Abstract
The Candida albicans cell wall is the immediate point of contact with the host and is implicated in the host-fungal interaction and virulence. To date, a number of cell wall proteins have been identified and associated with virulence. Analysis of the C. albicans genome has identified the IFF gene family as encoding the largest family of cell wall-related proteins. This family is also conserved in a range of other Candida species. Iff11 differs from other family members in lacking a GPI anchor, and we have demonstrated it to be O glycosylated and secreted in C. albicans. A null mutant lacking IFF11 was hypersensitive to cell wall-damaging agents, suggesting a role in cell wall organization. In a murine model of systemic infection the null mutant was highly attenuated in virulence, and survival-standardized infections suggest it is required to establish an infection. This work provides the first evidence of the importance of this gene family in the host-fungal interaction and virulence.
Candida albicans is the most common opportunistic fungal pathogen of humans. It is carried as a commensal in a significant proportion of individuals and can cause superficial epithelial infections and, in the immunocompromised, life-threatening systemic infections (5, 21, 24). A number of different virulence factors are involved with C. albicans pathogenicity; of these, the cell wall and secreted proteins are proposed to play a key role (15, 26, 27). The cell surface of C. albicans is the first point of contact with the host and plays a role in adhesion, colonization, and immunomodulation (6, 20, 25, 28). The cell wall is comprised of an inner layer consisting of glucans and chitin, which provide mechanical strength, and an outer layer enriched in mannoproteins (7, 15, 20, 26) that determine cell surface properties and play a vital role in the host-fungal interaction.
The major class of cell surface mannoproteins is the covalently attached glycosylphosphatidylinositol (GPI)-linked cell wall proteins (GPI-CWPs) (15, 25). These have a common structure consisting of an N-terminal signal sequence, conserved domain, variable-length Ser/Thr-rich region, and C-terminal GPI-anchor. Genomic approaches have identified 104 open reading frames in the C. albicans genome encoding potential GPI-CWPs (11). To date, only 12 GPI-CWPs have been shown to be expressed by proteomic analysis of cell walls derived from exponentially growing yeast cells (10). These include five carbohydrate-active enzymes (Cht2, Crh11, Pga4, Phr1, and Scw1), potential adhesion proteins of the ALS family (Als1 and Als4), a superoxide dismutase (Sod4), and others (Ywp1, Ecm33, Rbt5, and Ssr1). It is proposed that the majority of GPI-CWPs are differentially expressed under different growth conditions.
The IFF gene family was initially identified during the annotation of the C. albicans genome (9) and includes the previously reported hypha-specific GPI-CWP HYR1 (1). In the current annotation of the genome there are 12 members of the family (IFF1 to IFF11 and HYR1): it is therefore the largest family of cell wall and related proteins. A similar family of varying number is also present in a range of other pathogenic Candida species. We chose to target IFF11 from the family for experimental investigation. Uniquely among IFF family members it lacked a potential GPI-anchor or transmembrane domain, so its analysis was therefore expected to avoid the problem of functional redundancy commonly associated with gene families. Moreover, we were intrigued by the possibility that, without a GPI-anchor, the protein may play a role both inside and outside the cell. Analysis of the localization of Iff11 showed it to be secreted and not associated with the cell wall. Deletion of IFF11 resulted in a cell wall defect identified through hypersensitivity to cell wall stress. In a murine model of infection the iff11Δ null mutant was highly attenuated in virulence. We provide here the first example of this conserved gene family's role in virulence.
MATERIALS AND METHODS
Strains, media and culture conditions.
All strains constructed and used in the present study are detailed in Table 1. Strains were grown as yeasts at 30°C in YEPD medium (1% yeast extract, 2% mycological peptone [Oxoid, Basingstoke, United Kingdom], 2% glucose), SD medium (0.67% yeast nitrogen base, 2% glucose), or SC-U (0.67% yeast nitrogen base, 2% glucose, 0.077% complete supplement mixture minus uracil [Qbiogene, Cambridge, United Kingdom]). Uridine (50 μg/ml) was added to media as required. Pseudohyphal cells were induced by subculture of stationary-phase cells into fresh YEPD at 37°C. Hyphal cells were induced in YEPD plus 20% (vol/vol) newborn calf serum, RPMI 1640, Lee's medium (pH 6.5) (16) at 37°C, or solid spider medium at 30°C (17). Prior to virulence testing strains were grown at 30°C in NGY medium (0.1% neopeptone [Becton Dickinson, Cowley, Oxford, United Kingdom], 0.1% yeast extract, 0.4% glucose) for 16 to 18 h.
TABLE 1.
C. albicans strains
| Strain | Parent strain | Genotype | Source or reference |
|---|---|---|---|
| CAI-4 | ura3Δ::imm434/ura3Δ::imm434 | 11 | |
| NGY152 | CAI-4 | Same as CAI-4 but RPS1/rps1Δ::CIp10 | 4 |
| F3 | CAI-4 | Same as CAI-4 but IFF11/iff11Δ::hisG-URA3-hisG | This study |
| F3A | F3 | Same as CAI-4 but IFF11/iff11Δ::hisG | This study |
| JMCA6 | F3A | Same as CAI-4 but iff11Δ::hisG/iff11Δ::hisG-URA3-hisG | This study |
| SBC101 | JMCA6 | Same as CAI-4 but iff11Δ::hisG/iff11Δ::hisG | This study |
| SBC102 | SBC101 | Same as CAI-4 but iff11Δ::hisG/iff11Δ::hisG, RPS1/rps1Δ::CIp10 | This study |
| SBC104 | SBC101 | Same as CAI-4 but iff11Δ::hisG/iff11Δ::hisG, RPS1/rps1Δ::CIp10-IFF11 | This study |
| SCB119 | CAI-4 | Same as CAI-4 but RPS1/rps1Δ::CIp10-IFF11-V5 | This study |
| SCB121 | CAI-4 | Same as CAI-4 but RPS1/rps1Δ::CIp10-ENO1-IFF11-V5 | This study |
| SBC134 | CAI-4 | Same as CAI-4 but RPS1/rps1Δ::CIp10-ENO1-IFF11 | This study |
| SBC136 | SCB101 | Same as CAI-4 but iff11Δ::hisG/iff11Δ::hisG, RPS1/rps1Δ::CIp10-ENO-IFF11 | This study |
| SBC137 | SBC101 | Same as CAI-4 but iff11Δ::hisG/iff11Δ::hisG, RPS1/rps1Δ::CIp10-ENO-IFF11-V5 | This study |
Construction of iff11Δ null mutant and reintegrant strains.
