google Scholar]
- 2.Cameron, C. E. 2003. Identification of a Treponema pallidum laminin-binding protein. Infect. Immun. 71:2525-2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Centurion-Lara, A., C. Castro, J. M. Shaffer, W. C. Van Voorhis, C. M. Marra, and S. A. Lukehart. 1997. Detection of Treponema pallidum by a sensitive reverse transcriptase PCR. J. Clin. Microbiol. 35:1348-1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Champion, C. I., D. R. Blanco, and M. A. Lovett. 2005. Quantitative assessment of protection in experimental syphilis. Infect. Immun. 73:5923-5927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chesney, A. M., and C. J. Schipper. 1950. The effect of the method of inoculation upon the course of experimental syphilis in the rabbit. Am. J. Syph. 34:18. [PubMed] [Google Scholar]
- 6.Fitzgerald, T. J. 1991. Syphilis vaccine: up-regulation of immunogenicity by cyclophosphamide, Ribi adjuvant, and indomethacin confers significant protection against challenge infection in rabbits. Vaccine 9:266-272. [DOI] [PubMed] [Google Scholar]
- 7.LaFond, R. E., A. Centurion-Lara, C. Godornes, A. M. Rompalo, W. C. Van Voorhis, and S. A. Lukehart. 2003. Sequence diversity of Treponema pallidum subsp. pallidum tprK in human syphilis lesions and rabbit-propagated isolates. J. Bacteriol. 185:6262-6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.LaFond, R. E., and S. A. Lukehart. 2006. Biological basis for syphilis. Clin. Microbiol. Rev. 19:29-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leslie, D. E., F. Azzato, T. Karapanagiotidis, J. Leydon, and J. Fyfe. 2006. Development of a real-time PCR assay to detect Treponema pallidum in clinical specimens and assessment of the assay's performance by comparison with serological testing. J. Clin. Microbiol. 45:93-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lukehart, S. A. 2004. Syphilis, p. 977-985. In D. Kasper, E. Braunwald, A. S. Fauci, S. L. Hauser, D. L. Longo, and J. L. Jameson (ed.), Harrison's principles of internal medicine, 16th ed. McGraw Hill, New York, NY.
- 11.Lukehart, S. A., S. A. Baker-Zander, R. M. Cheri Lloyd, and S. Sell. 1980. Characterization of lymphocyte responsiveness in early experimental syphilis. II. Nature of cellular infiltration and Treponema pallidum distribution in testicular lesions. J. Immunol. 124:461-467. [PubMed] [Google Scholar]
- 12.Lukehart, S. A., S. A. Baker-Zander, and S. Sell. 1980. Characterization of lymphocyte responsiveness in early experimental syphilis. I. In vitro response to mitogens and Treponema pallidum antigens J. Immunol. 124:454-460. [PubMed] [Google Scholar]
- 13.Lukehart, S. A., and J. N. Miller. 1978. Demonstration of the in vitro phagocytosis of Treponema pallidum by rabbit peritoneal macrophages. J. Immunol. 121:2014-2024. [PubMed] [Google Scholar]
- 14.Magnuson, H. J., H. Eagle, and R. Fleischman. 1948. The minimal infectious inoculum of Spirochaeta pallida (Nichols strain), and a consideration of its rate of multiplication in vivo. Am. J. Syph. Gon. Vener. Dis. 32:1-19. [PubMed] [Google Scholar]
- 15.Magnuson, H. J., and B. J. Rosenau. 1948. The rate of development and degree of acquired immunity in experimental syphilis. Am. J. Syph. Gon. Vener. Dis. 32:418-436. [PubMed] [Google Scholar]
- 16.Marfin, A. A., H. Liu, M. Y. Sutton, B. Steiner, A. Pillay, and L. E. Markowitz. 2001. Amplification of the DNA polymerase I gene of Treponema pallidum from whole blood of persons with syphilis. Diagn. Microbiol. Infect. Dis. 40:163-166. [DOI] [PubMed] [Google Scholar]
- 17.Miller, J. N. 1971. Spirochetes in body fluids and tissues. Charles C Thomas, Springfield, IL.
