Abstract
Human monocytic ehrlichiosis, one of the most frequent life-threatening tick-borne zoonoses, is caused by Ehrlichia chaffeensis that lacks endotoxin and peptidoglycan. While sequence polymorphisms in several genes in E. chaffeensis strains have been reported, global genomic divergence and biological differences among strains are unknown. The objectives of the present study were to compare the genome sequences of strains of E. chaffeensis and to examine the virulence potentials of the strains with defined genome sequences. Genomic DNA was extracted from purified E. chaffeensis strains Wakulla and Liberty, and comparative genome hybridization was performed using a densely tiled microarray of 147,027 chromosome positions of the E. chaffeensis strain Arkansas genome. The results revealed that 4,663 and 5,325 positions in the chromosomes of strains Wakulla and Liberty, respectively, were different from those in the chromosome of strain Arkansas, including three common major polymorphic chromosomal regions. Of various functional categories, the differences were most concentrated in genes predicted to encode cell envelope proteins. Of all the open reading frames (ORFs), 21 omp-1 (p28 gene) paralogs, nine genes encoding hypothetical proteins, two genes encoding ankyrin repeat proteins, and hemE contained the most differences. Several highly polymorphic ORFs were confirmed by sequencing. When the E. chaffeensis strains were inoculated into severe combined immunodeficiency mice, the order of the severity of clinical signs and the bacterial burden detected in mice was Wakulla > Liberty > Arkansas. Severe diffuse inflammation and granulomatous inflammation were evident in the livers of mice infected with strains Wakulla and Arkansas, respectively, but not in the livers of mice infected with strain Liberty. These results revealed distinct virulence phenotypes of E. chaffeensis strains with defined genome sequences.
Human monocytotropic ehrlichiosis (HME) is an emerging tick-borne zoonosis that was discovered in the United States in 1986 (22). HME is caused by infection with Ehrlichia chaffeensis, an obligatory intracellular gram-negative bacterium in the order Rickettsiales that was isolated in 1990 (7). By 1997, 47 states had reported a total of 742 probable or confirmed cases of HME to the Centers for Disease Control and Prevention (25). The average reported annual incidence of HME during 2001 and 2002 was 0.6 case per million people (8). Several prospective epidemiologic studies have documented an increasing incidence of human ehrlichiosis (4, 13, 30). HME also has been reported in other parts of the globe, including Europe, Mexico, Argentina, Mali, Israel, and Thailand. E. chaffeensis has been identified most commonly in the Lone Star tick (Amblyomma americanum) (2), and white-tailed deer is considered to be the major reservoir in the United States (10).
HME generally is characterized by fever, headache, myalgia, anorexia, and chills and frequently is accompanied by leukopenia, thrombocytopenia, anemia, and elevated levels of serum hepatic aminotransferases (32). The manifestations of HME are broad, however, ranging from asymptomatic infection to death (32). Especially in patients with AIDS or other immunosuppressive conditions, E. chaffeensis can cause overwhelming infection (23, 33, 39). The clinical signs are not induced by endotoxin, since E. chaffeensis, like other sequenced members of the family Anaplasmataceae, lacks genes for biosynthesis of lipid A and lipopolysaccharide and thus is incapable of producing endotoxin (17, 20).
Genetic diversity among E. chaffeensis human isolates initially was found in the sequence of the p28 (p28-19) gene encoding the 28-kDa major outer membrane protein (49). Further comparison of the p28-19 gene in other strains of E. chaffeensis led to proposal of three distinct groups of E. chaffeensis isolates (21). Cheng et al. (5) placed the E. chaffeensis isolates into three distinct genetic groups, groups I, II, and III, on the basis of restriction enzyme cleavage analysis and sequence data for the partial 28-kDa outer membrane protein multigene locus. E. chaffeensis isolates also differ in their numbers of repeat sequences within the p120 gene and the variable-length PCR target gene (33, 45). For example, the Wakulla strain has a six-repeat version of the variable-length PCR target gene (45), whereas the Arkansas and Liberty strains have four repeats. Although the genome of E. chaffeensis strain Arkansas has been sequenced (17), the global genomic divergence among E. chaffeensis strains has not been determined.
Pathogenic differences between strains have not been noted in patients with HME, since the present diagnostic methods cannot distinguish between strains. Furthermore, clinical outcomes are influenced by patient factors (age, underlying illness, and immunosuppressive conditions) and therapeutic interventions. As only a single strain (Arkansas) has been used so far for experimental infection of animals, there are no experimental data on biological or pathogenetic differences among E. chaffeensis strains. We thought that biological diversity among E. chaffeensis strains might be demonstrated clearly if the pathogenesis of E. chaffeensis strains with distinct genomic sequences could be compared under controlled conditions in an experimental animal model.
Immunocompetent mice clear E. chaffeensis Arkansas infection within 2 weeks and do not develop clinical signs (47). Consequently, severe combined immunodeficiency (SCID) mice have been used for studies of E. chaffeensis infection and pathogenesis. The SCID mouse model recapitulates several features of clinical signs and histopathology of the human disease (HME), including wasting, tissue and cell tropism, extensive tissue inflammation, splenomegaly, lymphadenopathy, and liver granulomas and necroses (47). Hence, we used SCID mice to compare the virulence potentials of the three E. chaffeensis human isolates with defined genome sequences. The phenotype and the genotype data for a given strain were obtained from bacteria having the same passage history. The results provide novel insights into the pathogenesis of E. chaffeensis strains and suggest new investigation targets for potential virulence factors.
MATERIALS AND METHODS
Bacteria and cell lines.
E. chaffeensis strains Arkansas (from a patient residing in Arkansas) and Wakulla and Liberty (from patients residing in Florida) were kindly provided by C. Paddock and W. L. Nicholson at the Centers for Disease Control and Prevention, Atlanta, GA. E. chaffeensis strains were propagated in DH82 cells as previously described (38). Infected cells were lysed by nitrogen cavitation, and liberated bacteria were purified by Percoll density gradient centrifugation as previously described (27, 28).
DNA tiling microarray and sequencing.
Genomic DNA was extracted from the purified organisms using a QIamp DNA blood mini kit (QIAGEN, Valencia, CA). The contamination level of host cell DNA in the purified bacterial DNA preparation was less than 1% as determined by PCR using host-specific glyceraldehyde-3-phosphate dehydrogenase primers as previously described (17). The high-density E. chaffeensis DNA microarray was constructed at NimbleGen Systems (Madison, WI). Cy3- and Cy5-labeled probes were synthesized from genomic DNA (1 μg) of Arkansas, Wakulla, and Liberty using a randomly primed reaction as previously described (1). Labeled genomic DNA was hybridized to arrays in 1× NimbleGen hybridization buffer (NimbleGen); the arrays were scanned at a 5-μm resolution using a Genepix 4000b scanner (Axon Instruments, Union City, CA); and pixel intensities were extracted using NimbleScan software (NimbleGen). The genomic loci concerned were amplified by PCR with several primer sets designed based on conserved sequences for each locus. After purification with a QIAquick gel extraction kit (QIAGEN), the PCR products were used for direct sequencing.
Infection of mice.