The IFF11 gene was disrupted by “ura-blasting” (12). The regions of homology were amplified by PCR (upstream primer pair 5′-ATTTGTATGTTGCTCTAGGC-3′ and 5′-GTCGACTAGTGTTGAGCTGAAG-3′, with the SalI restriction site underlined; downstream primer pair 5′-AGATCTATCTTCAAAAATCACCAC-3′ and 5′-GAGCTCCTCGTCAGTGTTTGTCA-3′, with the BglII and SacI restriction sites, respectively, underlined) and cloned into pMB-7 (12). The disruption cassette was released by digestion with HindIII and SacI and was flanked by 468 bp upstream and 500 bp downstream to target IFF11. IFF11 was disrupted in strain CAI-4 by sequential rounds of gene replacement and recycling of the URA3 marker by selection on SD medium plus 5-fluoroorotic acid (1 mg/ml) and uridine (50 μg/ml). The ura-null mutant was transformed with StuI-digested CIp10 (19) to avoid potential problems associated with the level of URA3 expression (4). As a control a reintegrant strain was constructed in which the wild-type gene was introduced into the null mutant. The IFF11 open reading frame plus 491-bp promoter and 476-bp terminator were amplified by PCR (primer pair 5′-ATTTGTATGTTGCTCTAGGC-3′ and 5′-GAGCTCCTC GTCAGTGTTTGTCA-3′) and cloned into pGEM-T Easy (Promega, Ltd., Southampton, United Kingdom). The plasmid inserted was subcloned into the NotI site of CIp10. The resulting plasmid was linearized with StuI and transformed into the iff11Δ null mutant.
Construction of IFF11 tagged and overexpressing strains.
For overexpression studies the enolase (ENO1) promoter was utilized. The ENO1 promoter (−983 to −1) was amplified by PCR (primer pair, 5′-GAGCTCCATTTGTATCTTTAGTAGACATG-3′ and 5′-GCGGCCGCTGTTGTAATATTCCTGAATTATC-3′, SacI and NotI restriction sites underlined, respectively) and cloned into pGEM-T Easy. The plasmid insert was subcloned into the relevant sites of CIp10 to create CIp10-ENO a convenient vector for constructing overexpressing strains.
To internally tag IFF11 with the V5 epitope, the open reading frame plus 540-bp promoter and 496-bp terminator was amplified by PCR (primer pair, 5′-GAAAATCAATATCGTTAGTAGTG-3′ and 5′-CAAAATACATAGACAATGACTC-3′) and cloned into pGEM-T Easy. The resulting vector was amplified by inverse PCR (primer pair, 5′-GAAGGATCCGGTAAGCCTATCCCTAACCCTCTCCTCGGTCTCGATTCTACGGCTATCACAAGCACCCAATC-3′ and 5′-GAAGGATCCGGTGGCACTTGATTAATAAC-3′, with the BamHI sites underlined), and the product was digested with BamHI and self-ligated to introduce the V5 epitope tag in frame. The internally tagged open reading frame plus promoter and terminator were then subcloned on a NotI fragment into CIp10 to create CIp10-IFF11-V5. For overexpression studies either the wild-type or V5 tagged open reading frame plus 496-bp terminator were amplified by PCR (primer pair, 5′-AAGATGTTATTGTCAAACCTTG-3′ and 5′-CAAAATACATAGACAATGACTC-3′, with the start codon underlined) and cloned into pGEM-T Easy. The plasmid inserts were then cloned on a NotI fragment into CIp10-ENO to create CIp10-ENO-IFF11 and CIp10-ENO-IFF11-V5, respectively. Orientation of the open reading frame in relation to the ENO1 promoter was confirmed by PCR. To construct IFF11 overexpressing and tagged strains, CIp10-IFF11-V5, CIp10-ENO-IFF11 and CIp10-ENO-IFF11-V5 were linearized with StuI and independently transformed into both CAI-4 and SBC101 (iff11Δ ura-null mutant).
Preparation of protein extracts and Western blot analysis.
Secreted protein fractions from cells grown in SC-U were filter sterilized (0.45-μm pore diameter) and then concentrated, and the buffer was exchanged to phosphate-buffered saline (PBS) by ultrafiltration (Amicon Ultra-10-kDa Cutoff; Millipore, Watford, United Kingdom). Samples were then further concentrated by ethanol precipitation and reconstituted in PBS. Whole cells were extracted with β-mercaptoethanol according to published protocols (29). Briefly, washed cells were incubated in ammonium carbonate buffer (pH 8) containing 1% β-mercaptoethanol for 30 min at 37°C. The supernatant was then collected by centrifugation, the buffer was exchanged, and then the supernatant was concentrated by ultrafiltration and ethanol precipitation as before.