- 18.Pahl, A., U. Kühlbrandt, K. Brune, M. Röllinghoff, and A. Gessner. 1999. Quantitative detection of Borrelia burgdorferi by real-time PCR. J. Clin. Microbiol. 37:1958-1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Palmer, H. M., S. P. Higgins, A. J. Herring, and M. A. Kingston. 2003. Use of PCR in the diagnosis of early syphilis in the United Kingdom. Sex. Transm. Infect. 79:479-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pillay, A., H. Liu, C. Y. Chen, B. Holloway, A. W. Sturm, B. Steiner, and S. A. Morse. 1998. Molecular subtyping of Treponema pallidum subspecies pallidum. Sex. Transm. Dis. 25:408-414. [DOI] [PubMed] [Google Scholar]
- 21.Pillay, A., H. Liu, S. Ebrahim, C. Y. Chen, W. Lai, G. Fehler, R. C. Ballard, B. Steiner, A. W. Sturm, and S. A. Morse. 2002. Molecular typing of Treponema pallidum in South Africa: cross-sectional studies. J. Clin. Microbiol. 40:256-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pope, V., K. Fox, H. Liu, A. A. Marfin, P. Leone, A. C. Sena, J. Chapin, M. B. Fears, and L. Markowitz. 2005. Molecular subtyping of Treponema pallidum from North and South Carolina. J. Clin. Microbiol. 43:3743-3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Radolf, J. D., K. R. O. Hazlett, and S. A. Lukehart. 2006. Pathogenesis of syphilis, p. 197-236. In J. D. Radolf and S. A. Lukehart (ed.), Pathogenic treponemes: cellular and molecular biology. Caister Academic Press, Norfolk, United Kingdom.
- 24.Radolf, J. D. 1993. PCR detection of Treponema pallidum, p. 224-229. In D. H. Persing, T. F. Smith, F. C. Tenover, and T. J. White (ed.), Diagnostic molecular microbiology. Principles and applications. American Society for Microbiology, Washington, D.C.
- 25.Radolf, J. D., and S. A. Lukehart. 2006. Immunology of syphilis, p. 285-322. In J. D. Radolf and S. A. Lukehart (ed.), Pathogenic treponemes: cellular and molecular biology. Caister Academic Press, Norfolk, United Kingdom.
- 26.Radolf, J. D., M. V. Norgard, and W. W. Schulz. 1989. Outer membrane ultrastructure explains the limited antigenicity of virulent Treponema pallidum. Proc. Natl. Acad. Sci. USA 86:2051-2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salazar, J. C., K. R. Hazlett, and J. D. Radolf. 2002. The immune response to infection with Treponema pallidum, the stealth pathogen. Microbes. Infect. 4:1133-1140. [DOI] [PubMed] [Google Scholar]
- 28.Sánchez, P. J., G. D. Wendel, Jr., E. Grimprel, M. Goldberg, M. Hall, O. Arencibia-Mireles, J. D. Radolf, and M. V. Norgard. 1993. Evaluation of molecular methodologies and rabbit infectivity testing for the diagnosis of congenital syphilis and neonatal central nervous system invasion by Treponema pallidum. J. Infect. Dis. 167:148-157. [DOI] [PubMed] [Google Scholar]
- 29.Sell, S., D. Gamboa, S. A. Baker-Zander, S. A. Lukehart, and J. N. Miller. 1980. Host response to Treponema pallidum in intradermally infected rabbits: evidence for persistence of infection at local and distant sites. J. Investig. Dermatol. 75:470-475. [DOI] [PubMed] [Google Scholar]
- 30.Sell, S., and S. J. Norris. 1983. The biology, pathology, and immunology of syphilis. Int. Rev. Exp. Pathol. 24:203-276. [PubMed] [Google Scholar]
- 31.Sutton, M. Y., H. Liu, B. Steiner, A. Pillay, T. Mickey, L. Finelli, S. Morse, L. E. Markowitz, and M. E. St. Louis. 2001. Molecular subtyping of Treponema pallidum in an Arizona county with increasing syphilis morbidity: use of specimens from ulcers and blood. J. Infect. Dis. 183:1601-1606. [DOI] [PubMed] [Google Scholar]
- 32.Sykes, J. A., and J. N. Miller. 1971. Intracellular location of Treponema pallidum (Nichols strain) in the rabbit testis. Infect. Immun. 4:307-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tantalo, L. C., S. A. Lukehart, and C. M. Marra. 2005. Treponema pallidum strain-specific differences in neuroinvasion and clinical phenotype in a rabbit model. J. Infect. Dis. 191:75-80. [DOI] [PubMed] [Google Scholar]
- 34.Turner, T. B., P. H. Hardy, and B. Newman. 1969. Infectivity tests in syphilis. Br. J. Vener. Dis. 45:183-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Turner, T. B., and D. H. Hollander. 1957. Biology of the treponematoses. World Health Organization, Geneva, Switzerland.
- 36.Wicher, K., F. Abbruscato, V. Wicher, D. N. Collins, I. Auger, and H. W. Horowitz. 1998. Identification of persistent infection in experimental syphilis by PCR. Infect. Immun. 66:2509-2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wicher, V., and K. Wicher. 1984. Studies of rabbit testes infected with Treponema pallidum. III. Immunosuppressive activity of infiltrating mononuclear cells. Br. J. Vener. Dis. 60:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Woznicova, V., D. Smajs, D. Wechsler, P. Matejkova, and M. Flasarova. 2007. Detection of Treponema pallidum subsp. pallidum from skin lesions, serum, and cerebrospinal fluid in an infant with congenital syphilis after clindamycin treatment of the mother during pregnancy. J. Clin. Microbiol. 45:659-661. [DOI] [PMC free article] [PubMed] [Google Scholar]