Three- to 4-week-old male ICR-scid mice were obtained from Taconic (Tarrytown, NY) and infected intraperitoneally with 106 E. chaffeensis-infected DH82 cells (typically >95% infected) containing approximately the same number of bacteria. The number of bacteria was estimated as previously described (48). Determination of the viability of organisms in infected DH82 cells was performed with a LIVE/DEAD BacLight bacterial viability kit (Molecular Probes, Eugene, OR). Infected DH82 cells were harvested, washed, and resuspended in phosphate-buffered saline (137 mM NaCl, 2.68 mM KCl, 1.47 mM KH2PO4, 8.06 mM Na2HPO4), and 0.5 ml was injected into the peritoneal cavity with a 26-gauge needle. Heparinized blood specimens were collected from the submandibular sinus of mice anesthetized by inhalation with CO2. The experiment was terminated at day 15 postinfection (PI) according to the Institutional Animal Care and Use Committee regulations, since mice inoculated with the Wakulla strain became moribund by this time. The tissues were harvested and fixed in Histochoice (Amresco, Solon, OH) for histopathologic examination or were stored in RNAlater (QIAGEN) for PCR analyses. The levels of total serum albumin and serum aminotransferases (aspartate and alanine aminotransferase) were measured with a 911 automatic analyzer (Hitachi, Tokyo, Japan) at 15 days PI.
Real-time PCR and reverse transcriptase PCR.
DNA was extracted from blood, spleen, and liver specimens with a QIAamp blood kit (QIAGEN). To determine the numbers of E. chaffeensis bacteria, a real-time PCR assay was performed in triplicate to amplify the E. chaffeensis 16S rRNA gene as previously described (6). Total RNA was extracted from the spleen and liver using an RNeasy kit (QIAGEN). Total cellular RNA (2 μg) was treated with DNase I (Invitrogen, Carlsbad, CA) and was reverse transcribed with SuperScript III reverse transcriptase (Invitrogen), and the cDNA was used in the PCR as previously described (6).
Statistical analysis.
The unpaired two-tailed t test was performed to test the significance of differences between different groups. A P value of <0.05 was considered significant.
RESULTS
CGH.
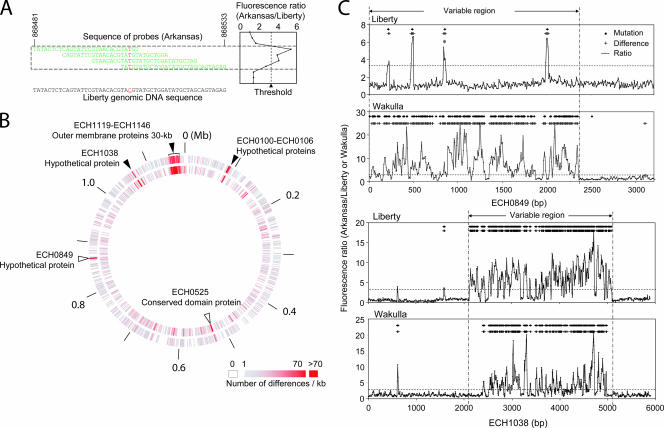
Based on the group designation of Cheng et al. (5) and the availability of strains, we selected the genomic sequences of the Wakulla (group II) and Liberty (group III) strains of E. chaffeensis for comparison with the genomic sequence of the Arkansas strain (group I). For comparative genome hybridization (CGH) analysis, a high-density DNA microarray was constructed by selecting 29-mer oligonucleotides every eight bases in the genome sequence of E. chaffeensis Arkansas (GenBank accession no. CP000236), resulting in a total of 147,027 chromosome positions on each strand (GEO accession no. GPL4360). With this approach, each nucleotide in the genome was covered with a total of six or eight overlapping oligonucleotides in the array (Fig. 1A [oligonucleotides for only one DNA strand are shown]). We purified bacteria from infected host cells as previously described (17) so that we could isolate E. chaffeensis genomic DNA that was almost free of host DNA. Arkansas genomic DNA was labeled with Cy3. Wakulla and Liberty DNA probes were labeled with Cy5, and each of the probes was hybridized with the Arkansas oligonucleotides on the microarray. Cy3/Cy5 ratios for each pair (Arkansas/Wakulla and Arkansas/Liberty) were determined across the entire 147,027 chromosome positions on both DNA strands, and the means of the two strands at each position were plotted (Fig. 1A) (GEO accession no. GSM137280 and GSM137281). Overall, the gene content was highly conserved (>99%) compared to the conservation observed in similar studies done with Helicobacter pylori (40) and Staphylococcus aureus (14). The median plus two times the standard deviation of the Cy3/Cy5 ratios for each pair of all chromosome positions was used as a threshold to identify differences at each nucleotide position in the E. chaffeensis sequence in the array. The threshold was 2.78 for Arkansas/Wakulla and 3.29 for Arkansas/Liberty. Wakulla differed from Arkansas at 4,663 positions in the genome (3.2%), and Liberty differed from Arkansas at 5,325 positions (3.6%) (Fig. 1B). These differences were distributed in 361 open reading frames (ORFs) and 133 intergenic regions of Wakulla and in 357 ORFs and 200 intergenic regions of Liberty. The numbers of differences in various functional categories divided by the total length of the ORFs in the categories are shown in Table 1. In the genomes of both Wakulla and Liberty, genes encoding cell envelope proteins (total length, ∼46 kb) were most different from the genes in the Arkansas strain, whereas genes for housekeeping and metabolic functions had fewer differences. In particular, a 30-kb contiguous region containing 22 OMP-1 (p28) gene paralogs, including the six OMP-1 (p28) gene paralogs reported previously by Cheng et al. (5), had many mutations when Wakulla and Arkansas were compared and when Liberty and Arkansas were compared (Fig. 1B and Table 1). Of all the ORFs, 21 omp-1 (p28 gene) paralogs, nine genes encoding hypothetical proteins, two genes encoding ankyrin repeat proteins, and hemE contained the most differences (Table 2).
FIG. 1.
CGH of E. chaffeensis strains. (A) Multiple overlapping probes cover each nucleotide. A mutation at a site, indicated by red, perturbs hybridization of the sample to all probes. The representative example is a mutation in Liberty ECH_0849, indicated by an asterisk in panel C. The sequence of probes on one of the strands is represented. (B) E. chaffeensis strains Wakulla and Liberty were compared with the Arkansas strain using an array containing a total of 294,050 29-mer probes spaced, on average, every 8 bp throughout the genome of E. chaffeensis Arkansas. Inner circle, Arkansas versus Liberty; outer circle, Arkansas versus Wakulla. The open and solid arrowheads indicate remarkably different regions. (C) ECH_0849 and ECH_1038: CGH results revealed different positions and actual mutations. Sequencing was used to validate the sensitivity and specificity of CGH. The horizontal dotted line indicates the threshold of the Cy3/Cy5 ratio used to indicate a difference. The diamonds and plus signs indicate positions of mutated bases and probes with differences, respectively. The variable regions of ECH_0849 and ECH_1038 in the Wakulla and Liberty strains compared with strain Arkansas are between two vertical dashed and dotted lines.
TABLE 1.