Cell walls were prepared from β-mercaptoethanol-extracted cells by a method modified from De Groot et al. (10). Cells were resuspended in 10 mM Tris-HCl (pH 7.5) plus protease inhibitor (Roche Applied Science) and disrupted by glass beads in a FastPrep machine (Qbiogene). Cell walls were washed extensively with 1 M NaCl and water and then extracted twice for 5 min at 100°C in 2% (wt/vol) sodium dodecyl sulfate (SDS)-100 mM EDTA-40 mM β-mercaptoethanol-50 mM Tris-HCl (pH 7.5) to remove nonconvalently attached proteins and cytoplasmic contaminants. Cell walls were then washed in water and freeze-dried. Mild alkali extracts were prepared by treating cell walls with 30 mM NaOH for 16 h at 4°C. The reaction was stopped by neutralization with 30 mM acetic acid, followed by the addition of 0.4% SDS and incubation at 100°C for 5 min. GPI-linked cell wall proteins were released by resuspending cell walls in HF-pyridine (Sigma-Aldrich, Gillingham, United Kingdom) at 4°C for 3 h. The reaction was quenched by adding an equal volume of water prior to dialysis to remove HF-pyridine. Mild alkali and HF-pyridine extracts were freeze-dried and resuspended in PBS. All secreted and cell wall extracts were prepared sequentially from the same set of cells, and extracts were resuspended at a volume to represent the proportion of original culture volume. Soluble protein extracts were prepared according to standard protocols (3), and 50 μg of protein was used in subsequent analysis.
Protein extracts were separated on a 4 to 12% NuPAGE bis-Tris gel (Invitrogen, Paisley, United Kingdom) and blotted onto a polyvinylidene difluoride membrane. The membrane was blocked with 5% bovine serum albumin in TBS-T (Tris-buffered saline containing 0.1% Tween 20) for 2 h at room temperature and subsequently incubated with the primary antibody, mouse anti-V5 (Invitrogen), at a 1:5,000 dilution in TBT-T for 1 h. The membrane was then washed twice for 5 min and twice for 15 min in TBT-T before incubation with the horseradish peroxidase-conjugated secondary antibody, anti-mouse immunoglobulin G-horseradish peroxidase (Invitrogen), at a 1:10,000 dilution in TBS-T for 1 h. After the final washes, twice for 5 min and twice for 15 min in TBT-T, the membrane was processed and exposed to film according to manufacturer's instructions (Lumiglo; New England Biolabs, Hitchin, United Kingdom).
Phenotypic analysis.
Strains were screened for hypersensitivity to cell wall-perturbing agents by the microdilution method and for phosphomannan content by Alcian Blue binding as described previously (2, 3, 13). Briefly, for cell wall sensitivity, standardized inocula were prepared from 24-h YEPD cultures by washing cells with water and resuspending them at an A600 of 1. YEPD medium was inoculated with the strains at an A600 of 0.01, and 95-μl volumes were dispensed into 96-well microtiter plates. Cell wall stressing agents were added in a 5-μl volume across a range of doubling dilutions. Plates were incubated for 16 h at 30°C, and the A600 was determined. The agents tested included Calcofluor White (500 μg/ml), Congo red (500 μg/ml), SDS (0.1%), hygromycin B (500 μg/ml), NaCl (1 M), KCl (1 M), caffeine (50 mM), vanadate (80 mM), and tunicamycin (100 μg/ml). The concentrations listed are the maximum concentration tested for each agent.
Cell surface hydrophobicity was screened in an octane partitioning assay. Washed stationary-phase cells (2 ml) were thoroughly mixed with octane (1 ml) and allowed to separate for 5 min. Once separated the change in A600 of the aqueous phase was calculated, and the proportion of cells that were hydrophobic and partitioned with the octane layer was determined. Adherence of yeast cells to buccal epithelial cells (BEC) was assessed as described previously (3, 23); at least 150 BEC were scored for adherence. A range of enzyme activities was screened for in concentrated secreted protein extracts (10-kDa ultrafiltration residues of membrane-sterilized culture filtrates) and intact cells. For determination of the exoglucanase activity a range of glucans (cellulose, glycogen, laminarin, and pustulan) was incubated with concentrated secreted protein extracts, and the glucose released was measured enzymically (glucose [GO] assay kit; Sigma-Aldrich). The protease activity was measured as previously described (8). API ZYM kits (bioMérieux) were used according to the manufacturer's instructions to detect the activities of acid and alkaline phosphatases, C4 esterase, C8 esterase-lipase, C14 lipase, leucine, valine and cysteine aminopeptidases, trypsin, chymotrypsin, phosphoamidase, α- and β-glucosidases, α- and β-galactosidases, β-glucuronidase, N-acetyl-β-d-glucosaminidase, α-mannosidase, and α-fucosidase. API ZYM tests were done with the wild type, the iffΔ null mutant strain, and the overexpressing strain, as well as with concentrated secreted protein extract.
To study the possible effects in trans of Iff11 on cell wall hypersensitivity, a concentrated culture filtrate was used in rescue experiments both by addition to the Calcofluor White sensitivity assay and by addition to the preculture used to grow the inocula of wild-type and iff11Δ cells.
Virulence studies.
For virulence tests female immunocompetent BALB/c mice were challenged intravenously. Challenge strains were grown in NGY medium at 30°C for 16 to 18 h, and cells were then washed in water and resuspended in physiological saline. Groups of six mice were challenged with 2 × 104 CFU/g (body weight) and monitored over 28 days. Mice showing signs of illness or distress were humanely terminated and recorded as dying on the following day. Mice surviving the course of the experiment were humanely terminated on day 28. Kidneys and brains were removed postmortem and homogenized, and tissue burdens were calculated by viable counting. Doses for a survival-standardized infection were determined by challenging pairs of mice intravenously with various doses of the test strains and monitoring the outcome. For survival-standardized infection studies (22), groups of three mice were infected with challenge strains and then humanely terminated on days 2, 4, and 7 postinfection; the spleen, kidney, and brain tissue burdens were then determined. All animal experimentation was done in accordance with United Kingdom Home Office regulations and was approved by both the Home Office and an institutional ethical review committee. The data from virulence studies were analyzed statistically by Kaplan-Meier/log-rank tests for survival and by Mann-Whitney U tests for burden differences.
RESULTS AND DISCUSSION
IFF gene family.
The IFF (for IPF family F) gene family was initially identified in the annotation of the C. albicans genome, CandidaDB (9). In the release of the genome called assembly 19 there are 12 members of the gene family, IFF1 to IFF11 and the previously reported hypha-specific gene HYR1 (1) (Table 2; assembly 20 was released after the present study was completed). The majority of the family displays a structure characteristic of GPI-anchored cell wall proteins, consisting of signal sequence, conserved domain, variable length Ser/Thr-rich region, and potential GPI-anchor (Table 2). The homology between family members is mainly found in the conserved N-terminal domain of 350 amino acids (Table 3). Multiple sequence alignments of the conserved domain demonstrate clustering of some family members, suggesting recent duplication. The conserved domain does not display significant homology to proteins of known function. In addition to the Ser/Thr-rich region that is potentially O glycosylated, all family members except Iff11 also contain potential N glycosylation sites. Iff7 and Iff11 also deviate from the general structure displayed by family members since both lack a potential GPI-anchor site. In the case of Iff7 the protein contains a potential C-terminal transmembrane domain, suggesting that it is membrane bound. Since Iff11 lacks both a potential GPI-anchor signal and a potential transmembrane domain it is likely either to be secreted or attached to the cell wall by other means. Iff11 also displays the lowest level of homology to the majority of other members (30 to 39% identity), except Iff10 (56.6% identity). IFF11 is directly downstream of IFF10 on chromosome 3, suggesting they resulted from tandem duplication.
TABLE 2.
Properties of IFF family
| IFF gene | Alias | ORF19a | Length (aa) | Signal sequenceb | GPI anchorc | TMDd | No. of N-glycosylation sitese | Ser/Thr (%) |
|---|---|---|---|---|---|---|---|---|
| IFF1 | RBR3 | ORF19.5124 | 1,632 | + (20-21) | + (1609) | - | 5 | 42 |
| IFF2 | HYR3 | ORF19.575 | 1,249 | + (20-21) | + (1231) | - | 39 | 36 |
| IFF3 | ORF19.4361 | 942 | + (20-21) | + (918) | - | 3 | 33 | |
| IFF4 | ORF19.7472 | 1,526 | + (20-21) | + (1502) | - | 3 | 35 | |
| IFF5 | ORF19.2879 | 1,308 | + (20-21) | + (1284) | - | 10 | 36 | |
| IFF6 | ORF19.4072 | 1,086 | + (19-20) | + (1053) | - | 31 | 38 | |
| IFF7 | HYR4 | ORF19.3279 | 1,225 | + (20-21) | − | + (1205-1224) | 11 | 27 |
| IFF8 | ORF19.570 | 714 | + (19-20) | + (690) | - | 6 | 35 | |
| IFF9 | ORF19.465 | 941 | + (20-21) | + (917) | - | 4 | 32 | |
| IFF10f | FLO9 | ORF19.5404 | 1,244 | + (19-20) | + (1219) | - | 7 | 33 |
| IFF11 | ORF19.5399 | 511 | + (19-20) | − | - | 0 | 25 | |
| HYR1 | ORF19.4975 | 919 | + (20-21) | + (985) | - | 13 | 32 |
ORF19 numbers refer to the C. albicans database assembly 19 (http://www.candidagenome.org).
Signal sequences were predicted by using SignalP (http://www.cbs.dtu.dk/services/SignalP/).
The GPI anchor attachment was predicted by using DGPI (http://129.194.185.165/dgpi).
Potential transmembrane domains (TMDs) were predicted by using TMHHH (http://www.cbs.dtu.dk/services/TMHMM-2.0/).
Potential N-glycosylation sites were predicted by using NetNGlyc (http://www.cbs.dtu.dk/services/NetNGlyc/).
The sequence for IFF10 contains a potential frameshift mutation joining ORF19.5404 to ORF19.5401.
TABLE 3.
Homology of IFF gene family productsa
| Gene type | % Identity
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Iff1 | Iff2 | Iff3 | Iff4 | Iff5 | Iff6 | Iff7 | Iff8 | Iff9 | Iff10 | Iff11 | Hyr1 | |
| Iff1 | 100 | |||||||||||
| Iff2 | 59.4 | 100 | ||||||||||
| Iff3 | 55.6 | 63.7 | 100 | |||||||||
| Iff4 | 55.1 | 62.3 | 76.3 | 100 | ||||||||
| Iff5 | 53.5 | 60.9 | 85.4 | 74 | 100 | |||||||
| Iff6 | 45.6 | 50.1 | 46.5 | 48.3 | 46.5 | 100 | ||||||
| Iff7 | 48 | 53.1 | 55.1 | 53.2 | 51.4 | 39.1 | 100 | |||||
| Iff8 | 45.6 | 54.1 | 50.1 | 49.4 | 47.5 | 46.8 | 41.8 | 100 | ||||
| Iff9 | 55.1 | 62.9 | 98 | 74.9 | 83.7 | 45.6 | 54.3 | 49 | 100 | |||
| Iff10 | 30.2 | 35.8 | 35.4 | 35.5 | 34.2 | 33.8 | 34.2 | 31.9 | 34.6 | 100 | ||
| Iff11 | 30 | 38.5 | 34.2 | 34.1 | 33.6 | 34.6 | 34.7 | 35.1 | 33.3 | 56.6 | 100 | |
| Hyr1 | 35.4 | 45.5 | 42.2 | 41.6 | 41.9 | 34.4 | 37.5 | 40.8 | 41.9 | 29.3 | 30.9 | 100 |
Protein homology was calculated over the first 350 amino acids incorporating the conserved domain.