Distribution of differences from strain Arkansas by functional category
| Category | Total length (bp) | Difference from strain Arkansas in strain:
|
Avg rate (kb−1) | |||
|---|---|---|---|---|---|---|
| Wakulla
|
Liberty
|
|||||
| No. | Rate (kb−1) | No. | Rate (kb−1) | |||
| Cell envelope | 45,936 | 1,342 | 29.21 | 1,511 | 32.89 | 31.05 |
| rRNA | 4,382 | 89 | 20.31 | 0 | 0.00 | 10.16 |
| Hypothetical proteins | 134,832 | 1,132 | 8.40 | 1,031 | 7.65 | 8.02 |
| Disrupted reading frame | 171 | 0 | 0.00 | 2 | 11.70 | 5.85 |
| Conserved hypothetical proteins | 88,142 | 364 | 4.13 | 642 | 7.28 | 5.71 |
| Intergenic regions | 230,774 | 592 | 2.57 | 1,108 | 4.80 | 3.68 |
| Unknown function | 93,622 | 251 | 2.68 | 318 | 3.40 | 3.04 |
| Central intermediary metabolism | 1,698 | 3 | 1.77 | 4 | 2.36 | 2.06 |
| Protein synthesis | 91,797 | 165 | 1.80 | 195 | 2.12 | 1.96 |
| Mobile and extrachromosomal element functions | 4,763 | 9 | 1.89 | 9 | 1.89 | 1.89 |
| Biosynthesis of cofactors, prosthetic groups, and carriers | 51,912 | 119 | 2.29 | 53 | 1.02 | 1.66 |
| tRNA | 2,860 | 4 | 1.40 | 5 | 1.75 | 1.57 |
| Transcription | 28,137 | 58 | 2.06 | 22 | 0.78 | 1.42 |
| Cellular processes | 29,157 | 36 | 1.23 | 46 | 1.58 | 1.41 |
| Energy metabolism | 88,473 | 148 | 1.67 | 98 | 1.11 | 1.39 |
| Transport and binding proteins | 34,129 | 32 | 0.94 | 57 | 1.67 | 1.30 |
| Regulatory functions | 12,315 | 9 | 0.73 | 23 | 1.87 | 1.30 |
| Fatty acid and phospholipid metabolism | 15,012 | 23 | 1.53 | 15 | 1.00 | 1.27 |
| Amino acid biosynthesis | 24,183 | 31 | 1.28 | 29 | 1.20 | 1.24 |
| Protein fate | 96,477 | 131 | 1.36 | 82 | 0.85 | 1.10 |
| DNA metabolism | 61,412 | 83 | 1.35 | 52 | 0.85 | 1.10 |
| Purines, pyrimidines, nucleosides, and nucleotides | 38,767 | 50 | 1.29 | 31 | 0.80 | 1.04 |
| Structural RNA | 746 | 0 | 0.00 | 0 | 0.00 | 0.00 |
TABLE 2.
Most different ORFs in CGH analyses of E. chaffeensis strains
| ORF | Gene | Direction | Length (bp) | Protein | No. of differences in CGH
|
||
|---|---|---|---|---|---|---|---|
| Wakulla | Liberty | Total | |||||
| ECH_1038 | − | 5,892 | Hypothetical protein | 201 | 281 | 482 | |
| ECH_0106 | + | 2,142 | Hypothetical protein | 176 | 184 | 360 | |
| ECH_0849 | + | 3,210 | Hypothetical protein | 234 | 13 | 247 | |
| ECH_0525 | + | 2,001 | Conserved domain protein | 98 | 135 | 233 | |
| ECH_0653 | − | 12,942 | Ankyrin repeat protein | 75 | 115 | 190 | |
| ECH_1127 | omp-1V | + | 840 | Major outer membrane protein OMP-1V | 78 | 95 | 173 |
| ECH_1142 | omp-1F | + | 843 | Major outer membrane protein OMP-1F | 88 | 84 | 172 |
| ECH_1126 | omp-1U | + | 888 | Major outer membrane protein OMP-1U | 64 | 104 | 168 |
| ECH_1123 | omp-1Q | + | 894 | Major outer membrane protein OMP-1Q | 74 | 86 | 160 |
| ECH_1133 | omp-1H | + | 897 | Major outer membrane protein OMP-1H | 71 | 86 | 157 |
| ECH_1125 | omp-1T | + | 819 | Major outer membrane protein OMP-1T | 61 | 91 | 152 |
| ECH_1129 | omp-1W | + | 852 | Major outer membrane protein OMP-1W | 58 | 93 | 151 |
| ECH_1132 | omp-1S | + | 876 | Major outer membrane protein OMP-1S | 71 | 80 | 151 |
| ECH_1131 | omp-1Y | + | 858 | Major outer membrane protein OMP-1Y | 75 | 75 | 150 |
| ECH_1124 | omp-1P | + | 858 | Major outer membrane protein OMP-1P | 68 | 77 | 145 |
| ECH_1135 | omp-1A | + | 882 | Major outer membrane protein OMP-1A | 45 | 95 | 140 |
| ECH_1134 | omp-1Z | + | 903 | Major outer membrane protein OMP-1Z | 44 | 94 | 138 |
| ECH_1143 | p28 | + | 846 | Major outer membrane protein P28 | 68 | 70 | 138 |
| ECH_1130 | omp-1X | + | 828 | Major outer membrane protein OMP-1X | 57 | 79 | 136 |
| ECH_1136 | omp-1B | + | 852 | Major outer membrane protein OMP-1B | 47 | 77 | 124 |
| ECH_1137 | omp-1C | + | 843 | Major outer membrane protein OMP-1C | 84 | 39 | 123 |
| ECH_1121 | omp-1N | + | 855 | Major outer membrane protein Omp-1N | 50 | 64 | 114 |
| ECH_1128 | + | 591 | Conserved hypothetical protein | 38 | 70 | 108 | |
| ECH_0684 | − | 4,392 | Ankyrin repeat protein | 59 | 44 | 103 | |
| ECH_1035 | − | 548 | Conserved hypothetical protein, Deg | 33 | 61 | 94 | |
| ECH_1140 | omp-1E | + | 837 | Major outer membrane protein OMP-1E | 75 | 9 | 84 |
| ECH_1144 | p28-1 | − | 816 | Major outer membrane protein P28-1 | 51 | 26 | 77 |
| ECH_1119 | omp-1M | + | 897 | Major outer membrane protein OMP-1M | 28 | 30 | 58 |
| ECH_1044 | − | 357 | Conserved hypothetical protein | 31 | 21 | 52 | |
| ECH_0150 | − | 2,019 | Hypothetical protein | 35 | 13 | 48 | |
| ECH_0282 | + | 675 | Hypothetical protein | 28 | 18 | 46 | |
| ECH_0030 | hemE | − | 1,005 | Uroporphyrinogen decarboxylase | 27 | 9 | 36 |
| ECH_1139 | omp-1D | + | 861 | Major outer membrane protein OMP-1D | 23 | 13 | 36 |
The following three major genomic regions were consistently different when Arkansas was compared with Wakulla or Liberty: the 30-kb region from ECH_1119 to ECH_1146 encoding 22 OMP-1 paralogs mentioned above; ECH_1038 (GenBank accession no. YP_507823), which encodes a hypothetical 222,638-Da protein; and the region from ECH_0100 to ECH_0106 encoding hypothetical proteins (Fig. 1B and Table 2). Wakulla had a deletion of ECH_0100 to ECH_0104. The ECH_0849 ORFs (GenBank accession no. YP_507645), which encode hypothetical 121,401-Da proteins, differed at many positions in Wakulla and Arkansas but were similar in Liberty and Arkansas (Fig. 1C). Both ECH_1038 and ECH_0849 were transcribed by the Wakulla and Liberty strains in DH82 cell cultures and in the spleens of SCID mice (data not shown), implying that these ORFs are functional genes.