A similar gene family with varying numbers of members can be identified in a range of related fungi, including the Candida species C. dubliniensis, C. guilliermondii, C. lusitaniae, C. tropicalis, C. glabrata, Kluyveromyces lactis, and Debaryomyces hansenii. However, the family is not present in Saccharomyces cerevisiae or in the genome sequences currently available for a range of fungi pathogenic to humans, including Aspergillus fumigatus, A. terreus, Coccidioides immitis, Cryptococcus neoformans, and Histoplasma capsulatum: our search extended to these and other fungal genomes currently available (http://www.broad.mit.edu/annotation/fgi/). The C. albicans genome also carries the ALS gene family of cell wall proteins (14). The IFF family is of comparable size, and these are the largest families of cell wall related proteins in the genome as currently annotated.
Since IFF11 is a family member that differs in lacking a GPI-anchor, and hence may be localized differently, it was chosen for further analysis as it may not display functional redundancy—a problem commonly associated with the analysis of gene families. The previously identified member of the family, HYR1, is known to display hypha-specific expression, but its inactivation generated no obvious phenotype (1). We therefore analyzed the expression of IFF11. IFF11 mRNA was found to be constitutively expressed in yeast, pseudohyphal and hyphal growth forms, and compared to positive control EFB1 was expressed at a low level (data not shown).
Localization of Iff11.
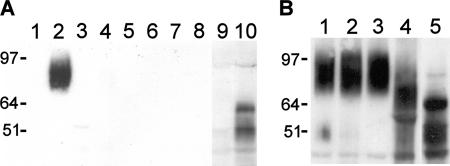
The bioinformatics analysis of Iff11 suggested that it lacks a potential GPI-anchor signal. We therefore tested whether it is secreted or cell wall associated. In order to localize Iff11, an internal epitope tag approach was chosen to avoid potential problems with alteration of the N- or C-terminal signals. An internal V5 epitope tag was inserted into an IFF11 cassette through inverse PCR and self-ligation. This was targeted to insert the V5 tag in frame at amino acid position 349 between the conserved domain and the Ser/Thr-rich region. To determine the cellular localization of Iff11, secreted and cell wall fractions were analyzed by Western blotting. When the V5-tagged version of IFF11 was expressed under its native promoter no signal could be detected, either in secreted, cell wall, or total protein extracts. This is consistent with the finding that IFF11 was constitutively expressed at a low level. Therefore, for localization studies Iff11-V5 was overexpressed under the control of the ENO1 (enolase) promoter. Western blot analysis of total soluble protein extracts identified three specific bands associated with Iff11-V5 overexpression (Fig. 1A, lanes 9 and 10) present at 53, 64, and 90 kDa. The 53-kDa protein band matched the predicted size of Iff11 after processing of the N-terminal signal sequence. The presence of the specific 64-kDa band on Western blot analysis is suggestive of an intermediate in posttranslational modification. The 90-kDa band is presumably fully modified (see below), mature Iff11 present in the secretory pathway. To determine the localization of Iff11, secreted and cell wall fractions were analyzed (Fig. 1A). No specific bands could be detected in a range of cell wall fractions, including nonconvalently attached proteins (whole-cell β-mercaptoethanol- and SDS-treated cell wall extracts) and covalently attached proteins (mild alkali, HF-pyridine, and β-1,3-glucanase extracts). In contrast, the secreted protein fraction contained a specific band at ∼90 kDa. Therefore, Iff11 appears to be a secreted protein and is not found associated with the cell wall. However, it is not possible to discount that a small, nondetectable, level of Iff11 may be associated with the cell wall.
FIG. 1.
Localization and modification of Iff11. (A) Secreted, cell wall, and cytoplasmic protein extracts were analyzed by Western blotting with an anti-V5 antibody. Fractions were secreted proteins (lanes 1 and 2), cell wall extracts from treatments with β-mercaptoethanol (lanes 3 and 4), mild alkali (lanes 5 and 6), or HF-pyridine (lanes 7 and 8) and soluble proteins (lanes 9 and 10). The extracts were prepared from wild-type NGY152 (lanes 1, 3, 5, 7, and 9) and SBC121 (overexpressing V5-tagged Iff11; lanes 2, 4, 6, 8, and 10). All secreted and cell wall extracts were prepared sequentially from the same set of cells, and extracts were resuspended at a volume to represent the proportion of original culture volume. For soluble protein extracts, 50 μg of protein was analyzed. The faint low-molecular-weight bands in lanes 9 and 10 are nonspecific. (B) Glycosylation status of Iff11. The secreted protein extracts from SBC121 (overexpressing Iff11-V5) were untreated (lanes 1 and 3) or treated with endo H (lane 2) or jack bean mannosidase (lane 4); soluble cytoplasmic extracts were included as a control (lane 5). Note that the low-molecular-weight band in each lane is nonspecific.
Modification by glycosylation is common for cell wall and secreted proteins. To access the nature of Iff11 modification the Iff11-V5 secreted protein extracts were treated with endoglycosidase H (endo H) and jack bean mannosidase (Fig. 1B). Consistent with bioinformatics analysis, Iff11 was found not to be modified by N glycosylation since there was no apparent shift in mobility after treatment with endo H. However, after treatment with jack bean mannosidase there was a clear increase in the mobility of Iff11-V5. This, in combination with the absence of any effect of endo H, implies that Iff11 is modified by O glycosylation. The increase in molecular mass from the predicted (53 kDa) to that observed (∼90 kDa) suggests extensive O glycosylation. C. albicans O-mannan comprises one to five α-1,2-linked mannose residues (18), the first added in the endoplasmic reticulum (ER) and subsequent residues in the Golgi. Jack bean mannosidase treatment would not release the first residue of O-mannan; hence, the mass of Iff11-V5 did not return to that predicted for the unmodified form. The mobility of Iff11-V5 after jack bean mannosidase treatment was close to that of the modification intermediate present in the soluble protein extract. This is suggestive of a potential block in the secretory pathway at the point of trafficking between the ER and Golgi in cells overexpressing Iff11-V5. No difference in the localization or modification of Iff11-V5 was apparent in yeast or hyphal cells (data not shown). In summary, localization studies suggest that Iff11 is an extensively O-glycosylated secreted protein.
Deletion of IFF11.
Both alleles of IFF11 were disrupted in strain CAI-4 by sequential gene deletion with the ura-blaster protocol (12), resulting in the deletion of the entire open reading frame. URA3 was introduced into the iff11Δ null mutant at the RPS1 locus to avoid potential problems with the level of URA3 expression (4). IFF11 was also reintroduced into the iff11Δ null mutant to act as a reintegrant control. In addition, IFF11-V5 was introduced into the null mutant to confirm function of V5-tagged Iff11, and IFF11 was also overexpressed under the control of the ENO1 (enolase) promoter.
Deletion of IFF11 had no effect on growth rate in complete (YEPD) or minimal (SD) medium. There was also no obvious effect on hyphal morphogenesis in response to serum, Lee's medium at pH 6.5 or RPMI 1640 medium. However, there was a delay in developing the classic colony morphology displayed on spider medium, and this was restored to wild type in the reintegrant strain. There was also no change in antifungal susceptibility or the carbon assimilation profile. Since secreted proteins normally express an enzymatic function, we screened the iff11Δ null mutant and IFF11-overexpressing strains and secreted fractions for a range of activities. No change was identified in protease activity, exoglucanase activity, or hydrolytic enzyme activities screened by API-ZYM.
Cell wall sensitivity.
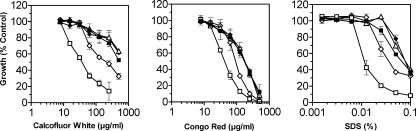
To determine the effect of deleting IFF11 on cell wall integrity, the sensitivity to a range of compounds associated with cell wall defects was determined. The iff11Δ null mutant was hypersensitive to the cell wall perturbing agents Calcofluor White, Congo red, and SDS (Fig. 2). The sensitivity was returned to wild-type levels in the iff11Δ+IFF11 reintegrant. There was no change in sensitivity to high-salt conditions, indicating that the mutant was not osmotically fragile. There was also no change in sensitivity to hygromycin B, vanadate, or tunicamycin, which would have been indicative of glycosylation defects, or to caffeine, which is usually associated with cell signaling defects. Overexpression of IFF11 in the null mutant background rescued cell wall sensitivity to wild-type levels and did not affect the overall level of resistance. The overexpression of IFF11-V5 in the null mutant background also rescued cell wall sensitivity although it was not fully restored to wild-type levels, indicating that insertion of the V5 tag affected Iff11 function. However, the rescue of the null mutant was sufficient to provide confidence in the localization studies.
FIG. 2.
Sensitivity of the iff11Δ null mutant to cell wall perturbing agents. Sensitivity was tested quantitatively by serial dilution of agents in multiwell plates. Agents to which iff11Δ displayed hypersensitivity (Calcofluor White, Congo red, and SDS) are shown. Strains tested were wild type (▪), iff11Δ (□), reintegrant (▵), iff11Δ-overexpressing Iff11 (⧫) and iff11Δ-overexpressing Iff11-V5 (⋄). Error bars indicate the means ± the standard deviation.
Since Iff11 is secreted we wanted to test whether it could function in trans on the iff11Δ null mutant strain. The null mutant was grown in the presence of wild-type Iff11 from concentrated spent medium and tested for cell wall sensitivity. No rescue of the iff11Δ null mutant was apparent after we provided Iff11 in this form (data not shown). The role of Iff11 in cell wall structure is therefore likely to be effected as the protein passes through the secretory pathway.
The iff11Δ null mutant had no defect in phosphomannan content, hydrophobicity, or adhesion to BEC. However, when IFF11 was overexpressed a decrease in phosphomannan content coupled with an increase in both hydrophobicity and adhesion to BEC was observed. This could suggest that Iff11 has a potential role in altering cell surface properties. However, since Iff11, an O-glycosylated protein, was being overexpressed this could also be an artifact of depleting the GDP-mannose pool required for glycosylation. This would lead to a phosphomannan defect that would be directly linked to an increase in hydrophobicity. Therefore, studies of cell wall and secreted proteins utilizing overexpression systems must be interpreted with care.
Attenuated virulence of iff11Δ cells.
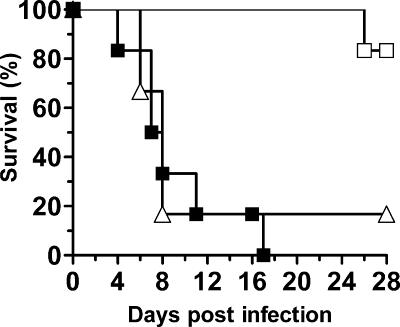
The virulence of the iff11Δ null mutant compared to wild-type and reintegrant controls was assessed in a murine model of systemic infection. The iff11Δ null mutant was clearly attenuated in virulence (Fig. 3 and Table 4, log-rank test; P < 0.001) with a mean survival time of 27.7 days compared to 9 days for the wild-type control. The virulence of the reintegrant control was restored in terms of both mean survival time and organ burdens. In mice infected with the iff11Δ null mutant there were a significant number of Candida-negative organs (58% kidneys, 83% brains). However, where infection was detectable the organ burdens were comparable to those seen in mice infected with the wild-type control strain (Mann-Whitney U test, P = 0.052). When survival-standardized infections were carried out, based on a challenge dose that results in similar mean survival times for all strains tested (22), tissue burdens of mice infected with the iff11Δ null mutant were comparable to those infected with the wild-type control strain. This suggests that once an infection is established Iff11 is not required, suggesting that it acts in the early stages of the infection process.