To confirm the CGH data, the ECH_0849, ECH_1038, and hemE genes of Wakulla and Liberty were sequenced (GenBank accession no. DQ888319, DQ912824, DQ915166, DQ915979, DQ924562, and DQ924563). The results revealed by CGH for different chromosome positions and actual mutations detected by sequencing are similar for ECH_0849 and ECH_1038 of strains Wakulla and Liberty (Fig. 1C). A comparison of the sequences indicated that the sensitivities of probes on the array for detection of differences between strains Wakulla and Arkansas and between strains Liberty and Arkansas were 81.0 and 87.8%, respectively (see Table S1 in the supplemental material). The specificities of probes on the array for detection of differences between strains Arkansas and Wakulla and between strains Arkansas and Liberty were 98.0 and 99.4%, respectively. The sensitivity and specificity of probes for strains Wakulla and Liberty combined were 83.6 and 98.7%, respectively (see Table S1 in the supplemental material). The net sensitivity of detection of mutations (percent detection) in the sequences in Wakulla and Liberty combined was 97.3% (see Table S1 in the supplemental material). CGH and sequencing results revealed the presence of polymorphic and conserved regions within ECH_0849 and ECH_1038 (Fig. 1C).
ECH_1038 is paralogous to ECH_1037 and ECH_1036 in the same orientation, and these three ORFs and their synteny are conserved among sequenced Ehrlichia species, including Ehrlichia ruminantium Welgevonden (Erum7970, Erum7960, Erum7950) and Ehrlichia canis Jake (Ecaj_0838, Ecaj0_835, Ecaj_0834). The ECH_0849 locus also is conserved among sequenced Ehrlichia species.
Clinical signs and bacterial burden.
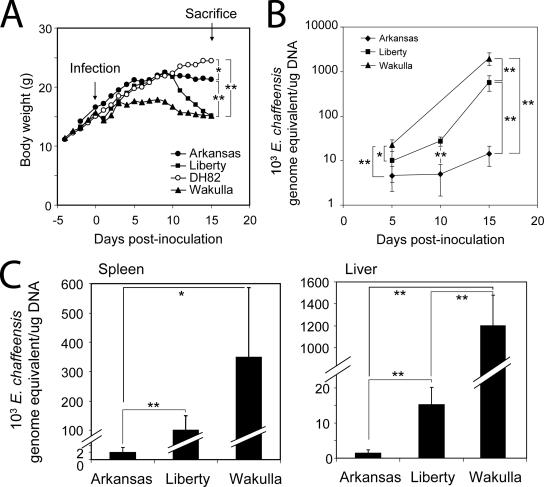
To compare the levels of pathogenesis of E. chaffeensis Liberty, Wakulla, and Arkansas, for each strain, five SCID mice were intraperitoneally inoculated with the same number of freshly harvested DH82 cells containing approximately the same number of viable bacteria. The numbers of Wakulla, Arkansas, and Liberty cells were 55 ± 7, 57 ± 2, and 68 ± 6 bacteria per DH82 cell, respectively. Retrospectively, regression analysis with Microsoft Excel showed that the correlation coefficient (r) was 0.932 for the bacterial genome equivalent (as determined by the real-time PCR) versus the bacterial count (see Fig. S1 in the supplemental material). The viabilities of bacteria used for inoculation were >99% as determined with the LIVE/DEAD BacLight bacterial viability kit. Although this kit is prescribed for determining the viability of extracellular bacteria, we found that it also is useful for determination of the viability of obligatory intracellular bacteria; live bacteria were stained green with SYTO9, and dead bacteria were stained red with propidium iodide (an example of a cell containing both live and dead bacteria is shown in Fig. S2 in the supplemental material). The y-axis intercepts (day 0) of the fitted curves for the Wakulla, Liberty, and Arkansas strains in blood were 2,430, 892, and 2,279 genome equivalents/μg DNA, respectively (Fig. 2B).
FIG. 2.
E. chaffeensis strains differ in virulence. (A) Body weights of mice inoculated with DH82 cells infected with E. chaffeensis Arkansas, Liberty, or Wakulla or with uninfected DH82 cells (control). The values are means (n = 5; on day 6 one mouse inoculated with Wakulla died and therefore the data for four mice in this group were used after day 7). One asterisk, P < 0.05 at day 15 PI (as determined by an unpaired two-tailed t test); two asterisks, P < 0.01 at day 15 PI (as determined by an unpaired two-tailed t test). (B) Bacterial loads in the blood of mice as determined by real-time PCR based on the 16S rRNA gene of E. chaffeensis (n = 5, except for mice inoculated with Wakulla after day 6 [n = 4]). One asterisk, P < 0.05; two asterisks, P < 0.01 (as determined by an unpaired two-tailed t test). (C) Bacterial loads on day 15 PI in the spleens and livers of mice inoculated with E. chaffeensis, as determined by real-time PCR of the 16S rRNA gene of E. chaffeensis (n = 5, except for mice inoculated with Wakulla [n= 4]). One asterisk, P < 0.05; two asterisks, P < 0.01 (as determined by an unpaired two-tailed t test).
Mice inoculated with Wakulla and Liberty rapidly lost body weight starting on day 10 PI (Fig. 2A). In contrast, the body weight loss of mice inoculated with Arkansas occurred more slowly. Anorexia and wasting were evident with Wakulla-inoculated mice even earlier, as they stopped gaining body weight starting 4 days PI, while Liberty- and Arkansas-inoculated mice gained amounts of body weight similar to the amounts gained by mock-infected mice until day 10 PI (Fig. 2A).
As determined by real-time PCR, the numbers of Wakulla and Liberty bacteria in the blood increased exponentially (Fig. 2B). The numbers of Wakulla and Liberty bacteria in the blood increased about 90- and 60-fold, respectively, from day 5 to day 15 PI, whereas the number of Arkansas bacteria increased only 3-fold as determined by quantitative PCR (Fig. 2B). The modest increase in the bacterial load with the Arkansas strain in the present study was similar to the previous observation for C.B-17-scid mice inoculated with the Arkansas strain (47). On day 15 PI, the numbers of Wakulla and Liberty bacteria in the blood were approximately 140- and 40-fold greater than the number of Arkansas bacteria, respectively (Fig. 2B). The differences in bacterial loads between Liberty- and Wakulla-inoculated mice were more pronounced in the liver than in the spleen or blood. The livers of Wakulla- and Liberty-inoculated mice had approximately 900- and 10-fold more bacteria, respectively, than the livers of mice inoculated with Arkansas (Fig. 2C). The spleens of Wakulla- and Liberty-inoculated mice had 170- and 50-fold more bacteria, respectively, than the spleens of mice inoculated with Arkansas (Fig. 2C).
Liver pathology and clinical pathology.