FIG. 3.
Attenuation of virulence in the iff11Δ null mutant. The wild-type (▪), iff11Δ (□), and reintegrant (▵) strains were tested for virulence in a mouse model of systemic infection. Six mice per strain were intravenously infected with 2 × 104 CFU/g (body weight).
TABLE 4.
Mean survival times and organ burdens for groups of six BALB/c mice infected with the iff11Δ null mutant and control strains
| Genotype (strain) | Mean ± SD
|
||
|---|---|---|---|
| Survival time (days) | Kidney burden (log10 CFU/g) | Brain burden (log10 CFU/g) | |
| IFF11 (NGY152) | 9.0 ± 4.5 | 6.3 ± 0.6 | 2.1 ± 0.7 |
| iff11Δ (SBC102) | 27.7 ± 0.8 | 3.2 ± 2.1 | 1.5 ± 0.4 |
| iff11Δ + IFF11 (SBC104) | 10.7 ± 8.5 | 6.0 ± 0.3 | 2.2 ± 0.8 |
Concluding remarks.
The IFF gene family is the largest family of cell wall related proteins encoded by C. albicans. Here we have demonstrated one member of the family, Iff11, is an O-glycosylated secreted protein. The other family members are presumably attached to the cell wall or membrane since they contain potential GPI-anchor signals or transmembrane domains. Deletion of IFF11 resulted in a defective cell wall, highlighted by hypersensitivity to cell wall-perturbing agents. That Iff11 is secreted and not maintained in the cell wall and the null mutant has a defective wall suggests that the protein carries out an enzymatic rather than a structural role, although we were unable to determine the nature of the enzyme activity despite tests with an extensive range of substrates. In addition, since Iff11 cannot be provided in trans its function is exerted as it passes through the secretory pathway or cell wall upon release. Understanding the function and role of cell wall-modifying enzymes is critical to the future understanding of cell wall assembly.
The iff11Δ null mutant was clearly highly attenuated in virulence in a mouse model of systemic infection. Further studies, using a survival-standardized infection model, suggest that the defect in the null mutant is in initially establishing an infection. Iff11 therefore does not display functional redundancy with the other family members and was the ideal choice to characterize the role of the gene family. The importance of Iff11 for virulence, coupled with the presence of the gene family in C. albicans and other pathogenic Candida species, also raises the possibility of this family as a potential antifungal or vaccine target.
Acknowledgments
This study was supported by grants 063204 and 080088 from the Wellcome Trust and PG01 from the British Society for Antimicrobial Chemotherapy to F.C.O., A.J.P.B., and N.A.R.G.
We thank Helen Gordon for skilled technical assistance.
Editor: A. Casadevall
Footnotes
Published ahead of print on 19 March 2007.
REFERENCES
- 1.Bailey, D. A., P. J. Feldmann, M. Bovey, N. A. Gow, and A. J. Brown. 1996. The Candida albicans HYR1 gene, which is activated in response to hyphal development, belongs to a gene family encoding yeast cell wall proteins. J. Bacteriol. 178:5353-5360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bates, S., H. B. Hughes, C. A. Munro, W. P. Thomas, D. M. MacCallum, G. Bertram, A. Atrih, M. A. Ferguson, A. J. Brown, F. C. Odds, and N. A. Gow. 2006. Outer chain N-glycans are required for cell wall integrity and virulence of Candida albicans. J. Biol. Chem. 281:90-98. [DOI] [PubMed] [Google Scholar]
- 3.Bates, S., D. M. MacCallum, G. Bertram, C. A. Munro, H. B. Hughes, E. T. Buurman, A. J. Brown, F. C. Odds, and N. A. Gow. 2005. Candida albicans Pmr1p, a secretory pathway P-type Ca2+/Mn2+-ATPase, is required for glycosylation and virulence. J. Biol. Chem. 280:23408-23415. [DOI] [PubMed] [Google Scholar]
- 4.Brand, A., D. M. MacCallum, A. J. Brown, N. A. Gow, and F. C. Odds. 2004. Ectopic expression of URA3 can influence the virulence phenotypes and proteome of Candida albicans but can be overcome by targeted reintegration of URA3 at the RPS10 locus. Eukaryot. Cell 3:900-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calderone, R. A. (ed.). 2002. Candida and candidiasis. ASM Press, Washington, DC.
- 6.Calderone, R. A., and N. A. R. Gow. 2002. Host recognition by Candida species, p. 67-86. In R. A. Calderone (ed.), Candida and candidiasis. ASM Press, Washington, DC.
- 7.Chauhan, N., D. Li, P. Singh, R. A. Calderone, and M. Kruppa. 2002. The cell wall of Candida spp., p. 159-175. In R. A. Calderone (ed.), Candida and candidiasis. ASM Press, Washington, DC.
- 8.Copping, V. M. S., C. J. Barelle, B. Hube, N. A. R. Gow, A. J. P. Brown, and F. C. Odds. 2005. Exposure of Candida albicans to antifungal agents affects expression of SAP2 and SAP9 secreted proteinase genes. J. Antimicrob. Chemother. 55:645-654. [DOI] [PubMed] [Google Scholar]
- 9.d'Enfert, C., S. Goyard, S. Rodriguez-Arnaveilhe, L. Frangeul, L. Jones, F. Tekaia, O. Bader, A. Albrecht, L. Castillo, A. Dominguez, J. F. Ernst, C. Fradin, C. Gaillardin, S. Garcia-Sanchez, P. de Groot, B. Hube, F. M. Klis, S. Krishnamurthy, D. Kunze, M. C. Lopez, A. Mavor, N. Martin, I. Moszer, D. Onesime, J. Perez Martin, R. Sentandreu, E. Valentin, and A. J. Brown. 2005. CandidaDB: a genome database for Candida albicans pathogenomics. Nucleic Acids Res. 33:D353-D357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Groot, P. W., A. D. de Boer, J. Cunningham, H. L. Dekker, L. de Jong, K. J. Hellingwerf, C. de Koster, and F. M. Klis. 2004. Proteomic analysis of Candida albicans cell walls reveals covalently bound carbohydrate-active enzymes and adhesins. Eukaryot. Cell 3:955-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Groot, P. W., K. J. Hellingwerf, and F. M. Klis. 2003. Genome-wide identification of fungal GPI proteins. Yeast 20:781-796. [DOI] [PubMed] [Google Scholar]
- 12.Fonzi, W. A., and M. Y. Irwin. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134:717-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hobson, R. P., C. A. Munro, S. Bates, D. M. MacCallum, J. E. Cutler, S. E. Heinsbroek, G. D. Brown, F. C. Odds, and N. A. Gow. 2004. Loss of cell wall mannosylphosphate in Candida albicans does not influence macrophage recognition. J. Biol. Chem. 279:39628-39635. [DOI] [PubMed] [Google Scholar]
- 14.Hoyer, L. L. 2001. The ALS gene family of Candida albicans. Trends Microbiol. 9:176-180. [DOI] [PubMed] [Google Scholar]
- 15.Klis, F. M., P. de Groot, and K. Hellingwerf. 2001. Molecular organization of the cell wall of Candida albicans. Med. Mycol. 39(Suppl. 1):1-8. [PubMed] [Google Scholar]
- 16.Lee, K. L., H. R. Buckley, and C. C. Campbell. 1975. An amino acid liquid synthetic medium for the development of mycelial and yeast forms of Candida albicans. Sabouraudia 13:148-153. [DOI] [PubMed] [Google Scholar]
- 17.Liu, H., J. Kohler, and G. R. Fink. 1994. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science 266:1723-1726. [DOI] [PubMed] [Google Scholar]
- 18.Munro, C. A., S. Bates, E. T. Buurman, H. B. Hughes, D. M. Maccallum, G. Bertram, A. Atrih, M. A. Ferguson, J. M. Bain, A. Brand, S. Hamilton, C. Westwater, L. M. Thomson, A. J. Brown, F. C. Odds, and N. A. Gow. 2005. Mnt1p and Mnt2p of Candida albicans are partially redundant alpha-1,2-mannosyltransferases that participate in O-linked mannosylation and are required for adhesion and virulence. J. Biol. Chem. 280:1051-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murad, A. M., P. R. Lee, I. D. Broadbent, C. J. Barelle, and A. J. Brown. 2000. CIp10, an efficient and convenient integrating vector for Candida albicans. Yeast 16:325-327. [DOI] [PubMed] [Google Scholar]
- 20.Netea, M. G., N. A. R. Gow, C. A. Munro, S. Bates, C. Collins, G. Ferwerda, R. P. Hobson, G. Bertram, H. B. Hughes, T. Jansen, L. Jacobs, E. T. Buurman, K. Gijzen, D. L. Williams, R. Torensma, J. W. M. Van der Meer, A. McKinnon, F. C. Odds, A. J. P. Brown, and B.-J. Kullberg. 2006. Immune sensing of Candida albicans: cooperative recognition of mannans and glucan by lectin and Toll-like receptors. J. Clin. Investig. 6:1642-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Odds, F. C. 1988. Candida and candidosis, 2nd ed. Bailliere-Tindall, Ltd., London, United Kingdom.
- 22.Odds, F. C., L. Van Nuffel, and N. A. Gow. 2000. Survival in experimental Candida albicans infections depends on inoculum growth conditions as well as animal host. Microbiology 146:1881-1889. [DOI] [PubMed] [Google Scholar]
- 23.Odds, F. C., and C. E. Webster. 1988. Effects of azole antifungals in vitro on host/parasite interactions relevant to Candida infections. J. Antimicrob. Chemother. 22:473-481. [DOI] [PubMed] [Google Scholar]
- 24.Pappas, P. G., J. H. Rex, J. Lee, R. J. Hamill, R. A. Larsen, W. Powderly, C. A. Kauffman, N. Hyslop, J. E. Mangino, S. Chapman, H. W. Horowitz, J. E. Edwards, and W. E. Dismukes. 2003. A prospective observational study of candidemia: epidemiology, therapy, and influences on mortality in hospitalized adult and pediatric patients. Clin. Infect. Dis. 37:634-643. [DOI] [PubMed] [Google Scholar]
- 25.Romani, L. 2002. Immunology of invasive candidiasis, p. 223-242. In R. A. Calderone (ed.), Candida and candidiasis. ASM Press, Washington DC.
- 26.Ruiz-Herrera, J., M. V. Elorza, E. Valentin, and R. Sentandreu. 2006. Molecular organization of the cell wall of Candida albicans and its relation to pathogenicity. FEMS Yeast Res. 6:14-29. [DOI] [PubMed] [Google Scholar]
- 27.Schaller, M., C. Borelli, H. C. Korting, and B. Hube. 2005. Hydrolytic enzymes as virulence factors of Candida albicans. Mycoses 48:365-377. [DOI] [PubMed] [Google Scholar]
- 28.Sundstrom, P. 2002. Adhesion in Candida spp. Cell. Microbiol. 4:461-469. [DOI] [PubMed] [Google Scholar]
- 29.Vediyappan, G., J. Bikandi, R. Braley, and W. L. Chaffin. 2000. Cell surface proteins of Candida albicans: preparation of extracts and improved detection of proteins. Electrophoresis 21:956-961. [DOI] [PMC free article] [PubMed] [Google Scholar]