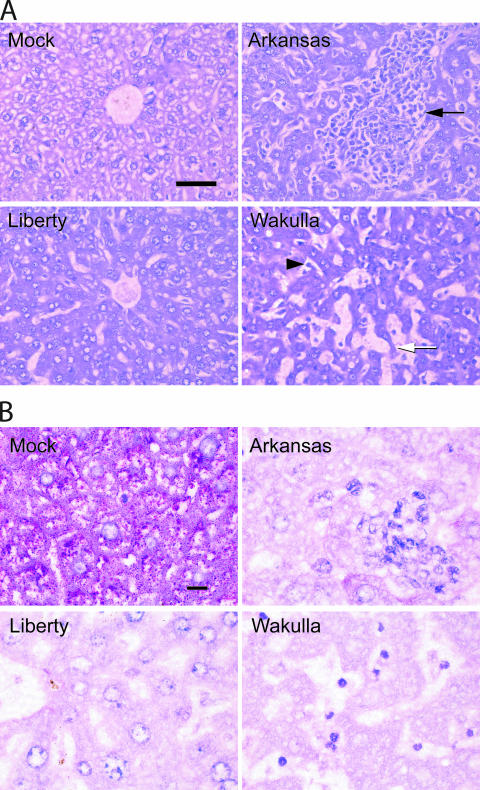
The liver is one organ where prominent pathological changes were demonstrated in patients with HME, in SCID mice infected with E. chaffeensis Arkansas, and in immunocompetent mice infected with the HF strain (Ixodes ovatus ehrlichia isolated from the I. ovatus tick in Japan [42]) (29, 31, 43, 47). The HF strain is an ehrlichial isolate that is most closely related to E. chaffeensis (the 16S rRNA sequence identity with E. chaffeensis Arkansas was 98.21% when 1,449 bp aligned base sequences were compared [42]). Hence, we compared the liver histopathologies in the four groups of mice. Compared with the livers of mock-inoculated control mice, parenchymal cells were much smaller and the sinusoidal space was larger in the livers of mice infected with any of the three E. chaffeensis strains (Fig. 3A). The sinusoidal irregular dilation was most prominent with the Wakulla strain, distorting liver cell cord arrangements (Fig. 3A). Severe granulomatous inflammatory leukocyte infiltration in the liver was evident with Arkansas infection, disorganizing liver cell cords, but coagulation necrosis was not observed (Fig. 3A). This is similar to the previous observations made with C.B-17-scid mice inoculated with the Arkansas strain (47) (Fig. 3A). Wakulla infection induced profound and diffuse leukocyte infiltration, but a large granulomatous aggregation of leukocytes was rare (Fig. 3A). Apoptotic cells with small nuclei and atrophy and degeneration of parenchymal cell cords were evident in the livers of mice inoculated with Wakulla (Fig. 3A). In contrast, the livers of mice inoculated with Liberty were almost free from leukocyte infiltration, granuloma, or degenerating parenchymal cells (Fig. 3A).
FIG. 3.
(A) Liver histopathology of mice inoculated with E. chaffeensis. Note the diffuse inflammatory cell infiltration in the sinusoids (arrowhead) in the mouse infected with the Wakulla strain, the severe granulomatous inflammation (arrow) in the mouse infected with the Arkansas strain, and the absence of lesions in the mouse infected with the Liberty strain. Severe and irregular dilatation of sinusoids and atrophy and degeneration of liver cell cords (arrow) were evident in the mouse infected with the Wakulla strain. Hematoxylin and eosin stain was used. Bar = 50 μm. (B) Liver glycogen depletion. The control mouse liver has massive glycogen accumulation, which is depleted in the mice infected with the three strains. PAS stain was used. Bar = 10 μm.
Since the cytoplasm of liver parenchymal cells was much smaller in three groups of infected mice, carbohydrate-specific periodic acid-Schiff (PAS) staining was performed to compare the glycogen amounts in the cytoplasm. PAS staining showed abundant granular glycogen accumulation in the cytoplasm of the parenchymal cells of the control mice. In contrast, glycogen was almost completely depleted in the parenchymal cells of mice infected with any of the three strains (Fig. 3B). The aspartate transaminase activities at day 15 PI were significantly elevated (575 ± 29 U/liter; n = 4) in the sera from mice infected with Wakulla but not in the sera from mice infected with Arkansas or Liberty compared to the activities in the sera from the control mice (100 to 200 ± U/liter; five mice in each group). The levels of serum aspartate transaminase in mice infected with Wakulla were similar to those reported for mice inoculated with the HF strain at days 7 and 9 PI just prior to death (43).
Anemia is one of clinical signs observed in human HME patients (32). Although anemia was not reported in any previous studies of SCID mice inoculated with the E. chaffeensis Arkansas strain, blood specimens taken from Wakulla-inoculated mice on day 15 PI had a lower hematocrit value (19.6% packed cell volume) and an increased number of reticulocytes, indicating severe anemia. In contrast, mice inoculated with the Arkansas or Liberty strain did not show significant anemia or reticulocytes in their blood. In addition, Wakulla-inoculated mice developed poor hemostasis, perhaps due to thrombocytopenia. One Wakulla-inoculated mouse died on day 6 PI, likely due to poor hemostasis after blood sample collection. Therefore, blood specimens could not be collected from the remaining Wakulla-inoculated mice until day 15 PI. Wakulla- and Arkansas-inoculated mice had six- to sevenfold-greater splenomegaly than mock-inoculated control mice, whereas Liberty-inoculated mice had two- to threefold-greater splenomegaly than mock-inoculated control mice (data not shown). These results indicated that the virulence and pathogenesis features of E. chaffeensis Wakulla and Liberty in SCID mice were distinct from each other and from those of Arkansas.
DISCUSSION
The present study demonstrated the utility of CGH using the densely tiled DNA microarray for comparison of E. chaffeensis strains. Like the genomic contents of other intracellular bacteria (3, 16, 17, 19), the genomic contents were found to be fairly conserved among E. chaffeensis strains. Thus, this type of microarray is more suitable than a gene content-based microarray for discerning diversity among E. chaffeensis strains. The high sensitivity and specificity demonstrated in the present study ensured the reliability of the data from CGH. Of the various functional categories, differences among the three E. chaffeensis strains were most concentrated in genes predicted to encode cell envelope proteins. Bacterial envelope proteins are at the host-bacterium interface and are involved in binding, internalization, signal transduction, nutrient transport, and immunoavoidance. Strain variation among envelope proteins of E. chaffeensis is expected to influence some of these functions and, consequently, the ability of bacteria to infect and proliferate in the host cells and cause diseases. Therefore, comparative functional studies of the products of the polymorphic envelope genes may shed light on E. chaffeensis pathogenesis in the future. Other polymorphic genes in E. chaffeensis strains are genes encoding ankyrin repeat proteins. Genomes of Ehrlichia and Anaplasma species encode several ankyrin repeat proteins per genome (17), one of which has been reported to interact with host cell chromosomes (34) and a host cell phosphatase (18). Strain polymorphisms of ankyrin repeat proteins may alter the host cell transcription or signaling process and, therefore, the survival of these obligatory intracellular bacteria.
Several genes highly polymorphic in E. chaffeensis strains, such as ECH_0100 to ECH_0106, ECH_0849, and ECH_1038, encode hypothetical proteins that have not been studied previously. PSORTB analysis (http://psort.ims.u-tokyo.ac.jp/) showed that proteins encoded by ECH_0849 and ECH_1038 are predicted to be inner and outer membrane proteins, respectively. Our study showed that genes encoding these proteins were expressed in infected cell cultures, as well as in infected mice, and thus are functional rather than degenerating on the way to elimination. This finding may have a broader implication, since ECH_0849 and ECH_1038 homologs (also encoding hypothetical proteins) were found in the genomes of E. canis and E. ruminantium, which are responsible for canine ehrlichiosis and heartwater of ruminants (37), respectively, suggesting that the gene products have functions unique to the genus Ehrlichia. In the future, when larger numbers of E. chaffeensis strains are compared using this type of CGH, a more distinct E. chaffeensis genomic pattern is expected to emerge.
The Arkansas strain was isolated from a young man with relatively mild clinical signs, including fever, headache, pharyngitis, nausea, vomiting, and dehydration. Cervical lymphadenopathy, splenomegaly, mild thrombocytopenia, and leukopenia also were observed. The patient defervesced within 24 to 48 h after tetracycline treatment and quickly recovered from the illness (7). Unfortunately, information on clinical signs of the patients from whom Wakulla and Liberty strains were isolated (45) is not available. Thus, it is not feasible to relate the present findings to patients' clinical signs or outcomes. Nonetheless, the diverse clinical spectra observed in the SCID mice infected with three strains of E. chaffeensis in the present study are similar to those seen in patients with HME. The clinical spectrum of HME ranges from mild to life threatening, with a case fatality rate of 2 to 3%; approximately 60 to 70% of infected patients have been hospitalized (9, 11, 12, 32, 35).
One of the most common laboratory abnormalities found in humans with HME is the elevation in levels of serum hepatic enzymes. Mildly or moderately elevated hepatic transaminase levels are noted in approximately 80 to 90% of HME patients at some point during their illness, suggesting that there is hepatic injury (32). However, there are relatively few histopathologic data describing lesions in tissues and organs of persons with HME. The previous effort to investigate the pathology of injury in liver tissues from seven patients with laboratory-confirmed HME revealed scattered lobular lymphohistiocytic foci and diffuse lymphohistiocytic infiltration and Kupffer cell hyperplasia with increased phagocytosis (41). In contrast, the injury in the livers of immunocompetent mice infected with the HF strain progressed rapidly from day 7 to day 9 PI and included Kupffer cell hyperplasia, ballooning cell injury, apoptosis, poorly formed granulomas, erythrophagocytosis, microvesicular fatty metamorphosis, and confluent severe necrosis (29, 43). Cholestasis was observed in 6/7 HME cases, sometimes along with bile duct epithelial injury (41), which was not evident in the previous SCID or SCID/bg models (47), in the present study, or in the immunocompetent mice infected with the HF strain. The causes of the cholestasis associated with HME (26, 41) are unknown.
The pathogenetic mechanisms of hepatic inflammation in HME are unknown. The livers of immunocompetent mice infected with the HF strain are heavily loaded with ehrlichiae (29, 42). However, 4/6 liver tissue samples from humans with HME were not heavily infected with E. chaffeensis (41), suggesting that host inflammatory or immune responses contribute to the liver injury seen in HME. The present study extended this notion by showing diverse responses of the liver tissue to E. chaffeensis strains with distinct genomic sequences. Leukocyte infiltration in the SCID mice did not correlate directly with bacterial load. For example, the Liberty strain did not induce severe hepatitis despite a higher bacterial load than the Arkansas strain. However, since the Liberty strain retained activity to deplete liver glycogen, it may not be completely inert in the liver. It is tempting to speculate that the hepatitis seen with Wakulla and Arkansas infections may involve a specific factor encoded by an ORF that is absent or mutated in strain Liberty. In this regard, a total of 143 ORFs, including 43 ORFs encoding hypothetical but unknown proteins, were conserved in Wakulla and Arkansas but were different in Liberty. In addition, although both the Arkansas and Wakulla strains induced severe infiltration of inflammatory cells into liver sinusoids, there was a distinct difference in the patterns: Arkansas induced severe granulomatous infiltration, as previously reported (47), whereas Wakulla induced homogeneous and diffuse infiltration disseminating throughout the liver tissue. Focal atrophy and necrosis of liver cell cords were much more pronounced in Wakulla-infected mice than in Arkansas-infected mice. SCID mice retain innate immunity (44). As granuloma formation generally is considered a part of the host innate defense system that can be modified by acquired immunity (46), the Wakulla strain may be able to overcome or suppress this host innate defense.
Glycogen depletion has not been reported in previous studies in patients with HME or in animal models. Glycogen loss in Wakulla- and Liberty-inoculated mice may be due in part to anorexia. However, mice inoculated with Arkansas did not appear to be anorexic at day 15 PI. Glycogen loss in the liver is a typical response to endotoxic shock (24). Since E. chaffeensis lacks lipopolysaccharide (20), other ehrlichial components may have contributed to this metabolic disorder in the mice.
As it is impossible for clinical isolates to have the same passage history, one concern is the possibility of mutation and genetic drift during passage of isolates in cell culture or experimental animals. It was reported previously that cell culture passage numbers of Anaplasma phagocytophilum influence disease severity in outbred horses but that incubation time, hematological changes, PCR detection, ehrlichial load, seroconversion time, and titer range do not (36). While this study did not analyze genomic differences between high- and low-passage cultures (36), the present study clearly demonstrated global genomic differences between E. chaffeensis strains. Unlike with A. phagocytophilum strains with different culture passage numbers (36), the present study revealed striking differences (more than 100-fold) in the bacterial burdens and growth rates among the three E. chaffeensis strains. The modest growth of bacteria in mice inoculated with the Arkansas strain was also consistent with the previous data (47). In addition, despite similar low-passage levels in cell culture, the Wakulla and Liberty strains had distinct genomic sequences and induced distinct liver pathologies. Our attempt to revive the Arkansas strain from an early passage received from the CDC was not successful. Nonetheless, the clinical signs and the histopathology of SCID mice in our study were remarkably similar to the previous data for the Arkansas strain reported 9 years ago by others (47). Thus, it is unlikely that the passage history difference is the primary cause of the genomic and phenotypic differences between the three E. chaffeensis strains. Regardless of the presence of mutations or drift, in the present study the phenotype and genotype of a given strain are the phenotype and genotype of bacteria with the same passage history.
While the cell-cultured bacteria used in the present study provide a more consistent inoculum than infected tissue (used for uncultivable Ehrlichia, such as the HF strain), blood (used for Anaplasma marginale, Ehrlichia ewingii, and Anaplasma platys and for some A. phagocytophilum studies), or ticks (used for A. marginale, A. phagocytophilum, E. canis, and E. ruminantium), another concern is variations in inocula. In this regard, a previous study showed that the inoculum dose of the Arkansas strain cultured in DH82 cells had a minimal effect on the bacterial load in SCID mice (47). Identification of the molecular basis of these phenotypic differences is perhaps the only way to unequivocally define the nature of virulence of E. chaffeensis strains. The present study is the first effort toward this goal.
Use of SCID mice imposes a limitation on the study of acquired immune responses. Recently, E. chaffeensis Arkansas cultured in ISE6 tick cells was reported to persist 9 days longer in immunocompetent mice than the same bacteria cultured in DH82 cells persisted (15). Therefore, by finding more suitable culture conditions and E. chaffeensis strains more virulent than the Arkansas strain, we may be able to develop an immunocompetent mouse model of E. chaffeensis infection that allows investigation of the acquired immune response in the future.
In summary, we demonstrated differences in virulence potential between three E. chaffeensis clinical isolates with genomic sequence divergence. Information gained from this study should facilitate many hypothesis-driven studies and characterization of additional strains to help elucidate HME pathogenesis, if all of the tools and advantages of the E. chaffeensis genome sequence data and mouse models are used.
Supplementary Material
Acknowledgments
We thank C. Paddock and W. L. Nicholson at the Centers for Disease Control and Prevention for sending us the Wakulla and Liberty stocks. We also thank Zhihui Cheng and Qingming Xiong for their technical assistance.
This work was supported by National Institutes of Health grant R01 AI30100 and by a David White Research Award from The OSU College of Veterinary Medicine. The E. chaffeensis genome sequence project was supported by National Institutes of Health grant R01 AI47885.
Editor: V. J. DiRita
Footnotes
Published ahead of print on 16 April 2007.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Albert, T. J., D. Dailidiene, G. Dailide, J. E. Norton, A. Kalia, T. A. Richmond, M. Molla, J. Singh, R. D. Green, and D. E. Berg. 2005. Mutation discovery in bacterial genomes: metronidazole resistance in Helicobacter pylori. Nat. Methods 2:951-953. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, B. E., K. G. Sims, J. G. Olson, J. E. Childs, J. F. Piesman, C. M. Happ, G. O. Maupin, and B. J. Johnson. 1993. Amblyomma americanum: a potential vector of human ehrlichiosis. Am. J. Trop. Med. Hyg. 49:239-244. [DOI] [PubMed] [Google Scholar]
- 3.Caldwell, H. D., H. Wood, D. Crane, R. Bailey, R. B. Jones, D. Mabey, I. Maclean, Z. Mohammed, R. Peeling, C. Roshick, J. Schachter, A. W. Solomon, W. E. Stamm, R. J. Suchland, L. Taylor, S. K. West, T. C. Quinn, R. J. Belland, and G. McClarty. 2003. Polymorphisms in Chlamydia trachomatis tryptophan synthase genes differentiate between genital and ocular isolates. J. Clin. Investig. 111:1757-1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carpenter, C. F., T. K. Gandhi, L. K. Kong, G. R. Corey, S. M. Chen, D. H. Walker, J. S. Dumler, E. Breitschwerdt, B. Hegarty, and D. J. Sexton. 1999. The incidence of ehrlichial and rickettsial infection in patients with unexplained fever and recent history of tick bite in central North Carolina. J. Infect. Dis. 180:900-903. [DOI] [PubMed] [Google Scholar]
- 5.Cheng, C., C. D. Paddock, and R. Reddy Ganta. 2003. Molecular heterogeneity of Ehrlichia chaffeensis isolates determined by sequence analysis of the 28-kilodalton outer membrane protein genes and other regions of the genome. Infect. Immun. 71:187-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng, Z., Y. Kumagai, M. Lin, C. Zhang, and Y. Rikihisa. 2006. Intra-leukocyte expression of two-component systems in Ehrlichia chaffeensis and Anaplasma phagocytophilum and effects of the histidine kinase inhibitor closantel. Cell. Microbiol. 8:1241-1252. [DOI] [PubMed] [Google Scholar]
- 7.Dawson, J. E., B. E. Anderson, D. B. Fishbein, J. L. Sanchez, C. S. Goldsmith, K. H. Wilson, and C. W. Duntley. 1991. Isolation and characterization of an Ehrlichia sp. from a patient diagnosed with human ehrlichiosis. J. Clin. Microbiol. 29:2741-2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Demma, L. J., R. C. Holman, J. H. McQuiston, J. W. Krebs, and D. L. Swerdlow. 2005. Epidemiology of human ehrlichiosis and anaplasmosis in the United States, 2001-2002. Am. J. Trop. Med. Hyg. 73:400-409. [PubMed] [Google Scholar]
- 9.Dumler, J. S., J. E. Dawson, and D. H. Walker. 1993. Human ehrlichiosis: hematopathology and immunohistologic detection of Ehrlichia chaffeensis. Hum. Pathol. 24:391-396. [DOI] [PubMed] [Google Scholar]
- 10.Ewing, S. A., J. E. Dawson, A. A. Kocan, R. W. Barker, C. K. Warner, R. J. Panciera, J. C. Fox, K. M. Kocan, and E. F. Blouin. 1995. Experimental transmission of Ehrlichia chaffeensis (Rickettsiales: Ehrlichieae) among white-tailed deer by Amblyomma americanum (Acari: Ixodidae). J. Med. Entomol. 32:368-374. [DOI] [PubMed] [Google Scholar]
- 11.Fichtenbaum, C. J., L. R. Peterson, and G. J. Weil. 1993. Ehrlichiosis presenting as a life-threatening illness with features of the toxic shock syndrome. Am. J. Med. 95:351-357. [DOI] [PubMed] [Google Scholar]
- 12.Fishbein, D. B., J. E. Dawson, and L. E. Robinson. 1994. Human ehrlichiosis in the United States, 1985 to 1990. Ann. Intern. Med. 120:736-743. [DOI] [PubMed] [Google Scholar]
- 13.Fishbein, D. B., A. Kemp, J. E. Dawson, N. R. Greene, M. A. Redus, and D. H. Fields. 1989. Human ehrlichiosis: prospective active surveillance in febrile hospitalized patients. J. Infect. Dis. 160:803-809. [DOI] [PubMed] [Google Scholar]
- 14.Fitzgerald, J. R., D. E. Sturdevant, S. M. Mackie, S. R. Gill, and J. M. Musser. 2001. Evolutionary genomics of Staphylococcus aureus: insights into the origin of methicillin-resistant strains and the toxic shock syndrome epidemic. Proc. Natl. Acad. Sci. USA 98:8821-8826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ganta, R. R., C. Cheng, E. C. Miller, B. L. McGuire, L. Peddireddi, K. R. Sirigireddy, and S. K. Chapes. 2007. Differential clearance and immune responses to tick cell-derived versus macrophage culture-derived Ehrlichia chaffeensis in mice. Infect. Immun. 75:135-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ge, H., Y. Y. Chuang, S. Zhao, M. Tong, M. H. Tsai, J. J. Temenak, A. L. Richards, and W. M. Ching. 2004. Comparative genomics of Rickettsia prowazekii Madrid E and Breinl strains. J. Bacteriol. 186:556-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hotopp, J. C., M. Lin, R. Madupu, J. Crabtree, S. V. Angiuoli, J. Eisen, R. Seshadri, Q. Ren, M. Wu, T. R. Utterback, S. Smith, M. Lewis, H. Khouri, C. Zhang, H. Niu, Q. Lin, N. Ohashi, N. Zhi, W. Nelson, L. M. Brinkac, R. J. Dodson, M. J. Rosovitz, J. Sundaram, S. C. Daugherty, T. Davidsen, A. S. Durkin, M. Gwinn, D. H. Haft, J. D. Selengut, S. A. Sullivan, N. Zafar, L. Zhou, F. Benahmed, H. Forberger, R. Halpin, S. Mulligan, J. Robinson, O. White, Y. Rikihisa, and H. Tettelin. 2006. Comparative genomics of emerging human ehrlichiosis agents. PLoS Genet. 2:e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ijdo, J. W., A. C. Carlson, and E. L. Kennedy. 2007. Anaplasma phagocytophilum AnkA is tyrosine-phosphorylated at EPIYA motifs and recruits SHP-1 during early infection. Cell. Microbiol., 9:1284-1296. [DOI] [PubMed] [Google Scholar]
- 19.Kato-Maeda, M., P. J. Bifani, B. N. Kreiswirth, and P. M. Small. 2001. The nature and consequence of genetic variability within Mycobacterium tuberculosis. J. Clin. Investig. 107:533-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin, M., and Y. Rikihisa. 2003. Ehrlichia chaffeensis and Anaplasma phagocytophilum lack genes for lipid A biosynthesis and incorporate cholesterol for their survival. Infect. Immun. 71:5324-5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Long, S. W., X. F. Zhang, H. Qi, S. Standaert, D. H. Walker, and X. J. Yu. 2002. Antigenic variation of Ehrlichia chaffeensis resulting from differential expression of the 28-kilodalton protein gene family. Infect. Immun. 70:1824-1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maeda, K., N. Markowitz, R. C. Hawley, M. Ristic, D. Cox, and J. E. McDade. 1987. Human infection with Ehrlichia canis, a leukocytic rickettsia. N. Engl. J. Med. 316:853-856. [DOI] [PubMed] [Google Scholar]
- 23.Martin, G. S., B. W. Christman, and S. M. Standaert. 1999. Rapidly fatal infection with Ehrlichia chaffeensis. N. Engl. J. Med. 341:763-764. [DOI] [PubMed] [Google Scholar]
- 24.McCallum, R. E., and L. J. Berry. 1973. Effects of endotoxin on gluconeogenesis, glycogen synthesis, and liver glycogen synthase in mice. Infect. Immun. 7:642-654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McQuiston, J. H., C. D. Paddock, R. C. Holman, and J. E. Childs. 1999. The human ehrlichioses in the United States. Emerg. Infect. Dis. 5:635-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moskovitz, M., R. Fadden, and T. Min. 1991. Human ehrlichiosis: a rickettsial disease associated with severe cholestasis and multisystemic disease. J. Clin. Gastroenterol. 13:86-90. [PubMed] [Google Scholar]
- 27.Niu, H., Y. Rikihisa, M. Yamaguchi, and N. Ohashi. 2006. Differential expression of VirB9 and VirB6 during the life cycle of Anaplasma phagocytophilum in human leucocytes is associated with differential binding and avoidance of lysosome pathway. Cell. Microbiol. 8:523-534. [DOI] [PubMed] [Google Scholar]
- 28.Ohashi, N., A. Unver, N. Zhi, and Y. Rikihisa. 1998. Cloning and characterization of multigenes encoding the immunodominant 30-kilodalton major outer membrane proteins of Ehrlichia canis and application of the recombinant protein for serodiagnosis. J. Clin. Microbiol. 36:2671-2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okada, H., T. Tajima, M. Kawahara, and Y. Rikihisa. 2001. Ehrlichial proliferation and acute hepatocellular necrosis in immunocompetent mice experimentally infected with the HF strain of Ehrlichia, closely related to Ehrlichia chaffeensis. J. Comp. Pathol. 124:165-171. [DOI] [PubMed] [Google Scholar]
- 30.Olano, J. P., E. Masters, W. Hogrefe, and D. H. Walker. 2003. Human monocytotropic ehrlichiosis, Missouri. Emerg. Infect. Dis. 9:1579-1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Olano, J. P., G. Wen, H. M. Feng, J. W. McBride, and D. H. Walker. 2004. Histologic, serologic, and molecular analysis of persistent ehrlichiosis in a murine model. Am. J. Pathol. 165:997-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paddock, C. D., and J. E. Childs. 2003. Ehrlichia chaffeensis: a prototypical emerging pathogen. Clin. Microbiol. Rev. 16:37-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paddock, C. D., J. W. Sumner, G. M. Shore, D. C. Bartley, R. C. Elie, J. G. McQuade, C. R. Martin, C. S. Goldsmith, and J. E. Childs. 1997. Isolation and characterization of Ehrlichia chaffeensis strains from patients with fatal ehrlichiosis. J. Clin. Microbiol. 35:2496-2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park, J., K. J. Kim, K. S. Choi, D. J. Grab, and J. S. Dumler. 2004. Anaplasma phagocytophilum AnkA binds to granulocyte DNA and nuclear proteins. Cell. Microbiol. 6:743-751. [DOI] [PubMed] [Google Scholar]
- 35.Petersen, L. R., L. A. Sawyer, D. B. Fishbein, P. W. Kelley, R. J. Thomas, L. A. Magnarelli, M. Redus, and J. E. Dawson. 1989. An outbreak of ehrlichiosis in members of an Army Reserve unit exposed to ticks. J. Infect. Dis. 159:562-568. [DOI] [PubMed] [Google Scholar]
- 36.Pusterla, N., J. E. Madigan, K. M. Asanovich, J. S. Chae, E. Derock, C. M. Leutenegger, J. B. Pusterla, H. Lutz, and J. S. Dumler. 2000. Experimental inoculation with human granulocytic Ehrlichia agent derived from high- and low-passage cell culture in horses. J. Clin. Microbiol. 38:1276-1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rikihisa, Y. 1991. The tribe Ehrlichieae and ehrlichial diseases. Clin. Microbiol. Rev. 4:286-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rikihisa, Y., S. A. Ewing, and J. C. Fox. 1994. Western immunoblot analysis of Ehrlichia chaffeensis, E. canis, or E. ewingii infections in dogs and humans. J. Clin. Microbiol. 32:2107-2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Safdar, N., R. B. Love, and D. G. Maki. 2002. Severe Ehrlichia chaffeensis infection in a lung transplant recipient: a review of ehrlichiosis in the immunocompromised patient. Emerg. Infect. Dis. 8:320-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Salama, N., K. Guillemin, T. K. McDaniel, G. Sherlock, L. Tompkins, and S. Falkow. 2000. A whole-genome microarray reveals genetic diversity among Helicobacter pylori strains. Proc. Natl. Acad. Sci. USA 97:14668-14673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sehdev, A. E., and J. S. Dumler. 2003. Hepatic pathology in human monocytic ehrlichiosis. Ehrlichia chaffeensis infection. Am. J. Clin. Pathol. 119:859-865. [DOI] [PubMed] [Google Scholar]
- 42.Shibata, S., M. Kawahara, Y. Rikihisa, H. Fujita, Y. Watanabe, C. Suto, and T. Ito. 2000. New Ehrlichia species closely related to Ehrlichia chaffeensis isolated from Ixodes ovatus ticks in Japan. J. Clin. Microbiol. 38:1331-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sotomayor, E. A., V. L. Popov, H. M. Feng, D. H. Walker, and J. P. Olano. 2001. Animal model of fatal human monocytotropic ehrlichiosis. Am. J. Pathol. 158:757-769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stephan, J. L., V. Vlekova, F. Le Deist, S. Blanche, J. Donadieu, G. De Saint-Basile, A. Durandy, C. Griscelli, and A. Fischer. 1993. Severe combined immunodeficiency: a retrospective single-center study of clinical presentation and outcome in 117 patients. J. Pediatr. 123:564-572. [DOI] [PubMed] [Google Scholar]
- 45.Sumner, J. W., J. E. Childs, and C. D. Paddock. 1999. Molecular cloning and characterization of the Ehrlichia chaffeensis variable-length PCR target: an antigen-expressing gene that exhibits interstrain variation. J. Clin. Microbiol. 37:1447-1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Velayudham, A., I. Hritz, A. Dolganiuc, P. Mandrekar, E. Kurt-Jones, and G. Szabo. 2006. Critical role of toll-like receptors and the common TLR adaptor, MyD88, in induction of granulomas and liver injury. J. Hepatol. 45:813-824. [DOI] [PubMed] [Google Scholar]
- 47.Winslow, G. M., E. Yager, K. Shilo, D. N. Collins, and F. K. Chu. 1998. Infection of the laboratory mouse with the intracellular pathogen Ehrlichia chaffeensis. Infect. Immun. 66:3892-3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yoshiie, K., H. Y. Kim, J. Mott, and Y. Rikihisa. 2000. Intracellular infection by the human granulocytic ehrlichiosis agent inhibits human neutrophil apoptosis. Infect. Immun. 68:1125-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu, X. J., J. W. McBride, and D. H. Walker. 1999. Genetic diversity of the 28-kilodalton outer membrane protein gene in human isolates of Ehrlichia chaffeensis. J. Clin. Microbiol. 37:1137-1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.